Abstract
The IQSEC2 gene is located on chromosome Xp11.22 and encodes a guanine nucleotide exchange factor for the ADP-ribosylation factor family of small GTPases. This gene is known to have a significant role in cytoskeletal organization, dendritic spine morphology and synaptic organization. Variants in IQSEC2 cause moderate to severe intellectual disability in males and a variable phenotype in females because this gene escapes from X-chromosome inactivation. Here we report on the first splicing variant in IQSEC2 (g.88032_88033del; NG_021296.1) that co-segregates in a family diagnosed with an X-linked form of ID. In a percentage of the cells, the variant activates an intraexonic splice acceptor site that abolishes 26 amino acids from the highly conserved PH domain of IQSEC2 and creates a premature stop codon 36 amino acids later in exon 13. Interestingly, the percentage of aberrant splicing seems to correlate with the severity of the disease in each patient. The impact of this variant in the target tissue is unknown, but we can hypothesize that these differences may be related to the amount of abnormal IQSEC2 transcript. To our knowledge, we are reporting a novel mechanism of IQSEC2 involvement in ID. Variants that affect splicing are related to many genetic diseases and the understanding of their role in disease expands potential opportunities for gene therapy. Modulation of aberrant splicing transcripts can become a potent therapeutic approach for many of these diseases.
Introduction
Intellectual disability (ID) affects 1–3% of the general population being highly heterogeneous and involving both genetic and environmental factors. Approximately, half of the ID cases have a familial origin and in ~50% the genetic defect remains unknown. X-linked ID (XLID) represents an important proportion of the patients affected by ID; indeed, >100 genes have been involved in syndromic and non-syndromic forms of XLID, making molecular diagnosis and genetic counseling difficult. Recently, in an X-chromosome exome resequencing project, Tarpey et al.1 identified three non-synonymous variants in the IQSEC2 gene (Sec7 domain of, and IQ-like domain; MIM*300522) in patients with XLID. Shoubridge et al.2 identified another mutation and demonstrated the implication of these variants in the etiology of ID. This gene is located in the X-chromosome and escapes X-inactivation. It codes for a guanine nucleotide exchange factor (GEF) for the small GTP-binding proteins. The protein has a critical role in cytoskeletal and synaptic organization by activating diverse ARF substrates, such as Arf6 that is localized in the postsynaptic density (PSD) of excitatory synapses.3 The IQSEC2 protein has three functional domains and one PDZ-binding motif. The IQ domain, including amino acids from 337 to 366, serves as a binding site for different proteins including the myosin light chains, calmodulin (CaM) and CaM-like proteins.4 The Sec7 domain, including amino acids from 736 to 929, catalyses GDP–GTP exchange on Arf GTPases such as Arf1 and Arf6. The PH domains, which include amino acids from 941 to 1045, are small modular domains that occur in a large variety of proteins and have a role in recruiting proteins to different membranes, thus targeting them to appropriate cellular compartments or enabling them to interact with other components of the signal transduction pathways. Finally, the PDZ motif, including amino acids from 1484-1488, mediates many cellular and biological functions, especially those involving signal transduction complexes.2, 5, 6, 7, 8 The first variants described in the IQSEC2 gene were located in the IQ-like and sec7 domains2 and the loss of GEF activity was the most likely underlying molecular mechanism for the ID.2, 9, 10 Almost all male patients carrying IQSEC2 variants showed moderate to severe ID, whereas females were variably affected.9, 11 In the present study, we have identified a novel splicing variant in the IQSEC2 gene that activates an alternative splicing, producing a mosaic of normal and aberrant IQSEC2 mRNA.
