Abstract
We report two brothers from a consanguineous couple with spondyloepimetaphyseal dysplasia (SEMD), multiple joint dislocations at birth, severe joint laxity, scoliosis, gracile metacarpals and metatarsals, delayed bone age and poorly ossified carpal and tarsal bones, probably representing a yet uncharacterized SEMD with laxity and dislocations. This condition has clinical overlap with autosomal dominantly inherited SEMD with joint laxity, leptodactylic type caused by recurrent missense variants in the kinesin family member 22 gene (KIF22). Single-nucleotide polymorphism array analysis and whole-exome sequencing in the two affected siblings revealed a shared homozygous nonsense variant [c.906T>A/p.(Tyr302*)] in EXOC6B as the most likely cause. EXOC6B encodes a component of the exocyst complex required for tethering secretory vesicles to the plasma membrane. As transport of vesicles from the golgi apparatus to the plasma membrane occurs through kinesin motor proteins along microtubule tracks, the function of EXOC6B is linked to KIF22 suggesting a common pathogenic mechanism in skeletal dysplasias with joint laxity and dislocations.
Introduction
Skeletal dysplasias with multiple joint dislocations is a heterogeneous group of disorders comprising ~10 distinct entities.1 Spondyloepimetaphyseal dysplasia (SEMD) with joint laxity type 2 (leptodactylic type, Hall type or SEMD-JL2; MIM 603546) is an autosomal dominant skeletal dysplasia with short stature, multiple joint dislocations and/or laxity, delayed ossification of carpals, small carpus, severe epiphyseal and mild metaphyseal changes, mild vertebral body deformities and gracile metacarpals.2 Missense variants affecting one of two amino acids in the motor domain of the kinesin family member KIF22 have been identified to cause SEMD-JL2.3, 4 Key features of SEMD with joint laxity, type 1, with or without fractures (SEMD-JL1; MIM 271640), which is caused by biallelic variants in B3GALT6,5, 6 are joint laxity and dislocations and SEMD. Affected individuals show facial dysmorphism, kyphoscoliosis at birth, talipes equinovarus, cleft palate and congenital heart disease.7 Another SEMD-JL2-like phenotype is associated with a homozygous missense variant in NIN.8 Here, we report two brothers with a new type of autosomal recessive SEMD with laxity and joint dislocations. We used single-nucleotide polymorphism (SNP) array analysis and whole-exome sequencing (WES) to identify a homozygous nonsense variant in EXOC6B in the affected siblings.
Patients and methods
Patients
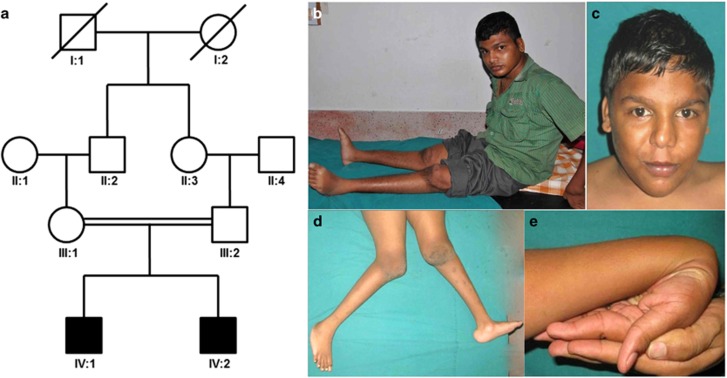
Clinical data and samples were obtained with informed consent, including consent to use photographs. Consent procedures were approved by the Ethics Committees of the Medical Chamber of Hamburg (reference number PV3802) and Kasturba Hospital, Manipal. The two brothers were born to consanguineous healthy parents from South India (Figure 1a) and delivered by vaginal delivery after uncomplicated pregnancy.
Figure 1.
(a) Pedigree of the family shows that the two affected brothers (IV:1 and IV:2) were born to first cousins. Clinical photographs show normal face of subject 1 (b) and subject 2 (c), dislocated knee joints and flat feet (d) and joint laxity (e) in subject 2.
