Abstract
Cancer stem‐like cells (CSC) or cancer‐initiating cells are now considered to be an important cell population related to cancer recurrence and the resistance to anti‐cancer therapy. Tumor‐associated macrophages (TAM) are a main component of stromal cells and are related to cancer progression in clear cell renal cell carcinoma (ccRCC). Because the detailed mechanisms allowing the maintenance of CSC in cancer tissues remain unclear, we investigated the relationship between TAM and CD44‐expressing cancer cells in ccRCC. CD44 was used as a marker for CSC, and CD163 and CD204 were used as markers for TAM. CD44‐positive cancer cells were detected in 37 of the 103 cases. Although statistical analysis showed no relationship between CD44‐positive cancer cells and the clinical course, the distribution of CD44‐positive cancer cells was significantly associated with a high density of TAM. Our in vitro study using RCC cell lines and human macrophages demonstrated that CD44 expression was upregulated by direct co‐culture with macrophages. Silencing of TNF‐alpha on macrophages abrogated the upregulation of CD44 expression in cancer cells. Macrophage‐induced CD44 overexpression was also suppressed by NF‐κB inhibitors. These results suggest that TNF‐alpha derived from TAM is linked to CD44 overexpression via NF‐κB signaling in ccRCC.
Keywords: CD163, CD44, macrophage, RCC, TAM
Kidney cancer is one of the main malignant tumors found in human urogenital organs, and clear cell renal cell carcinoma (ccRCC) is the most common histological subtype. Approximately 30–40% of patients with ccRCC have metastatic lesions at diagnosis or during the follow‐up period.1, 2, 3 Although novel tyrosine kinase inhibitors, including sorafenib and sunitinib, are reported to improve the survival period of patients with RCC, the median overall survival for patients with metastatic RCC is approximately 3 years.4
Cancer stem‐like cells (CSC) are now considered to be an important cell population related to the recurrence and progression of cancer.5, 6 Cancer tissue cells can be divided into CSC and non‐CSC, and CSC are known to be closely involved in cancer recurrence and metastasis.7, 8 However, it remains unclear why and how the stemness of CSC is maintained in cancer tissues.
CD44 is a receptor for hyaluronic acid and is widely expressed on normal cells, including immune cells. Recent findings suggested that standard and variant isoforms of CD44 could be used as markers of CSC in several malignant tumors, and the presence of cancer cells positive for the standard isoform of CD44 (CD44s) has been reported to be a factor of poor clinical prognosis in ccRCC and several other malignant tumors.9, 10, 11, 12, 13 Inhibition of CD44 signaling significantly abrogated the migration, invasion, proliferation and progression of RCC cell lines.14
Recent findings have demonstrated the role of stromal cells present in the cancer microenvironment in cancer progression and the maintenance of CSC.15 We previously demonstrated that tumor‐associated macrophages (TAM) are a main component of stromal cells in ccRCC, that a high number of TAM is significantly associated with a worse clinical course, and that direct cell–cell interaction between cancer cells and macrophages play an important role in cancer progression.16, 17 Therefore, in the present study, we investigated whether TAM are involved in CD44 expression on ccRCC cells.
Materials and Methods
Tissue samples
Paraffin‐embedded tissue samples from patients who had undergone curative surgery and were pathologically diagnosed as ccRCC between 1998 and 2008 at the University Hospital of Occupational and Environmental Health and Kumamoto University Hospital were included in the analysis.18 Tissue samples with massive necrosis were not included. In total, 103 cases were included in this study. Because CD44 expression is heterogeneous in cancer tissues, routine tissue sections were used in this study, and no tissue array sections were used. Data on progression‐free survival (PFS) and cancer‐specific overall survival (OS) were obtained for 103 and 91 patients, respectively. All samples were obtained with informed consent from patients in accordance with the study protocols approved by the review board of each university. Tissue samples were routinely fixed in 10% neutral buffered formalin and were embedded in paraffin.
