Abstract
Recently WAC was reported as a candidate gene for intellectual disability (ID) based on the identification of a de novo mutation in an individual with severe ID. WAC regulates transcription-coupled histone H2B ubiquitination and has previously been implicated in the 10p12p11 contiguous gene deletion syndrome. In this study, we report on 10 individuals with de novo WAC mutations which we identified through routine (diagnostic) exome sequencing and targeted resequencing of WAC in 2326 individuals with unexplained ID. All but one mutation was expected to lead to a loss-of-function of WAC. Clinical evaluation of all individuals revealed phenotypic overlap for mild ID, hypotonia, behavioral problems and distinctive facial dysmorphisms, including a square-shaped face, deep set eyes, long palpebral fissures, and a broad mouth and chin. These clinical features were also previously reported in individuals with 10p12p11 microdeletion syndrome. To investigate the role of WAC in ID, we studied the importance of the Drosophila WAC orthologue (CG8949) in habituation, a non-associative learning paradigm. Neuronal knockdown of Drosophila CG8949 resulted in impaired learning, suggesting that WAC is required in neurons for normal cognitive performance. In conclusion, we defined a clinically recognizable ID syndrome, caused by de novo loss-of-function mutations in WAC. Independent functional evidence in Drosophila further supported the role of WAC in ID. On the basis of our data WAC can be added to the list of ID genes with a role in transcription regulation through histone modification.
Introduction
Intellectual disability (ID) is a heterogeneous disorder, both clinically and genetically. To date, >650 genes have been associated with ID and novel genes are still being identified. The introduction of trio-based whole-exome sequencing (WES) in individuals with ID has proven to be a valuable approach for the identification of novel ID genes, especially for those individuals who do not show a clinical recognizable syndrome.1, 2 In addition to the identification of mutations in known disease genes, WES has facilitated the identification of candidate ID genes. To establish the pathogenicity of mutations in such candidate ID genes, it is essential to identify additional individuals with an overlapping phenotype and a mutation in the same gene.3, 4, 5 With increasing availability of WES in routine diagnostics6 as well as technological advances facilitating targeted resequencing of candidate ID genes in larger cohorts of samples,7 chances of finding such additional individuals are increasing. Furthermore, supporting evidence and insights into underlying mechanisms can be obtained from functional studies in cell or animal models.8, 9
Previously, we and others separately reported on an individual with a de novo mutation in the 'WW domain-containing adapter with coiled-coil' (WAC) gene using trio-based exome sequencing.1, 10 The mutations were reported as potential cause of disease, based on mutation severity, protein function, its expression in fetal stages and high expression in adult brain.1, 10, 11 WAC encodes a protein-regulating transcription-coupled histone H2B ubiquitination and contains two evolutionary conserved domains, including an N-terminal WW domain interacting with RNA polymerase II and a C-terminal coiled-coil domain promoting the RNF20/RNF40's E3 ligase activity for ubiquitination at active transcription sites.12, 13 Furthermore, the RNF20/40/WAC complex may have a role in cell cycle checkpoint activation upon genotoxic stress.12
In addition, WAC has previously been implicated in ID based on the finding that deletions of chromosome 10p12p11 result in a contiguous gene deletion syndrome, for which the shortest deleted region contains two genes, WAC and BAMBI. All individuals with a deletion of at least these two genes were reported to have a similar phenotype including ID, behavior problems and dysmorphic features, supporting a disease cause of WAC heterozygous loss-of-function.14, 15, 16, 17
Although it may well be hypothesized that WAC haploinsuffiency may explain the ID phenotype observed in the 10p12p11 contiguous gene syndrome, and ID in individuals with mutations in this gene, detailed evidence to support this hypothesis is lacking. In the present study we aimed to identify additional individuals with de novo mutations in WAC by using different sequencing strategies to define the clinical spectrum associated with WAC haploinsufficiency. Finally, to address the role of WAC in cognition, we investigated the role of the Drosophila WAC orthologue in habituation, a form of non-associative learning.
Materials and methods
Diagnostic exome sequencing
Individuals 1, 3, 4, 5 and 6 were ascertained through family-based WES in a diagnostic setting using techniques as described before.1 All clinically relevant candidate de novo mutations were validated using Sanger sequencing, and subsequently tested for absence in parental DNA samples. Individual 1 was previously reported as part of a large study assessing the clinical utility of WES, in which she was also identified to have a second de novo mutation in MIB1.1 Individuals 7 and 8 were identified in a large multicenter study to establish the contribution of de novo coding mutations to autism spectrum disorder.18, 19 Apart from the de novo mutation in WAC these two patients were not reported to have additional de novo mutations.19
Database searches for copy number variations disrupting WAC
We systematically searched for individuals with small deletions including WAC in our in-house database and international databases such as the database of the European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA) and the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER).20, 21
Targeted resequencing of WAC
Upon identification of the de novo mutation in Individual 1, targeted resequencing was performed on a cohort of 2326 patients with unexplained ID using molecular inversion probes (MIPs) as described previously.7 This cohort was selected from the in-house collection of the Department of Human Genetics of Radboud University Medical Center (Nijmegen, The Netherlands) containing patients with unexplained ID. Candidate loss-of-function mutations as well as highly conserved missense mutations (PhyloP>5) were validated by standard Sanger sequencing approaches on DNA extracted from peripheral blood. For assessing the de novo occurrence of validated mutations, DNA from the parents was tested. This study was approved by the institutional review board ‘Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen'.