Materials and methods
Patients
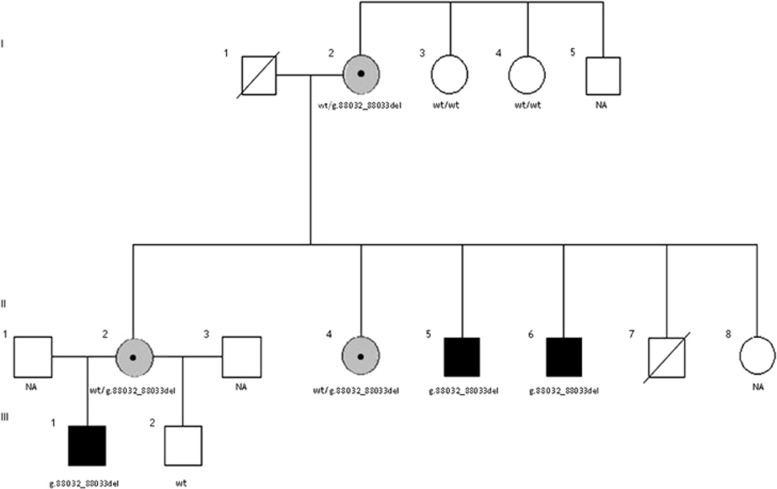
We studied a non-consanguineous family with five affected individuals with unexplained mild to severe ID and one with learning disabilities (Figure 1). Previous routine studies discarded fragile X syndrome and copy-number changes by aCGH. Exome sequencing was performed in two affected (I2 and II5) and two non-affected (I3 and I4) individuals of the family. All participants have provided written consent and the study was approved by the Institutional Review Boards of the Hospital Clinic of Barcelona.
Figure 1.
Pedigree of the family. The individuals who could be tested are indicated as carrying the 2-bp deletion or not (wt). NA, not available; wt, wild type.
Exome sequencing
Exome sequencing was performed in individuals I2, I3, I4 and II5 by the SNP&SEQ Technology Platform in Uppsala, Sweeden (www.sequencing.se) as described previously.12 Common variants (MAF >1%), based on frequencies in dbSNP13 and the 1000 Genomes Project,14 were discarded. Putative candidate variants were selected according to their potential effect on the phenotype as predicted by the tools polyphen2 (ref. 15) and SIFT16 and the function of the gene and segregation in the family. The variant detected by exome sequencing was confirmed by Sanger sequencing in DNA of the patients and their relatives (primers' sequences available upon request). PCR products were bidirectionally sequenced using Big Dye Terminator in an ABI3130 automated sequencer (Life Technologies, Madrid, Spain). The variant has been submitted into the public database ClinVar (submission ID SCV000245768; http://www.ncbi.nlm.nih.gov/clinvar/submitters/505524/).
Fibroblast culture
Skin biopsies from individuals I2 and II5 were obtained using a 3-mm punch without local anesthesia. The biopsy was diced under sterile conditions and then plated in T25 flasks in Minimum Essential Media (MEM) 13% containing 500 ml MEM (Gibco, Thermo Fisher Scientific, Madrid, Spain) and 75 ml Newborn Calf Serum (Gibco) supplement with 0.30 ml Penicillin (Gibco) and 0.30 ml Streptomycin (Gibco) at 37 °C with a 5% CO2 atmosphere. After 10–15 days in culture, one of the cultures is supplemented with cycloheximide (10 μg/ml) for 5 h. Cycloheximide is the most common protein synthesis inhibitor, and is believed to specifically inhibit the cytoplasmic protein synthesis. It has been previously reported that cycloheximide pretreatment increases the level of aberrant transcripts due to splice-site variants.17
RNA extraction and cDNA synthesis
For individuals I2, I3, I4, II4, II5 and II6, 2.5 ml of peripheral venous blood was collected in 5 ml PAXgene tubes (Qiagen, Hilden, Germany). Whole-blood RNA was isolated with the PAXgene Blood RNA Kit (Qiagen) according to the manufacturer's instructions. RNA extraction from fibroblasts was automatically performed using the QIAsymphony RNA Kit (Qiagen). RNA was quantified with a ND-1000 spectrophotometer (Thermo Fisher Scientific). cDNA was generated starting from 0.15 μg of total RNA using the High cDNA capacity RNA following manufacturer's instructions (Life Technologies) and was amplified by Sanger PCR (primers' sequences available upon request). Isolation of the 374-bp band was performed using the High Pure PCR Product Purification Kit, following the manufacturer's recommendation (Roche Diagnostics, Barcelona, Spain) and PCR products were bidirectionally sequenced using Big Dye Terminator in an ABI3130 automated sequencer (Life Technologies).
Expression analysis by PCR and qPCR
Real-time quantitative PCR was performed using the Power Master Mix PCR SYBR Green on an automated ABI 7300 PCR System by standard curve method for quantification (Applied Biosystems, CA, USA). A pair of primers was designed to amplify only the wild-type transcript of exon 12 and a second pair of primers was designed to amplify both the wild-type and the aberrant mRNA transcript (primers' sequences available upon request). Relative quantification of aberrant transcript of the IQSEC2 gene was determined based on the amplification of the wild-type transcript with respect the total product. Individuals I3 and I4 were considered as controls.
