Abstract
This study investigated whether the expression of CD44 variant 9 (CD44v9) might be a functional marker of tumor‐initiating stem‐like cells in primary hepatocellular carcinomas (HCCs) of hepatitis C virus (HCV)+ patients and provide an indicator of patient survival, as well as associated mechanisms. A total of 90 HCV + HCC patients who underwent surgery from 2006 to 2011 were enrolled and monitored for 2–8 years. Expression of CD44v9 was validated immunohistochemically in all HCCs, followed by comparative proteome, survival, and clinicopathological analyses. CD44 variant 8–‐10 was further evaluated in diethylnitrosamine‐induced HCCs of C57Bl/6J mice. Focally localized CD44v+ cells with a membranous staining pattern were detected in human HCV + and mouse HCCs. CD44v9+ cells of HCCs were predominantly negative for Ki67 and P‐p38, indicating decrease of cell proliferation in the CD44v9+ tumor cell population, likely to be related to suppression of intracellular oxidative stress due to activation of Nrf2‐mediated signaling, DNA repair, and inhibition of xenobiotic metabolism. CD44v9 IHC evaluation in 90 HCV + HCC cases revealed that positive expression was significantly associated with poor overall and recurrence‐free survival, a younger age, poor histological differentiation of HCCs, and high alkaline phosphatase levels compared with patients with negative expression. CD44v9 is concluded to be a potential biomarker of tumor‐initiating stem‐like cells and a prognostic marker in HCV + HCC patients associated with Nrf2‐mediated resistance to oxidative stress.
Keywords: Biomarker, CD44v9, hepatocellular carcinoma, oxidative stress, TISC
Hepatocellular carcinoma is the most common primary malignant tumors in liver, accounting for approximately 80% of all hepatic cancer cases worldwide. Surgical resection and liver transplantation are generally applied for early‐stage HCCs, but the prognosis of HCC patients is generally poor due to high levels of tumor invasion and resistance to chemotherapy.1 Accumulating evidence suggests that subpopulations of cancer cells with stem‐like cell properties, so‐called CSCs or TISCs, play a critical role in HCC development and progression.2 Recent studies reported that dedifferentiated hepatocytes, hepatic oval cells, and bone marrow cells are the three major types of liver stem cells and CD133, CD44, CD90, OV6, epithelial cell adhesion molecule, and CD13 have been identified as specific antigenic markers for CSCs isolated from different human HCC cell lines or specimens.2, 3, 4 Wnt, Hedgehog, Notch, TGF‐β, and angiogenic signaling are main pathways that regulate HCC stem cell self‐renewal, differentiation, and survival, and may be potential targets for novel therapeutic strategies of HCC.2, 4
CD44 is a single‐pass type I transmembrane protein that serves as a cell surface receptor and functions as a cellular adhesion molecule for hyaluronic acid, a major component of the ECM.5 It is involved in a wide variety of physiological processes, such as leukocyte homing and activation, wound healing, and cell migration.1, 5 It has been shown that the existence of alternative splicing of CD44 variant exons generates many CD44 variant isoforms (CD44v), which function differentially.5, 6 Furthermore, CSCs with an epithelial phenotype predominantly express isoforms containing variant exons, whereas CSCs that have undergone an epithelial–mesenchymal transition downregulate these variant isoforms and upregulate expression of the standard CD44 isoform that contains no variant exons.7 Expression of CD44v in some tissues appears to relate to tumor progression, and in particular to metastatic potential of some cancers.1, 8, 9, 10
Expression of the CD44v6 isoform has been found to be significantly correlated with a poor prognosis and more aggressive stages of disease in patients with high‐grade non‐Hodgkin's lymphoma,11 as well as stomach, colorectal, breast, and cervical cancers, with suggested potential as a CSC marker.12, 13, 14 However, significant correlations between CD44v6 expression and clinicopathological factors in patients appear to be lacking. Recently, it was reported that interaction of CD44v8‐10 with xCT (SLC7A11), a subunit of the glutamate–cystine transporter system xc(−), stabilizes the latter and thereby potentiates the ability of cancer cells to promote glutathione synthesis and control the cell redox potential, making TISCs resistant to induction of ROS, this being important for CSC survival.15, 16 CD44 variant 9 has been reported to feature an xCT interaction site.17 Furthermore, it was recently proposed as a potential predictive marker for recurrence in early gastric cancers.18 Overexpression was found in metaplastic cells in gastric cancer under chronic inflammation conditions, suggesting induction in response to inflammation and possible promotion of metaplasia of the gastric epithelium.19 To establish whether CD44v9 might be a novel reliable TISC marker for human HCC, in the present study we assessed its expression in HCV+ human HCCs and DEN‐induced mouse hepatomas. Proteome analysis of CD44v9+ and CD44v9− HCCs and adjacent liver tissues was used for the investigation of CD44v9‐related proteins, upstream regulators, molecular functions, and signaling and canonical pathways. Furthermore, the possible clinical significance of CD44v9 was investigated in 90 HCV+ HCC patients by IHC analysis.
Materials and Methods
Chemicals
All reagents were obtained from Sigma (St. Louis, MO, USA) or Wako Pure Chemicals Industries (Osaka, Japan). N‐nitrosodiethylamine was purchased from Sakai Research Laboratory (Fukui, Japan).
Institutional review board approval and informed consent
The present study was approved by the Osaka City University Graduate School of Medicine ethics committee (Osaka, Japan). Written informed consent was obtained from the patients prior to the study. The study was carried out in accordance with the principles of the Declaration of Helsinki.
Human tissue specimens, patients, and treatment
This prospective study included tissue specimens obtained from 90 HCV+ patients with primary HCC undergoing surgery at Osaka City University Hospital (Osaka, Japan) from January 2006 to December 2011. All of these patients were histologically proven to have HCC and were enrolled in this study to assess the clinical significance of expression levels of CD44v9 in resected liver specimens of primary HCCs. Information on the features of the patients such as gender, age, body mass index, smoking history, tumor volume, and tumor differentiation were obtained from medical records (Table S1). There were 62 male and 28 female patients, with a median age of 72 years (range, 45–84 years) at the time of surgery. Histopathological analysis and staging were carried out in accordance with the Japanese classification of hepatocellular carcinoma.20 The pathological diagnoses were made by at least two pathologists from the pathology department in our hospital according to the criteria of the general guidelines for primary liver cancer of the American Joint Committee on Cancer/International Union Against Cancer Staging Systems21 and the Liver Cancer Study Group of Japan.20, 22 Fifty‐four of the 90 patients were diagnosed with recurrence.
Animals, experimental design, and histopathology
A total of 32 male 7‐day‐old C57BL/6J mice (CLEA, Tokyo, Japan) were quarantined for 1 week before the start of the experiment. They were housed in an animal facility maintained on a 12:12 h (07:00–19:00) light : dark cycle, at a constant temperature of 23 ± 1°C and relative humidity of 44 ± 5%, and were given free access to tap water and food (Oriental MF pellet diet; Oriental Yeast Co., Tokyo, Japan). All experimental procedures were carried out following approval of the Animal Care and Use Committee of the Osaka City University Graduate School of Medicine.
A total of 32 male 14‐day‐old C57Bl/6J mice were divided into two experimental groups. Twenty‐four animals underwent an i.p. injection of DEN (10 mg/kg b.w.) and eight mice (control) were injected with saline. At 38 weeks the mice were killed. Livers were macroscopically examined and separate portions fixed in 10% phosphate‐buffered formalin for histopathological assessment and IHC analyses.
