1. Disease characteristics
1.1 Name of the disease (synonyms)
Familial platelet disorder with associated myeloid malignancies, FPDMM.
Alternative names
Familial platelet syndrome with associated myeloid malignancies.
Aspirin-like platelet disorder.
Familial thrombocytopenia with propensity to acute myeloid leukaemia.
FPD/AML syndrome.
FPS/AML syndrome.
1.2 OMIM# of the disease
601399.
1.3 Name of the analysed genes or DNA/chromosome segments
RUNX1, RUNT-related transcription factor 1.
Alternative titles; symbols
AML1, acute myeloid leukaemia 1 gene.
CBFA2, core-binding factor, RUNT-domain, alpha subunit 2.
PEBP2AB, PEBP2-Alpha-B.
1.4 OMIM# of the gene(s)
151385.
1.5 Mutational spectrum
Point mutations, intragenic deletions, and large deletions affecting 21q22.
1.6 Analytical methods
Sequencing; array-based comparative genomic hybridisation (aCGH), if available custom-made arrays with high-resolution of RUNX1 including non-coding regions; multiplex ligation-dependent probe amplification (MLPA); in case of large genomic aberrations, cytogenetic investigations and fluorescence in situ hybridisation (FISH) using RUNX1-specific probes.
1.7 Analytical validation
Confirmation of identified sequence variants in an independent biological sample of the index case or an affected relative. In case of intragenic deletions, an independent technique or probes should be used for confirmation purposes, for example, breakpoint spanning long-distance PCR; MLPA following aCGH or vice versa.
Due to known somatic sequence variants acquired during leukaemogenesis, DNA of non-haematopoietic origin needs to be investigated to prove the constitutional origin of sequence variants in leukaemia patients.
1.8 Estimated frequency of the disease (incidence at birth (‘birth prevalence') or population prevalence. If known to be variable between ethnic groups, please report)
<1 in 1 000 000 (approximate), there are no available data on the incidence of FPDMM.
1.9 Diagnostic setting

Comment: Familial platelet disorder with associated myeloid malignancies (FPDMM) is a rare autosomal dominant inherited disease exhibiting incomplete penetrance and variable intra and interfamilial expressivity. Patients typically present with mild–moderate thrombocytopenia which often coincides with a bleeding history due to impaired platelet aggregation. However, some patients have asymptomatic thrombocytopenia. Overt leukaemia, which is mainly of myeloid origin, can arise from early childhood to late adulthood. Clinically, some families present with familial leukaemia rather than familial platelet disorder. However, there are obligate adult carriers displaying neither thrombocytopenia nor leukaemia.1,2
Heterozygous constitutional disease-causing genetic variants of RUNX1 or larger deletions affecting its locus in the long arm of chromosome 21 are the only known causes of FPDMM. Point mutations are mainly located in the N-terminal conserved Runt homology domain (RHD), and lead to haploinsufficiency and/or dominant negative effects. Leukaemogenesis requiring secondary genetic aberrations co-occurs with somatic variants affecting the second RUNX1 allele.3, 4, 5, 6, 7, 8, 9
FPDMM can be part of a contiguous gene syndrome with thrombocytopenia, dysmorphisms, mental retardation, and increased risk of leukaemia due to genomic deletions affecting large proportions of the long arm of chromosome 21.10, 11
There is no publically available RUNX1-specific database of known pathogenic variants. However, FPDMM-associated variants previously reported are, for example, listed in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php, 17 November 2015) and the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/, 17 November 2015).
2. Test characteristics
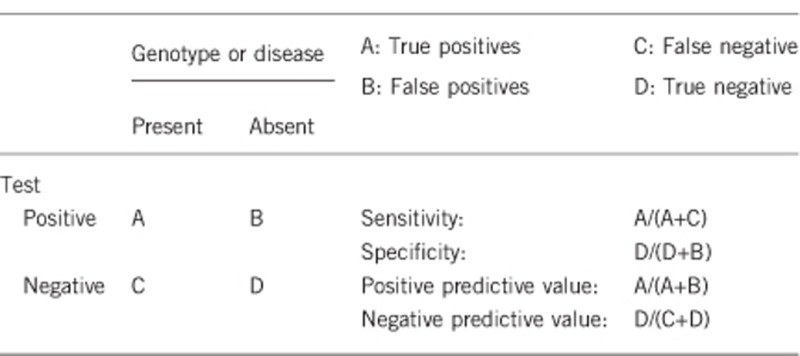
2.1 Analytical sensitivity
(proportion of positive tests if the genotype is present)
If sequencing is complemented by investigations allowing the detection of large deletions, examples by custom-made CGH arrays or MLPA, analytical sensitivity will be above 95%. Causative genetic variants in non-coding regions, for example, in introns or regulatory elements in the non-transcriped region, which are not covered by sequencing, will be missed. In addition, causative genetic variants might be missed due to allelic dropout caused, for example, by single nucleotide polymorphisms or variants within primer binding sites.
