Abstract
Transcriptional enhancers are short segments of genomic DNA (50 bp to 1 kb in length) that can work over long distances (≥1 Mb) to regulate gene expression in specific cells and tissues. Genomic assays have identified on the order of 400,000 to one million putative enhancers in the human genome (e.g., ENCODE Consortium). This suggests that a typical gene is regulated by tens of enhancers, ensuring stringent regulation of gene expression in response to a variety of intrinsic and external signals. Despite the discovery of the first transcriptional enhancer more than 30 years ago, we know surprisingly little about how enhancers regulate gene expression. In particular, the relationship between primary DNA sequence and enhancer specificity remains obscure. Here we summarize recent high-throughput studies in whole embryos aimed at the systematic identification of the sequence and organizational constraints underlying enhancer function and specificity.
A detailed understanding of how enhancers mediate tissue-restricted gene expression requires the functional analysis of thousands or even millions of enhancer variants in all of the cell types of an organism. This has not been possible in the context of whole animals because the methods of creating transgenic organisms are slow and tedious. For example, it takes weeks for a single enhancer variant to be incorporated in the Drosophila germline and then examined in F1 and F2 descendants. Similar experiments take even longer, several months, in vertebrate systems. These low-throughput methods have hindered comprehensive structure–function studies of transcriptional enhancers.
To circumvent this bottleneck we have exploited the potential of the model chordate, Ciona intestinalis, commonly known as sea squirts, which are a member of the tunicates, the closest living relatives of vertebrates (Delsuc et al. 2006). Despite the modest appearance of adults, the embryos and larvae look like simplified vertebrate tadpoles with a head and a tail that possess a prominent notochord and dorsal neural tube. These tadpoles share many common properties with vertebrates, including simplified rudiments for vertebrate-specific innovations such as second heart field (Stolfi et al. 2010), neural crest (Abitua et al. 2012; Stolfi et al. 2015), and neurogenic placodes (Abitua et al. 2015). In addition to these vertebrate features, Ciona also possesses a particularly compelling property for high-throughput structure–function studies of developmental enhancers. Namely, thousands of synchronously developing embryos can be readily transformed with transgenic DNA using simple electroporation methods (Corbo et al. 1997; Christiaen et al. 2009).
Ciona has become a laboratory workhorse because of its worldwide distribution, the ease with which it can be experimentally manipulated, and its durability in culture. Adults are hermaphrodites and thereby contain both sperm and eggs. These are easily extracted from a few adults, mixed to produce fertilized eggs, combined with transgenic DNA, and then electroporated en masse to generate thousands of synchronously developing transformed embryos (Corbo et al. 1997; Christiaen et al. 2009). These methods have been successfully used to identify a number of tissue-specific enhancers, including those mediating expression in the central nervous system (Bertrand et al. 2003; Stolfi et al. 2011) and notochord (Corbo et al. 1997).
Typically one or just a handful of transgenic DNAs are electroporated into thousands of fertilized Ciona eggs. We sought to multiplex the facile electroporation method to conduct a high-throughput analysis of the best-characterized developmental enhancer in Ciona, the Otx-a enhancer, which mediates expression in the neural plate (Fig. 1; Bertrand et al. 2003; Rothbacher et al. 2007; Khoueiry et al. 2010). The enhancer is only 69 bp in length and contains five well-defined DNA binding motifs: two ETS sites that mediate activation by fibroblast growth factor (FGF) signaling and three GATA binding sites that restrict expression to the ectoderm (Rothbacher et al. 2007; Khoueiry et al. 2010). Combinatorial activation of the Otx-a enhancer by pleiotropic FGF signaling and the tissue-determinant GATA ensure restricted expression in the neural plate (Rothbacher et al. 2007; Khoueiry et al. 2010). The Otx-a enhancer is not normally activated in any of the other tissues that receive FGF signals, such as the notochord, hindbrain, anterior endoderm, and heart (Hudson and Lemaire 2001; Bertrand et al. 2003; Yasuo and Hudson 2007; Shi and Levine 2008; Stolfi et al. 2011; Wagner and Levine 2012).
Figure 1.
