Abstract
Circulating adiponectin is involved in the atherosclerotic process and has been associated with cardiovascular disease as well as obesity, insulin resistance, metabolic syndrome, and type 2 diabetes. The adiponectin gene (ADIPOQ) encodes the circulating protein adiponectin and affects its expression. Only a small proportion of all known ADIPOQ polymorphisms have been investigated in relation to circulating adiponectin concentrations. Using data from 3,355 African-American and white men and women aged 33–45 at the year 15 examination from the Coronary Artery Development in Young Adults (CARDIA) Study the association between 10 single-nucleotide polymorphisms (SNPs) within ADIPOQ and serum adiponectin was examined using linear regression. SNPs were chosen based on a tagSNP approach. Models were stratified by self-reported race to control for population stratification, and Bonferroni corrected for multiple comparisons. ADIPOQ SNPs rs17300539 (P < 0.0001), rs182052 (P = 0.0013), rs822393 (P = 0.0005), rs9882205 (P = 0.0001), and rs3774261 (P = 0.0001) were strongly associated with serum adiponectin concentrations in whites. In general, there was a dose–response relationship of adjusted mean adiponectin concentrations across genotypes. Only one SNP, rs17300539 was marginally associated with serum adiponectin concentrations (P = 0.0087) in African Americans. Significant interactions were found between waist and rs182052 (P = 0.0029) and between rs9882505 and smoking (P = 0.001) in whites. Many ADIPOQ SNPs have not yet been examined, and additional studies are needed to determine whether these may be functional variants.
INTRODUCTION
Adiponectin is a 247-amino acid, 30 kDa protein secreted by adipocytes that circulates at high concentrations in the blood (1). Adiponectin is involved in the atherosclerotic process by stimulating the production of nitric oxide in endothelial cells (2) and suppressing the expression of cellular adhesion molecules by inhibiting endothelial nuclear factor-κB signaling through the activation of cAMP protein kinase C (3–6). It also inhibits the proliferation and migration of smooth muscle cells by binding to platelet derived growth factor and promotes plaque stability in more advanced stages of atherosclerosis by increasing the expression of a tissue inhibitor of matrix metal-loproteinases in macrophages (1). A number of studies have evaluated the association between circulating adiponectin and coronary heart disease or cardiovascular disease outcomes with some showing higher levels of adiponectin associated with decreased risk (7–11) whereas other studies show an increased risk (12–14). There is some evidence that adiponectin concentrations may relate to the stage of cardiovascular disease; higher adiponectin levels may be an attempt to slow progression of the disease (15) and may account for discrepant findings of these studies. It is well established that adiponectin has also been linked to obesity, insulin resistance, type 2 diabetes, and the metabolic syndrome which are risk factors for cardiovascular disease (16).
The adiponectin gene (ADIPOQ) encodes the circulating protein adiponectin and is expressed primarily in the adipose tissue of various organs, but it is expressed in vascular tissue as well (17). Only a small proportion of all known ADIPOQ polymorphisms have been investigated in relation to circulating adiponectin concentrations and it is likely that functional variants responsible for these observed associations have yet to be discovered or studied (18). Several studies have examined the association of single-nucleotide polymorphisms (SNPs) and haplotypes of ADIPOQ with circulating adiponectin concentrations, but these studies have had mixed results and have concentrated on a small number of SNPs, usually within one ethnic group (19–24). A review and meta-analysis by Menzaghi et al. (18) note that although evidence for the association of a few ADIPOQ SNPs with circulating adiponectin is strong, there is still much variation in ADIPOQ that has not been examined.
Thus, this study examined the association of 10 SNPs in ADIPOQ chosen by a tagSNP method with serum adipo nectin in whites and African Americans from Coronary Artery Risk Development in Young Adults (CARDIA) study. Given circulating adiponectin's likely involvement in the pathophysio logy of adiposity, interactions of ADIPOQ SNPs with BMI, waist circumference, as well as age, sex, and smoking status were evaluated.