Deposition of genotypes and phenotypes in a locus-specific database
All mutations and phenotypes reported in this publication are deposited in the locus-specific database for WAC, under the realm of the Leiden Open Variation Database (LOVD; http://databases.lovd.nl/shared/genes/WAC). Variant information and phenotypes can be retrieved using the following submission entries: Individual 1: #00054835; Individual 2: #00054836; Individual 3: #00054837; Individual 4: 00054838; Individual 5: #00054839; Individual 6: #00054848; Individual 7: #00054849; Individual 8: #00054850; Individual 9: #00054851; and Individual 10: #00054852.
Drosophila lines and maintenance
Fly stocks were kept on standard Drosophila diet (cornmeal/sugar/yeast) at 25 °C and 45–60% humidity at 12 h:12 h light/dark cycle. Inducible RNAi lines targeting the Drosophila WAC orthologue CG8949 (vdrc48307, vdrc107328) and corresponding genetic background control lines (vdrc60000, vdrc60100) were obtained from the Vienna Drosophila RNAi Center.22 The s19 value, which refers to the specificity of the dsRNA hairpin construct,23 is 1.00 for vdrc48307 with no off-target and 0.99 for vdrc107328 (two possible off-targets, CG11122 and CG11354).
RNAi was induced using the UAS-Gal4 system and the panneuronal driver lines: elav-Gal4 w1118; 2xGMR-wIR; elav-Gal4, UAS-Dicer-2 or nSyb-Gal4 w1118, UAS-Dicer-2; nSyb-Gal4..9, 24 Flies were reared and tested at 25 °C (elav-Gal4) and 28 °C (nSyb-Gal4) and 70% humidity. The ubiquitous actin-Gal4 driver w1118; P(w[+mC]=Act5c-Gal4)/CyO obtained from Bloomington Drosophila Stock Center,25 was used to generate RNAi-mediated knockdown for quantitative PCR (qPCR).
Analysis of CG8949 mRNA levels from larval brains by qPCR
RNA isolations (three biological replicates) from third instar larvae brains were performed using Arcturus Picopure RNA Isolation Kit (Life Technologies, Bleiswijk, The Netherlands). RNA was treated with DNase (DNAfree Kit, Ambion, Bleiswijk, The Netherlands). First-strand cDNA synthesis was performed using iScript cDNA Synthesis Kit (Biorad, Veenendaal, The Netherlands). Gene expression was analysed by real-time PCR (7900HT Fast Real-Time PCR system, Applied Biosystems, Bleiswijk, The Netherlands). PCR reactions were performed in a volume of 25 μl containing 150 nM primers and GoTag Green Mastermix (Promega, Leiden, The Netherlands). Primer sequences used for amplification of CG8949: 5′-TGGAATTACGACAACGATGG-3′ and 5′-TAACTGGCTTCCGAGGTAGG-3′. BetaCop was used as reference gene, primer sequences: 5′-AACTACAACACCCTGGAGAAGG-3′, 5′-ACATCTTCTCCCAATTCCAAAG-3′.
Light-off jump reflex habituation
The light-off jump reflex habituation assay was performed as previously described.26 Habituation of the startle jump response towards repeated light-off stimuli of 3–7-day-old individual male flies was tested in two independent 16-unit light-off jump habituation systems. A total of 32 flies (16 flies/system) were simultaneously exposed to series of 100 short (15 ms) light-off pulses with a 1 s inter-pulse interval. The noise amplitude of wing vibrations following every jump response was recorded for 500 ms after the start of light-off pulse and an appropriate threshold was applied to filter out the background noise. Data were collected and analysed by a custom-made Labview Software (National Instruments). High initial jumping responses to light-off pulse decreased with the growing number of trials and flies were considered habituated when they failed to jump in five consecutive trials (non-jump criterion). Habituation was scored as the mean number of trials required to reach the non-jump criterion (trials to criterion, TTC). Main effects of genotype (mutant vs control), day and test system on log-transformed TTC values were tested using linear model regression analysis (lm) in R statistical software (R version 3.0.0 (2013-04-03)).27