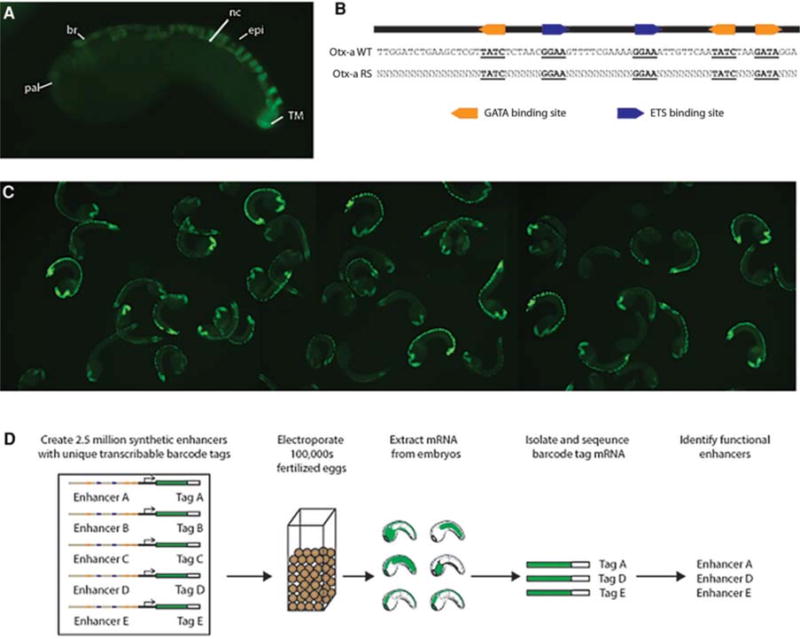
High-throughput characterization of the Otx-a enhancer. (A) Endogenous Otx-a expression pattern. Expression can be seen in the anterior neural plate: anterior brain (br) and palps (pal), dorsal nerve chord (nc), dorsal epidermis (epi), and two tail muscle cells (TM). (B) Sequence of the Otx-a enhancer and schematic showing randomized variants. (C) Small proportion of an electroporation. (D) Schematic of experimental approach. Electroporation of library of Otx-a enhancer variants into Ciona embryos to identify functional enhancer variants. Analysis of functional variants enables identification of principles governing tissue-specific gene expression patterns. (Adapted from Farley et al. 2015.)
To understand the basis for tissue-specific induction in the neural plate, we created millions of synthetic variants of the Otx-a enhancer (Fig. 1B). These were systematically electroporated into thousands of embryos, which were grown to the stage when the Otx gene is normally activated. RNA was extracted from these embryos, reverse transcribed to produce cDNAs, and then systematically sequenced to identify active enhancers using a barcoding scheme that links specific 3′ transcribed barcodes with particular enhancer variants (Fig. 1D).
SUBOPTIMAL AFFINITY BINDING SITES ARE ESSENTIAL FOR RESTRICTED EXPRESSION
Altogether, our analysis identified approximately 20,000 functional enhancer variants. This collection represents, by far, the largest data set of active enhancers identified in developing embryos. Computational methods were used to identify sequence features overrepresented in the active enhancers. This analysis led to a clear-cut finding: Short dinucleotide sequences flanking each of the five core ETS and GATA binding sites are necessary and sufficient for full enhancer activity in the neural plate (Fig. 2A). The consensus flanking sequences that were identified, CCGGAARY and SMGATAAS, possess 0.98 and 0.96 Pearson coefficient correlations with the highest-affinity position weight matrices identified by high-throughput binding assays for orthologous ETS and GATA transcription factors in flies, mice, and humans (Badis et al. 2009; Wei et al. 2010; Jolma et al. 2013; Nitta et al. 2015).
Figure 2.