METHODS AND PROCEDURES
Participants
The CARDIA study is a prospective cohort study examining the determinants and trends in risk factors of heart disease development starting in young adulthood. Between 1985 and 1986 5,115 black and white men and women aged 18–30 years were recruited from four US centers—Birmingham, AL; Chicago, IL; Minneapolis, MN; and Oakland, CA. Details of inclusion and exclusion criteria can be found elsewhere (25). Follow-up examinations occurred during 1987–1988 (year 2), 1990–1991 (year 5), 1992–1993 (year 7), 1995–1996 (year 10), and 2000–2001 (year 15), with 74% of the surviving cohort participating in the year 15 examination. The CARDIA participants attending the year 15 clinic visit consented to isolation of genomic DNA from a blood sample obtained at the year 10 and year 15 examinations. A total of 3,355 year 15 participants had serum adiponectin assayed as part of the Young Adult Longitudinal Trends in Antioxidants (YALTA) ancillary study and comprised the total sample size for this study.
ADIPOQ SNP selection and genotyping
Polymorphisms spanning ADIPOQ were selected from the International HapMap Project database (HapMap Phase I release) based on the estimated pairwise linkage disequilibrium (LD), r2, between all common SNPs in HapMap, i.e., SNPs with minor allele frequency >10%, as described by Carlson et al. (26). Briefly, the algorithm bins SNPs for which pairwise LD exceeds a threshold r2 (0.80 in this case). For each racial group (Centre d'Etude du Polymorphisme Humain and Yoruban populations), tagSNPs were selected from each bin so as to constitute a minimal set of highly informative markers while minimizing redundant data. In addition, tagSNPs with potential functional relevance were included where available. When a full set of tagSNPs was not available due to poor coverage in HapMap, additional SNPs were selected from the literature. Through this process, ADIPOQ SNPs rs17300539, rs182052, rs822393, rs822395, rs2036373, rs9882205, rs2241766, rs1501299, rs3774261, rs17366743, rs4686804, and rs1063539 were chosen to be genotyped. Two of these SNPs, rs1501299 and rs4686804, could not be designed in the multiplex assays, leaving 10 total SNPs that were genotyped.
The selected ADIPOQ tagSNPs were genotyped using a multiplex matrix-assisted laser desorption/ionization time-of-flight mass spectrometry method (Sequenom, San Diego, CA). Polymorphism genotyping in the CARDIA study adheres to a rigorous quality control program, which includes barcode identification of samples, robotic sample handling, and blind replicate genotype assessment on 5% of the total sample. The geno-type call rate was 96.7%, with a maximum missing per SNP of 8.62%.
Data collection
Information on gender, race, age, and smoking status was collected via interview and questionnaire at the year 15 examination. Smoking was categorized into current vs. never/former. Weight was measured to the nearest 0.2 pound, rounding down, using a calibrated balance beam scale. BMI was calculated as weight in kilograms divided by height in meters squared. Waist circumference was measured once to the nearest 0.5 cm with a Gulick Plus II measurement tape, in a location laterally midway between the iliac crest and the lowest lateral portion of the rib cage and anteriorly midway between the xiphoid process of the sternum and the umbilicus. Blood was drawn by venipuncture and stored as serum at −70 °C. Serum adiponectin was measured via radioimmunoassay using a polyclonal rabbit antibody and purified recombinant adiponectin standards (Linco Research, St Charles, MO). The assay had a range of 0.20–40 mg/l and an interassay coefficient of variation of 7–9%.
Statistical analysis
Pairwise LD (r2) was calculated among the SNPs and LD plots were generated using Haploview V4.0 (27). Hardy–Weinberg equilibrium tests were conducted using the exact test within each race group, and SNPs with substantial deviation from Hardy–Weinberg equilibrium (P < 0.001) were excluded. Baseline characteristics for each ethnic group were compared across groups by t-test, χ2-test or Wilcoxon test as appropriate.
Linear regression models were used to assess the association of SNPs with natural log transformed serum adiponectin. Analyses were stratified by self-reported race to control for population stratification. Associations of each SNP with serum adiponectin were evaluated first by two degree of freedom tests in models adjusting for age, sex, and field center. Additional adjustment for smoking, waist circumference, and BMI was performed. African-American models were also additionally adjusted for African ancestry to further control for population stratification. Unadjusted and adjusted geometric means for each genotype of the SNP were examined for individual SNPs with two degree of freedom P < 0.05. A Bonferroni correction was used to adjust for multiple comparisons within each race group, α = 0.05/10 = 0.005.
Interactions of ADIPOQ SNPs with age, sex, BMI, waist circumference, and smoking status were examined on an additive scale. Interactions were not corrected for multiple testing. With the exception of sex and smoking status, all covariates in interactions with ADIPOQ were modeled as continuous variables. All analyses were conducted using SAS V9.1 (SAS Institute, Cary, NC).