Immunohistochemistry and scoring
Single IHC for CD44v and double IHC for Ki67, P‐p38, Nrf2, p62‐SQSTM1, Keap1, and CD44v were carried out using the ABC method as described previously.23 In double IHC, CD44v was visualized with 3,3′‐diaminobenzidine tetrahydrochloride solution (DAKO, Tokyo, Japan) or alkaline phosphatase (Vectastain ABC‐AP kit, Vector red; Vector Laboratories, Burlingame, CA, USA), and Ki‐67, P‐p38, Nrf2, p62‐SQSTM1, and Keap1 with alkaline phosphatase (Vectastain ABC‐AP kit, Vector blue; Vector Laboratories, Burlingame, CA, USA). CD44v was detected with rat mAbs against human CD44v9 (RV3; 1:500) and mouse CD44v8‐10 (CD44v10‐e16 [RM1]; 1:300), kindly provided by Prof. H. Saya (Keio University, Tokyo, Japan), generated as previously described.17 Ki67 was detected with rabbit mAbs (1:300; Abcam, Tokyo, Japan) and P‐p38 with mouse mAbs (1:150; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Rabbit polyclonal antibodies for Nrf2 (1:100; Abcam, Tokyo, Japan), p62‐SQSTM1 (1:1000; MBL Co., Osaka, Japan), and Keap1 (1:200; Abcam, Tokyo, Japan) were applied. The membranous CD44v expression levels in cancer cells of the tumors were interpreted using guidelines published in previous studies.24 For human CD44v9, the results were graded from 0 to 3+ as follows: 0, no staining; 1+, 1–25% staining; 2+, 26–50% staining; 3+, >50% of the specimen was stained. Ki67 and P‐p38 IHC scores were calculated in CD44v9+ and CD44v9− tumor areas as the averages of the percentage of Ki67+ or P‐p38+ cells in each of three high‐power fields of areas containing tumor cells.
QSTAR Elite LC‐MS/MS
Of the 90 HCV+ HCC patients, five cases each of CD44v9+ and CD44v9− HCCs were randomly selected for proteome analysis. Samples from dissected 10% phosphate‐buffered formalin‐fixed HCCs and adjacent normal‐appearing tissues were prepared in liquid tissue buffer (AMR, Tokyo, Japan), treated with heat at 90°C for 90 min and digested with trypsin at 37°C for 16–18 h. Proteome analysis was carried out with the selected CD44v9+ and CD44v9− HCCs and adjacent normal tissue samples (20 μg each) with a DiNa‐AI nano LC System (KYA Technologies, Tokyo, Japan) coupled to a QSTAR Elite hybrid mass spectrometer (AB Sciex, Concord, ON, Canada) through a NanoSpray ion source (AB Sciex), as previously described.25 For the quantitative MS/MS analysis, labeling was carried out with 4‐plex iTRAQ reagents.25 The LC‐MS/MS and ProteinPilot analyses results of five CD44v9+ and five CD44v9− patients were averaged and further underwent IPA (Ingenuity Systems, Mountain View, CA, USA) to investigate protein molecular functions, localization, and identification of networks of interacting proteins, as well as functional groups and pathways and altered upstream regulators.
Statistical analyses
All statistical analyses were carried out using SPSS statistics version 19.0 (SPSS, Chicago, IL, USA). Statistical significance of any associations between CD44v9 immunohistochemical staining and various clinicopathological variables was evaluated using χ2‐ and Fisher's tests. Survival curves were calculated from the day of surgery to relapse, death, or the last follow‐up observation using the Kaplan–Meier method, and differences in overall and recurrence‐free survival curves were assessed with the log–rank test. In all applied analyses, P‐values < 0.05 were considered statistically significant. Univariate and multivariate Cox proportional hazard model analyses were carried out to calculate HRs and to determine associations between clinicopathologic variables and disease‐specific mortality. Univariate analysis describes survival with respect to the factor under investigation, but necessarily ignores the impact of any others. We therefore undertook a multivariate survival analysis to compare prognostic factors of survival after adjustment for the impact of other factors. Different potential predictive variables were evaluated in the univariate analysis: CD44v9 (positive vs negative), age (>70 vs ≤70 years), diabetes (positive vs negative), fibrosis (stages 3 and 4 vs stages 1 and 2), aspartate transaminase value (>34 vs 13–33 IU/L), alanine aminotransferase value (>28 vs 6–27 IU/L), ALP value (>360 vs 115–359 IU/L), tumor size (≥2 vs <2 cm3), vessel invasion (positive vs negative), infiltration to capsule by tumor cells (positive vs negative), tumor differentiation (poor vs well and moderate), T category (T2–4 vs T1), stage (stages 2–4 vs stage 1), and recurrence (positive vs negative). A Cox proportional hazard regression model was used to estimate prognostic factors associated with survival in the multivariate analysis.
Results
Representative results of CD44v9 IHC in HCV+ human and mouse HCCs
Representative CD44v9 immunohistochemical staining is illustrated in Fig. 1(A). In the specimens of 90 patients examined, CD44v9 was strongly elevated in 24 cases (26.7%; score 3+), moderately expressed in 16 cases (17.8%; score 2+), weakly expressed in 25 cases (27.8%: score 1+), and negative in 25 cases (27.7%; score 0). Although CD44v9 was not detectable in HCC patients of the CD44v9− group and non‐tumor areas of all patients, it was overexpressed in several well, moderately, and most strongly, poorly differentiated HCCs with focal heterogeneous expression patterns (CD44v9+ group). In poorly differentiated tumors we observed single giant multinuclear cells with eosinophilic cytoplasm, which were positive for CD44v9. Significant correlations were identified between CD44v9 positivity with younger patient age and poor tumor differentiation (Table 1). In the livers of colorectal cancer patients, used as positive control for CD44v9, 100% of metastatic lesions were stained, while in the normal‐appearing liver area, no CD44v9+ cells were observed.
Figure 1.
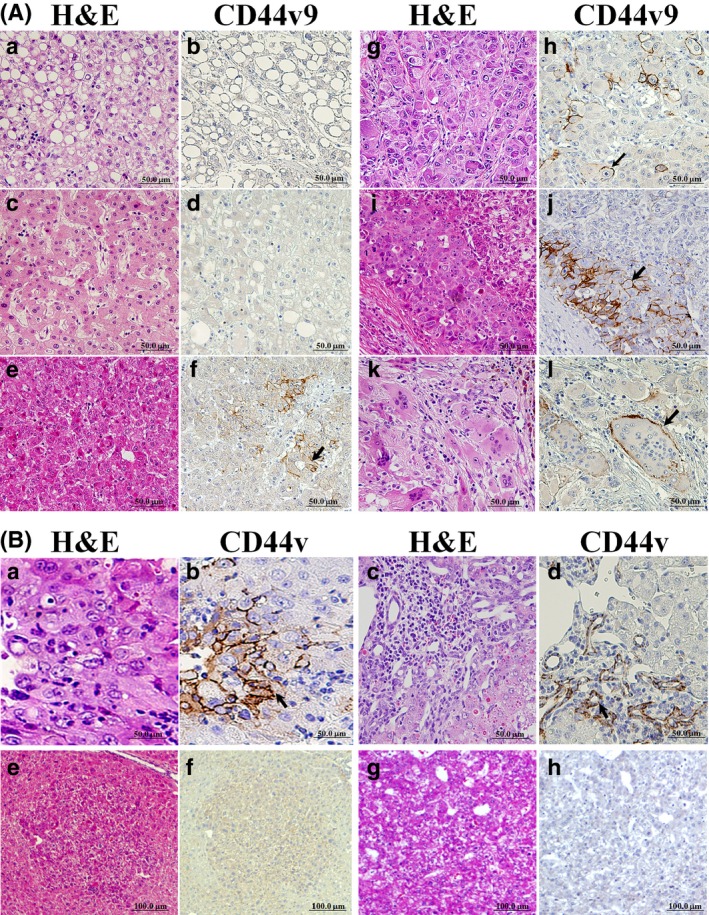
Immunohistochemistry for CD44 variant 9 (CD44v9) in hepatitis C virus‐positive human (A) and mouse (B) hepatocellular carcinomas (HCCs). (A) CD44v9− non‐recurrent HCC (a,b), normal‐appearing liver (c,d), and well (e,f), moderately (g,h), and poorly (i,j and k,l) differentiated CD44v9+ recurrent HCCs. Staining in human HCC used H&E (a,c,e,g,i,k) and CD44v9 (b,d,f,h,j,l). Note CD44v9+ cells (arrows) and positive multinuclear cells (l) in HCCs. (B) In mouse HCCs, note CD44v+ cells (a,b) and positive proliferative ductular epithelial cells in the mixed type tumor (c,d) (arrows). CD44v− basophilic foci (e,f) and hepatocellular adenoma (HCA) (g,h) in mouse liver (serial sections).