Birth weight of subject 1 (S1) was 3.25 kg (normal). At age 20 years, his height was 120 cm (−8 SD) and weight 42 kg (−3 SD; Figure 1b). Subject 2 (S2) is the younger brother and presented to us at age 14 years (Figure 1c). He had short stature (height 116 cm; −6 SD) and weighed 40 kg (normal). His head circumference was 52 cm (normal). No intrauterine growth retardation was observed. Both subjects had dislocations of hip and knee joints at birth, joint laxity (Figures 1d and e) and underwent surgical operations for correction of deformities. Delayed walking due to the joint problems was noted in both. They had normal facial features (Figures 1b and c). Both had left lower thoracic scoliosis (Figure 2) and short neck. Skin, nails, hair and teeth were normal. They had normal intellectual functioning. Both did not have cutaneous hyperelasticity, bruisability, fragility, scar formation or any bleeding diathesis. There was no laryngeal stenosis (as observed during general anesthesia for surgery). Ophthalmological, otorhinolaryngological, cardiological evaluations (including echocardiography) and routine blood and urine examinations were unremarkable. There was no evidence of nephropathy.
Figure 2.
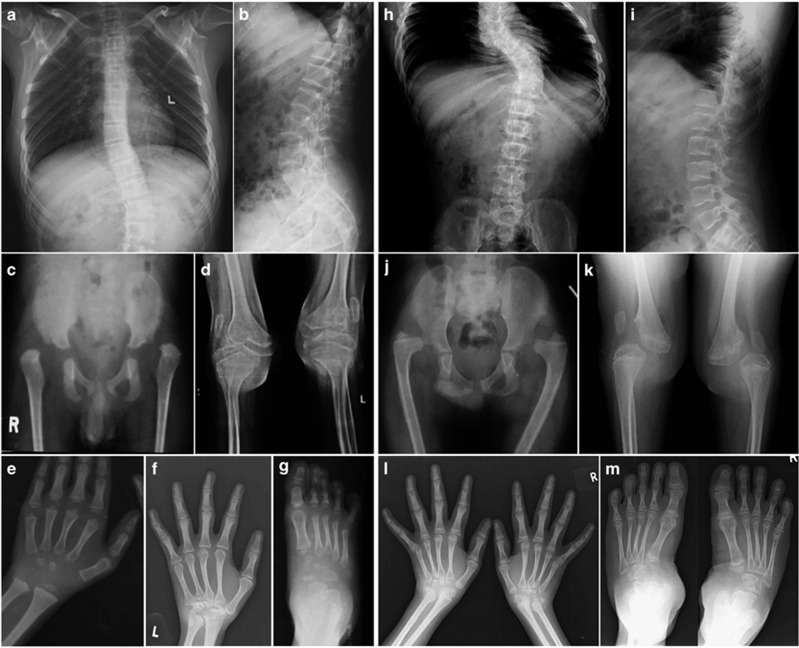
Key radiological features in spine, large joints, hands and feet of the two affected brothers. Spine shows thoracolumbar scoliosis and narrow interpedicular distance in lumbar vertebrae (a, h). Vertebral end plates are irregular (b, i). Dislocations are seen in hip and knee joints (c, d, j, k). Patellae were also dislocated (d, k). Slender metacarpals and metatarsals are seen in both individuals (e–g, l, m). Carpal bone ossification is delayed at younger age (e). Proximal row of carpal bones are not ossified and other carpal bones are small, irregular and disorganized (f, l). Similar changes are seen in tarsal bones too (g, m). Epiphyseal dysplasia is evident in long bones and carpal and tarsal bones. Lower ends of ulna are small (f, l). (Subject 1: a and b at 21 years, c at 2 years, d at 18 years, e at 4 years, f at 21 years and g at 4 years; Subject 2: h and i at 14 years, j at 9 years and k–m at 14 years).
Radiographic examination revealed SEMD (Figure 2). Spine showed mild platyspondyly, scoliosis and narrow interpedicular distance (Figures 2a, b, h and i). Ribs were slender. Epiphyses showed severe dysplasia in long tubular bones, which was marked at the upper ends of humerus and femur (Figures 2c, d, j and k and Supplementary Figures 1 and 2). Dislocation of hip and knee joints and patella were noted on both sides (Figures 2c, d, j and k). Proximal row of carpal bones showed absent ossification with an overall short carpus. Distal carpal bones and tarsal bones showed delayed ossification, irregularity and poor morphology (Figures 2e–g, l and m). Metacarpals and metatarsals were slender (gracile or leptodactylic). Cupped metaphyses of radius and ulna with short sclerotic striations, flattened epiphyses of lower ends of radius and ulna and distal ulnar hypoplasia were noted. Mild osteopenia was observed. Bone mineral density was not determined. Radiograph of skull was normal and there was no instability of cervical spine and no wormian bones (Supplementary Figures 1a and b and Supplementary Figure 2a). S2 showed dislocation of radial heads and flat feet (Figure 1 and Supplementary Figure 2b). Clinical details were submitted to the Phenome Central database at https://phenomecentral.org/ (Patient ID: GR1805859).