RESULTS
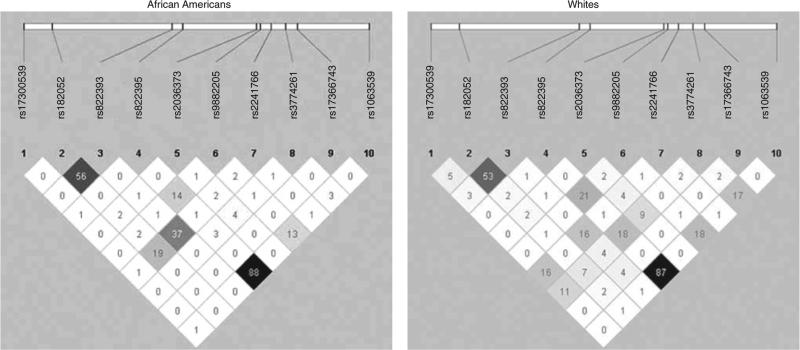
All SNPs were in Hardy–Weinberg equilibrium (P > 0.001) except for rs9882205 among African Americans with a P < 0.0001 (Supplementary Table S1 online), so this SNP was eliminated from further analysis in African Americans. The LD plots showed little LD among the SNPs in either race group (Figure 1). As estimation of haplotypes in the presence of low LD can result in high levels of haplotype inaccuracy (28), estimation of haplotypes was not attempted. African Americans and whites significantly differed on age, gender, smoking status, waist circumference, BMI, and serum adipo nectin concentrations at the year 15 examination (Table 1).
Figure 1.
Pairwise linkage disequilibrium (r2) for adiponectin gene SNPs. Linkage disequilibrium plots by race group. These plots show the pairwise correlation (r2) for each tag single-nucleotide polymorphism (tagSNP) chosen in this study.
Table 1.
Coronary Artery Risk in Young Adults (CARDIA) characteristics from the year 15 (2000–2001) examination by race group
| Characteristics | White (n = 1,772) | African Americans (n = 1,583) | P valuea |
|---|---|---|---|
| Age, years | 40.5 ± 3.4 | 39.5 ± 3.8 | <0.001 |
| Gender | |||
| Male | 838 (47.3) | 652 (41.2) | <0.001 |
| Female | 934 (52.7) | 931 (58.8) | |
| Field center | |||
| Birmingham | 372 (21.0) | 465 (29.4) | <0.001 |
| Chicago | 441 (24.9) | 364 (23.0) | |
| Minneapolis | 533 (30.1) | 299 (18.9) | |
| Oakland | 426 (24.0) | 455 (28.7) | |
| Current smoking | 287 (16.2) | 441 (27.9) | <0.001 |
| BMI, kg/m2 | 27.2 ± 5.9 | 30.6 ± 7.4 | <0.001 |
| Waist circumference, cm | 87.5 ± 14.7 | 92.0 ± 15.7 | <0.001 |
| Serum adiponectin, mg/lb | 11.0 (7.2, 16.0) | 7.2 (5.0, 11.0) | <0.001 |
P values by χ2, Wilcoxon or t-test as appropriate.
Median (quartile 1, quartile 3).