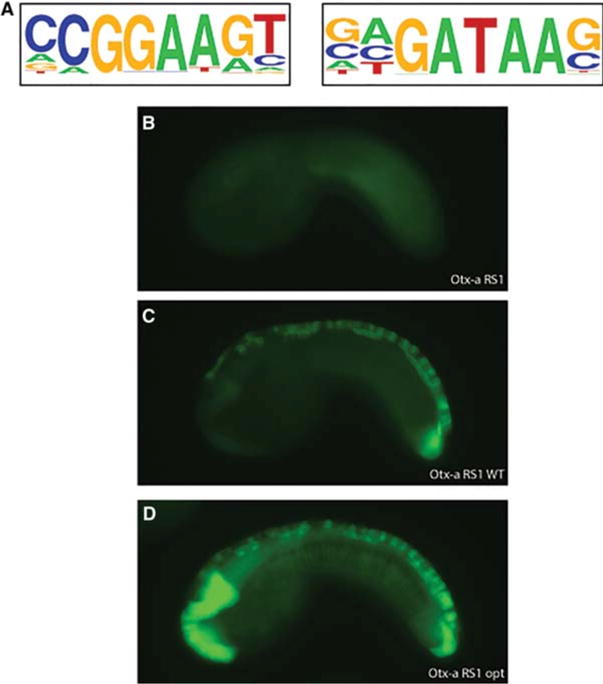
Suboptimal affinity binding sites are necessary and sufficient for tissue-specific enhancer activity. (A) Position weight matrices (PWMs) of the motifs enriched in functional enhancers. These PWMs match those identified by high-throughput in vitro binding assays (Badis et al. 2009; Wei et al. 2010; Jolma et al. 2013; Nitta et al. 2015). (B) Otx-a RS1—an inert enhancer variant that drives no expression in reporter assays. (C) Otx-a RS1 WT, in which the dinucleotide flanking of the binding sites has been altered so the affinity matches that of the native Otx-a enhancer. Changes to the dinucleotides create a functional tissue-specific neural enhancer indistinguishable from the Otx-a enhancer. (D) Embryo electroporated with Otx-a RS1 opt, a variant in which the dinucleotides flanking the binding sites have been changed so these sites are the highest-affinity sites. Expression is seen in the native neural location as well as ectopic expression in the notochord, endoderm, and posterior brain—all regions of FGF signaling. All images are taken at the same exposure with no manipulation. (Adapted from Farley et al. 2015.)
The sufficiency of these dinucleotide flanking sequence motifs was showed by “Lazarus” experiments, whereby inactive enhancer variants were restored by simply altering these sequences (Fig. 2). Changing the flanking dinucleotide sequences was sufficient to restore the activities of an inactive enhancer that essentially lacks sequence homology with the native Otx-a enhancer, except for the five core ETS (GGAA) and GATA binding sites (Fig. 2B,C). In this first experiment the flanking dinucleotides were modified to mimic the sequences present in the native Otx-a enhancer. This produces normal levels of expression and specific expression within the neural plate (Fig. 2C).
To our surprise, the native Otx-a enhancer contains mediocre matches to the consensus flanking sequences identified by our high-throughput functional assays. Converting the flanking sequences in the inactive Otx-a variant (Fig. 2A) to produce perfect matches to the consensus dinucleotide motifs, and thus creating the highest-affinity binding motifs, causes both an increase in the levels of green fluorescent protein (GFP) reporter gene expression and a loss of tissue specificity (Fig. 2D). Significant expression is now observed at additional sites of FGF signaling, including the notochord. These results suggest that the native Otx-a enhancer contains binding sites with suboptimal affinity, and these are essential for restricted activity in the neural plate (Farley et al. 2015). Other studies have highlighted the importance of suboptimal binding motifs for tissue-specific patterns of gene expression in both Drosophila and differentiating T cells (Small et al. 1991; Hentsch et al. 1992; Jiang and Levine 1993; Swanson et al. 2011; Ramos and Barolo 2013; Crocker et al. 2015).
SUBOPTIMAL SPACING OF LINKED SITES IS FOUND IN THE ENDOGENOUS ENHANCER
Having identified the sequence constraints sufficient for tissue-specific expression, we next asked whether the spacing between binding sites was important for enhancer activity. Our reasoning is that cooperative interactions of GATA and ETS, or cooperative recruitment of secondary “coactivators,” might be important for restricted expression in the neural plate. Manipulations of the native Otx-a enhancer and synthetic variants showed that the spacing between ETS and GATA sites influences the levels of gene expression (Farley et al. 2015). In the native enhancer, the 5′ pair of GATA and ETS binding sites is separated by 10 bp, whereas the 3′ ETS and GATA sites are separated by 13 bp. In both the endogenous enhancer and synthetic variants, changing the spacing of the 5′ pair from 10 to 13 bp caused a significant increase in expression. Conversely, changing the spacing of both pairs of ETS and GATA to 10 bp (10–10 spacing) results in very poor expression (Farley et al. 2015). These findings suggest that the native Otx-a enhancer contains mediocre ETS and GATA binding motifs that are poorly organized for optimal transcriptional output. This suboptimization appears to be important for tissue-specific expression in the neural plate.