2.2 Analytical specificity
(proportion of negative tests if the genotype is not present)
In the absence of genetic variants and by performing appropriate analytical validation, the probability of negative test results is close to 100%.
2.3 Clinical sensitivity
(proportion of positive tests if the disease is present)
The clinical sensitivity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if a quantification can only be made case by case.
Due to the broad clinical spectrum that includes clinically healthy carriers, patients with bleeding histories, and individuals with early onset familial leukaemia, the clinical sensitivity is variable and depends on the past medical history of the index case, the expressivity of his/her disease and the family history.
2.4 Clinical specificity
(proportion of negative tests if the disease is not present)
The clinical specificity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if a quantification can only be made case by case.
Due to the incomplete penetrance and the chance to detect variants of unknown significance (VUS), one can expect a clinical sensitivity above 95%. Detection of VUS may hamper final conclusions.
Please note that in the context of familial leukaemia, constitutive disease-causing genetic variants in other genes need to be considered, for example, CEBPA, GATA2, DDX41 and ETV6. In families with thrombocytopenia and leukaemia, ETV6, which is also associated with thrombocytopenia (OMIM #616216), might be of special interest.
2.5 Positive clinical predictive value
(lifetime risk to develop the disease if the test is positive)
Data allowing an estimation of the penetrance of platelet disorder are not available. However, the lifetime risk of developing leukaemia is estimated to be 30–40%.
2.6 Negative clinical predictive value
(probability not to develop the disease if the test is negative)
Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
If the index case of the family carries a disease-causing RUNX1 gene variant and predictive testing including appropriate analytical validation was negative in a relative, the likelihood of FPDMM is close to 0%. Regarding known allelic heterogeneity and potential locus heterogeneity in FPDMM, there remains a theoretical chance to develop the disease due to de novo mutations, the presence of more than one causative genetic variant of RUNX1 in a family or the presence of causative genetic variants in other genes associated with platelet disorders or familial leukaemia.
Index case in that family had not been tested:
If no causative genetic variant is known in the index case, it is not recommended to test non-affected relatives.
3. Clinical utility
3.1 (Differential) diagnostics: The tested person is clinically affected
(To be answered if in 1.9 ‘A' was marked)
3.1.1 Can a diagnosis be made other than through a genetic test?

3.1.2 Describe the burden of alternative diagnostic methods to the patient
Not applicable.
3.1.3 How is the cost effectiveness of alternative diagnostic methods to be judged?
Not applicable.
3.1.4 Will disease management be influenced by the result of a genetic test?

3.2 Predictive Setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.9 ‘B' was marked)
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe):
Yes. Due to the present risk of developing thrombocytopenia and/or leukaemia, biannual/annual clinical evaluation including a differential blood cell count should be offered. Responsible physicians should be informed to allow prompt investigations in case of suspicious symptoms. However, there is no general consensus statement on preventive measures and the necessity of routine bone marrow investigations for surveillance. In families with frequent and childhood onset of leukaemia, regular bone marrow aspirates can be discussed. In future, it might be worth evaluating whether deep or targeted-sequencing approaches can be used to detect evolving leukaemic clones8, 9, 14 and warrant haematopoietic stem cell transplantation prior to overt leukaemia.
Regarding the autosomal dominant inheritance, lifestyle and family planning might be affected by positive test results. Predictive genetic testing can only be offered after genetic counselling. Since there is no general consensus on the surveillance of individuals at risk, predictive testing of minors is debatable, but is obligate, if minors are potential-matched stem cell donors for affected relatives.
If the test result is negative (please describe):
Yes. Relief of the familial risk for the counsellee and his/her offspring. There is no need for routine investigations.
3.2.2 Which options in view of lifestyle and prevention does a person at risk have if no genetic test has been done (please describe)?