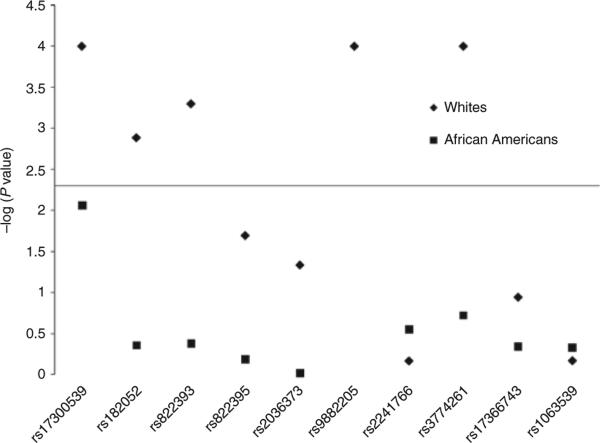
Figure 2 shows the results of single SNP association tests for both race groups and all outcomes adjusted for age, sex, and center. In some cases (SNPs rs2241766 and rs1063539 in African Americans, and rs17366743 in whites) where the rare homozygote group had <10 participants, this group was combined with the heterozygote group in order to obtain more stable estimates of association. Among whites, rs17300539, rs182052, rs822393, rs9882205, and rs3774261 were strongly significantly associated with serum adiponectin concentrations with P < 0.005 or –log(P value) of 2.3 (Figure 2). There was some indication of associations of rs822395 and rs2036373 with serum adiponectin, although these associations did not reach the P < 0.005 Bonferroni threshold of significance (Figure 2). Unadjusted and adjusted geometric mean adiponectin concentrations among whites are shown in Table 2. For most of the significant SNPs, there appeared to be a dose– response relationship across the genotypes. In particular, rs17300539, rs9882205, and rs3774261 had the largest between genotype differences in mean adiponectin concentrations. Further adjustment for waist circumference, BMI, and smoking did not substantially change these results (data not shown). The amount of variance explained (R2) with only age, sex, and center in the model was 21%. Adding the five SNPs that were significantly associated with adiponectin in whites, 24% of the variance in adiponectin was accounted for.
Figure 2.
Results of two degree of freedom tests for adiponectin gene SNPs and serum adiponectin. The –log(P values) for each two degree of freedom test of each single-nucleotide polymorphism (SNP) with serum adiponectin concentrations by race. Each SNP is along the x axis with the corresponding –log(P value) on the y axis. The solid horizontal line represents the –log(0.0050) Bonferroni significance threshold, and the black diamonds represent the P values for whites whereas black squares show the results for African Americans.
Table 2.
Associations of single adiponectin gene (ADIPOQ) SNPs with serum adiponectin concentrations (mg/l) among whites (n = 1,772)
| Unadjusteda |
Demographic adjustedb |
|||
|---|---|---|---|---|
| Means (95% CI) | P value | Means (95% CI) | P value | |
| rs17300539 | ||||
| GG (n = 1,429) | 10.3 (10.0, 10.6) | ref | 10.3 (10.0, 10.5) | ref |
| AG (n = 264) | 12.8 (11.9, 13.6) | <0.0001 | 12.8 (12.1, 13.6) | <0.0001 |
| AA (n = 20) | 15.3 (12.0, 19.4) | 0.0014 | 15.7 (12.7, 19.5) | 0.0001 |
| rs182052 | ||||
| GG (n = 750) | 11.2 (10.7, 11.6) | ref | 11.1 (10.8, 11.6) | ref |
| AG (n = 780) | 10.2 (9.8, 10.6) | 0.0016 | 10.2 (9.9, 10.6) | 0.0008 |
| AA (n = 201) | 9.9 (9.2, 10.7) | 0.0082 | 10.1 (9.4, 10.8) | 0.01 |
| rs822393 | ||||
| CC (n = 951) | 11.1 (10.7, 11.5) | ref | 11.1 (10.8, 11.5) | ref |
| CT (n = 656) | 10.3 (9.8, 10.7) | 0.0066 | 10.2 (9.8, 10.6) | 0.0005 |
| TT (n = 101) | 9.7 (8.7, 10.8) | 0.02 | 9.8 (8.9, 10.8) | 0.02 |
| rs822395c | ||||
| AA (n = 696) | 10.5 (10.1, 11.0) | ref | 10.5 (10.1, 10.9) | ref |
| AC (n = 700) | 10.7 (10.3, 11.2) | 0.11 | 10.7 (10.3, 11.1) | 0.44 |
| CC (n = 197) | 11.5 (10.6, 12.4) | 0.05 | 11.8 (11.0, 12.6) | 0.02 |
| rs2036373d | ||||
| GG/GT (n = 231) | 10.5 (10.2, 10.8) | ref | 10.5 (10.3, 10.8) | ref |
| TT (n = 1,518) | 11.3 (10.5, 12.2) | 0.07 | 11.3 (10.6, 12.1) | 0.05 |
| rs9882205c | ||||
| GG (n = 874) | 11.1 (10.7, 11.5) | ref | 11.1 (10.7, 11.5) | ref |
| AG (n = 635) | 10.0 (9.6, 10.5) | 0.001 | 10.1 (9.7, 10.5) | 0.0002 |
