Abstract
Genome-wide association studies (GWAS) have long relied on proposed statistical significance thresholds to be able to differentiate true positives from false positives. Although the genome-wide significance P-value threshold of 5 × 10−8 has become a standard for common-variant GWAS, it has not been updated to cope with the lower allele frequency spectrum used in many recent array-based GWAS studies and sequencing studies. Using a whole-genome- and -exome-sequencing data set of 2875 individuals of European ancestry from the Genetics of Type 2 Diabetes (GoT2D) project and a whole-exome-sequencing data set of 13 000 individuals from five ancestries from the GoT2D and T2D-GENES (Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples) projects, we describe guidelines for genome- and exome-wide association P-value thresholds needed to correct for multiple testing, explaining the impact of linkage disequilibrium thresholds for distinguishing independent variants, minor allele frequency and ancestry characteristics. We emphasize the advantage of studying recent genetic isolate populations when performing rare and low-frequency genetic association analyses, as the multiple testing burden is diminished due to higher genetic homogeneity.
Introduction
In genetic association analyses of complex traits, determining the correct P-value threshold for statistical significance is critical to control the number of false-positive associations. Although the genome-wide significance (WGS) P-value threshold of 5 × 10−8 has become a standard for genome-wide association studies (GWAS),1, 2 it has not been updated to account for the lower allele frequency spectrum used in many recent array-based GWAS studies3 and sequencing studies. Different statistical procedures accounting for multiple testing have been used in the genome-wide setting, including the naive Bonferroni correction,4 which can be overly conservative due to the assumption that every genetic variant tested is independent of the rest; false discovery rate procedures,5 permutation based-approaches2 and Bayesian approaches.6
Here, we set out to perform an updated evaluation of the significance threshold for genome-wide genetic association studies designed to discover loci associated with complex traits using a multiple testing approach to control the number of false-positive associations. Guidelines developed in this paper can be useful for researchers using human sequence data (for either direct association testing or as an imputation panel) to evaluate variants in the lower frequency spectrum of their samples. In 2005 the International HapMap Consortium1 used permutation testing of genotypes in 10 densely genotyped Encyclopedia of DNA Elements genomic regions to estimate the number of common independent variants (minor allele frequency (MAF)≥5%) to be 150 per 500 kilobase pairs (kb) in European population. Extrapolating to all the genome (~3.3 Gb) suggested a significance threshold of 5 × 10−8. Since then, this WGS threshold became a standard for reporting genome-wide association significance hits at MAF≥5% for European ancestry populations.2, 3 Moreover, the HapMap variation catalog1, 7 established most of the variation that one could test for association and set a P-value threshold for WGS that was invariant to a study's sample size at MAF≥5%. More recently, whole-exome- and -genome-sequencing projects greatly expanded the number of genetic variants that one could use in association studies. In the 1000 Genomes sequencing project,8 it was observed that ~50% of observed genetic variants were novel, even in the well-characterized Encyclopedia of DNA Elements regions. Sequencing studies lead to an increased number of low-frequency (0.5%<MAF<5%) and rare (MAF<0.5%) variants, arguing for a more stringent statistical threshold for association testing in studies utilizing sequence data.