Results
Identification of individuals with de novo CNVs affecting WAC
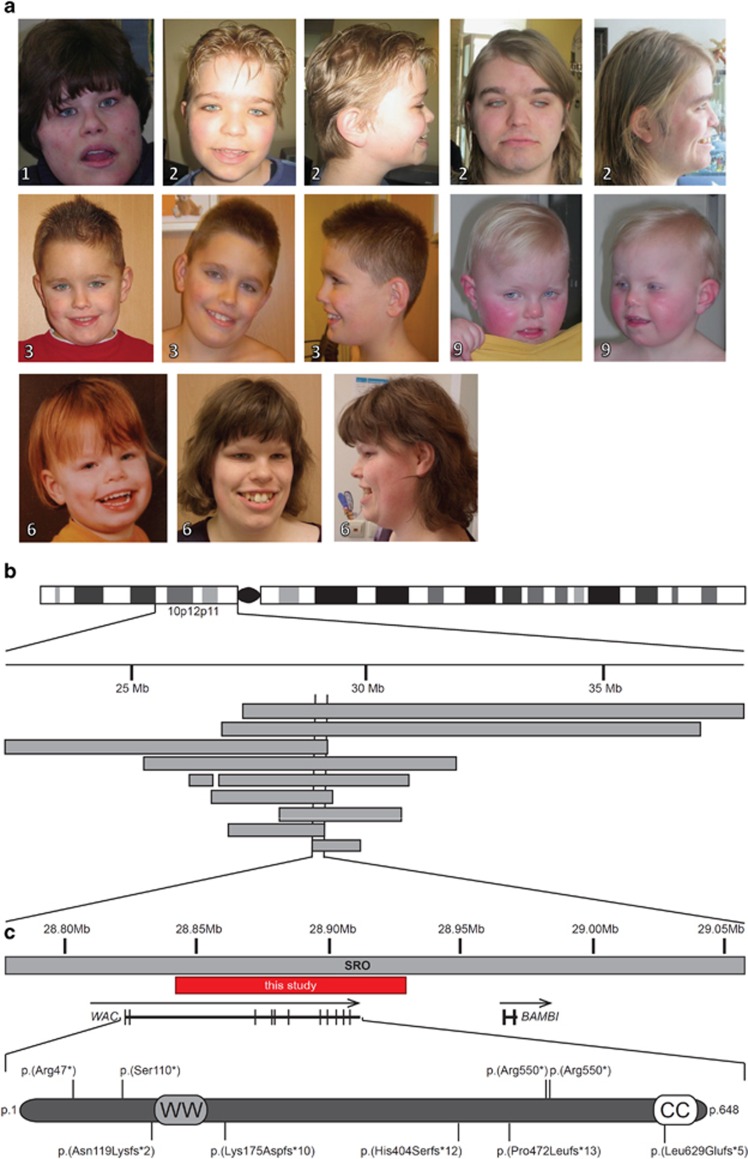
After the identification of the de novo mutation c.139C>T (NM_016628.3) leading to nonsense mutation p.(Arg47*) in Individual 1 (as reported before1), we set out to find additional individuals with mutations affecting WAC to obtain more evidence for its involvement in ID. Systematic analysis of DECIPHER and ECARUCA, two databases collecting clinically relevant copy number variants (CNVs), for CNVs affecting WAC yielded one small deletion (Individual 2; Figure 1). This deletion, hg19 chr10:g.(?_288422777)_(28929097_?)del, disrupted the coding sequence of WAC by deletion of exons 5-14 (NM_016628.3). The shortest region of deletion overlap of the chromosome 10p12p11 contiguous gene deletion syndrome was previously determined by nine deletions ranging in size between 0.99 and 10.66 Mb.14, 15, 16, 17 Comparison of the deletion in Individual 2 to the shortest region of deletion overlap indicates WAC as a only remaining candidate gene for the ID phenotype (Figure 1b).
Figure 1.
Individuals with WAC mutations. (a) Frontal and lateral photographs of individuals with de novo mutations in WAC. All individuals shared overlapping facial dysmorphisms including a square-shaped face, long palpebral fissures, broad mouth and broad chin. Additional features included deep set eyes, epicanthal folds and short philtrum in Individual 1 (photograph at the age of 19 years); low posterior hairline, broad forehead, simple ears, hypertelorism, deep set eyes, low-set full eyebrows, synophrys, deep nasal bridge, flat nose, bifid tongue and broad gums in Individual 2 (photographs at the age of 12 years and 23 years); brachycephaly, posterior ear creases, broad forehead, prominent antihelix, low-set full eyebrows, synophrys and prominent teeth in Individual 3 (photographs at the age of 4 years and 9 years); prominent antihelix, frontal bossing, dental crowding, broad teeth and high palate in Individual 6 (photographs at the age of 3 years and 20 years) and prominent antihelix and deep set eyes in Individual 9 (photographs at the age of 3 years). Photographs were published with consent. (b) The genomic region involved in the 10p12p11 contiguous gene deletion region with the previously published microdeletions, represented by gray horizontal bars.14, 15, 16 (c) Detailed view of the smallest region of overlap (SRO) and the deletion described in this study, represented by a red horizontal bar. In addition, de novo mutations in WAC (NM_016628.3) reported in this study are shown according to their relative position at protein level.
Diagnostic exome sequencing in individuals with neurodevelopmental disorders to identify de novo point mutations in WAC
In routine diagnostic trio-based exome sequencing for individuals with unexplained ID, performed as described before,1 we identified four additional individuals with de novo loss-of-function mutations in WAC (NM_016628.3): c.329C>A, p.(Ser110*) in Individual 3; c.1885_1886del, p.(Leu629fs) in Individual 4; c.356dup, p.(Asn119fs) in Individual 5 and c.1648C>T, p.(Arg550*) in Individual 6 (Figure 1c). In addition, two more individuals with de novo mutations were identified by trio-based exome sequencing of a large cohort of individuals with autism spectrum disorder:18, 19 c.523_524del, p.(Lys175fs) in Individual 7 and c.1209_1212del, p.(His404fs) in Individual 8. Whereas exome sequencing had identified a second de novo mutation in Individual 1 (MIB1; NM_020774.2:c.521G>A; p.(Arg174His)),1 no further clinically relevant de novo mutations were identified in Individuals 3–8, leaving WAC haploinsufficiency as the most likely candidate to explain disease.