Methods
Homozygosity mapping was performed by SNP array analysis, using DNA samples of both siblings, followed by WES. Sanger sequencing was performed to confirm functional relevant variants. Detailed description of methods is provided in Supplementary Information. The EXOC6B variant has been submitted to the LOVD database (http://databases.lovd.nl/shared/variants/EXOC6B).
Results and Discussion
Patients S1 and S2 clinically show some resemblance to both SEMD-JL1 and -JL2 (Supplementary Table 1). However, as other features were absent, such as airway obstruction due to laryngeal stenosis, facial dysmorphism, kyphoscoliosis at birth, talipes equinovarus, cleft palate and congenital heart disease, they most likely present with a new type of autosomal recessive multiple joint dislocation syndrome resembling SEMD-JL2. Consanguinity of the parents suggests a variant affecting function within a shared region of homozygosity (ROH).
SNP array revealed 10 ROHs in the siblings (Supplementary Table 2). Visual examination of WES reads did not identify any rare variant in genes within the ROHs, and/or which had previously been related to skeletal dysplasias (Supplementary Table 2), KIF22 (MIM 603213), NIN (MIM 608684), B3GALT6 (MIM 615291), MMP2 (MIM 120360) and MAFB (MIM 608968; data not shown).
After filtering of WES data, we obtained five private and five rare (MAF<0.5%) variants that were absent in the homozygous state in public databases: a nonsense, six non-synonymous and three synonymous variants (Supplementary Table 3). All variants were validated by Sanger sequencing; the parents of the siblings carried each in the heterozygous state (data not shown). No X-chromosomal variant passed the filter (Supplementary Table 4).
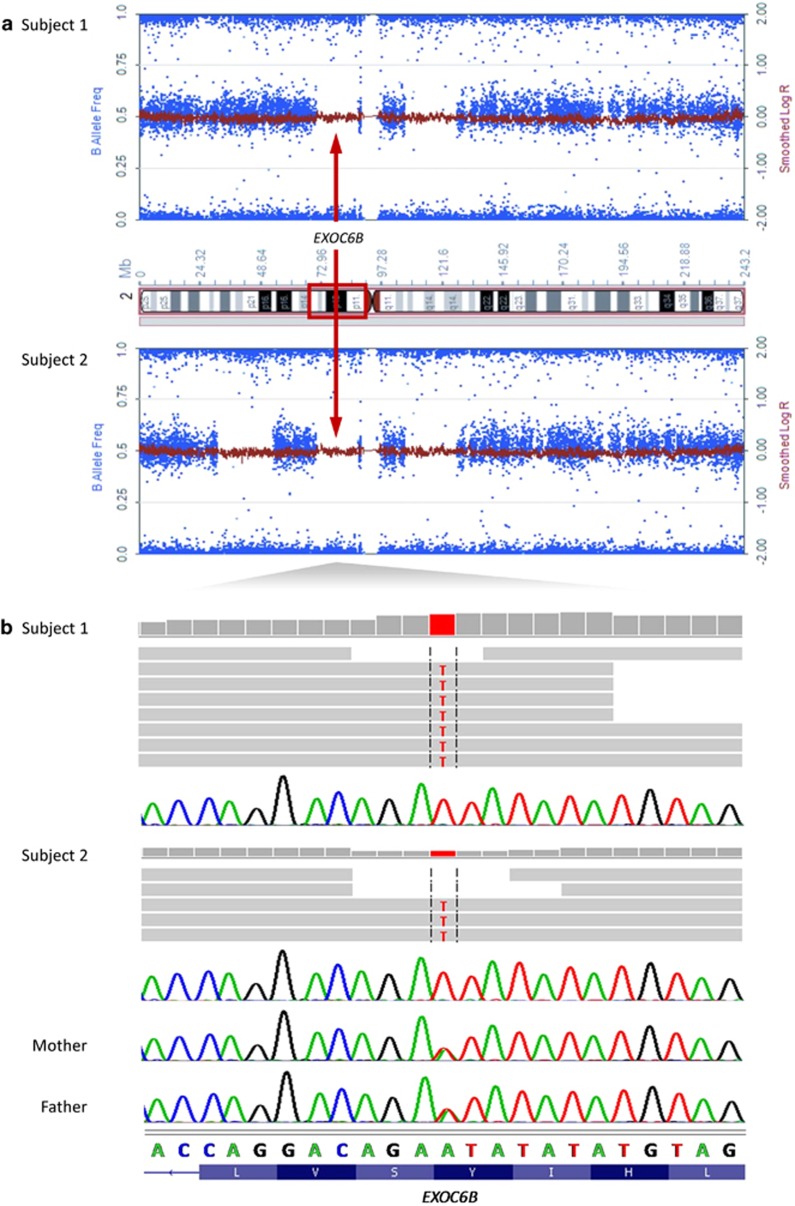
The patients' phenotype was uncommon fitting with a private, not yet described variant. We considered four of the five private variants as unlikely to affect function (Supplementary Table 3), only one had a predicted damaging effect: c.906T>A [p.(Tyr302*)] in EXOC6B (Figure 3 and Supplementary Table 3). This nonsense variant leads to loss of the last 509 amino acids of the encoded protein or nonsense-mediated mRNA decay. EXOC6B became the most probable causative gene as no other homozygous deleterious EXOC6B variant has been reported in ExAC, and EXOC6B is predicted to be intolerant to functional genetic variation (data not shown).