Materials and methods
For genome-wide (WGS) and exome-wide (WES) significance threshold calculations the Genetics of Type 2 Diabetes (GoT2D) genome-wide integrated SNP panel data freeze v.20120804 and GoT2D.exomes.2760.qc_plus.86_swap_fixed.vcf were used, respectively. The integrated SNP panel contains QC genotypes from low-coverage (4 ×) whole-genome sequencing (4 ×), deep (70 ×)-exome sequencing and 2.5 m SNP genotyping of 2875 samples from four European cohorts: FUSION (Finland), DGI (Sweden and Finland), WTCCC (UK) and KORA (Germany). For the exome ancestry analysis, we sampled exome sequencing from Europeans (Finland and Ashkenazi cohorts), African-Americans (JHS cohort), South Asians (LOLIPOP cohort), East Asians (KARE cohort) and Hispanic (FHS cohort) from the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) consortium. Exome-sequencing target capture was performed with the Agilent SureSelect Human All Exon platform. The function snpgdsLDpruning from the SNPrelate R package9 was used to calculate the number of biallelic tag SNPs using the correlation coefficient (r2) linkage disequilibrium (LD) metric at different LD thresholds (in autosomes plus chromosome X). Tag SNPs are the SNPs selected by the LD pruning algorithm to be kept on the pruned subset. The estimate of the number of independent variants is consistent if the snpgdsLDpruning algorithm is re-run. We repeated the snpgdsLDpruning command 10 times for the whole-genome-sequencing variants on chromosome 21, and we found that the SD of the number of independent variants at each LD threshold was always <0.1% of the mean number of independent variants. The vcftools package10 was used to subset the data at different MAFs. The P-value needed to reach genome- and exome-wide significance at different MAFs and LD thresholds was calculated as 0.05/number of tag SNPs. SNPs below a defined LD threshold are considered independent. For simplicity, the comparison between different WGS ancestry groups (UK, Sweden and Finland) and LD reliability based on sample size were done only on chromosome 21 at MAF≥5%, MAF≥1% and MAF≥0.5%. The results were then extrapolated to all genome based on the genome and chromosome 21 sizes taken from Ensembl browser.11 Only 512 samples taken at random from each population were chosen for the WGS ancestry comparison as this was the minimum number of samples for one of our ancestry groups (Sweden). The same was applied for the WES ancestry comparison (minimum number of samples=861). We also used the D′ LD metric, implemented in the snpgdsLDpruning algorithm, to calculate the number of biallelic tag SNPs at different LD thresholds. D′ measures the evolutionary genealogy of a pair of variants – and is influenced by the amount of recombination that has occurred between the two loci as the appearance of the more recent variant. When used with the binning approach described in the paper, D′ would create bins of SNPs that do not tag the same association signal – as the power of LD mapping (observing an association in a non-causal SNP that is linked to the causal SNP) is a function of r2 and not D′. Furthermore, D′ has the disadvantage that the estimate can be biased upward if allele frequencies between loci are very different, the sample size is small, or the frequency of the variants are low.
Results
For this analysis, we used a WGS and WES data set of 2875 individuals of European ancestry from the GoT2D and a whole-exome-sequencing data set of 13 000 individuals from five ancestries from the T2D-GENES projects (Flannick et al, Teslovich et al, submitted). For each scenario of data type, MAF, LD threshold and ancestry, we estimate the number of independent genetic variants and calculate a statistical significance threshold to maintain a family-wise type I error rate of 5%: 0.05/number of independent variants (Materials and methods).
We found that the genome- and exome-wide association significance P-value thresholds needed for association testing depend upon the LD cut-off chosen for defining independence between variants, MAF and ancestry (Figure 1). As expected, for both genome- and exome-wide significance thresholds in the GoT2D data set, as lower LD is considered, more variants are considered dependent, relaxing the required P-value significance threshold (Materials and methods). In addition, as the minimum MAF of the variants included in a study decreases, more stringent significant thresholds are needed due to the increasing number of variants and the lower LD between less frequent variants (Figure 1; Supplementary Table S1; Supplementary Table S2).
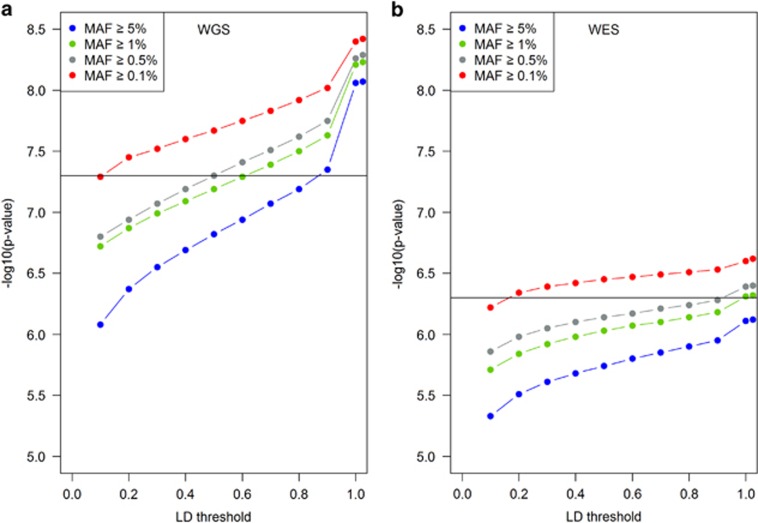
Figure 1.