Immunohistochemistry
The anti‐CD44s antibody was purchased from Abcam (clone IM7; Cambridge, UK). The anti‐CD163 antibody (10D6; Novocastra, Newcastle, UK) and anti‐CD204 antibody (SRA‐E5; Transgenic, Kumamoto, Japan) were used for detecting TAM.16 Secondary antibodies were purchased from Nichirei (Tokyo, Japan) and reactions were visualized using a diaminobenzidine substrate system (Nichirei). Two investigators, who were blinded to any information about the samples, evaluated the infiltration of CD163+, CD204+ and CD44+ cancer cells under microscopic observation. The numbers of CD163+ or CD204+ TAM in all samples were counted and the averages of the results were used. In the cases in which cancer cells are positive for CD44, the numbers of TAM in CD44+ areas and CD44− areas were additionally counted in the serial sections.
Cell lines
The human RCC cell lines, ACHN and 786‐O, were purchased from ATCC (Manassas, VA, USA). The MAMIYA cells had previously been kindly provided by Professor K. Itoh.14 Cells were maintained in DMEM/Ham F‐12 supplemented with 10% FBS. A mycoplasma test was performed using a PCR detection kit (Takara Bio, Otsu, Japan). Cell lines stably expressing green fluorescence protein (GFP) were established using GFP‐expressing lentivirus (Santa Cruz Biotechnology, Santa Cruz, CA, USA), as described in the manufacturer's protocol. The cell numbers were tested using a WST assay kit (Donjindo, Kumamoto, Japan).
Macrophage culture
Peripheral blood mononuclear cells were obtained from healthy volunteer donors, who had all provided written informed consent for the use of their cells in accordance with the study protocols approved by the Kumamoto University Hospital Review Board.16 CD14+ monocytes were isolated using CD14‐microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). These monocytes were plated in 6‐well plates (2 × 105 cells/well) and were cultured in 2% human serum, 1 ng/mL granulocyte macrophage‐colony stimulating factor (GM‐CSF; WAKO, Tokyo, Japan), and 50 ng/mL macrophage‐colony stimulating factor (M‐CSF, WAKO) for 7 days to induce differentiated macrophages. Then medium was changed and cells were cultured with DMEM/Ham F‐12 supplemented with 10% FBS (without any cytokines) until the usage of cells.
Flow cytometry
Cells were detached using cell dissociation buffer (Thermo Fisher Scientific, Waltham, MA, USA), and stained using APC‐labeled anti‐CD44s antibody (BioLegend, San Diego, CA, USA) with Fc receptor blocking solution (BioLegend).
Reverse transcriptase‐PCR and real‐time PCR
Total RNA was isolated using RNAiso Plus (Takara Bio). RNA was reverse‐transcribed by means of a PrimeScript RT Reagent Kit (Takara Bio). Quantitative real‐time PCR was performed using TaqMan polymerase with SYBR Green Fluorescence (Takara Bio) and an ABI PRISM 7300 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The primers used were as follows: for tumor necrosis factor (TNF)‐α, sense 5′‐TGG.AGA.AGG.GTG.ACC.GAC.TC‐3′, antisense 5′‐TCC.TCA.CAG.GGC.AAT.GAT.CC‐3′; for β‐actin, sense 5′‐ATT.CCT.ATG.TGG.GCG.ACG.AG‐3′, antisense 5′‐AAG.GTG.TGG.TGC.CAG.ATT.TTC‐3′. Reverse transcriptase‐PCR (RT‐PCR) was performed by using a TaKaRa Ex Taq Kit (TaKaRa Bio), and electrophoresed by Mupid electrophoresis system (Mupid, Tokyo, JP). The primers used were as follows: for TNFR1 (TNFrsf1a, 243 bp), sense 5′‐ACC.TCC.AGC.TCC.ACC.TAT.ACC‐3′, antisense 5′‐GAA.TTC.CTT.CCA.GCG.CAA.C‐3′; for TNFR2 (TNFrsf1b, 242 bp), sense 5′‐AAA.CAT.CAG.ACG.TGG.TGT.GC‐3′, antisense 5′‐AGT.GCT.GGG.TTC.TGG.AGT.TG‐3′.