Results
Clinical examination
Patient 1
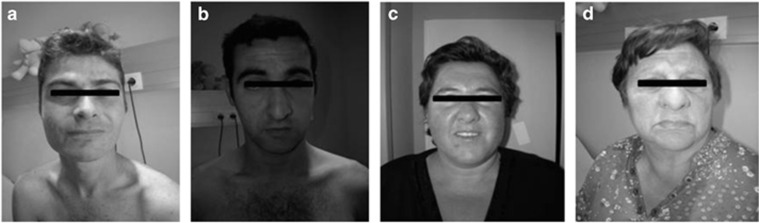
The index patient (Figure 1, II5) was born to unrelated parents and he was diagnosed with severe ID. At the time of evaluation he was 32 years old, OFC was 53.5 cm (3rd percentile; −1.94 SD) with a weight of 57 kg (7th percentile; −1.55SD) and a height of 169.5 cm (11th percentile, −1.24 SD). On physical examination, the patient showed some minor dysmorphic facial features such as prominent superciliary arch, wide and smooth philtrum, and thin upper lip (Figure 2a), multiple lentiginosis on his back and macroorchidism. He also showed poorly marked palmar flexion creases and a hypoplastic nail on the fifth toe. The patient has a severe language- (only few words) and motor-development delay. Although no IQ tests have been performed, a disability of 80% was established by the Elderly and Social System Institute of Spain.
Figure 2.
Images of four of the individuals carrying the mutation in the IQSEC2 gene. All present similar facial features with bulbous nose, smooth wide philtrum and thin upper lip. (a) Patient II5 at the age of 32 years. He presents with severe ID. (b) Patient II6 at the age of 29 years. He presents with moderate ID. (c) Patient II.4 at the age of 39 years. She presents with mild ID. (d) Patient I2 at the age of 62 years. She presented mild ID.
Patient 2
This patient (II6) presents with moderate ID. He is less affected than his brother (II5) and is nowadays working in a supermarket. At time of evaluation, he was 29 years old. OFC was 55.5 cm (26th percentile, −0.67 SD), weight was 67 kg (26th percentile; −0.67 SD) and length was 170 cm (13th percentile, −1.17 SD). As his brother he presented with minor facial features (Figure 2b): findings may include thin upper lip, narrow nasal root, slanted and wide nasal tip, and wide and smooth philtrum. He presents with a frontal angioma (3 × 3 cm). Unlike his brother, he presents normal testes. Upper extremity exam showed short fingers and the lower extremity exam showed a bilateral hypoplastic nail in the second toe. This patient presented discrete motor involvement and had a slight speech delay. A disability of 33% was established by the Elderly and Social System Institute of Spain.
Patient 3
This patient (II4) presented mild ID, obesity and short stature. At the time of examination she was 39 years old. The OFC was 53.5 cm (8th percentile, −1.45 SD), her weight was 113 kg (99th percentile; +6SD) and her height was 151 cm (2nd percentile, −2.2 SD). The facial characteristics included a wide nasal tip, wide and smooth philtrum, and a thin upper lip (Figure 2c). She presented speech delay and discrete motor involvement.
Patient 4
This patient (II2) is the oldest sibling of patients 1, 2 and 3 (II5, II6 and II4) and the mother of patient 6 (III1). She presented mild learning disabilities.
Patient 5
This patient (I2) is the 62-year-old mother patients 1, 2, 3 and 4. At the time of examination she presented with mild ID, obesity (73 kg; 96th percentile) and short stature (147 cm, −2.86 SD). Her facial characteristics included a round face with a narrow nasal root and wide nasal tip, a smooth wide philtrum and thin upper lip (Figure 2d). She also had short fingers and a single palmar crease in the right hand and dysplastic nails in both feet.
Patient 6
This patient (III1) presented severe ID and postnatal microcephaly (OFC: 51 cm; 1st percentile,−3.26 SD). At the time of evaluation he was 15 years old, weighed 41 kg (4th percentile; −1.81 SD) and was 153-cm tall (2nd percentile, −2.30 SD). He had smooth wide philtrum, thin upper lip and high arched palate. Unfortunately, RNA sample from this patient was not available.