Impact of linkage disequilibrium pruning on P-value significance threshold for whole-genome and -exome association studies of European ancestries (N=2875). Variants with an LD between each other bigger or equal than a certain threshold are pruned, so only randomly chosen tagging SNPs are used for the multiple testing P-value threshold calculation. (a) GoT2D WGS integrated SNP panel with horizontal line showing the commonly used 5 × 10−8 genome-wide significance P-value threshold. (b) WES SNP panel with horizontal line showing the commonly used 5 × 10−7 exome-wide significance Bonferroni corrected P-value threshold for MAF≥0.5%. The plotted P-values were calculated as 0.05/number of tag SNPs at each described MAF and LD pruning thresholds (Materials and methods; Supplementary Table S1; Supplementary Table S2). The data points with LD>1 refer to no LD pruning.
Interestingly, the widely used genome-wide P-value threshold of 5 × 10−8 is valid for common variants (MAF≥5%) only if a LD r2<0.8 is applied (see Supplementary Table S1 for other thresholds). Under a model using this LD threshold for tagging SNPs, then we would have a P-value threshold of 3 × 10−8, 2 × 10−8 and 1 × 10−8, for analyses of variants with MAF≥1%, MAF≥0.5% and MAF≥0.1%, respectively (Figure 1a; Supplementary Table S1). For exome-wide significance (also at LD r2<0.8), we would have a P-value threshold of 1 × 10−6 at MAF≥5%, 7 × 10−7 at MAF≥1%, 5 × 10−7 at MAF≥0.5% and 3 × 10−7 at MAF≥0.1% (Figure 1b; Supplementary Table S2), roughly consistent with the WES threshold commonly used that is based on a Bonferroni correction for 100 000 variants with MAF≥0.5% (P-value=5 × 10−7).
Importantly, if no LD threshold is applied, that is, including all variants even if they are in perfect LD (LD r2=1), this naive Bonferroni correction will lead to unnecessary testing. For instance at MAF≥0.1%, you end up testing 833 000 variants that are in perfect LD (Figure 1a; Supplementary Table S1). Of note, 92% of biallelic SNPs from our GoT2D exome-sequencing data set of European ancestry are also captured by the general population (ExAC database—http://exac.broadinstitute.org/).
We also examined the significance P-value threshold for various sample sizes to determine its effect at different allele frequencies. We observed clear evidence that for studies that include variants with MAF≥0.5%, the statistical significance P-value threshold calculated in a European sample of N=2875 is reliable for smaller studies of the same ancestry when N>500 (Supplementary Figure S1; Supplementary Table S4). When we evaluated the D′, we observed that the number of tag SNPs was much lower and less dependent on allele frequency than the using r2 as the LD measure (Supplementary Figure S2).
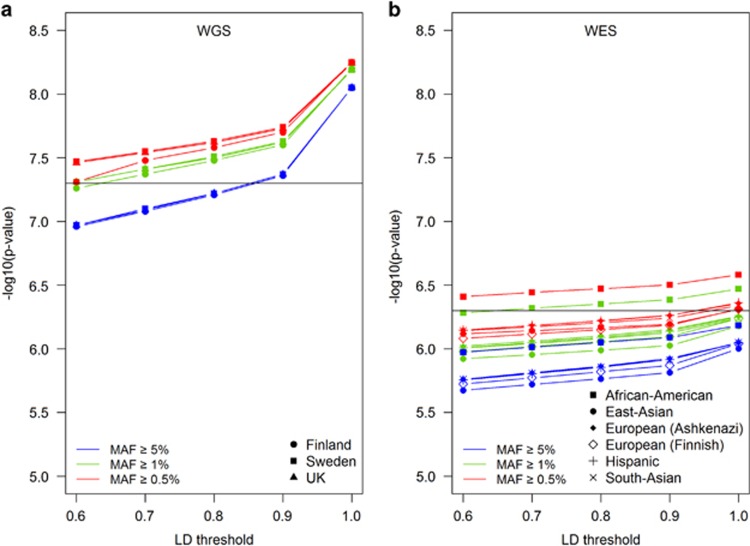
As the GoT2D whole-genome data set included >500 individuals from each of three ancestries: UK (n=660), Sweden (n=512) and Finland (n=1442) and the T2D-GENES exome-sequencing data set included >861 individuals from each of five diverse ancestries (South Asian, East Asian, European, Hispanic and African-American), we questioned if the statistical significance threshold controlling the false-positive rate for low-MAF variants changes with ancestry characteristics. We hypothesized that due to the Finnish population history shaped by relative few founders and recent rapid expansion,12, 13 we would have an advantage when performing rare/low-frequency variant analysis in this population in comparison with the UK and Swedish populations. In fact, for the Finnish population there are a lower number of independent variants among low-frequency variants, requiring a less stringent correction for statistical association testing and increased power (Figure 2a; Supplementary Table S3). For instance, at a LD threshold of r2<0.8 and MAF ≥0.5%, we would have a WGS significance P-value threshold of 2.6 × 10−8 in Finns, whereas for the Swedish and British ancestries it would be 2.3 × 10−8, which would require testing >200 000 extra variants in the latter two populations (Supplementary Table S3). Likewise, when considering the ancestries represented in the T2D-GENES whole-exome-sequence data set, we observe an increased testing burden for ancestry groups with a greater genetic diversity, in particular the African-Americans (Figure 2b; Supplementary Figure S3). This emphasizes the advantage of performing rare/low-frequency variant association studies in isolated populations with a relative lower effective population size, as the length of shared haplotypes is greater with lower allele frequency, in line with what has been previously reported.7
Figure 2.