Targeted resequencing of WAC in an ID cohort identifies additional de novo mutations
On the identification of multiple de novo loss-of-function mutations in WAC, we performed targeted resequencing of this gene in a cohort of over 2300 individuals with unexplained ID using MIPs as described before.7 This cohort was selected from the in-house collection of the Department of Human Genetics of Radboud University Medical Center containing individuals with unexplained ID. All candidate loss-of-function mutations as well as highly conserved missense mutations (PhyloP>5) were validated by standard Sanger sequencing approaches. For assessing the de novo occurrence of validated mutations, DNA from the parents was tested. This targeted screen identified two additional de novo truncating mutations: c.1415del, p.(Pro472fs) in Individual 9 and c.1648C>T, p.(Arg550*) in Individual 10 (Figure 1c).
WAC mutation spectrum in control individuals
Of all ten de novo mutations identified, nine are predicted to directly result in nonsense-mediated decay of the RNA transcripts; the de novo frameshift in Individual 4 is located in the last exon, suggesting it may skip nonsense-mediated RNA decay. None of the de novo mutations are reported in our in-house variant database containing exome sequencing variants detected in 5031 individuals, nor in ExAC, a large database collecting NGS variants in over 60 000 exomes as proxy for variant allele frequencies in the general population.28 The latter, however, does contain three other, presumable loss-of-function, variants (by insertion–deletion events), each observed only once in ~100 000 alleles. Whereas these three variants have not been validated by Sanger sequencing, thereby possible being sequencing artefacts rather than true mutations, this observation may also reflect the very mild end of the ID spectrum in the general population.
Clinical spectrum associated with WAC haploinsuffiency
Clinical evaluation of all individuals with de novo loss-of-function mutations in WAC showed distinct phenotypic overlap (Table 1; Supplementary Information (clinical descriptions); and Supplementary Table S1). All, but one individual, had ID. The range of ID observed ranged from mild-to-severe and was accompanied by language and motor delay. In addition, individuals showed a variety of neurological problems including hypotonia (6/9), with remarkable manifestation in the oral region resulting in dysarthria, and behavioral problems (10/10). The latter recurrently included autism (4/9), anxiety (3/10), concentration disorder (4/10) and/or sleep disturbance (6/10). Other overlapping features consisted of unexplained reduced vision (3/9) and respiratory problems (7/9) with recurrent respiratory infections reported most often (5/7). Notably, all individuals had overlapping facial dysmorphisms consisting of a square shape of the face, deep set eyes, long palpebral fissures, broad mouth and broad chin (Figure 1a).
Table 1. Clinical details of individuals with WAC mutations.
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | Individual 5 | Individual 6 | Individual 7 | Individual 8 | Individual 9 | Individual 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Female | Male | Male | Male | Female | Female | Female | Female | Female | Male |
| Age at last visit | 22 years | 12.5 years | 9 years | 17 months | 3 years | 20 years | 6 years | 9 years | 12 years | 7 years |
| Mutation WAC (NM_016628.3) | ||||||||||
| cDNA change | c.139C>T | Partial gene | c.329C>A | c.1885_1886del | c.356dup | c.1648C>T | c.523_524del | c.1209_1212del | c.1415del | c.1648C>T |
| Amino acid change | p.(Arg47*) | Deletion | p.(Ser110*) | p.(Leu629fs) | p.(Asn119fs) | p.(Arg550*) | p.(Lys175fs) | p.(His404fs) | p.(Pro472fs) | p.(Arg550*) |
| Chromosome position (Hg19) | Chr10:28824551 | chr10:g.(?_288422777)_(28929097_?)del | Chr10:28872382 | Chr10:28908476_ 28908477 | Chr10:28872409 | Chr10:28905193 | Chr10:28879674_ 28879675 | Chr10:28899671_ 28899674 | Chr10:28900827 | Chr10:28905193 |
| Growth | ||||||||||
| Birth weight (g) | NR | 3600 (>+2 SD) | 3600 (0 SD) | NR | 2920 (0 SD) | 3070 (−1 SD) | NR | NR | 3380 (0 SD) | 4500 (+2.5 SD) |
| Height (cm) | NR | 149.7 (−1.1 SD) | 131.3 (−1.5 SD) | 75.4 (−2 SD) | 87 (−3 SD) | 156.5 (−2.5 SD) | +0.86 SD | + 2.88 SD | 161.0 (+0.75 SD) | 114.8 (−1.75 SD) |