Molecular studies
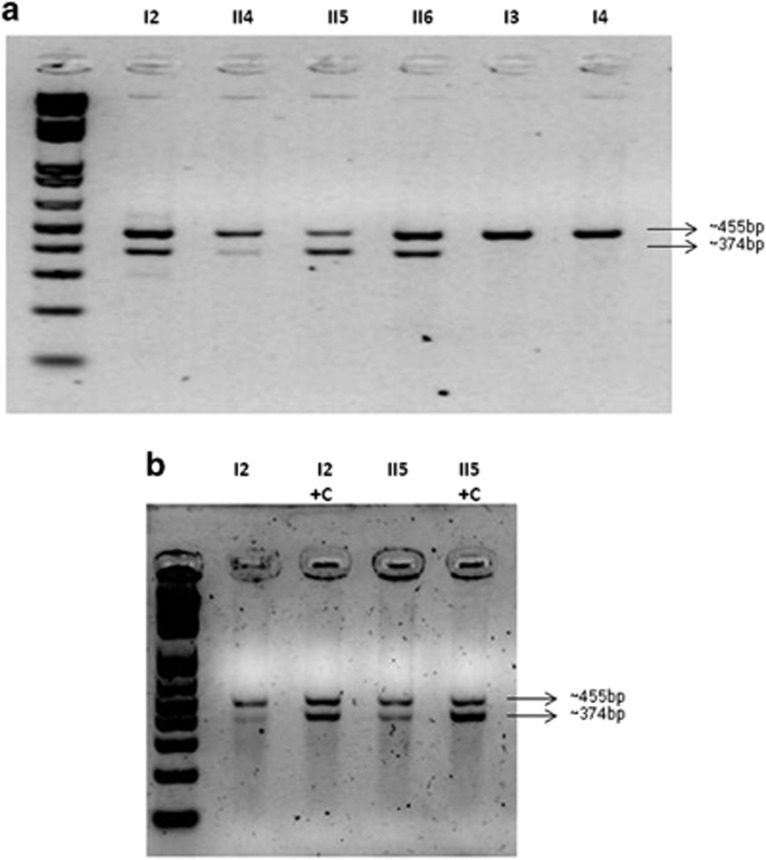
Exome sequencing identified a novel splicing variant (g.88032_88033del) in the IQSEC2 gene (NG_021296.1) in cases I2 and II5. Familial study by direct Sanger sequencing of exon 12 and boundaries confirmed the presence of the variant in four other affected family members (Figure 1). The variant is an intronic deletion of 2 bp. The amplification of IQSEC2 mRNA with primers spanning exons 9–12 showed two separate products in variant carriers: a ~455bp band (wild-type size product) and an extra ~374-bp band (Figure 3a). The same patterns were observed in mRNA from fibroblasts treated with and without cycloheximide (Figure 3b).
Figure 3.
Analysis of IQSEC2 cDNA. The amplified products encompassing exons 10–13 are shown. (a) The first six bands correspond to mRNA IQSEC2 products obtained from lymphocytes. Two bands were visualized in mutation carrier samples, whereas only one band was visualized in samples from healthy relatives. The first column corresponds to 1 kb plus DNA ladder. Note the different intensity patterns of the bands in the patients. (b) Amplification pattern in mRNA from fibroblasts from patients I2 and II5. +C indicates pretreatment with cycloheximide.
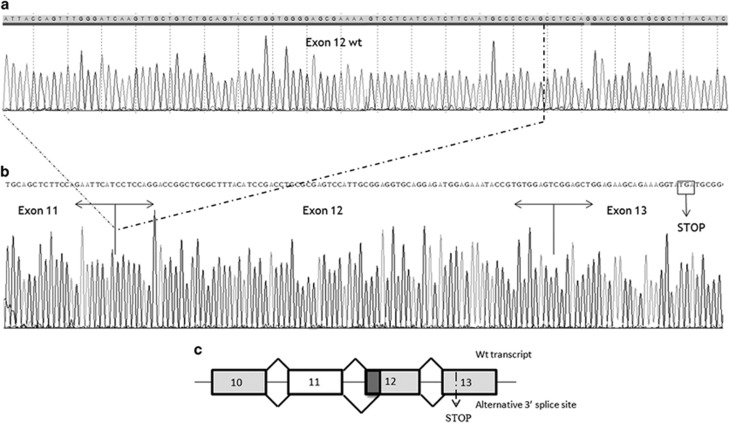
Isolation and sequencing of the 374-bp band showed that the variant activated an exonic splice acceptor site producing a deletion spanning 79 nucleotides in exon 12 (r.3116_3194del). This deletion results in aberrant mRNA transcript product with the abolishment of 26 amino acids from the highly conserved pleckstrin homology (PH) domain in the IQSEC2 gene and the creation of a premature stop codon 36 amino acids later in exon 13 (Figure 4). Quantitative PCR analysis of cDNA showed that the percentage of aberrant transcript and wild-type transcript varied in the different individuals and increased with the clinical severity. In individuals I3 and I4, the observed ratio of wild-type transcript versus total transcript was 0.98 and 1.03, respectively. In patient II5, the most affected, the ratio was ~0.5, whereas in his brother, the ratio was around 0.6. The carrier mother (I2) and sister (II4) presented ratios of 0.82 and 0.90, respectively.