OPTIMIZATION OF THE Otx-a ENHANCER RESULTS IN LOSS OF TISSUE SPECIFICITY
To test this idea of suboptimization, we created a synthetic Otx-a enhancer that contains optimized binding motifs and spacing. Each ETS and GATA recognition sequence corresponds to the highest-affinity binding site, and both pairs of sites are separated by 13 bp. The resulting enhancer mediates intense expression in the neural plate, as well as significant expression in most of the other tissues receiving FGF signals, including the notochord, hindbrain, and anterior endoderm (Fig. 3B; cf. 3A). These results suggest that the evolution of developmental enhancers is constrained by the need for tissue specificity. Design features that allow optimal biochemical interactions cause a loss of specificity.
Figure 3.
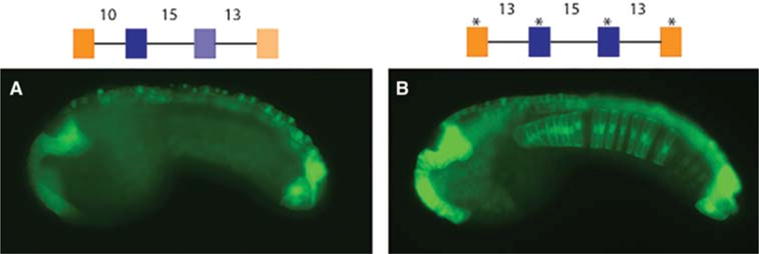
Suboptimization of spacing and binding affinity is required for tissue specificity. (A) Embryo electroporated with WT enhancer with suboptimal spacing and suboptimal affinity binding sites mediates weak expression restricted to the neural tissue. (B) Embryo electroporated with a synthetic enhancer with high-affinity binding sites and spacing optimized for transcriptional output. This “optimized” enhancer shows strong expression in the entire nervous system, notochord, and endoderm. Schematics of the enhancers are shown above the images. Blue boxes depict ETS binding sites, orange GATA binding sites. Asterisks above binding sites represent high-affinity sites. Numbers describe distance in base pairs between binding sites. All images are taken at the same exposure with no manipulation. (Adapted from Farley et al. 2015.)
TRADE-OFF IN LEVELS AND SPECIFICITY OF GENE EXPRESSION
Enhancers with suboptimal binding affinities and suboptimal organization produce specific but weak patterns of expression. It is possible that this trade-off in specificity and expression levels is circumvented by clusters of enhancers with overlapping activities. To test this idea, we multimerized a suboptimized Otx-a enhancer. A single copy of this enhancer directs only very weak expression of the GFP reporter gene in the neural plate, whereas two tandem copies generate strong and specific expression (Farley et al. 2015).
The preceding results suggest that there are two ways to obtain strong expression in the neural plate: (1) optimized binding motifs and spacing or (2) multimers of suboptimized enhancers. Optimization results in ectopic sites of expression in response to pleiotropic FGF signaling, whereas suboptimal multimers retain tissue specificity. We suggest that this principle of suboptimization is the basis for many cases of enhancer clusters such as “super-enhancers” in vertebrate genomes. It might also explain why so many of the patterning genes in the Drosophila embryo contain pairs of primary and shadow enhancers (Hong et al. 2008; Perry et al. 2010; Cannavo et al. 2016).