Table 1.
Correlation between CD44 variant 9 expression and clinicopathological variables
| Factors | No. of patients (%) | |||
|---|---|---|---|---|
| Total | Positive (n = 64) | Negative (n = 26) | P‐value† | |
| Age | ||||
| >70 | 68 | 44 (65) | 24 (35) | 0.018 |
| ≤70 | 22 | 20 (91) | 2 (9) | |
| Gender | ||||
| Male | 62 | 44 (71) | 18 (29) | 0.964 |
| Female | 28 | 20 (71) | 8 (29) | |
| Smoking | ||||
| Smoker | 50 | 33 (66) | 16 (64) | 0.232 |
| Non‐smoker | 40 | 31 (77) | 9 (23) | |
| Drinking | ||||
| Drinker | 36 | 25 (69) | 11 (31) | 0.776 |
| Non‐drinker | 54 | 39 (72) | 15 (28) | |
| Diabetes | ||||
| Positive | 20 | 17 (85) | 3 (15) | 0.120 |
| Negative | 70 | 47 (67) | 23 (33) | |
| Tumor size, cm3 | ||||
| ≤2 | 55 | 39 (71) | 16 (29) | 0.871 |
| >2 | 35 | 25 (71) | 10 (29) | |
| T category (pT) | ||||
| T1 | 25 | 16 (64) | 9 (36) | 0.356 |
| T2–T4 | 65 | 48 (74) | 17 (26) | |
| Im | ||||
| Positive | 14 | 10 (71) | 4 (29) | 0.977 |
| Negative | 76 | 54 (71) | 22 (29) | |
| pM | ||||
| Positive | 1 | 0 (0) | 1 (100) | 0.115 |
| Negative | 89 | 64 (72) | 25 (28) | |
| pB | ||||
| Positive | 1 | 1 (100) | 0 (0) | 0.522 |
| Negative | 89 | 63 (71) | 26 (29) | |
| Venous invasion | ||||
| Positive | 25 | 20 (80) | 5 (20) | 0.187 |
| Negative | 65 | 44 (68) | 21 (32) | |
| Fc inf | ||||
| Positive | 54 | 41 (76) | 13 (24) | 0.159 |
| Negative | 36 | 23 (64) | 13 (36) | |
| Differentiation‡ | ||||
| Well | 8 | 5 (62) | 3 (38) | 0.033 |
| Moderate | 35 | 21 (60) | 14 (40) | |
| Poor | 47 | 38 (81) | 9 (19) | |
| Clinical stage§ | ||||
| I | 25 | 16 (64) | 9 (36) | 0.251 |
| II | 41 | 30 (73) | 11 (27) | |
| III | 21 | 16 (76) | 5 (24) | |
| IV | 3 | 2 (67) | 1 (33) | |
| Recurrence | ||||
| Positive | 54 | 41 (76) | 13 (24) | 0.159 |
| Negative | 36 | 23 (64) | 13 (44) | |
| Cirrhosis | ||||
| Stages 1 and 2 | 41 | 28 (68) | 13 (32) | 0.589 |
| Stages 3 and 4 | 49 | 36 (73) | 13 (27) | |
| AST, IU/L | ||||
| 13–33 | 23 | 16 (70) | 7 (30) | 0.850 |
| >34 | 67 | 48 (72) | 19 (28) | |
| ALT, IU/L | ||||
| 6–27 | 31 | 21 (68) | 10 (32) | 0.609 |
| >28 | 59 | 43 (73) | 16 (27) | |
| ALP, IU/L | ||||
| 115–359 | 60 | 39 (65) | 21 (35) | 0.049¶ |
| >360 | 30 | 25 (83) | 5 (17) | |
†Pearson's χ2‐test. ‡Well and moderately differentiated versus poorly differentiated. §Stage I versus stages II–IV. No lymph node metastases were detected in any patient. ¶Fisher's test. ALP, alkaline phosphatase; ALT, alanine transferase; AST, aspartate transferase; Fc inf, infiltration to capsule; im, intrahepatic metastasis; pM factor, pathological M factor; pT factor, pathological T factor, pB factor, pathological B factor (bile duct invasion).
Histopathological analysis and assessment of CD44v positivity in C57Bl/6J mice
During this study, no animals died and no significant changes in body weight, food, or water intake were observed. The number of putative preneoplastic foci observed in C57Bl/6J mice treated with DEN was 1.24 ± 0.90 no/cm2. The incidence of HCA and HCC of mice observed at week 38 after DEN initiation were 50% (12 mice) and 12.5% (3 mice), respectively, and multiplicities were 1.13 ± 1.36 and 0.13 ± 0.35 no./mouse. A mixed‐type tumor, hepatocholangiocarcinoma (1 mouse, 4%) was found in the DEN group. Representative CD44v IHC staining is illustrated in Fig. 1(B). Assessment of CD44v showed that HCCs but not HCAs or putative preneoplastic foci of mice contained focal regions or single cells positive for CD44v (Fig. 1Ba,b,e–h). No CD44v overexpression was obvious in adjacent non‐tumor areas, and normal biliary ductular cells were negative for CD44v. Almost all proliferating ductular epithelial cells (>90%) within the mixed‐type tumor were positive for CD44v (Fig. 1Bc,d). In this experiment, no bile duct proliferative lesions in the normal‐appearing liver tissue were found.
Observed correlations between Ki67, P‐p38, Nrf2, p62‐SQSTM1, Keap1, and CD44v in human and mouse HCCs
Next, we examined the expression of Ki67, Nrf2, p62‐SQSTM1, Keap1, and activation of p38MAPK, a major target of ROS, in cancer tissues to assess the role of CD44v in the regulation of intracellular oxidative stress. The representative pictures of double immunostaining for target proteins and CD44v are presented in Fig. 2. Ki67 immunopositive nuclei were barely detectable in CD44v9+ regions of human HCCs (3.41 ± 2.06%, P < 0.05), CD44v9+ colorectal cancer metastases to the liver, and mouse CD44v+ HCC regions (4.24 ± 7.35%, P < 0.05), but were apparent in human (17.63 ± 9.48%) and mouse (30.00 ± 12.00% CD44v− HCC areas (Fig. 2Aa–d,Bb,Ca,Db). Similarly, P‐p38 staining was not obvious in human (1.80 ± 2.65%, P < 0.05) or mouse (1.52 ± 2.62%, P < 0.05) CD44v+ HCC regions but was detected in human (45.92 ± 38.77%) and mouse (24.00 ± 12.12%) CD44v− HCC areas (Fig. 2Ae,Bc,Cc,Dc). Furthermore, it was found that CD44v positivity was correlated with Nrf2, p62‐SQSTM1, and Keap1 overexpression in both human and mouse HCCs (Fig. 2Bd–f,Dd–f).
Figure 2.