Figure 4.
The top panel (a) shows an IQSEC2 wild-type PCR product in which the normal exon structure from exons 10–13 is maintained. The bottom panel (b) shows the isolated 374-bp PCR product that corresponds to the aberrant spliced form in which 79 nucleotides of exon 12 are deleted. (c) The splicing patterns identified in the patients are represented schematically. Exons and introns are indicated by boxes and horizontal lines, respectively. The diagonal lines indicate the splicing events that were observed in lymphocytes and fibroblasts. The figure is not drawn to scale.
Discussion
A novel splicing variant in intron 11 of the IQSEC2 gene was identified in a family with several members affected with ID. The variant caused two molecular events both in lymphocytes and fibroblasts: (1) it created a new splice acceptor site, producing an aberrantly spliced transcript in a percentage of the cells, and (2) a premature termination codon (PTC) was introduced into the alternate spliced mRNA product. The PTC produces a shorter mRNA with the deletion of the PH domain and the PDZ-binding motif. In most cases, the use of unnatural splice sites introduces PTCs into the mRNA. These PTCs lead either to nonsense-mediated mRNA decay (NDM) or production of a truncated protein lacking a number of functional domains.18 As suggested by the presence of a PDZ-binding motif, the IQSEC2 protein is anchored at PSD via its interaction with PSD-95, which also interacts with NMDA receptors.19, 20 At the PSD, the IQSEC2 protein activates the ARF6 substrate that has been shown to regulate the formation and maintenance of dendritic spines,21 the branching of axons and dendrites,22, 23 exocytosis and endocytosis of synaptic vesicles,24, 25 and receptor internalization.25, 26
Disease-related variants directly modify the DNA sequence or can influence mRNA splicing leading to the lack of the correct gene product or to the formation of truncated proteins. The mechanism of exon recognition involves identification of a complex code of cis-acting elements within genes, located at the 5′ and 3′ ends of exon–intron–exon junctions (known as donor and acceptor splice sites, respectively).27 Pre-mRNA transcripts may be differentially spliced, depending basically on tissue localization and/or stage of development. This different processing allows several transcripts and different proteins to be synthesized from the same gene. The different use of a novel splice acceptor site suggests that trans-elements besides cis-elements regulate the activation of the novel splice acceptor site, that is, nuclear proteins expressed in tissue-specific patterns. We have evaluated the splicing pattern in two tissues with different embryonic origins: fibroblasts and lymphocytes. The splicing pattern observed was the same in both cultures and the use of cycloheximide in fibroblasts did not reveal any other transcripts at low levels. The clinical examination of affected members of the present family revealed that the phenotype differed among variant carriers. The analysis of the aberrant/wild-type transcript ratio in patients 1 and 2, has shown that the most clinically affected patient (patient 1) seems to have less wild-type transcript (50%), whereas the patient less clinically affected (patient 2) has more wild-type transcript (60%). We hypothesize that these differences may be related to the percentage of cells showing an abnormal splicing pattern. The IQSEC2 protein is mainly expressed in nervous system (fetal and adult human brain and frontal cortex), kidney and small intestine. As the target tissue was not available, it is unknown how this variant can affect the structure of the IQSEC2 protein. The most likely option is the degradation of the mRNAs containing PTCs by the NDM pathway (Leeds et al.;28 for a review see Kervestin and Jacobson29). Regarding clinical features in females, these are less severe due to the presence of a normal X-chromosome, which has been previously described.9, 11
To date, around 25 families with variants in IQSEC2, both deletions/duplications and point variants have been described.2, 5, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 Tables 1 and 2 summarize the clinical manifestations of patients with deletions or duplications in the IQSEC2 gene and Table 3 summarizes the clinical manifestations of patients with point variants in this gene. It has been proposed that non-synonymous variants in this gene are responsible for non-syndromic ID, whereas truncating variants may generate a more severe neurodevelopmental phenotype related to the loss of function of IQSEC2.5 In agreement with our hypothesis of a dosage effect of the IQSEC2 protein, Moey et al.34 have recently published a review of cases with submicroscopic copy-number gains on Xp11.22 in which they suggest that IQSEC2 gene is a dosage-sensitive gene contributing to ID and behavioral disturbances.34 These authors speculate that increased dosage of IQSEC2 gene may lead to an inappropriate response to glutamate signaling with similar phenotypic consequences as those observed in patients with duplications involving glutamate receptors.