P-value needed to reach WGS at different MAF and LD thresholds for a whole-genome analysis of Finnish, Swedish and British populations (a) and whole-exome-sequencing analysis of diverse ancestries (b) using the same sample sizes across ancestries (N=512 for WGS, N=861 for WES). The horizontal lines show the commonly used significance P-value threshold for WGS (5 × 10−8) and WES (5 × 10−7). Supplementary Figure S3 presents the multiple ancestry data divided into separate plots.
Discussion
Taken together, this study provides guidelines for genome- and exome-wide association P-value thresholds needed to correct for multiple testing, explaining the impact of LD thresholds for distinguishing independent variants, MAF and ancestry characteristics.
We confirm the 5 × 10−8 P-value threshold for WGS to be valid for common (MAF>5%) genetic variation in the European population. However, for lower frequency variants, the genome-wide P-value threshold needs to be more stringent for studies with European ancestry (3 × 10−8 for MAF≥1%, 2 × 10−8 for MAF≥0.5% and 1 × 10−8 for MAF≥0.1% at LD r2<0.8). For exome-sequencing studies, exome-wide-significant thresholds should also be agreed and adopted by the scientific community; for studies with European ancestry, P-value threshold of 1 × 10−6, 7 × 10−7, 5 × 10−7 and 3 × 10−7, for MAF≥5%, MAF≥1%, MAF≥0.5% and MAF≥0.1%, respectively, are reasonable. Studies of other ancestry groups should consider the degree of genetic variation when considering the appropriate statistical significance threshold.
We also demonstrate the advantage of studying isolated young populations with a relative lower effective population size, for analysis of rare variants, since their lower genetic diversity translates into fewer independent rare variants and therefore, less multiple testing burden and consequent increased power in rare variant analysis.
We acknowledge that the frequentist approach of using P-value thresholds as a measure of statistical evidence has important limitations, as it does not take into account the power of the tests, as it is a threshold suggested for all sample sizes and allele frequencies.14, 15 Although in a Bayesian setting, one can incorporate these parameters as prior odds of belief, it needs prior distributions to be defined for model parameters, involving intensive computation to incorporate likelihoods over the defined parameter space. By doing so, if different studies adopt different priors, comparability of findings between studies remain problematic. Nevertheless, we believe that the Bayesian approach has its most value for region fine mapping to identify the true causal variant(s).16
Acknowledgments
We gratefully acknowledge the members of T2D-GENES and GoT2D consortia for sharing prepublication data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- International HapMap Consortium: A haplotype map of the human genome. Nature 2005; 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ: Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008; 32: 381–385. [DOI] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J et al: The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014; 42: D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE: Il calcolo delle assicurazioni su gruppi di teste. In Studi in Onore del Professore Salvatore Ortu Carboni. Bardi: Rome, Italy, 1935, pp 13–60. [Google Scholar]
- Storey JD, Tibshirani R: Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium: Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature 2007; 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium, Frazer KA, Ballinger DG et al: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis GR, Auton A et al: An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS: A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012; 28: 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G et al: The variant call format and VCFtools. Bioinformatics 2011; 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D et al: Ensembl 2014. Nucleic Acids Res 2014; 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T: Molecular genetics of the Finnish disease heritage. Hum Mol Genet 1999; 8: 1913–1923. [DOI] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M et al: Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 2006; 38: 556–560. [DOI] [PubMed] [Google Scholar]
- Wacholder S, Chanock S, Garcia-Closas M et al: Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 2004; 96: 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell SM: Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet 2014; 15: 335–346. [DOI] [PubMed] [Google Scholar]
- Stephens M, Balding DJ: Bayesian statistical methods for genetic association studies. Nat Rev Genet 2009; 10: 681–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.