| Weight (kg) | NR | 48.8 (+2.1 SD) | 31.5 (+0.75 SD) | 10.0 (+2 SD) | 13 (+0.7 SD) | 75.5 (+3.5 SD) | +1.7 SD | −2.9 SD | 61.2 (+2 SD) | 26.2 (+3 SD) |
| Head circumference (cm) | NR | 56.3 (+1.1 SD) | 52.0 (−0.5 SD) | 48.5 (0 SD) | 50.4 (−0.6 SD) | 54 (−1 SD) | +2.9 SD | + 0.53 SD | 56.0 (+1.5 SD) | 54.9 (+2 SD) |
| Development | ||||||||||
| Intellectual disability | Severe | Mild (IQ 58) | Mild (IQ 65) | Not assessed owing to young age | Mild | Moderate (IQ 44) | No (IQ 89) | Mild (IQ 61) | Mild (IQ 65) | Mild (IQ 61) |
| Adaptive ability | 70 (−2 SD) | 69 (−2 SD) | ||||||||
| Speech delay | + No speech | + Dysarthria | + 2.5 years: first words Dysarthria | + 1.5 years: no words, only babbles | + Dysarthria | + 2 years: first words Dysarthria | − 20 months: first words | + 5 years: first words | + 5 years: first words | + 1.5 years: first words |
| Motor delay | + 34 months: first steps | + 24 months: first steps | + 24 months: first steps | + 1.5 years: walks with support | + 36 months: walks unsupported | + 2 years: first steps | + | NR | + 22 months: first steps | + 27 months: first steps |
| Behaviour | ||||||||||
| Anxiety | − | + | − | − | − | + | − | − | + | − |
| Concentration problems/hyperactivity | − | − | + | − | + | + | − | − | − | + |
| Sleep disturbances | + | + | − | + | + | + | − | − | + | − |
| Autism | − | + | − | − | NR | + | + | + | − | − |
| Other | Self-mutilation | − | Age 1 year: lack of eye contact | − | − | − | − | − | − | |
| Neurological | ||||||||||
| Epilepsy | + | − | − | − | − | − | − | − | − | − |
| Oral hypotonia | NR | + | + | + | + | + | − | − | + | + |
| Hypotonia | NR | + | + | + | + | − | − | − | + | + |
| Toe walking | + | − | + | − | NR | − | NR | NR | − | + |
| Ocular | ||||||||||
| Vision | Normal | Normal | Unexplained reduced vision | To further establish: lack of eye contact and staring | Hypermetropia (+3.75 Dpt) | Unexplained reduced vision | Normal | Hyper/hypometropia | CVI | Normal |
| Strabismus | − | − | + | − | − | + | − | − | − | + |
| Pulmonal | ||||||||||
| Abnormal breathing pattern | + | + | − | − | NR | − | NR | ±a | − | − |
| Recurrent respiratory infections | NR | + | + | + | − | + | NR | ±a | + | − |
| Asthma | − | + | + | − | − | − | NR | ±a | − | − |
| Extremities | ||||||||||
| Brachydactyly fingers | NR | − | + | − | − | + | NR | NR | + | − |
| Pedes plano valgi | NR | − | + | + | − | + | NR | NR | + | + |
| Fetal finger pads | NR | − | + | − | − | − | NR | NR | + | + |
| Other | − | Broad first digits | Sandal gap Simian crease | − | Short hands and feet | Tapering fingers | − | − | Hip dysplasia Simian crease | − |
| Other | ||||||||||
| Neonatal feeding difficulties | + | − | + | − | + | − | − | − | + | − |
| Other clinical features | Hernia diafragmatica, pubertas tarda | Laryngomalacy Hoarse voice | Fever convulsions, fatigue, phimosis, hypertrichosis back | Large frontal fontanel | Low IgG class 2 and 3 | Fatigue, paroxysmal atonia | − | Kidney problems | − | Inverted nipples |
| Additional investigations | ||||||||||
| MRI/CT | Normal | Normal | Megacisterna magna | Normal | Normal | Normal | NR | Abnormal (no details available) | Mild asymmetrical right frontotemporal hemisphere | Mild enlarged ventricles |
| Muscle biopsy | − | − | Low mitochondrial activity | − | − | Normal | NR | NR | − | − |
| Genetic analysis | 250k SNP array:− MECP2:− RAI1:− TCF4:− Angelman methylation:− | FMR1:− RAI1: − | Subtelomeric regions: − 250k SNP array: − OPA1: − POLG: − RYR1:− | Cytoscan HD array: 9p22.1 gain and Xp22.31 gain, maternally inherited | FMR1:− RAI1:− EHMT1:− Cytoscan HD array: 6 Mb region of homozygosity chromosome 6q14.2q15 | Angelman methylation: − FMR1:− CACNA1A:− array CGH:− | NR | NR | FMR1: − Subtelomeric regions: − | 250k SNP array: − |
| Other (clinically relevant) de novo variant(s) by exome sequencing | MIB1(NM_020774.2)c.521G>A; p.(Arg174His) | Not tested | None | None | None | None | None | None | Not tested | Not tested |
Abbreviations: CVI, cerebral visual impairment; Dpt, diopter; IQ, intelligence quotient; NR, not reported.
± indicates that Individual 8 is reported to have pulmonal problems, but this is not further specified.