Figure 3.
Identification of a homozygous nonsense variant in EXOC6B by whole-exome sequencing. (a) Plot of B allele frequency (BAF) and log R ratio on chromosome 2. The x axis shows positions of SNPs in subjects 1 and 2 on chromosome 2 and the y axis shows the BAF. A BAF value of around 0.5 indicates a heterozygous genotype, whereas 0 and 1 indicate homozygous genotypes. The shared homozygous region of ∼17 Mb length harboring the EXOC6B gene is indicated on the ideogram of chromosome 2 (p13.3-p11.2; red rectangle) and in the plot of each subject (red arrows). (b) Exome sequence data are visualized using the Integrated Genomics Viewer. The horizontal gray arrows depict the paired-end reads aligned to the positive strand of the human genome (hg19). The negative strand with complement nucleotides and reverse complement translations is shown. Parts of the Sanger sequencing electropherograms are depicted and demonstrate homozygosity for the nonsense variant in subjects 1 and 2 and heterozygosity in mother and father. Encoded amino acids are indicated in the one-letter code.
Individuals with a microdeletion encompassing EXOC6B or a translocation disrupting EXOC6B, all with a neurodevelopmental phenotype have been reported (Supplementary Figure 3).9, 10, 11, 12 EXOC6B haploinsufficiency has been suggested as a cause for intellectual disability (ID).10 A homozygous loss-of-function EXOC6B variant in siblings with a skeletal disorder and heterozygosity of the same in healthy parents contrast with the literature. Possibly, another variant is causative for the SEMD type studied here. However, the other four private variants were not predicted to be damaging, and no effect on pre-mRNA splicing was predicted for the two synonymous changes (Supplementary Table 3 and data not shown). Alternatively, EXOC6B is not an ID-associated gene. As chromosomal abnormalities can delete/interrupt a coding region, but finally lead to disruption of cis-regulatory elements,13 we suggest EMX1 (MIM 600034) as candidate for intellectual development (Supplementary Figure 3). It encodes a transcription factor important for brain development.14, 15 We found the fetal brain enhancers 11 270 and 5866 to reside in EXOC6B and both are covered by the smallest ID-associated microdeletions; the translocation breakpoints are located between these brain enhancers and EMX1 (Supplementary Figure 3).9, 11 We hypothesize that deletion of enhancers or translocation to another region changed the regulatory environment of EMX1 and resulted in dysregulated EMX1 expression in the ID-affected individuals, providing further evidence for involvement of EXOC6B in SEMD.
EXOC6B is widely expressed in human tissues such as bone and central nervous system (https://www.proteomicsdb.org/ proteomicsdb/#human/proteinDetails/Q9Y2D4/expression). EXOC6B is a component of the exocyst complex required for tethering secretory vesicles to the plasma membrane; it likely plays a role in polarized exocytosis.16 Tethering and fusion of exocytic vesicles are required for ciliogenesis, which is important for skeletal development.17 A postulated role of KIF22 in cartilage biology by implication in intracellular transport and/or cilia-associated transport mechanisms3, 4 indicates a possible link between kinesins (eg KIF22) and the exocyst complex (eg EXOC6B). In line with this, NIN, another gene associated with an SEMD-JL2-like phenotype,8 encodes ninein required for centrosome maturation and architecture.18 As building and maintenance of a cilium depend on membrane vesicle trafficking, microtubule-dependent motor proteins and formation of basal bodies,19 we speculate that dysfunction of EXOC6B, KIF22 and ninein negatively affects ciliogenesis leading to overlapping SEMD forms.