Table 1. Clinical features of patients with deletions or duplications below 100 kb involving the IQSEC2 gene.
| Present report | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | I2 | II2 | II4 | II5 | II6 | III1 | Olson et al.30 | Gandomi et al.31 | Tran Mau-Them et al.5 | Tran Mau-Them et al.5 | Gilissen et al.32 |
| Mutation | g.88032_88033del (NG_021296.1) | c.273_282del | c.2052_2053del | 22-kb dup including exon 3 | 42-kb dup including exon 3–7 | 62 kb insertion of TENM3 sequence | |||||
| Database | ClinVar SCV000245768 | − | − | ClinVar SCV000147968 | ClinVar SCV000245768 | − | |||||
| Gender | F | F | F | M | M | M | 1 F | M | 1 M | 1 M | NR |
| Microcephaly | − | − | − | − | − | + | − | + | + | + | + |
| Motor development | Delay | No delay | Delay | Delay | Delay | Delay | − | Delay | Delay | Regresion | NR |
| Intellectual disability | Mild | Learning disabilities | Mild | Severe | Moderate | Severe | Mild | Severe | Severe | Severe | Severe |
| Language development | Delay | No delay | Delay | Severe | Delay | Delay | Delay | Not acquired | Not acquired | Not acquired | NR |
| Seizures | − | − | − | − | − | − | − | + | + | + | + |
| Behavioral disturbances | − | − | − | − | − | − | − | + | ASD | ASD | NR |
| Schizophrenia | − | − | − | − | − | − | − | − | − | − | NR |
| Strabismus | − | − | − | − | − | − | − | + | + | + | NR |
| Stereotypies | − | − | − | + | − | − | + | + | + | + | NR |
Abbreviations: ASD, autistic spectrum disorder; del, deletion; dup, duplication; F, female; M, male; NR, not reported; Ref., reference.
Table 2. Clinical features of patients with deletions or duplications above 100 kb involving the IQSEC2 gene.
| Ref. | Fieremans et al.33 | Moey et al.34 | Moey et al.34 | Moey et al.34 | Moey et al.34 | Froyen et al.35 | Froyen et al.35 | Froyen et al.35 Gedeon et al.36 | Froyen et al.37 | Froyen et al.37 | Froyen et al.37 | Froyen et al.37 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | A119 | A057 | A009/MRX17 | ON1 | AU88848 | FTD | F538 | ||
| Mutation | 400-kb del | 361-kb dup | 403-kb.dup | 579-kb dup | 1.141-kb dup | 837-kb dup | 725-kb dup | 762-kb dup | 730-kb dup | 931 kb dup | 1038 kb dup | 1032 kb dup |
| Gender | 1 F | 1 M | 1 M | 1 M | 1 M | 2 M | 2 M | 1 M | 1 M | 2 M | 1 M | 2 M |
| Microcephaly | − | − | − | − | + | − | ± | − | − | NR | − | − |
| Motor development | − | Delay | Delay | Delay | Delay | Delay | No delay | − | Delay | NR | Delay | − |
| Intellectual disability | Severe | NR | Moderate | Moderate | Moderate | Moderate | Moderate | Mild | Moderate | Mild | Mild | Moderate |
| Language development | Not acquired | Delay | No delay | Delay | Delay | Delay | Delay | NA | Delay | NR | Delay | Delay |
| Seizures | − | − | − | − | + | + | − | NA | − | NR | − | − |
| Behavioral disturbances | ASD | + | ASD | + | + | ADHD | + | NA | ADHD | NR | ADHD | − |
| Schizophrenia | − | − | − | − | − | − | − | NA | − | NR | − | − |
| Strabismus | + | − | − | − | − | − | − | NA | − | NR | − | − |
| Stereotypies | − | − | − | − | − | − | − | NA | − | NR | − | − |
Abbreviations: ASD, autistic spectrum disorder; ADHD, attention-deficit hyperactivity disorder; del, deletion; dup, duplication; F, female; M, male; NA, not available; NR, not reported.