Western blotting
Cells were lysed in ice‐cold lysis buffer (50 mM Tris pH 8.0, 1 mM EDTA, 150 mM NaCl, 1% NP‐40) and protease inhibitor cocktail (Sigma‐Aldrich, St. Louis, MO, USA). PVDF membrane was incubated with the anti‐TNF‐α or anti‐β‐actin antibodies (Santa Cruz Biotechnology) overnight at 4°C. HRP‐Goat Anti‐mouse IgG (Invitrogen, Camarillo, CA, USA) was performed as the second antibody. Immunoreactive bands were visualized using the Pierce Western Blotting Substrate Plus kit (Thermo Scientific, Rockford, IL, USA) and ImageQuant LAS‐4000 mini (Fuji Film, Tokyo, Japan).
Elisa
A high sensitivity ELISA Kit for human TNF‐α was purchased from eBioscience (San Diego, CA, USA), and was performed according to the manufacturer's protocol.
siRNA in human macrophages
Human macrophages were transfected with siRNA against human TNF‐α (Santa Cruz Biotechnology) using Lipofectamine RNAi MAX (Thermo Fisher). Control siRNA (Santa Cruz Biotechnology) was used as a negative control.
Inhibitors and neutralizing antibody
Nuclear factor (NF)‐κB inhibitors (BAY‐11‐7082 and parthenolide) were purchased from WAKO (Tokyo, Japan) and dissolved in DMSO. A neutralizing antibody against TNF‐α and an isotype‐matched control antibody were obtained from BioLegend.
Statistics
Statistical analysis of in vitro and in vivo data was carried out using JMP10 (SAS Institute, Chicago, IL, USA) and StatMate III (ATOMS, Tokyo, Japan). A P‐value <0.05 was considered to be statistically significant. The χ2‐test, Mann–Whitney U‐test, and Cox proportional hazard test were used to analyze associations with clinical factors. All data used in the cell‐culture study was representative of at least three independent experiments.
Results
The distribution of CD44+ cells in clear cell renal cell carcinoma
In the immunostaining analysis, CD44+ cancer cells were detected in 37 cases among the 103 primary ccRCC cases (Table 1). CD44 was mainly expressed on the cell surface membrane of cancer cells, and was also expressed on TAM and leukocytes (Fig. 1a). The CD44+ cancer cells and immune cells could be distinguished by morphological features (Fig. 1a). The CD44+ area showed a heterogeneous staining pattern and covered <25% of the area of the cancer tissue in approximately half of the cases (Fig. 1b). Analysis of the correlations between CD44 expression in cancer cells and clinicopathological factors showed that there were more CD44+ cancer cells in males than in females; however, there was no significant correlation between CD44 expression and any of the other factors (Table 1). In addition, the results showed no association between CD44 expression in cancer cells and PFS or OS (Table 2).
Table 1.
CD44 expression in cancer cells and clinicopathological parameters
| CD44 | |||
|---|---|---|---|
| Negative | Positive | P‐value | |
| Age | |||
| <60 | 28 | 13 | 0.47 |
| ≧60 | 38 | 24 | |
| Gender | |||
| Male | 16 | 31 | <0.001 |
| Female | 50 | 6 | |
| Nuclear grade | |||
| G1, G2 | 55 | 26 | 0.12 |
| G3, G4 | 11 | 11 | |
| T classification | |||
| T1 | 42 | 24 | 0.91 |
| T2, T3, T4 | 24 | 13 | |
Figure 1.