| AA (n = 133) | 9.7 (8.9, 10.7) | 0.01 | 9.6 (8.8, 10.4) | 0.0016 |
| rs2241766c | ||||
| TT (n = 1,306) | 10.5 (10.2, 10.9) | ref | 10.6 (10.3, 10.9) | ref |
| GT (n = 346) | 10.4 (9.8, 11.0) | 0.6 | 10.4 (9.8, 10.9) | 0.52 |
| GG (n = 22) | 12.8 (10.1, 16.1) | 0.11 | 11.2 (9.1, 13.8) | 0.57 |
| rs3774261 | ||||
| GG (n = 656) | 10.0 (9.5, 10.4) | ref | 10.0 (9.7, 10.4) | ref |
| CG (n = 783) | 10.8 (10.3, 11.2) | 0.0083 | 10.8 (10.4, 11.1) | 0.0086 |
| CC (n = 307) | 11.7 (10.9, 12.4) | <0.0001 | 11.6 (10.9, 12.2) | <0.0001 |
| rs17366743 | ||||
| TT (n = 1,645) | 10.6 (10.3, 10.8) | ref | 10.6 (10.3, 10.8) | ref |
| CT (n = 103) | 11.4 (10.2, 12.7) | 0.17 | 11.4 (10.4, 12.6) | 0.11 |
| rs1063539 | ||||
| GG (n = 1,349) | 10.7 (10.3, 11.0) | ref | 10.7 (10.4, 11.0) | ref |
| CG (n = 371) | 10.4 (9.8, 11.0) | 0.41 | 10.4 (9.9, 10.9) | 0.38 |
| CC (n = 26) | 12.1 (9.7, 15.0) | 0.27 | 10.5 (8.6, 12.7) | 0.83 |
Geometric means.
Geometric means, adjusted for age, sex, and center.
These SNPs were genotyped on ~200 fewer CARDIA participants than the remaining SNPs.
GG and GT genotypes combined due to rare homozygotes, i.e., < n = 10 in the rare homozygote group.
Rs17300539 was marginally associated with serum adipo nectin in African Americans in models adjusted for age, sex, and center, P = 0.0087. The geometric mean adiponectin concentration for those with the AG genotype was 9.5 mg/l (95% CI (7.8, 11.5)) whereas those with the GG genotype had a mean of 7.3 mg/l (95% CI (7.0, 7.5)). No African-American participants had the AA genotype of rs17300539. The amount of variance explained using age, sex, center, and percent African ancestry was 9%; adding rs17300539 resulted in 11% of the variation in adiponectin levels explained. Further adjustment for percent African ancestry, waist circumference, BMI, and smoking did not substantially change these results (data not shown). No other statistically significant SNP associations were observed among African Americans.
Among whites, there was a significant interaction of waist and rs182052 in their association with serum adiponectin, P = 0.0029. Those with the AA genotype had 15.6% lower (95% CI (−21.3%, −9.4%) in serum adiponectin per standard deviation increase in waist circumference, while those with the AG genotype had a 22.8% lower (−25.4%, −20.1%) and those with the GG genotype had a 16.9% lower (−19.7, −14.0) serum adiponectin concentrations. There was also a significant interaction of rs9882205 and smoking among whites (P = 0.001). Smokers with the GG genotype had a geometric mean adiponectin concentration of 11.1 mg/l (95% CI (10.7, 11.5)), whereas those with the AG and AA genotypes had significantly lower mean adiponectin concentrations, 8.7 mg/l (95% CI (7.9, 9.6)) and 6.5 mg/l (95% CI (5.1, 8.2)), respectively. No other interactions of ADIPOQ SNPs and age, sex, BMI, or waist circumference were significant.
DISCUSSION
In this study, SNPs rs17300539, rs182052, rs822393, rs9882205, and rs3774261 within ADIPOQ were strongly associated with serum adiponectin concentrations in whites. In general, there was a dose–response relationship of mean adiponectin concentrations across genotypes. This study adds to the growing body of literature suggesting that rs17300539 is strongly associated with serum adiponectin levels in whites (21,22), and confirms the likely importance of four other SNPs associated with adipo nectin levels in whites that had previously been identified (22). Confirmatory studies such as the present study are necessary to ensure that the associations observed are true rather than spurious.