Panneuronal knockdown of the Drosophila WAC orthologue results in learning deficit
To obtain independent evidence for the involvement of WAC in the ID phenotype of the described individuals, we decided to study the functional consequences of WAC knockdown using Drosophila as a model. The Drosophila genome contains a previously uncharacterized WAC orthologue named CG8949, not be confused with the unrelated Drosophila WAC (wee Augmin) gene. CG8949 codes for several protein isoforms, the longest one consisting of 876 amino acids, and shows the highest expression in adult ovaries and the larval central nervous system.29, 30 WAC is 26% identical and 39% conserved over the C-terminal 588 amino acids of the fly protein, with sequence similarity distributed over the whole protein, further characterized by clusters of short sequences of high conservation for the important functional motifs of the protein.13
We investigated the role of the Drosophila WAC orthologue in light-off jump reflex habituation paradigm. Habituation is a simple form of non-associative learning, in which an initial strong behavioral response towards a repeated, non-threatening stimulus gradually wanes. It provides a filtering mechanism, which is an important prerequisite for higher cognitive functioning.31, 32 Using the light-off jump reflex habituation, we have previously identified learning deficits in number of Drosophila ID models.9, 24, 26 Two independent inducible RNAi lines targeting the Drosophila WAC orthologue CG8949 (vdrc48307 and vdrc107328) and their corresponding genetic background control lines (vdrc60000 and vdrc60100) were obtained (Vienna Drosophila RNAi Center22) and fly stocks were kept under standard conditions. Expression of CG8949 was specifically downregulated in neurons using the UAS-Gal4 system.
The efficiency of ubiquitous RNAi knockdown was measured using qPCR on RNA isolated from Drosophila brains of third instar larvae, representing the tissue and developmental stage with the highest expression of CG8949 allowing for an efficient detection of expression differences. There was no significant CG8949vdrc107328-mediated knockdown on CG8949 expression levels (88% remaining gene expression; P=0.16, student's t-test; Supplementary Table S2). In contrast, CG8949vdrc48307-mediated RNAi reduced levels of CG8949 to 58% remaining gene expression (P<0.01, Student's t-test; Supplementary Table S2).
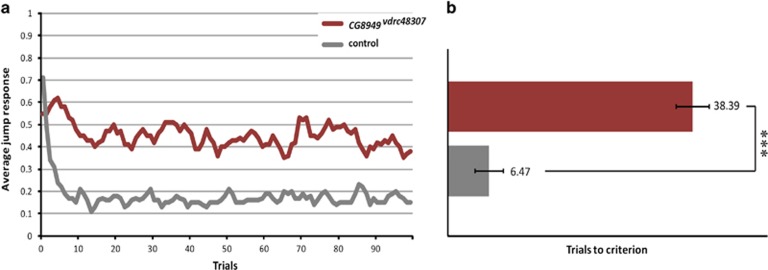
Flies were exposed to series of 100 short (15 ms) light-off stimuli with 1 s interval between stimuli. Both control and CG8949vdrc48307 knock-down flies showed good initial jump response; there was no significant difference between the initial startle response of CG8949vdrc48307 and control flies (t-test, P=0.469). Whereas, control flies quickly habituated to the repeated light-off stimuli, CG8949vdrc48307 knock-down animals failed to adapt their behavioral response and retained high average jump response throughout the whole experiment (Figure 2). This defect was statistically significant (fold-change=5.93; P=5.21 × 10−12). No habitation defects were seen in the CG8949vdrc107328 knock-down flies (data not shown), as was to be expected based on the insufficient mRNA knockdown.
Figure 2.
Knockdown of the Drosophila WAC orthologue CG8949 affects non-associative learning in the light-off jump reflex habituation paradigm. Jump responses of 3–7-day-old individual male flies were induced by repeated light-off pulses (100 trials) with a 1 s inter-trial interval. CG8949 knockdown flies (CG8949vdrc48307; genotype: 2xGMR-wIR/+ UAS-CG8949vdrc48307/elav-Gal4, UAS-Dicer-2) are plotted in red and genetic background control flies are plotted in dark gray. Habituation was scored as the mean number of trials required to reach the non-jump criterion (TTC). Main effects of genotype (mutant vs control), day and test system on log-transformed TTC values were tested using linear model regression analysis.27 (a) Average jump response across 100 light-off trials. (b) Mean TTC of CG8949vdrc48307 (TTC=38.39, n=54) vs mean TTC of control flies (TTC=6.47, n=49). Quantification of average jump responses revealed that flies with pan-neuronally induced CG8949 knockdown habituated significantly slower (***P<0.001, linear model regression analysis).
Discussion
Here, we report the identification of a novel clinically recognizable syndrome caused by haploinsufficiency of WAC. All but one patient showed mild ID, with speech and motor delay, whereas one had an overall more severe ID phenotype, epilepsy and an absence, rather than delay, of speech. All patients had neurological problems including hypotonia and a variety of behavioral problems including autism, anxiety, concentration problems, sleep disturbance and/or self-mutilation. Notably, all patients had overlapping facial dysmorphisms consisting of square shape of the face, deep set eyes, long palpebral fissures, broad mouth and broad chin. Complementary experimental evidence in Drosophila showed a role of the evolutionarily conserved WAC proteins in cognitive processes and a role for the Drosophila WAC orthologue in non-associative learning.