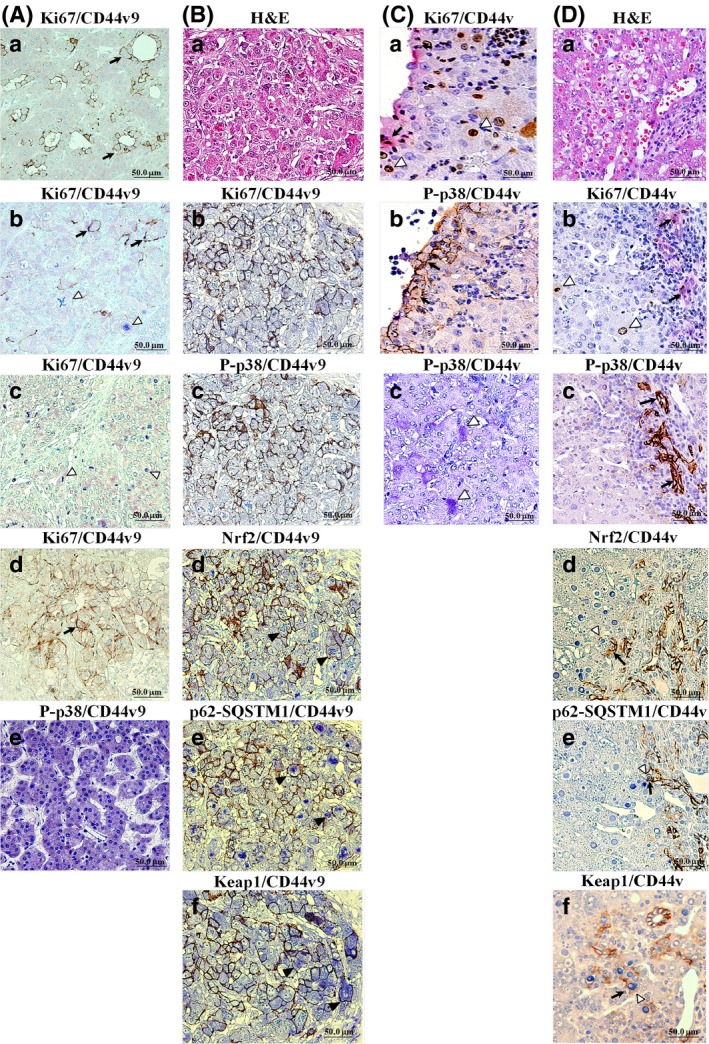
Double immunohistochemistry (IHC) for CD44v (brown/black) and Ki67, phospho‐p38 (P‐p38), nuclear factor (erythroid‐derived 2)‐like 2 (Nrf2), p62‐sequestosome 1 (SQSTM1), or Kelch‐like ECH‐associated protein 1 (Keap1) (blue) in representative cases of recurrent HCV + human and mouse hepatocellular carcinomas (HCCs). (A) IHC for human CD44 variant 9 (CD44v9; black arrows) and Ki67 or P‐p38 (white arrowheads). (a) CD44v9+ and Ki67− moderately differentiated HCC. (b) Independent CD44v9+ and Ki67+ cells in moderately differentiated HCC. (c) CD44v9− and Ki67+ moderately differentiated HCC. (d) CD44v9+ and Ki67− colorectal adenocarcinoma metastatic lesion in the liver (positive control). (e) CD44v9− and P‐p38+ human HCC. (B) H&E staining (a) and double IHC in serial sections for Ki67 (b), P‐p38 (c), Nrf2 (d), p62‐SQSTM1 (e), or Keap1 (f) and CD44v9 in human poorly differentiated HCC. (C) Double IHC for Ki67 (a,b), P‐p38 (c), and CD44v in mouse HCC. (D) H&E staining (a) and double IHC for Ki67 (b), P‐p38 (c), Nrf2 (d), p62‐SQSTM1 (e), Keap1 (f), and CD44v in mouse mixed‐type tumor. Note the inverse correlation between CD44v and Ki67 or P‐p38 and the positive correlations between Nrf2, p62‐SQSTM1, Keap1, and CD44v9 expression (black arrowheads).
Association of CD44v9 staining with clinicopathological variables
To evaluate the correlation between CD44v9 expression and clinicopathological outcome, its expression was assessed by IHC in 90 HCV+ HCC patients. The relationship between CD44v9 overexpression and various clinicopathological parameters including pathological stage, TNM factors, and tissue differentiation, venous and bile duct invasion of human HCCs was analyzed using the χ2‐test (Table 1). No statistically significant associations between increased CD44v9 expression (scores 0, 1+ to 3+) with gender, smoking, drinking, past history of diabetes, stage of cirrhosis, tumor size, the established prognostic factor of pT, pB, and pM status, or clinicopathological stage were found (Table 1). No lymph node metastases (pN status) were observed in any HCV+ HCC patients. Interestingly, positivity and negativity of membranous CD44v9 expression were significantly associated with younger age (≤70 vs >70 years; P = 0.018) and poor histological tumor differentiation (poorly vs well and moderately; P = 0.033) (Fig. S1). Furthermore, a positive correlation was found between ALP elevation in the blood and CD44v9 expression (>360 vs 115–359 IU/L, P = 0.049) (Table 1, Fig. S1). Moreover, positive CD44v9 expression in tumors tended to be associated with venous invasion (positive vs negative, P = 0.187), infiltration of the capsule (positive vs negative, P = 0.159), and recurrence (positive vs negative, P = 0.159) (Table 1).
Prognosis
Next, univariate survival analysis with recurrence‐specific survival curves was carried out, according to the Kaplan–Meier method, and differences in overall and recurrence‐free survival were assessed with the log–rank test (Fig. 3). Importantly, positive CD44v9 expression was associated with poorer overall (P = 0.022, log–rank test) and recurrence‐free (P = 0.049, log–rank test) survival compared with negative expression (Fig. 3A,B). Moreover, median recurrence‐free survival periods for patients with positive and negative CD44v9 expression were 1016 days and 1370 days, respectively. The overall 100‐month (3000‐day) survival rates for patients with positive and negative CD44v9 expression were 54% and 84%, respectively. Moreover, median recurrence‐free survival for patients with positive and negative CD44v9 expression was 736 days and 934 days, respectively. The recurrence‐free 2500‐day survival rates for patients with positive and negative CD44v9 expression were 38% and 44%, respectively. According to the results of univariate analysis, poor survival of patients was significantly associated with increased CD44v9 expression (HR, 2.781; 95% confidence interval, 1.155–6.694, P = 0.022), higher T category (T2–4 vs T1, P = 0.031), clinicopathological stage (II–IV vs I, P = 0.031), tumor size (>2 vs ≤2 cm3 , P = 0.009), venous invasion (positive vs negative, P < 0.0001), infiltration to capsule (positive vs negative, P = 0.047), poor tumor differentiation (poorly vs well and moderately, P = 0.037), and recurrence (positive vs negative, P = 0.015) (Table 2).
Figure 3.
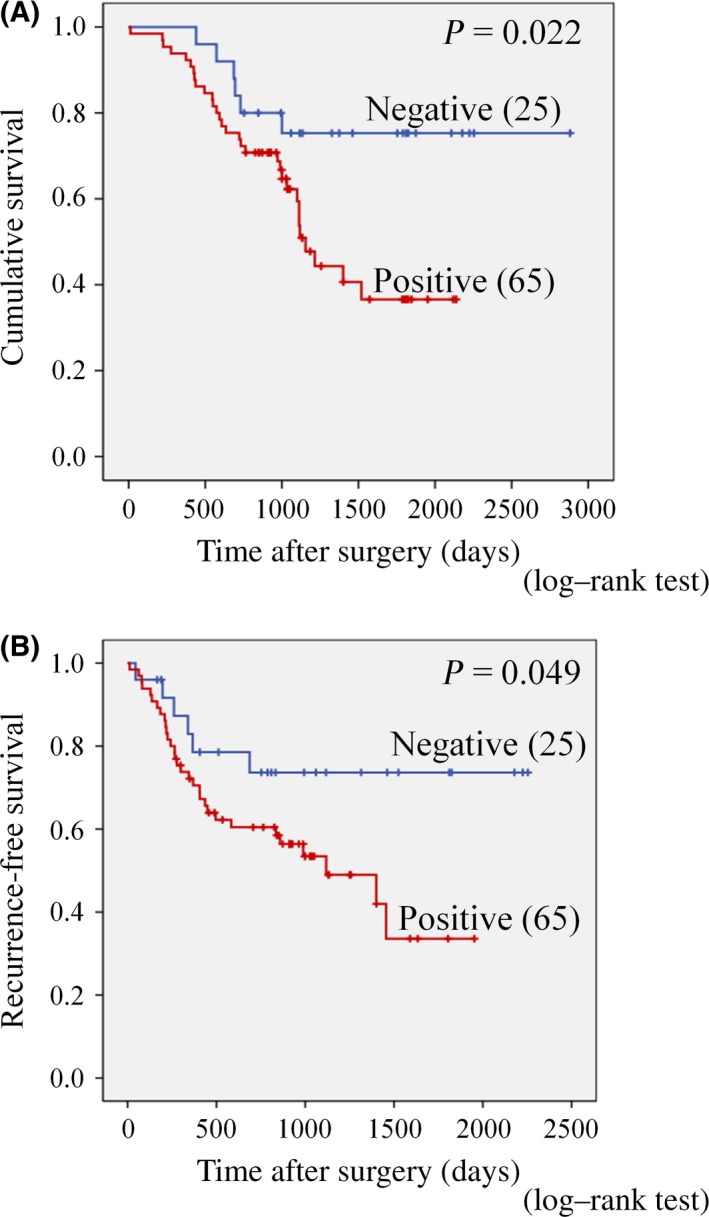
Kaplan–Meier curves evaluating differences in overall (A) and recurrence‐free (B) survival in hepatitis C virus‐positive patients with hepatocellular carcinoma depending on CD44 variant 9 expression. Significant differences in overall and recurrence‐free survival were observed for patients with CD44 variant 9 positive (n = 65) and negative (n = 25) expression (log–rank test).