We identified only one SNP, rs17300539, in African Americans that had a marginally significant association with serum adiponectin. This is the first large scale study, to our knowledge, to examine the association of ADIPOQ SNPs with serum adiponectin levels in African Americans. Most SNPs associated with serum adiponectin levels in whites were not associated with serum levels in African Americans, with the exception of rs17300539, which had a marginal association. It is possible that this SNP is a functional variant or is in LD with another SNP that is a functional variant among both African Americans and whites. Differences in SNP findings between the whites and African Americans in this study could be due to different patterns of LD within race, i.e., it could be that none of these SNPs are in LD with functional SNPs of ADIPOQ in African Americans, but may be in LD among the whites.
A significant interaction of rs182052 and waist circumference was found in the association with serum adiponectin in whites. Although this interaction makes sense biologically given the evidence that adiponectin plays a role in central adiposity (16), it should be interpreted with caution given the number of interactions examined, and should be confirmed in independent samples in future studies. A significant interaction of rs9882205 with smoking was also found in whites. Smoking is associated with lower adiponectin concentrations (29), and cessation of smoking results in increased serum adiponectin concentrations, both in healthy subjects (30) and those with existing coronary heart disease (31).
Many previous studies have examined the influence of ADIPOQ SNPs on circulating adiponectin concentrations, and as much as 70% of the variation in adiponectin concentrations is thought to be due to variants in ADIPOQ (18). Two studies most similar to ours in sample size and ADIPOQ variants genotyped were performed among 1,727 whites (22) and 922 French whites (21). Heid et al. (22) found 9 of 15 tagSNPs, including rs822387, rs17300539, rs822388, rs182052, rs9882205, rs2241766, rs1501299, rs3774261, and rs4686804, significantly associated with circulating adiponectin concentrations. In the Vasseur et al. study (21), those with the A allele of rs17300539 had significantly higher adiponectin concentrations as compared to those with the G allele, P < 0.0001. In addition, the T allele of rs2241766 was significantly associated with higher adiponectin levels. A meta-analysis of ADIPOQ SNPs from the literature with greater than 2,000 cumulative participants concluded that fairly strong evidence exists for rs17300539 and rs2241766 influencing adiponectin concentrations, but there is far less evidence for rs1501299 (18). Among whites in our study, rs17300539, rs182052, rs822393, rs9882205, and rs3774261 were strongly associated with serum adiponectin concentrations. These results are consistent in direction and magnitude with Heid et al. (22) for all SNPs genotyped in both studies except rs2241766. The study by Heid et al. was very similar to ours not only in sample size and ethnic composition, but also in the tagSNP method used to choose the ADIPOQ SNPs. Similar to Vasseur et al. (21), our study found a strong association of rs17300539 with serum adiponectin, with increasing number of A alleles associated with higher adiponectin levels.
The most significant associations observed were between ADIPOQ SNPs rs17300539, rs182052, rs822393, rs9882205, and rs3774261 and serum adiponectin in whites. SNP rs17300539 lies in the promoter region of ADIPOQ, whereas rs182052, rs822393, rs9882205, and rs3774261 lie in intron regions of the gene. Introns are non-coding regions of a gene; however, there is evidence that introns of the protein-coding gene transcripts can affect gene expression by repressing translation or cleaving RNA transcripts (32). In particular, rs3774261 is an intronic enhancer, and thus could affect protein levels via enhancing transcription. Variants in the promoter region of ADIPOQ also affect circulating adiponctin concentrations by enhancing ADIPOQ promoter activity(18). We might have expected that either rs2241766 in exon 2 or rs17366743 in exon 3 to be associated with serum adiponectin given that they both lie in coding regions of the gene. However, rs17366743 is a rare mis-sense mutation (His-Tyr) so there could have been a lack of power to detect any associations of this SNP and adiponectin concentrations unless effects were large. Although rs2241766 lies in a coding region, it is a synonymous (Gly/Gly) SNP, which would not likely affect the adiponectin protein.
There are several important strengths of this study, but also some limitations. Using the tagSNP approach to choose SNPs enables the capturing of the variation across the adiponectin gene. Very few of the previous ADIPOQ-serum adiponectin studies have used a tagSNP approach to examine associations or have examined these associations in multiple ethnicities. On the other hand, some SNPs that were chosen with the tagSNP approach were not able to be genotyped because they could not be designed in the multiplex assays. It is also possible that there are important SNPs with minor allele frequencies of less than 0.10 used to choose the tagSNPs in this study. Other regions of ADIPOQ where SNPs were not genotyped could affect serum adiponectin concentrations, particularly in the exon or promotor regions of the gene. Despite using a tagSNP approach, this still does not ensure all known gene variation was captured.