Previously, another individual was reported with a truncating mutation in WAC10 who shows a phenotype similar to the individuals reported in this study (Supplementary Table S1). Also, a large-scale study aiming at the identification of genetic causes underlying developmental disorders recently reported the identification of one de novo nonsense mutation in WAC but further clinical details of this individual were lacking, hampering detailed phenotypic comparison.33 Interestingly, three of our patients were negative tested for RAI1, known to cause Smith–Magenis syndrome.34 The coarse facial appearance as well as the ID with variable behavior problems of the individuals with WAC mutations have similarities with individuals reported with Smith–Magenis syndrome.35
Interstitial deletions including WAC were previously described and associated with ID.14, 15, 16, 17 Wentzel et al.14 presented six individuals with an interstitial deletion at 10p12p11, all sharing a region of overlap including two genes: BAMBI and WAC. All individuals were reported to have developmental delay, abnormal behavior and facial dysmorphic features including a bulbous nasal tip, deep set eyes, synophrys/thick eyebrows and full cheeks. This phenotype is highly similar to the phenotype observed in the current individuals and consistent with our finding that loss of WAC causes ID and the characteristic facial dysmorphisms in 10p12p11 microdeletion syndrome (Supplementary Table S1). In this 10p12p11 microdeletion syndrome, cardiac abnormalities have frequently been reported (7/9 individuals) and heterozygosity of two other genes, LYZL1 and SVIL, has been suggested to contribute to the development of these cardiac abnormalities.14 This is in line with the fact that in none of our individuals cardiac abnormalities were present. Epilepsy has been reported in two out of nine individuals with deletion of 10p12p11 and is present in only one of our individuals (Individual 1). This more severely affected individual carried also a de novo MIB1 mutation. This variant has been reported twice in ExAC, containing NGS variants in healthy controls of several ethnicities.28 Moreover, one missense and one nonsense mutations in MIB1 were identified previously and segregated each in two large dominant families with affected individuals with cardiomyopathy, but without ID.36 Therefore, a contribution of the second mutation in MIB1 as potential modifier of the more severe phenotype is unlikely. The more severe phenotype may be caused by other yet unknown potential genetic modifier(s) or reflects the severe end of the clinical spectrum caused by WAC haploinsufficiency.
Human WAC encodes a protein containing a WW domain-containing adapter and a coiled-coil region. WAC is an evolutionary conserved protein, but its exact function is still unknown. Protein–protein interaction studies suggest a role in the regulation of histone H2B ubiquitination and gene transcription.12 Interaction of WAC via the coiled-coil domains with RNF20 and RNF40 activates UBE2A-mediated H2B ubiquitination. On the basis of this interaction, RNF20 and RNF40 could be considered as candidate genes for ID. Interestingly, de novo mutations in both genes have been described in individuals with autism and unrelated unaffected siblings.19 A clear relation to a clinical phenotype, if any, remains to be established. RNA polymerase II recruits WAC to active transcription sites by binding to the WW domain. On the basis of sequence homology it has also been suggested that WAC is involved in RNA processing or transcription.13 Besides co-localization of WAC with splicing factor SC35 in nuclear speckles, there is, however, currently no further evidence for a role in RNA splicing. The localization of WAC in the nucleus supports its suggested function in gene transcription via UBE2A histone H2B ubiquitination. Although WAC and UBE2A lack a direct interaction, they are both likely to function within a protein complex important for histone H2B ubiquitination and transcription regulation.12 Interestingly, for UBE2A, microdeletions and point mutations have been associated in males with an X-linked inherited clinical syndrome characterized by ID, seizures, absent speech, urogenital and skin anomalies. Recent work on Drosophila has uncovered a novel role of UBE2A in clearance of defective mitochondria from the synaptic compartment and in synaptic plasticity.37 On the basis of the suggested interaction of WAC and UBE2A being part of the same complex, and their clinical phenotypes both including ID, we experimentally addressed whether CG8949, like fly UBE2A, is required for synaptic vesicle cycling, mitochondrial functioning and morphology.37 We found all these processes unperturbed in our Drosophila model (Supplementary Figure S1). Despite the unavailability of the second CG8949 knockdown fly as an independent confirmation of the habituation phenotype, our data supports the role of WAC in cognition by the deficit in non-associative learning.
In summary, we describe a clinically recognizable syndrome owing to loss-of-function mutations in WAC, which is characterized by mild ID, hypotonia, behavioral problems and facial dysmorphisms consisting of square shape of the face, deep set eyes, long palpebral fissures, broad mouth and broad chin. Complementary experimental evidence in Drosophila showed a role of the evolutionarily conserved WAC proteins in cognitive processes and a role for the Drosophila WAC orthologue in non-associative learning. WAC is a component of the evolutionary conserved histone H2B ubiquitination complex, regulating gene transcription. On the basis of our results, WAC can be added to the growing list of genes involved the H2B ubiquitination complex leading to cognitive defects.