Table 3. Clinical features of patients with point mutations in the IQSEC2 gene.
| Ref. | Shoubridge et al.2 | Shoubridge et al.2 | Shoubridge et al.2 | Rauch et al.38 Tran Mau-Them et al.5 | Gandomi et al.31 | Shoubridge et al.2 | Purcell et al.39 | Redin et al.40 | Allen et al.41 |
|---|---|---|---|---|---|---|---|---|---|
| Mutation | c.1075C>T p.R359C | c.2273G>A p.R758Q | c.2402A>C p.Q801P | c.2563C>T p.R855* | c.2582G>C p.S861T | c.2587C>T p.R863W | c.2975T>A p.L992* | c.3097C>T p.Q1033* | c.3322C>T p.Q1108* |
| Database | ClinVar SCV000031844 | ClinVar SCV000031843 | ClinVar SCV000031842 | ClinVar SCV000147969 | − | ClinVar SCV000031841 SCV000232601 | ExAC X:53271006A/T | − | − |
| Gender | 4M | 8M | 8M/1F | 1M | M | 12M | NR | 1M | 1F |
| Microcephaly | NR | NR | ± | + | −a | − | NR | − | NR |
| Motor development | NR | Delay (1/8) | Delay (1/6M) | Delay | Delay | NR | NR | Delay | Delay |
| Intellectual disability | Mild to moderate | Mild | Moderate to severe (8 M) Moderate (1 F) | Severe | Severe | Moderate | + | Severe | + |
| Language development | Delay (3) | Delay (1) | Delay (4/8 M) | Severe | Not acquired | NR | NR | Not acquired | Delay |
| Seizures | 2/4 | − | 2/8 M 1/6 F | − | + | 1/12 | NR | + | LGS |
| Behvioural distrubances | 1/4 | − | 4/8 M 1/6 F | − | ASD | − | NR | + | NR |
| Schizophrenia | − | − | − | − | − | − | + | − | NR |
| Strabismus | NR | NR | 1/8 M | + | + | NR | NR | + | NR |
| Stereotypies | NR | NR | NR | + | + | NR | NR | NR |
Abbreviations: ASD, autism spectrum disorder; F, female; LGS, Lennox–Gastaut syndrome; M, male; NR, not reported. *designates a translation termination codon.
The authors report plagiocephaly and a generalized cerebral and cerebellar volume loss.
To our knowledge, this is the first description of a splicing variant affecting the IQSEC2 gene that produces different grades of affectation in the carrier patients. A similar case has been previously described in Duchenne muscular dystrophy (DMD), in which the authors report a tissue-specific production of semi-functional dystrophin mRNA via activation of a nonsense variant-created intraexonic splice acceptor site.42 In this work, the authors postulate that the modulation of the splicing of muscle dystrophin mRNA in cases of DMD could be considered as a potential target for therapeutic strategies to generate a milder phenotype for this disease.
Variants that affect splicing are related to many genetic diseases and the understanding of its role in disease expands potential opportunities for therapeutic intervention. Therefore, modulation of splicing can become a potent therapeutic approach for many of these diseases.
Acknowledgments
This study was supported as a transnational access project at the European Sequenicng and Genotyping Infrastructure, which has received funding from the Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 262055 and by AGAUR from the Autonomous Catalan Government (2014-SGR603). The ‘CIBER de Enfermedades Raras' is an initiative of the ISCIII.
The authors declare no conflict of interest.
References
- Tarpey PS, Smith R, Pleasance E et al: A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge C, Tarpey PS, Abidi F et al: Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 2010; 42: 486–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE: Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 2007; 8: 1476–1485. [DOI] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F: Sequence motifs for calmodulin recognition. FASEB J 1997; 11: 331–340. [DOI] [PubMed] [Google Scholar]
- TranMau-Them F, Willems M, Albrecht B et al: Expanding the phenotype of IQSEC2 mutations: truncating mutations in severe intellectual disability. Eur J Hum Genet 2014; 22: 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Carrel L: Escape from X-chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci USA 2008; 105: 17055–17060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal I, Alvarez-Mora MI, Karlberg O et al: Efficient application of next-generation sequencing for the diagnosis of rare genetic syndromes. J Clin Pathol 2014; 67: 1099–10103. [DOI] [PubMed] [Google Scholar]
- Database of Single Nucleotide Polymorphisms (dbSNP). National Center for Biotechnology Information, National Library of Medicine. Bethesda, MD, USA. Available at http://www.ncbi.nlm.nih.gov/SNP/.