Table 2.
Univariate and multivariate analyses of survival in hepatitis C virus‐positive patients with resected hepatocellular carcinoma (n = 90)
| Clinicopathological features | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| CD44v9 | ||||||
| Positive versus negative | 2.781 | 1.155–6.694 | 0.022 | 2.536 | 0.964–6.672 | 0.049 |
| Age, years | ||||||
| ≤70 versus >70 | 1.650 | 0.688–3.955 | 0.2620 | NA | ||
| Tumor size, cm3 | ||||||
| >2 versus ≤2 | 2.433 | 1.244–4.756 | 0.0090 | NA | ||
| T category | ||||||
| T2–4 versus T1 | 2.831 | 1.102–7.270 | 0.0310 | NA | ||
| Intrahepatic metastasis | ||||||
| Positive versus negative | 2.237 | 1.082‐4.627 | 0.0300 | NA | ||
| Venous invasion | ||||||
| Positive versus negative | 1.909 | 0.630–5.789 | <0.0001 | 2.450 | 1.116–5.379 | 0.026 |
| Infiltration to capsule | ||||||
| Positive versus negative | 2.055 | 1.011–4.176 | 0.0470 | 2.399 | 1.010–5.698 | 0.047 |
| Differentiation | ||||||
| Poor versus well/moderate | 2.125 | 1.046–4.314 | 0.0370 | NA | ||
| Stage | ||||||
| II–IV versus I | 2.831 | 1.102–7.270 | 0.0310 | NA | ||
| Recurrence | ||||||
| Positive versus negative | 2.770 | 1.216–6.310 | 0.0150 | 3.383 | 1.357–8.434 | 0.009 |
| Cirrhosis | ||||||
| Stage 3/4 versus stage 1/2 | 1.286 | 0.516–3.206 | 0.3790 | NA | ||
| AST, IU/L | ||||||
| >34 versus 13–33 | 0.988 | 0.478–2.041 | 0.9740 | NA | ||
| ALT, IU/L | ||||||
| >28 versus 6–27 | 0.965 | 0.491–1.896 | 0.9170 | NA | ||
| ALP, IU/L | ||||||
| >360 versus 115–359 IU/L | 0.850 | 0.420–1.721 | 0.6520 | NA | ||
Both multivariate and univariate analyses used Cox proportional‐hazards regression. Variables were adopted in multivariate analysis for their prognostic significance by univariate analysis. ALP, alkaline phosphatase; ALT, alanine transferase; AST, aspartate transferase; CD44v9, CD44 variant 9; NA, not available. No lymph node metastases were detected in any patient.
Multivariate analysis was performed using the Cox proportional hazards model for all significant variables in the univariate analysis. Survival of HCV+ HCC patients was significantly associated with CD44v9 expression (HR, 2.536; 95% confidence interval (CI), 0.964–6.672, P = 0.049), venous invasion (positive vs negative, P = 0.026), infiltration of the capsule (positive vs negative, P = 0.047) and recurrence (positive vs negative, P = 0.009) (Table 2). Based on these results, CD44v9 expression was proven to be an independent prognostic factor for human HCV+ HCC survival.
Comparison of HCC proteomes of CD44v9+ and CD44v9− patients
The results of LC‐MS/MS and IPA analyses of differentially expressed proteins in CD44v9+ and CD44v9− HCCs as compared to adjacent liver tissues are presented in Table 3. In all CD44v9+ HCCs, we observed that many genes related to xenobiotic metabolism including various cytochrome P450 isoenzymes and epoxide hydrolase 2 were downregulated, while genes involved in Nrf2‐mediated signaling and degradation of superoxide radicals, such as NQO1 (8.22‐fold upregulation) and SOD2 (5.02‐fold upregulation) were strongly elevated. We also observed that β‐catenin, DNA repair proteins APEX nuclease 1 and poly(ADP ribose) polymerase 1, vimentin, and proteins involved in actin cytoskeleton organization and glucose metabolism were overexpressed, whereas keratins 8 and 18 were downregulated only in CD44v9+ HCCs (Table 3).
Table 3.
Differentially expressed proteins in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas, identified by liquid chromatography MS/MS
| Protein name | Symbol | GI number/Swiss‐Prot accession | Ratio | Location | Function(s) | |
|---|---|---|---|---|---|---|
| CD44v9 (−) | CD44v9 (+) | |||||
| NAD(P)H dehydrogenase, quinone 1 | NQO1 | 118 607 | – | 8.22 | C | XM, NO S |
| Superoxide dismutase 2, mitochondrial | SOD2 | 134 665 | – | 5.02 | C | RSR |
| Epoxide hydrolase 2, cytoplasmic | EPHX2 | 67 476 665 | – | 0.45 | C | XM, ROS M |
| Flavin containing monooxygenase 3 | FMO3 | 6 166 183 | 0.62 | 0.55 | C | XM |
| Flavin containing monooxygenase 5 | FMO5 | 1 346 021 | – | 0.70 | C | XM |
| Cytochrome P450, fam. 2, subfam. C, polypep. 8 | CYP2C8 | 117 225 | – | 0.28 | C | XM, AAM, ORP |
| Cytochrome P450, fam. 2, subfam. C, polypep. 9 | CYP2C9 | 6 686 268 | 0.60 | 0.27 | C | XM, AAM, ORP |
| Cytochrome P450, fam. 2, subfam. D, polypep. 6 | CYP2D6 | 84 028 191 | – | 0.50 | C | XM, ORP |
| Cytochrome P450, fam. 2, subfam. E, polypep. 1 | CYP2E1 | 117 250 | – | 0.66 | C | XM, EM, ORP |
| Cytochrome P450, fam. 3, subfam. A, polypep. 4 | CYP3A4 | 116 241 312 | 0.41 | 0.32 | C | XM, ORP |
| Cytochrome P450, fam. 3, subfam. A, polypep. 5 | CYP3A5 | 117 157 | – | 0.57 | C | XM, ORP |
| Cytochrome P450, fam. 4, subfam. A, polypep. 11 | CYP4A11 | 2 493 371 | – | 0.21 | C | XM, AAM, ORP |
| Cytochrome P450, fam. 4, subfam. F, polypep. 3 | CYP4F3 | 56 757 430 | – | 0.67 | C | XM, AAM, ORP |
| Cytochrome P450, fam. 8, subfam. B, polypep. 1 | CYP8B1 | G3V8J2 | – | 0.57 | C | XM, BAM, ORP |
| Carboxylesterase 1 | CES1 | 119 576 | – | 0.45 | C | XM |
| Catechol‐O‐methyltransferase | COMT | 116 907 | – | 0.56 | C | MET, XM, ESM |
| Glutathione S‐transferase alpha 1 | GSTA1 | 121 730 | 0.58 | 0.43 | C | XM, GM |
| Thioredoxin | TXN | 135 773 | 1.63 | 1.97 | C | ORP, NO R |
| Catenin (cadherin‐ass. protein), beta 1, 88 kDa | CTNNB1 | 461 854 | – | 1.65 | N | TRA |
| Prothymosin, alpha | PTMA | 135 834 | – | 1.70 | N | TRA |
| Tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein, eta | YWHAH | 1 345 593 | 1.32 | 1.68 | C | TRA, ST |