We found variants of ADIPOQ associated with serum adiponectin concentrations in whites. This study replicates findings of earlier studies such as Heid et al. and Vasseur et al. that found the same ADIPOQ SNPs associated with serum adiponectin concentrations in whites. This article makes the novel contribution of evaluating tagSNPs of ADIPOQ with serum adiponectin levels in African Americans. In contrast to whites, we found only one ADIPOQ variant to be associated with serum adiponectin concentrations in African Americans, an association that was of only modest strength. There are still many ADIPOQ variants that have not been examined; so far only a small proportion of them have been studied (18) even in relation to serum adiponectin. Further, studies are needed to determine whether SNPs that replicate well are functional variants or if other SNPs may in fact be the functional variants.
Supplementary Material
ACKNOWLEDGMENTS
C.L.W. was supported by the NIH training grant in cardiovascular genetic epidemiology (T32 HL097972) and cardiovascular epidemiology and prevention (T32HL07779) during this research. The authors thank the CARDIA participants for their participation. Measurement of serum adiponectin was funded by the Young Adult Longitudinal Trends in Antioxidants (YALTA) ancillary study to CARDIA (R01 HL53560).
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Szmitko PE, Teoh H, Stewart DJ, Verma S. Adiponectin and cardiovascular disease. Am J Physiol Heart Circ Physiol. 2007;292:H1655–H1663. doi: 10.1152/ajpheart.01072.2006. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Montagnani M, Funahashi T, Shimomura I, Quon MJ. Adiponectin stimulates production of nitric oxide in vascular endothelial cells. J Biol Chem. 2003;278:45021–45026. doi: 10.1074/jbc.M307878200. [DOI] [PubMed] [Google Scholar]
- 3.Ekmekci H, Ekmekci OB. The role of adiponectin in atherosclerosis and thrombosis. Clin Appl Thromb Hemost. 2006;12:163–168. doi: 10.1177/107602960601200203. [DOI] [PubMed] [Google Scholar]
- 4.Karaduman M, Sengul A, Oktenli C, et al. Tissue levels of adiponectin, tumour necrosis factor-alpha, soluble intercellular adhesion molecule-1 and heart-type fatty acid-binding protein in human coronary atherosclerotic plaques. Clin Endocrinol (Oxf) 2006;64:196–202. doi: 10.1111/j.1365-2265.2006.02448.x. [DOI] [PubMed] [Google Scholar]
- 5.Li CJ, Sun HW, Zhu FL, et al. Local adiponectin treatment reduces atherosclerotic plaque size in rabbits. J Endocrinol. 2007;193:137–145. doi: 10.1677/JOE-06-0173. [DOI] [PubMed] [Google Scholar]
- 6.Shimada K, Miyazaki T, Daida H. Adiponectin and atherosclerotic disease. Clin Chim Acta. 2004;344:1–12. doi: 10.1016/j.cccn.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Pischon T, Girman CJ, Hotamisligil GS, et al. Plasma adiponectin levels and risk of myocardial infarction in men. JAMA. 2004;291:1730–1737. doi: 10.1001/jama.291.14.1730. [DOI] [PubMed] [Google Scholar]
- 8.Koenig W, Khuseyinova N, Baumert J, Meisinger C, Löwel H. Serum concentrations of adiponectin and risk of type 2 diabetes mellitus and coronary heart disease in apparently healthy middle-aged men: results from the 18-year follow-up of a large cohort from southern Germany. J Am Coll Cardiol. 2006;48:1369–1377. doi: 10.1016/j.jacc.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto N, Kanda J, Nakamura T, et al. Association of hypoadiponectinemia in men with early onset of coronary heart disease and multiple coronary artery stenoses. Metab Clin Exp. 2006;55:1653–1657. doi: 10.1016/j.metabol.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Frystyk J, Berne C, Berglund L, et al. Serum adiponectin is a predictor of coronary heart disease: a population-based 10-year follow-up study in elderly men. J Clin Endocrinol Metab. 2007;92:571–576. doi: 10.1210/jc.2006-1067. [DOI] [PubMed] [Google Scholar]
- 11.Sattar N, Wannamethee G, Sarwar N, et al. Adiponectin and coronary heart disease: a prospective study and meta-analysis. Circulation. 2006;114:623–629. doi: 10.1161/CIRCULATIONAHA.106.618918. [DOI] [PubMed] [Google Scholar]