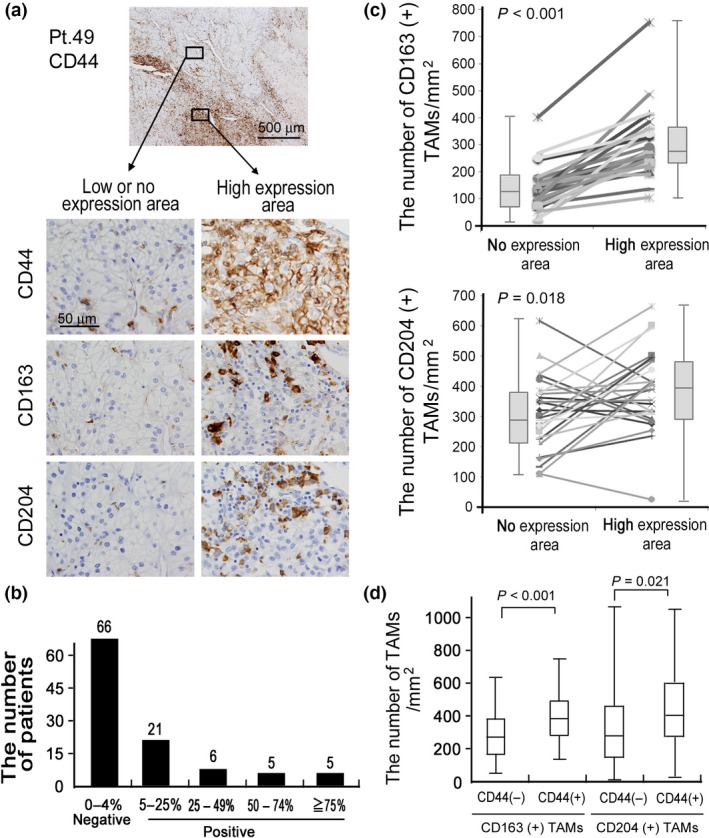
Immunostaining of CD44 CD163, and CD204 in clear cell renal cell carcinoma (ccRCC). (a) Cancer cells and macrophages are positive for CD44, and the distribution of CD44+ cancer cells is heterogeneous. CD163 and CD204 were used as macrophage markers. The results of two patients are shown. (b) Patients were divided into four groups according to the percentage of CD44+ cancer cells. (c) The numbers of CD163+ and CD204+ tumor‐associated macrophages (TAM) were counted in the CD44− and CD44+ areas, respectively. A paired Student t‐test was performed. (d) The numbers of CD163+ and CD204+ TAM were counted in all cases, and the numbers of TAM were compared between CD44− cases and CD44+ cases. A Mann–Whitney U‐test was performed.
Table 2.
Univariate cox regression analysis of PFS and OS
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| Age, <60 vs ≧60 | 1.3 | 0.6–2.9 | 0.46 | 1.4 | 0.5–3.9 | 0.54 |
| Gender, M vs F | 1.2 | 0.6–2.6 | 0.56 | 2.4 | 0.8–10.5 | 0.25 |
| Stage, T1 vs T2 + 3+4 | 3.4 | 1.6–7.5 | <0.001 | 2.8 | 1.1–8.2 | 0.037 |
| Nuclear grade, G1 + 2 vs G3 + 4 | 5.2 | 2.4–11.1 | <0.001 | 5.3 | 1.8–14.9 | 0.003 |
| CD44, negative vs positive | 1.9 | 0.8–4.2 | 0.14 | 1.3 | 0.4–3.6 | 0.68 |
CI, confidential interval; HR, hazard ratio; OS, overall survival; PFS, progression free survival.
An increased density of tumor‐associated macrophages was correlated to CD44 overexpression on clear cell renal cell carcinoma cancer cells
Next, serial sections were stained using anti‐CD163 and anti‐CD204 antibodies to evaluate the correlation between TAM and CD44‐expressing cancer cells. Areas of the tissue sections were divided into those that showed CD44 expression (CD44+ areas) and those that showed no or little CD44 expression (CD44− areas), and the numbers of CD163+ or CD204+ TAM in these areas were counted in the serial sections (Fig. 1a). Comparison of the numbers of TAM in the CD44+ areas and CD44− areas showed that there were significantly higher numbers of CD163+ and CD204+ TAM in the CD44+ areas than in the CD44− areas, and that there was a significant relationship between CD44 expression on cancer cells and the number of CD163+ TAM (Fig. 1c). In addition, increased density of TAM are detected in CD44+ RCC cases compared with CD44− RCC cases; moreover, strong correlation was observed between the number of CD163+ TAM and existence of CD44+ cancer cells (Fig. 1d).