| Heat shock 70‐kDa protein 5 (glucose‐reg.prot.,78 kDa) | HSPA5 | 14 916 999 | 1.31 | 1.69 | C | PF, A(−) |
| Heat shock 70‐kDa protein 9 (mortalin) | HSPA9 | 21 264 428 | 1.29 | 2.27 | C | PF, A(−) |
| Calnexin | CANX | 543 920 | 1.88 | 1.68 | C | PF |
| Transketolase | TKT | 1 729 976 | – | 3.60 | C | CM |
| Glucose‐6‐phosphate dehydrogenase | G6PD | 116 242 483 | – | 3.23 | C | CM, GLM, LM |
| Hexokinase domain containing 1 | HKDC1 | Q2 TB90 | – | 1.85 | C | CM, GLM |
| Enolase 1, (alpha) | ENO1 | 119 339 | – | 1.64 | C | CM, GLM, TRA |
| Carnitine O‐acetyltransferase | CRAT | 215 274 265 | – | 2.03 | C | LM, CAM, T |
| UDP‐glucose pyrophosphorylase 2 | UGP2 | 59 803 098 | 0.52 | 0.43 | C | CM, GLM, XM |
| Arginase 1 | ARG1 | 12 230 985 | 0.51 | 0.38 | C | RD, UC |
| Argininosuccinate synthase 1 | ASS1 | 20 141 195 | 0.32 | 0.36 | C | RD, UC |
| Carbamoyl‐phosphate synthase 1, mitochondrial | CPS1 | 4033 707 | 0.29 | 0.33 | C | RD, UC |
| Ornithine carbamoyltransferase | OTC | 84 028 235 | 0.70 | 0.60 | C | RD, UC |
| Vimentin | VIM | 55 977 767 | – | 1.61 | C | CO |
| Keratin 18 | KRT18 | 125 083 | – | 0.62 | C | CO |
| Keratin 8 | KRT8 | 90 110 027 | – | 0.58 | C | CO |
| Cofilin 1 (non‐muscle) | CFL1 | 116 848 | – | 1.65 | N | ACO |
| Solute carrier fam. 9, subfam. A, mem. 3 reg. 1 | SLC9A3R1 | 41 688 557 | – | 1.62 | PM | ACO, CP(−) |
| Transgelin 2 | TAGLN2 | 586 000 | 1.25 | 2.18 | C | ACO |
| Periostin, osteoblast specific factor | POSTN | 93 138 709 | – | 3.52 | ES | CA |
| Fibronectin 1 | FN1 | P04937 | 2.60 | 2.04 | ES | CA, EMO |
| APEX nuclease (multifunct. DNA repair enzyme) 1 | APEX1 | 113 984 | – | 1.67 | N | DNAR, ReOS |
| Poly (ADP‐ribose) polymerase 1 | PARP1 | 130 781 | – | 1.60 | N | DNAR, BER |
| Synaptophysin‐like 1 | SYPL1 | 48 474 786 | – | 2.90 | PM | T |
| GTPase activating protein binding protein 1 | G3BP1 | 14 916 572 | – | 3.14 | N | T, ST |
| RAB35, member RAS oncogene family | RAB35 | Q15286 | – | 1.71 | C | T, ST |
| Apolipoprotein E | APOE | 114 039 | 6.36 | 1.54 | ES | T, ST |
| Fatty acid binding protein 1, liver | FABP1 | 119 808 | 0.66 | 0.23 | C | T ReOS |
| DEAD (Asp‐Glu‐Ala‐Asp) box polypeptide 39A | DDX39A | 61 212 932 | 1.83 | 1.76 | N | mRNA SPT |
A(−), negative regulation of apoptosis; AAM, arachidonic acid metabolism; ACO, actin cytoskeleton organization; BAM, bile acid metabolism; BER, base excision repair; C, cytoplasm; CA, cell adhesion; CAM, carnitine metabolism; CM, carbohydrate metabolism; CO, cytoskeleton organization; CP(−), negative regulation of cell proliferation; DNAR, DNA repair; E, enzyme; EM, ethanol metabolism; EMO, ECM organization; ES, extracellular space; ESM, estrogen metabolism; GM, glutathione metabolism; GLM, glucose metabolism; MET, methylation; mRNA SPT, mRNA splicing, processing, and transport; N, nucleus; NO R, response to nitric oxide; NO S, nitric oxide synthesis; ORP, oxidation‐reduction process; PF, protein folding; PM, plasma membrane; RD, response to drug; ReOS, response to oxidative stress; ROS M, reactive oxygen species metabolism; RSR, removal of superoxide radicals; ST, signal transduction; T, transport; TRA, transcription; UC, urea cycle.
Comparison analysis of protein functions by IPA indicated that in CD44v9+ human HCCs, numerous proteins with altered expression were involved in activation of cellular invasion, migration, synthesis of nitric oxide, metabolism of nucleotides (z‐score ≥2.0), and suppression of generation and quantity of ROS, lipid oxidation, fatty acid metabolism, DNA damage, and cell death (z‐score ≤2.0) (Table S2). Furthermore, analysis of altered canonical pathways showed that most upregulated proteins were involved in Nrf2‐mediated oxidative stress responses and superoxide radicals degradation, protein kinase A signaling, glycolysis, gluconeogenesis, tricarboxylic acid cycle, fatty acid α‐oxidation, signaling by Rho family GTPases, hepatic fibrosis/hepatic stellate cell activation, gap junction signaling, DNA double‐strand break repair by non‐homologous end joining, and DNA repair of oxidative modifications (Table S3). In contrast, xenobiotic metabolism and p53 signaling pathways were suppressed in CD44v9+ HCCs.
Ingenuity pathway analysis of upstream regulators indicated that CD44v9 positivity resulted in significant activation of Nrf2, TGFB1, platelet‐derived growth factor BB, epidermal growth factor receptor, basic fibroblast growth factor 2, vascular endothelial growth factor A, IL1A and IL1B, and low‐density lipoprotein, and significant inhibition of hepatocyte nuclear factor 4A, transcription factor E2F1, WNT1‐inducible‐signaling pathway protein 2, and miR‐1‐3p miRNA (Fig. 4, Table S4). Furthermore, we found a tendency for inhibition of pregnane X receptor, constitutive androstane receptor (CAR), and CCAAT/enhancer binding protein (C/EBP).
Figure 4.
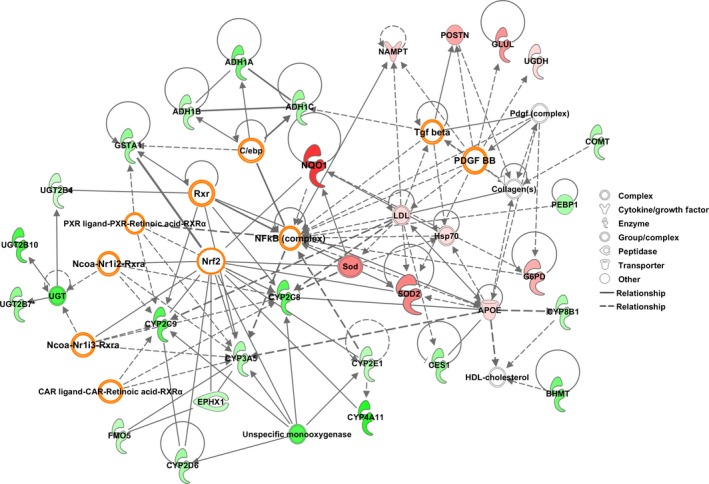
Signaling pathways involving differentially expressed proteins in CD44 variant 9‐positive hepatocellular carcinomas. Note activation of nuclear factor (erythroid‐derived 2)‐like 2 (Nrf2), with strong overexpression of its downstream proteins NAD(P)H quinone oxidoreductase (NQO1) and superoxide dismutase 2 (SOD2), as well as activation of transforming growth factor‐β (TGF ‐β) and platelet‐derived growth factor (PDGF) and inhibition of pregnane X receptor (PXR), constitutive androstane receptor (CAR), and CCAAT/enhancer binding protein (C/EBP). The illustration shows proteins with increased (red) and decreased (green) expression in CD44 variant 9‐positive hepatocellular carcinomas.