- 12.Kanaya AM, Wassel Fyr C, Vittinghoff E, et al. Health ABC Study. Serum adiponectin and coronary heart disease risk in older Black and White Americans. J Clin Endocrinol Metab. 2006;91:5044–5050. doi: 10.1210/jc.2006-0107. [DOI] [PubMed] [Google Scholar]
- 13.Laughlin GA, Barrett-Connor E, May S, Langenberg C. Association of adiponectin with coronary heart disease and mortality: the Rancho Bernardo study. Am J Epidemiol. 2007;165:164–174. doi: 10.1093/aje/kwk001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnabel R, Messow CM, Lubos E, et al. Association of adiponectin with adverse outcome in coronary artery disease patients: results from the AtheroGene study. Eur Heart J. 2008;29:649–657. doi: 10.1093/eurheartj/ehn009. [DOI] [PubMed] [Google Scholar]
- 15.Steffes MW, Gross MD, Lee DH, Schreiner PJ, Jacobs DR., Jr Adiponectin, visceral fat, oxidative stress, and early macrovascular disease: the Coronary Artery Risk Development in Young Adults Study. Obesity (Silver Spring) 2006;14:319–326. doi: 10.1038/oby.2006.41. [DOI] [PubMed] [Google Scholar]
- 16.Kadowaki T, Yamauchi T, Kubota N, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann TS, Li W, Dominguez H, et al. Quinapril treatment increases insulin-stimulated endothelial function and adiponectin gene expression in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91:1001–1008. doi: 10.1210/jc.2005-1231. [DOI] [PubMed] [Google Scholar]
- 18.Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:1198–1209. doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- 19.Jang Y, Lee JH, Chae JS, et al. Association of the 276G->T polymorphism of the adiponectin gene with cardiovascular disease risk factors in nondiabetic Koreans. Am J Clin Nutr. 2005;82:760–767. doi: 10.1093/ajcn/82.4.760. [DOI] [PubMed] [Google Scholar]
- 20.Qi L, Li T, Rimm E, et al. The +276 polymorphism of the APM1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes. 2005;54:1607–1610. doi: 10.2337/diabetes.54.5.1607. [DOI] [PubMed] [Google Scholar]
- 21.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 22.Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- 23.Hoefle G, Muendlein A, Saely CH, et al. The -11377 C>G promoter variant of the adiponectin gene, prevalence of coronary atherosclerosis, and incidence of vascular events in men. Thromb Haemost. 2007;97:451–457. [PubMed] [Google Scholar]
- 24.Filippi E, Sentinelli F, Romeo S, et al. The adiponectin gene SNP+276G>T associates with early-onset coronary artery disease and with lower levels of adiponectin in younger coronary artery disease patients (age <or=50 years). J Mol Med. 2005;83:711–719. doi: 10.1007/s00109-005-0667-z. [DOI] [PubMed] [Google Scholar]
- 25.Hughes GH, Cutter G, Donahue R, et al. Recruitment in the Coronary Artery Disease Risk Development in Young Adults (Cardia) Study. Control Clin Trials. 1987;8:68S–73S. doi: 10.1016/0197-2456(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 26.Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Avery CL, Martin LJ, Williams JT, North KE. Accuracy of haplotype estimation in a region of low linkage disequilibrium. BMC Genet. 2005;6(Suppl 1):S80. doi: 10.1186/1471-2156-6-S1-S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swarbrick MM, Havel PJ. Physiological, pharmacological, and nutritional regulation of circulating adiponectin concentrations in humans. Metab Syndr Relat Disord. 2008;6:87–102. doi: 10.1089/met.2007.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Efstathiou SP, Skeva II, Dimas C, et al. Smoking cessation increases serum adiponectin levels in an apparently healthy Greek population. Atherosclerosis. 2009;205:632–636. doi: 10.1016/j.atherosclerosis.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Otsuka F, Kojima S, Maruyoshi H, et al. Smoking cessation is associated with increased plasma adiponectin levels in men. J Cardiol. 2009;53:219–225. doi: 10.1016/j.jjcc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Ying SY, Lin SL. Intron-derived microRNAs–fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.