TRADE-OFF IN BINDING AFFINITIES AND SYNTAX
The organization of the native Otx-a enhancer raises the possibility of a balance between binding affinities and regulatory syntax (spacing and orientation of linked sites). The Otx-a enhancer contains two pairs of linked ETS and GATA sites (Fig. 4). The distal pair shows suboptimal spacing (10 bp) but possesses relatively high binding affinities (0.9 and 0.6 of the optimal motifs, respectively). In contrast, the proximal pair is separated by the optimal distance (13 bp) but possesses lower relative affinities (0.4 and 0.2 of the optimal motifs, respectively). This observation raises the possibility that there is a trade-off between affinity and spacing of sites, whereby optimal sites have suboptimal spacing to maintain restricted expression.
Figure 4.
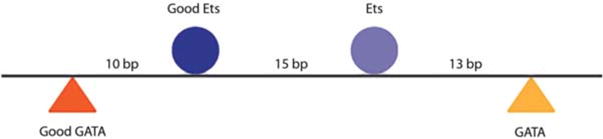
The Otx-a enhancer has a balance between affinity and regulatory syntax. Schematic of Otx-a enhancer. The 5′ pair of ETS and GATA have higher-affinity binding sites but spacing that is suboptimal for transcriptional output. In contrast, the second pair of ETS and GATA have lower affinities but optimal spacing for transcriptional output.
To test this idea we characterized the RS 6 synthetic Otx-a enhancer, which directs strong expression in the notochord, another tissue that receives FGF. This expression is mediated by Ets and a fortuitous ZicL binding site. These factors have previously been shown to be important for activation of the Ciona Brachyury gene (Ci-Bra), a T-box transcription factor that controls notochord differentiation (Imai et al. 2002; Yagi et al. 2004; Yasuo and Hudson 2007). A variety of manipulations suggest that the optimal spacing between this ZicL site and the distal ETS site is 11 bp. Moreover, orientation of linked ZicL and ETS sites also plays a role in enhancer function. Simple inversion of the ETS site abolishes expression of the synthetic enhancer in the notochord (Farley et al. 2016). The specificity of notochord expression, like neural expression, is determined by binding sites with suboptimal affinities and syntax (spacing, order, and orientation of sites). This finding suggests that enhancer suboptimization is a general mechanism for controlling tissue specificity.
We surveyed the Ciona genome for putative notochord enhancers containing linked ZicL and ETS binding sites with appropriate constraints on spacing and orientation. One of the sequences that we identified represents a particularly stringent test of the idea of compensation between binding affinities and regulatory syntax. It contains two ETS sites and a ZicL site showing perfect regulatory syntax (viz., 11-bp spacing and convergent orientation of the linked ETS site) (Fig. 5). However, this sequence contains very poor ETS sites that possess just 0.14 and 0.25 relative binding affinities compared with the optimal site. It nonetheless functions as a surprisingly strong notochord enhancer.
Figure 5.
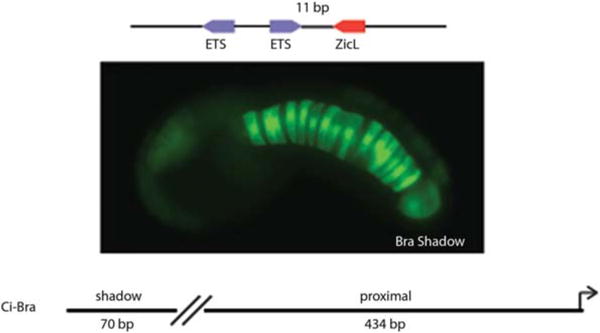
Regulatory syntax can compensate for poor binding affinity to mediate tissue-specific enhancer activity. A 70-bp enhancer with poor-affinity ETS binding sites but optimal grammar mediates restricted expression in the notochord. This enhancer is a shadow enhancer to the previously defined proximal Bra enhancer. (Adapted from Farley et al. 2016.)
A 70-bp genomic DNA segment encompassing this region was attached to the GFP reporter gene and expressed in electroporated embryos (Fig. 5). It directs robust and restricted expression in the notochord, suggesting that regulatory syntax can indeed compensate for low-affinity binding sites. This newly identified enhancer maps 300 bp upstream of a previously identified notochord-specific enhancer for the Ci-Bra gene. It appears to represent the first example of a shadow enhancer in the Ciona genome; such enhancers are common in Drosophila and vertebrates (Hong et al. 2008; Perry et al. 2010; Nolte et al. 2013).