Discussion
This study provided the first experimental evidence of focal overexpression of CD44v9 in human HCV+ HCCs and CD44v8‐10 in mouse HCCs with similar membranous staining patterns. The absence of CD44v+ cells in mouse preneoplastic foci or HCAs indicated that formation of CD44v+ TISCs is a late event in mouse hepatocarcinogenesis. Importantly, positive CD44v9 expression was associated with poorer overall and recurrence‐free survival, younger age, and poor histological tumor differentiation, as compared with negative expression, indicating that CD44v9 could be a novel potential TISC biomarker and independent prognostic factor for human HCV+ HCC. Furthermore, a positive correlation was found between ALP elevation in the blood and CD44v9 expression in HCCs, which could reflect increased invasion activity of CD44v9+ cells and might have diagnostic significance.
It was recently reported that expression of CD44v9 in primary gastric cancer tissue could serve as an indicator of recurrence in early gastric cancers patients.18 In addition, it has been reported that, in lung cancer patients, CD44v8‐10 mRNA and protein are predominantly expressed in non‐small‐cell lung carcinomas, while non‐tumorous tissues principally express CD44s and small‐cell lung carcinomas express either CD44s or no detectable CD44. Thus, CD44v8‐10 was concluded to be the dominant splicing isoform in non‐small‐cell lung carcinomas and can be practically utilized for diagnosis and as a therapeutic target.26
Interestingly, Nrf2 downstream proteins such as NQO1 and SOD2, participating in degradation and removal of ROS, were greatly overexpressed in human CD44v9+ HCCs, pointing to an important role for Nrf2 activation in TISCs. We further observed the concomitant overexpression of Nrf2, p62‐SQSTM1, Keap1, and CD44v9, being an indicator of their interrelation and activation of Nrf2 by p62‐SQSTM1. Nrf2 is a potent transcription activator that recognizes a unique DNA sequence known as the antioxidant response element, and xCT is one of its downstream genes.27 Furthermore, CD44v9 positivity was associated with suppressed expression of numerous proteins involved in xenobiotic metabolism, which are controlled by hepatocyte nuclear factor 4A, E2F1, constitutive androstane receptor, and PXR upstream regulators.28, 29 These data are in line with previously published results demonstrating involvement of CD44v8‐10 in potentiation of the ability of cancer cells to promote GSH synthesis30 due to its interaction with a glutamate‐cystine transporter xCT.17 CD44v controls the cell redox potential of TISCs, thus increasing their resistance to induction of ROS and maintaining CSC survival.15, 16 Important cross‐talk was recently identified between the Keap1–Nrf2 system and the autophagy‐adaptor protein p62–SQSTM1, which was found to compete with Keap1 for binding with Nrf2 by using the STGE motif, and through this competition promoting the stabilization of Nrf2 and upregulation of Nrf2 activity.31, 32 The present findings supported this idea and furthermore suggested the participation of CD44v9 in this pathway. Another protein that can bind to Nrf2 or Keap1, thereby disrupting their interaction is p21WAF1/Cip1, which may act together with p62‐SQSTM1.28 It was suggested that high ROS levels are generally detrimental to cells but they also may constitute a barrier to tumorigenesis.33 The persistent high levels of inflammation and ROS generated by HCV in the liver could result in the development of defense mechanisms to survive in the conditions of persistent oxidative stress, thus p62‐SQSTM1 and Nrf2 factors could be activated and drove the CD44v9 expression.
Tumor‐initiating stem‐like cells were recently suggested to feature enhanced mechanisms of protection from stress induced by ROS that render them resistant to chemotherapy and radiotherapy. As it is accepted that during tumor progression cancer cells are often exposed to high levels of ROS, the ability to escape the consequences of ROS exposure is extremely important for cancer cells to survive and propagate. Tumor‐initiating stem‐like cells, in which defense against ROS is enhanced by CD44v, are thus thought to drive tumor growth, chemoresistance, and metastasis.6 Our findings further supported previously published results, where xCT inhibition was shown to selectively induce apoptosis in CD44v‐expressing tumor cells without affecting CD44v− differentiated cells in the same squamous cell carcinomas,16 and that ablation of CD44 induced loss of xCT from the cell surface and suppressed tumor growth in a transgenic mouse model of gastric cancer.17
In our study, the focal pattern of CD44v9 expression and the reverse correlation with cell proliferation markers such as Ki67 and P‐p38 suggested that CD44v9 could be a marker of hepatic TISCs. Previously, inhibition of cell proliferation in terms of BrdU labeling was reported in expanded CD44+ glandular elements in gastric tumors of K19‐Wnt1/C2 mE mice.34 Moreover, ablation of CD44v was shown to induce activation of p38MAPK, a downstream target of ROS, and expression of the gene for the cell cycle inhibitor p21WAF1/Cip1.17 In the present study, the poorly differentiated HCCs contained higher percentages of CD44v9+ cells, suggesting that CD44v9 expression is mainly correlated with poor HCC differentiation, which is likely to become the best marker of tumor metastasizing capacity. Hepatocellular carcinomas contained both CD44v9+ and CD44v9− regions; the CD44v9+ cells that comprised only a small part of the tumor are likely to have high invasion activity, whereas the CD44v9− cells, which are the majority, have higher proliferation levels. Our data are in good correlation with previous results, indicating that in CD44v9+ cells proliferation could be inhibited through the antioxidant response element/Nrf2/glutathione pathway, characterized by development of resistance to oxidative stress and damage.17 In addition, ROS have been shown to induce the proliferation of cells that are not resistant to oxidative stress.23 In addition, actively proliferating cells are considered to be more sensitive to treatment compared to those that are not proliferating and could remain in a “sleeping state” for a long time.34
It is conceivable that CD44v9+ HCC overexpression of β‐catenin, enzymes involved in energy metabolism, ECM proteins controlled by TGFB1, vimentin, actin cytoskeleton members, and those involved in DNA repair of double‐strand breaks and oxidative base modifications, are likely to be mediated by Nrf2, TGFB1, epidermal growth factor receptor, platelet‐derived growth factor, basic fibroblast growth factor 2, endothelial growth factor A, IL1A, IL1B, and other factors, which may explain lowered apoptosis, survival maintenance, migration, and invasion activity of CD44v9+ TISCs. In line with our data, clinically, overexpression of CD44s was shown to be associated with high expression of vimentin, low expression of E‐cadherin, a high percentage of phospho‐Smad2‐positive nuclei, mesenchymal spindle‐like morphology, tumor invasiveness, and poor prognosis in HCC patients, and reduced disease‐free and overall survival. Thus, CD44s are likely to play a critical role in the TGF‐β‐mediated mesenchymal phenotype.24
In conclusion, in the present study, focal CD44v9 overexpression was found in human and CD44v8‐10 in mouse HCCs. In human HCV+ HCC cases it was correlated with poorer overall and recurrence‐free survival and clinicopathological factors including younger age, a poorly differentiated invasive phenotype of HCC, and increase of ALP levels in the blood, thus suggesting that CD44v9 could be a novel biomarker of liver TISCs and a prognostic factor for HCV+ HCC patients.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- ABC
avidin–biotin complex
- ALP
alkaline phosphatase
- CD44v
CD44 variant isoform
- CSC
cancer stem‐like cell
- DEN
diethylnitrosamine
- HCA
hepatocellular adenoma
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HR
hazard ratio
- IHC
immunohistochemistry
- IL1A
interleukin‐α
- IL1B
interleukin‐β
- IPA
Ingenuity pathway analysis
- Keap1
Kelch‐like ECH‐associated protein 1
- LC
liquid chromatography
- NQO1
NAD(P)H quinone oxidoreductase
- Nrf2
nuclear factor (erythroid‐derived 2)‐like 2
- P‐p38
phospho‐p38MAPK
- PXR
pregnane X receptor (PXR) ligand–PXR–retinoic acid–retinoid receptor α (RXRα)
- ROS
reactive oxygen species
- SOD2
superoxide dismutase 2
- SQSTM1
sequestosome 1
- TGF‐β
transforming growth factor‐β
- TGFB1
transforming growth factor‐β1
- TISC
tumor‐initiating stem‐like cell
Supporting information
Fig. S1. Immunohistochemical scores of CD44 variant 9 in hepatitis C virus‐positive patients with hepatocellular carcinoma.
Table S1. Patient clinicopathological characteristics, tumor characteristics, and treatment characteristics for hepatitis C virus‐positive patients with hepatocellular carcinoma (n = 90).