Even though there is no general statement on appropriate surveillance, annual clinical examinations can be offered to relatives at risk and responsible physicians should be informed about their risk of developing thrombocytopenia/thrombocyte malfunction and leukaemia.15
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.9 ‘C' was marked)
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
Yes. If a causative genetic variant is identified in an index patient, FPDMM can be diagnosed, and predictive testing following genetic counselling can be offered to all family members.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
Yes. If no causative genetic variant is identified, it is not worth screening unaffected family members for disease-causing RUNX1 variants. If a causative genetic variant is identified, by direct testing of the identified familial gene variant, surveillance can be restricted to relatives carrying the causative gene variant; regarding stem cell transplantation, healthy relatives carrying the familial mutation need no evaluation as potential stem cell donors.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
Yes. Predictive genetic analyses can be offered following genetic counselling. However, regarding predictive testing of minors, the legal framework may vary from nation to nation and it should be considered that there is no expert statement on recommended surveillance measures.
3.4 Prenatal diagnosis
(To be answered if in 1.9 ‘D' was marked)
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnosis?
Technically, prenatal or preimplantational genetic analyses are feasible, if the causative genetic alteration is known. However, the legal framework needs to be considered and may differ in various countries.
4. If applicable, further consequences of testing
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or his/her relatives? (Please describe)
Haematopoietic stem cell transplants using cells of HLA-matched relatives carrying the familial gene variant need to be avoided.2 In this respect, predictive testing of healthy relatives, even of minors, is indicated, although the test result might not be of immediate medical consequence, since surveillance of relatives at risk will not be recommended by all physicians.
Acknowledgments
This work was supported by EuroGentest2 (Unit 2: ‘Genetic testing as part of health care'), a Coordination Action under FP7 (Grant Agreement Number 261469), the European Society of Human Genetics, and by Bloodwise Programme Award UK, 14032.
The authors declare no conflict of interest.
References
- Song WJ, Sullivan MG, Legare RD et al: Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 1999; 23: 166–175. [DOI] [PubMed] [Google Scholar]
- Owen CJ, Toze CL, Koochin A et al: Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 2008; 112: 4639–4645. [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Renneville A, Bourdon V et al: High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood 2009; 113: 5583–5587. [DOI] [PubMed] [Google Scholar]
- Ripperger T, Steinemann D, Göhring G et al: A novel pedigree with heterozygous germline RUNX1 mutation causing familial MDS-related AML: can these families serve as a multistep model for leukemic transformation? Leukemia 2009; 23: 1364–1366. [DOI] [PubMed] [Google Scholar]
- Béri-Dexheimer M, Latger-Cannard V, Philippe C et al: Clinical phenotype of germline RUNX1 haploinsufficiency: from point mutations to large genomic deletions. Eur J Hum Genet 2008; 16: 1014–1018. [DOI] [PubMed] [Google Scholar]
- Michaud J, Wu F, Osato M et al: In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood 2002; 99: 1364–1372. [DOI] [PubMed] [Google Scholar]
- Ripperger T, Tauscher M, Ehlert K et al: Childhood onset of leukaemia in familial platelet disorder with propensity for myeloid malignancies due to an intragenic RUNX1 deletion. Haematologica 2012; 97 (s3): S14. [Google Scholar]
- Antony-Debré I, Duployez N, Bucci M et al: Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia 2015, e-pub ahead of print 28 August 2015;doi:10.1038/leu.2015.236. [DOI] [PubMed]
- Yoshimi A, Toya T, Kawazu M et al: Recurrent CDC25C mutations drive malignant transformation in FPD/AML. Nat Commun 2014; 27: 4770. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Erez A, Shardy DL et al: Syndromic thrombocytopenia and predisposition to acute myelogenous leukemia caused by constitutional microdeletions on chromosome 21q. Blood 2008; 112: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T, Tauscher M, Thomay K et al: No evidence for ITSN1 loss in a patient with mental retardation and complex chromosomal rearrangements of 21q21-21q22. Leuk Res 2013; 37: 721–723. [DOI] [PubMed] [Google Scholar]
- Iizuka H, Kagoya Y, Kataoka K et al: Targeted gene correction of RUNX1 in induced pluripotent stem cells derived from familial platelet disorder with propensity to myeloid malignancy restores normal megakaryopoiesis. Exp Hematol 2015; 43: 849–857. [DOI] [PubMed] [Google Scholar]
- Connelly JP, Kwon EM, Gao Y et al: Targeted correction of RUNX1 mutation in FPD patient-specific induced pluripotent stem cells rescues megakaryopoietic defects. Blood 2014; 124: 1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Steinemann D, Katsman-Kuipers JE et al: Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood 2014; 123: 2229–2237. [DOI] [PubMed] [Google Scholar]
- Ripperger T, Tauscher M, Haase D, Griesinger F, Schlegelberger B, Steinemann D: Managing individuals with propensity to myeloid malignancies due to germline RUNX1 deficiency. Haematologica 2011; 96: 1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]