CONCLUSION
We propose that regulatory syntax is an underappreciated property of developmental enhancers because of a systematic bias in the way that enhancers are identified in whole-genome surveys. The newly identified Ci-Bra shadow enhancer would fall below the cutoff values used by in silico detection methods because of poor matches with the ETS and ZicL position weight matrices. Conversely, the identification of enhancers with good matches to these matrices are likely to lack optimal syntax because this would be expected to trigger ectopic sites of gene activation (e.g., Fig. 3). These properties, high-affinity binding sites or optimal syntax, represent extreme ends of a spectrum of enhancer design. It is likely that a typical developmental enhancer has a mixture of optimal and suboptimal binding site affinities and syntax, as seen for the native Otx-a enhancer.
An important implication of enhancer suboptimization is that sequence polymorphisms might augment binding affinities and thereby cause gain-of-function expression in unwanted tissues. It is therefore possible that enhancer “optimization” could cause developmental disorders. Such a mechanism might pertain to certain forms of polydactyly in humans. The long-range ZRS (zone of polarizing activity regulatory sequence) enhancer regulates sonic hedgehog (Shh) expression in developing limb buds (Lettice et al. 2008). This enhancer normally regulates restricted expression within the ZPA (zone of polarizing activity) of the limb bud in response to FGF signaling, but mutations in the ZRS cause ectopic activation of Shh and polydactyly (Lettice et al. 2008, 2014). Indeed, sequence changes found in families with polydactyly occur in the critical dinucleotide sequences flanking core ETS binding sites, potentially augmenting their affinities and causing excessive activation by FGF. Such a scenario could underlie other diseases such as autoimmunity and cancer.
Acknowledgments
This work was supported by National Institutes of Health grants GM46638 and NS076542.
References
- Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abitua PB, Gainous TB, Kaczmarczyk AN, Winchell CJ, Hudson C, Kamata K, Nakagawa M, Tsuda M, Kusakabe TG, Levine M. The pre-vertebrate origins of neurogenic placodes. Nature. 2015;524:462–465. doi: 10.1038/nature14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115:615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, Ciglar L, Korbel JO, Furlong EE. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol. 2016;26:38–51. doi: 10.1016/j.cub.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L, Wagner E, Shi W, Levine M. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5345. pdb prot5345. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science. 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1605085113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentsch B, Mouzaki A, Pfeuffer I, Rungger D, Serfling E. The weak, fine-tuned binding of ubiquitous transcription factors to the Il-2 enhancer contributes to its T cell–restricted activity. Nucleic Acids Res. 1992;20:2657–2665. doi: 10.1093/nar/20.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100:189–203. doi: 10.1016/s0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129:2723–2732. doi: 10.1242/dev.129.11.2723. [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol. 2010;20:792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Hill AE, Devenney PS, Hill RE. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum Mol Genet. 2008;17:978–985. doi: 10.1093/hmg/ddm370. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Williamson I, Devenney PS, Kilanowski F, Dorin J, Hill RE. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141:1715–1725. doi: 10.1242/dev.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EE, et al. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife. 2015;4 doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C, Jinks T, Wang X, Martinez Pastor MT, Krumlauf R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev Biol. 2013;383:158–173. doi: 10.1016/j.ydbio.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AI, Barolo S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130018. doi: 10.1098/rstb.2013.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbacher U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and b-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134:4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135:931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Wagner E, Taliaferro JM, Chou S, Levine M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development. 2011;138:5429–5439. doi: 10.1242/dev.072108. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature. 2015;527:371–374. doi: 10.1038/nature15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Schwimmer DB, Barolo S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr Biol. 2011;21:1186–1196. doi: 10.1016/j.cub.2011.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, Levine M. FGF signaling establishes the anterior border of the Ciona neural tube. Development. 2012;139:2351–2359. doi: 10.1242/dev.078485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Satoh N. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development. 2004;131:1279–1288. doi: 10.1242/dev.01011. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Hudson C. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev Biol. 2007;302:92–103. doi: 10.1016/j.ydbio.2006.08.075. [DOI] [PubMed] [Google Scholar]