Table S2. Ingenuity pathway analysis of altered molecular functions in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.
Table S3. Ingenuity canonical pathway analysis in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.
Table S4. Ingenuity pathway analysis of upstream regulators in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.
Acknowledgments
We thank Kaori Nakakubo, Azusa Inagaki, Keiko Sakata, Yuko Hisabayashi, and Rie Onodera for their valuable technical assistance. This research was supported by grants from the Ministry of Health, Labor and Welfare of Japan and the Japan Science and Technology Corporation.
Cancer Sci 107 (2016) 609–618
Funding Information
Ministry of Health, Labor and Welfare of Japan; Japan Science and Technology Corporation.
References
- 1. Mima K, Okabe H, Ishimoto T et al The expression levels of CD44v6 are correlated with the invasiveness of hepatocellular carcinoma in vitro, but do not appear to be clinically significant. Oncol Lett 2012; 3: 1047–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pang RW, Poon RT. Cancer stem cell as a potential therapeutic target in hepatocellular carcinoma. Curr Cancer Drug Targets 2012; 12: 1081–94. [DOI] [PubMed] [Google Scholar]
- 3. Ma S. Biology and clinical implications of CD133(+) liver cancer stem cells. Exp Cell Res 2013; 319: 126–32. [DOI] [PubMed] [Google Scholar]
- 4. Shen Y, Cao D. Hepatocellular carcinoma stem cells: origins and roles in hepatocarcinogenesis and disease progression. Front Biosci (Elite Ed) 2012; 4: 1157–69. [DOI] [PubMed] [Google Scholar]
- 5. Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci 2004; 95: 930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagano O, Okazaki S, Saya H. Redox regulation in stem‐like cancer cells by CD44 variant isoforms. Oncogene 2013; 32: 5191–8. [DOI] [PubMed] [Google Scholar]
- 7. Biddle A, Gammon L, Fazil B, Mackenzie IC. CD44 staining of cancer stem‐like cells is influenced by down‐regulation of CD44 variant isoforms and up‐regulation of the standard CD44 isoform in the population of cells that have undergone epithelial‐to‐mesenchymal transition. PLoS ONE 2013; 8: e57314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saya H. Molecular mechanisms of brain tumor invasion. Nihon Rinsho 2005; 63(Suppl 9): 61–7. [PubMed] [Google Scholar]
- 9. Saya H, Nagano O. Role of CD44‐mediated signal in cancer invasion and metastasis. Tanpakushitsu Kakusan Koso 2008; 53: 1225–30. [PubMed] [Google Scholar]
- 10. Sugahara KN, Murai T, Nishinakamura H, Kawashima H, Saya H, Miyasaka M. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44‐expressing tumor cells. J Biol Chem 2003; 278: 32259–65. [DOI] [PubMed] [Google Scholar]
- 11. Stauder R, Eisterer W, Thaler J, Gunthert U. CD44 variant isoforms in non‐Hodgkin's lymphoma: a new independent prognostic factor. Blood 1995; 85: 2885–99. [PubMed] [Google Scholar]
- 12. Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res 2006; 66: 10233–7. [DOI] [PubMed] [Google Scholar]
- 13. Kainz C, Kohlberger P, Tempfer C et al Prognostic value of CD44 splice variants in human stage III cervical cancer. Eur J Cancer 1995; 31A: 1706–9. [DOI] [PubMed] [Google Scholar]
- 14. Wielenga VJ, van der Neut R, Offerhaus GJ, Pals ST. CD44 glycoproteins in colorectal cancer: expression, function, and prognostic value. Adv Cancer Res 2000; 77: 169–87. [DOI] [PubMed] [Google Scholar]
- 15. Tsugawa H, Suzuki H, Saya H et al Reactive oxygen species‐induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem‐like cells. Cell Host Microbe 2012; 12: 764–77. [DOI] [PubMed] [Google Scholar]
- 16. Yoshikawa M, Tsuchihashi K, Ishimoto T et al xCT inhibition depletes CD44v‐expressing tumor cells that are resistant to EGFR‐targeted therapy in head and neck squamous cell carcinoma. Cancer Res 2013; 73: 1855–66. [DOI] [PubMed] [Google Scholar]
- 17. Ishimoto T, Nagano O, Yae T et al CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(‐) and thereby promotes tumor growth. Cancer Cell 2011; 19: 387–400. [DOI] [PubMed] [Google Scholar]
- 18. Hirata K, Suzuki H, Imaeda H et al CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer 2013; 109: 379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wada T, Ishimoto T, Seishima R et al Functional role of CD44v‐xCT system in the development of spasmolytic polypeptide‐expressing metaplasia. Cancer Sci 2013; 104: 1323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minagawa M, Ikai I, Matsuyama Y, Yamaoka Y, Makuuchi M. Staging of hepatocellular carcinoma: assessment of the Japanese TNM and AJCC/UICC TNM systems in a cohort of 13,772 patients in Japan. Ann Surg 2007; 245: 909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vauthey JN, Lauwers GY, Esnaola NF et al Simplified staging for hepatocellular carcinoma. J Clin Oncol 2002; 20: 1527–36. [DOI] [PubMed] [Google Scholar]
- 22. Liver Cancer Study Group of Japan . General Rules for the Clinical and Pathological Study of Primary Liver Cancer. The 5th edition (Serial online). [Cited 2009.]
- 23. Kinoshita A, Wanibuchi H, Imaoka S et al Formation of 8‐hydroxydeoxyguanosine and cell‐cycle arrest in the rat liver via generation of oxidative stress by phenobarbital: association with expression profiles of p21(WAF1/Cip1), cyclin D1 and Ogg1. Carcinogenesis 2002; 23: 341–9. [DOI] [PubMed] [Google Scholar]
- 24. Mima K, Okabe H, Ishimoto T et al CD44s regulates the TGF‐beta‐mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res 2012; 72: 3414–23. [DOI] [PubMed] [Google Scholar]
- 25. Kakehashi A, Ishii N, Shibata T et al Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis. Toxicol Sci 2011; 119: 61–72. [DOI] [PubMed] [Google Scholar]
- 26. Sasaki JI, Tanabe KK, Takahashi K et al Expression of CD44 splicing isoforms in lung cancers: dominant expression of CD44v8‐10 in non‐small cell lung carcinomas. Int J Oncol 1998; 12: 525–33. [DOI] [PubMed] [Google Scholar]
- 27. Mitsuishi Y, Motohashi H, Yamamoto M. The Keap1‐Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol 2012; 2: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiyosawa N, Kwekel JC, Burgoon LD et al Species‐specific regulation of PXR/CAR/ER‐target genes in the mouse and rat liver elicited by o, p'‐DDT. BMC Genom 2008; 9: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maglich JM, Stoltz CM, Goodwin B, Hawkins‐Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 2002; 62: 638–46. [DOI] [PubMed] [Google Scholar]
- 30. Tanabe KK, Ellis LM, Saya H. Expression of CD44R1 adhesion molecule in colon carcinomas and metastases. Lancet 1993; 341: 725–6. [DOI] [PubMed] [Google Scholar]
- 31. Komatsu M, Kurokawa H, Waguri S et al The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010; 12: 213–23. [DOI] [PubMed] [Google Scholar]
- 32. Suzuki T, Yamamoto M. Molecular basis of the Keap1‐Nrf2 system. Free Radic Biol Med 2015; 88: 93–100. [DOI] [PubMed] [Google Scholar]
- 33. Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 2013; 12: 931–47. [DOI] [PubMed] [Google Scholar]
- 34. Ishimoto T, Oshima H, Oshima M et al CD44+ slow‐cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci 2010; 101: 673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Immunohistochemical scores of CD44 variant 9 in hepatitis C virus‐positive patients with hepatocellular carcinoma.
Table S1. Patient clinicopathological characteristics, tumor characteristics, and treatment characteristics for hepatitis C virus‐positive patients with hepatocellular carcinoma (n = 90).
Table S2. Ingenuity pathway analysis of altered molecular functions in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.
Table S3. Ingenuity canonical pathway analysis in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.
Table S4. Ingenuity pathway analysis of upstream regulators in CD44 variant 9 (CD44v9)+ and CD44v9− hepatocellular carcinomas.