Acknowledgments
We thank the patients and their parents for participating in this study. We thank Peter Meinecke, Jürgen Spranger and Andrea Superti-Furga for diagnostic advice, Inka Jantke, Frederike L Harms and Jelena Ljubković for resequencing and segregation analysis and Malte Spielmann for help with preparing Supplementary Figure 3. We are grateful to MedGenome Labs, India for the information on frequencies of rare alleles in the local population. This work was supported by grants from the Indian Council of Medical Research (BMS 54/2/2013) and the Deutsche Forschungsgemeinschaft (KO 4576/1-1 to F.K. and KU 1240/5-1 to K.K.).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bonafe L, Cormier-Daire V, Hall C et al: Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A 2015, ; e-pub ahead of print 23 September 2015; doi:10.1002/ajmg.a.37365. [DOI] [PubMed]
- Kim OH, Cho TJ, Song HR et al: A distinct form of spondyloepimetaphyseal dysplasia with joint laxity (SEMDJL)-leptodactylic type: radiological characteristics in seven new patients. Skelet Radiol 2009; 38: 803–811. [DOI] [PubMed] [Google Scholar]
- Boyden ED, Campos-Xavier AB, Kalamajski S et al: Recurrent dominant mutations affecting two adjacent residues in the motor domain of the monomeric kinesin KIF22 result in skeletal dysplasia and joint laxity. Am J Hum Genet 2011; 89: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BJ, Kim N, Chung T et al: Whole-exome sequencing identifies mutations of KIF22 in spondyloepimetaphyseal dysplasia with joint laxity, leptodactylic type. Am J Hum Genet 2011; 89: 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Mizumoto S, Miyake N et al: Mutations in B3GALT6, which encodes a glycosaminoglycan linker region enzyme, cause a spectrum of skeletal and connective tissue disorders. Am J Hum Genet 2013; 92: 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait F, Kariminejad A, Van Damme T et al: Defective initiation of glycosaminoglycan synthesis due to B3GALT6 mutations causes a pleiotropic Ehlers-Danlos-syndrome-like connective tissue disorder. Am J Hum Genet 2013; 92: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P: Spondyloepimetaphyseal dysplasia with joint laxity (SEMDJL). J Med Genet 1994; 31: 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosch M, Gruner B, Spranger S et al: Identification of a Ninein (NIN) mutation in a family with spondyloepimetaphyseal dysplasia with joint laxity (leptodactylic type)-like phenotype. Matrix Biol 2013; 32: 387–392. [DOI] [PubMed] [Google Scholar]
- Borsani G, Piovani G, Zoppi N et al: Cytogenetic and molecular characterization of a de-novo t(2p;7p) translocation involving TNS3 and EXOC6B genes in a boy with a complex syndromic phenotype. Eur J Med Genet 2008; 51: 292–302. [DOI] [PubMed] [Google Scholar]
- Evers C, Maas B, Koch KA et al: Mosaic deletion of EXOC6B: further evidence for an important role of the exocyst complex in the pathogenesis of intellectual disability. Am J Med Genet 2014; 164A: 3088–3094. [DOI] [PubMed] [Google Scholar]
- Frühmesser A, Blake J, Haberlandt E et al: Disruption of EXOC6B in a patient with developmental delay, epilepsy, and a de novo balanced t(2;8) translocation. Eur J Hum Genet 2013; 21: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lopes F, Soares G et al: Phenotypic and functional consequences of haploinsufficiency of genes from exocyst and retinoic acid pathway due to a recurrent microdeletion of 2p13.2. Orphanet J Rare Dis 2013; 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Kleinjan DA: Disruption of long-range gene regulation in human genetic disease: a kaleidoscope of general principles, diverse mechanisms and unique phenotypic consequences. Hum Genet 2014; 133: 815–845. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Garel S, Nakagawa Y, Rubenstein JL, O'Leary DD: Emx1 and Emx2 cooperate to regulate cortical size, lamination, neuronal differentiation, development of cortical efferents, and thalamocortical pathfinding. J Comp Neurol 2003; 457: 345–360. [DOI] [PubMed] [Google Scholar]
- Hong SM, Liu Z, Fan Y et al: Reduced hippocampal neurogenesis and skill reaching performance in adult Emx1 mutant mice. Exp Neurol 2007; 206: 24–32. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P: The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 1996; 15: 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Serra RA, Yang S: Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Ann N Y Acad Sci 2015; 1335: 78–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M: Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 2005; 118: 1565–1575. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW: Centrioles, centrosomes, and cilia in health and disease. Cell 2009; 139: 663–678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.