Acknowledgments
We thank the participating individuals and their families. We also thank Maartje van de Vorst and Jolanda de Gooyert for excellent technical assistance and Willy Nillesen and Sander Stegmann for expert clinical interpretation. This study was in part financially supported by a TOP-grant from the Netherlands Organization for Scientific Research (NWO), ZonMW (912-12-109 to AS, JV and BBAdV; 917-96-346 to AS; 916-12-095 to AH; 907-00-365 to TK), the European Union under the 7th framework program (Gencodys HEALTH-F4-2010-241995 to AS), the Jérôme Lejeune foundation (to AS), the National Institutes for Mental Health (#1R01MH101221 to E.E.E.) and an ERC Starting Grant (260678), the Research Foundation Flanders (FWO grants G053913N, G079013N and G094915N), the Hercules Foundation, the Instituut voor Wetenschap en Technologie (IWT grant 111352), the Interuniversity Attraction Pole program by BELSPO, the research fund KU Leuven, a Methusalem grant of the Flemish government and VIB. E.E.E. is an investigator of the Howard Hughes Medical Institute.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JH, Oh EC, Vissers LE et al: Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet 2012; 91: 1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Vissers LE, Willemsen MA et al: Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet 2012; 49: 179–183. [DOI] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O et al: De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet 2014; 94: 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Green RC: Diagnostic clinical genome and exome sequencing. N Engl J Med 2014; 370: 2418–2425. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W et al: Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulto-van Silfhout AT, Rajamanickam S, Jensik PJ et al: Mutations affecting the SAND domain of DEAF1 cause intellectual disability with severe speech impairment and behavioral problems. Am J Hum Genet 2014; 94: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Nijhof B, Fenckova M et al: GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet 2013; 50: 507–514. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi JM et al: De novo mutations in moderate or severe intellectual disability. PLoS Genet 2014; 10: e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O: Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 2001; 8: 85–95. [DOI] [PubMed] [Google Scholar]
- Zhang F, Yu X: WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 2011; 41: 384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GM, Arnaout MA: WAC, a novel WW domain-containing adapter with a coiled-coil region, is colocalized with splicing factor SC35. Genomics 2002; 79: 87–94. [DOI] [PubMed] [Google Scholar]
- Wentzel C, Rajcan-Separovic E, Ruivenkamp CA et al: Genomic and clinical characteristics of six patients with partially overlapping interstitial deletions at 10p12p11. Eur J Hum Genet 2011; 19: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Hayashi S, Masui A et al: Deletion at chromosome 10p11.23-p12.1 defines characteristic phenotypes with marked midface retrusion. J Hum Genet 2012; 57: 191–196. [DOI] [PubMed] [Google Scholar]
- Shahdadpuri R, de Vries B, Pfundt R, de Leeuw N, Reardon W: Pseudoarthrosis of the clavicle and copper beaten skull associated with chromosome 10p11.21p12.1 microdeletion. Am J Med Genet Part A 2008; 146A: 233–237. [DOI] [PubMed] [Google Scholar]
- Mroczkowski HJ, Arnold G, Schneck FX, Rajkovic A, Yatsenko SA: Interstitial 10p11.23-p12.1 microdeletions associated with developmental delay, craniofacial abnormalities, and cryptorchidism. Am J Med Genet Part A 2014; 164A: 2623–2626. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Stessman HA, Boyle EA et al: Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014; 5: 5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ et al: The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DECIPHER. Available at https://decipher.sanger.ac.uk/index.
- ECARUCA. Available at http://umcecaruca01.extern.umcn.nl:8080/ecaruca/ecaruca.jsp.
- Vienna Drosophila RNAi Center. Available at www.vdrc.at.
- Dietzl G, Chen D, Schnorrer F et al: A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007; 448: 151–156. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Oortveld MA, Nijtmans LG et al: CEP89 is required for mitochondrial metabolism and neuronal function in man and fly. Hum Mol Genet 2013; 22: 3138–3151. [DOI] [PubMed] [Google Scholar]
- Bloomington Drosophila Stock Center. Available at www.flystocks.bio.indiana.edu.
- Kramer JM, Kochinke K, Oortveld MA et al: Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol 2011; 9: e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria, 2014.
- Exome Aggregation Consortium (ExAC). Cambridge, MA, USA, 2015.
- Graveley BR, Brooks AN, Carlson JW et al: The developmental transcriptome of Drosophila melanogaster. Nature 2011; 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modMine. Available at http://www.modencode.org/Celniker.shtm.
- Wilson DA, Linster C: Neurobiology of a simple memory. J Neurophysiol 2008; 100: 2–7. [DOI] [PubMed] [Google Scholar]
- Typlt M, Mirkowski M, Azzopardi E, Ruettiger L, Ruth P, Schmid S: Mice with deficient BK channel function show impaired prepulse inhibition and spatial learning, but normal working and spatial reference memory. PloS One 2013; 8: e81270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders S: Large-scale discovery of novel genetic causes of developmental disorders. Nature 2014; 19: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH: Mutations in RAI1 associated with Smith-Magenis syndrome. Nature Genet 2003; 33: 466–468. [DOI] [PubMed] [Google Scholar]
- Elsea SH, Girirajan S: Smith-Magenis syndrome. Eur J Hum Genet 2008; 16: 412–421. [DOI] [PubMed] [Google Scholar]
- Luxan G, Casanova JC, Martinez-Poveda B et al: Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med 2013; 19: 193–201. [DOI] [PubMed] [Google Scholar]
- Haddad DM, Vilain S, Vos M et al: Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell 2013; 50: 831–843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.