- Abecasis GR, Auton A, Brooks LD et al: 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S: SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling TN, Yee C, Koh B et al: Cycloheximide facilitates the identification of aberrant transcripts resulting from a novel splice-site mutation in COL17A1 in a patient with generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 1998; 110: 165–169. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA: Pre-mRNA splicing and human disease. Genes Dev 2003; 17: 419–437. [DOI] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL: PDZ domains in synapse assembly and signalling. Trends Cell Biol 2000; 10: 274–280. [DOI] [PubMed] [Google Scholar]
- Harris BZ, Lim WA: Mechanism and role of PDZ domains in signalling complex assembly. J Cell Sci 2001; 114: 3219–3231. [DOI] [PubMed] [Google Scholar]
- Fan JS, Zhang M: Signaling complex organization by PDZ domain proteins. Neurosignals 2002; 11: 315–321. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wang W: Organization of signaling complexes by PDZdomain scaffold proteins. Acc Chem Res 2003; 36: 530–538. [DOI] [PubMed] [Google Scholar]
- Roh MH, Margolis B: Composition and function of PDZ protein complexes during cell polarization. Am J Physiol Renal Physiol 2003; 285: F377–F387. [DOI] [PubMed] [Google Scholar]
- Dosemeci A, Makusky AJ, Jankowska-Stephens E, Yang X, Slotta DJ, Markey SP: Composition of the synaptic PSD-95 complex. Mol Cell Proteomics 2007; 6: 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami H, Sanda M, Fukaya M et al: IQ-ArfGEF/BRAG1 is a guanine nucleotide exchange factor for Arf6 that interacts with PSD-95 at postsynaptic density of excitatory synapses. Neurosci Res 2008; 60: 199–212. [DOI] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR et al: ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci 2006; 26: 4811–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Deviez DJ, Casanova JE, Wilson JM: Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci 2002; 5: 623–624. [DOI] [PubMed] [Google Scholar]
- Hernández-Deviez DJ, Roth MG, Casanova JE, Wilson JM: ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell 2004; 15: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N, Chasserot-Golaz S, Bader MF: Regulated secretion in chromaffin cells: an essential role for ARF6-regulated phospholipase D in the late stages of exocytosis. Ann N Y Acad Sci 2002; 971: 193–200. [DOI] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V: ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol 2003; 162: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houndolo T, Boulay PL, Claing A: G protein-coupled receptor endocytosis in ADP-ribosylation factor 6-depleted cells. J Biol Chem 2005; 280: 5598–5604. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB: Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 2008; 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR: The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 1991; 5: 2303–2314. [DOI] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A: NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 2012; 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson HE, Tambunan D, LaCoursiere C et al: Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A 2015; 167A: 2017–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandomi SK, Farwell Gonzalez KD et al: Diagnostic exome sequencing identifies two novel IQSEC2 mutations associated with X-linked intellectual disability with seizures: implications for genetic counseling and clinical diagnosis. J Genet Couns 2014; 23: 289–298. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Fieremans N, Van Esch H, de Ravel T et al: Microdeletion of the escape genes KDM5C and IQSEC2 in a girl with severe intellectual disability and autistic features. Eur J Med Genet 2015; 58: 324–327. [DOI] [PubMed] [Google Scholar]
- Moey C, Hinze SJ, Brueton L et al: Xp11.2 microduplications including IQSEC2, TSPYL2 and KDM5C genes in patients with neurodevelopmental disorders. Eur J Hum Genet 2015, e-pub ahead of print 10 June 2015 doi:10.1038/ejhg.2015.123. [DOI] [PMC free article] [PubMed]
- Froyen G, Corbett M, Vandewalle J et al: Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet 2008; 82: 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gedeon A, Kerr B, Mulley J, Turner G: Pericentromeric genes for non-specific X-linked mental retardation (MRX). Am J Med Genet 1994; 51: 553–564. [DOI] [PubMed] [Google Scholar]
- Froyen G, Belet S, Martinez F et al: Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet 2012; 91: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M et al: A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Gérard B, Lauer J et al: Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014; 51: 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome ProjectEpi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS et al: De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VK, Takeshima Y, Zhang Z et al: A nonsense mutation-created intraexonic splice site is active in the lymphocytes, but not in the skeletal muscle of a DMD patient. Hum Genet 2007; 120: 737–742. [DOI] [PubMed] [Google Scholar]