CD44 expression in MAMIYA cells was increased by co‐culture with macrophages
We previously demonstrated that direct cell–cell interaction with human macrophages could induce the activation of RCC cell lines.16 Therefore, a co‐culture experiment with double immunostaining was performed to investigate whether macrophages influence CD44 expression on RCC cell lines (Fig. 2a). Because ACHN and 786‐O cells strongly expressed CD44 while the MAMIYA cells weakly expressed CD44 when cell lines are cultured alone (Fig. 2b), MAMIYA cells were used for the co‐culture assay. Comparison of the CD44 expression levels before and after co‐culture with macrophages showed that the co‐culture with macrophages significantly increased the level of CD44 expression on MAMIYA cells (Fig. 2c). Flow cytometric analysis performed to confirm the upregulation of CD44 in GFP‐transduced MAMIYA cells (Fig. 2d) showed a significant upregulation of CD44 in MAMIYA cells following direct co‐culture with macrophages (Fig. 2e). The level of CD44 expression on ACHN cells and 786‐O cells was not changed by co‐culture with macrophages (data not shown).
Figure 2.
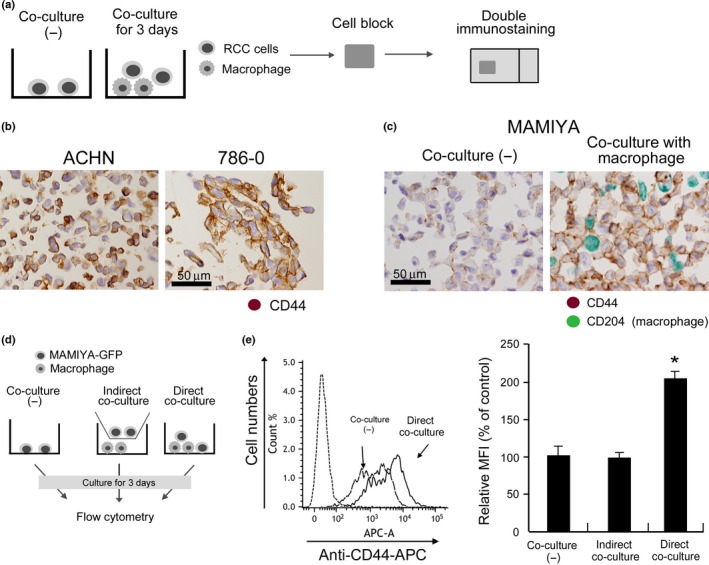
CD44 expression in cultured renal cell carcinoma (RCC) cell lines. (a) Cultured cells were prepared as cell‐block specimens and double immunostaining was performed. (b) CD44 expression on ACHN and 786‐O cells was evaluated by immunostaining. (c) Following co‐culture with macrophages for 3 days, CD44 expression in MAMIYA cells was evaluated by double immunostaining. Anti‐CD204 antibody was used to label macrophages (green), and we evaluated CD44 expression (brown) on CD204− cancer cells. (d) Following co‐culture with macrophages for 3 days, CD44s expression in RCC cell lines was evaluated by flow cytometry. (e) Following flow cytometry analysis, the mean fluorescence intensity (MFI) of CD44 was evaluated and statistically analyzed.
TNF‐α expressed on macrophages is involved in the upregulation of CD44 in co‐cultured MAMIYA cells
We previously reported that macrophage‐derived factors, such as C5a, TNF‐α, I‐309, growth‐related oncogene (GRO)‐α, and interleukin (IL)‐6, induced lymphoma cell proliferation,19 so we suspected that these molecules were involved in the upregulation of CD44 in the co‐cultured MAMIYA cells. Thus, MAMIYA cells were treated for 3 days with recombinant molecules of the macrophage‐derived factors mentioned above, then CD44 expression was evaluated by flow cytometry. The results showed that CD44 expression was induced by TNF‐α, but not by any of the other molecules (Fig. 3a). Although macrophages expressed a high level of TNF‐α, mRNA and protein expression of TNF‐α was scarcely observed in our RCC cell lines (Fig. 3b,c). TNFR1 expression was detected in RCC cell lines, whereas no or less expression of TNFR2 was seen (Fig. 3d). The expression of TNF‐α was detected in the cytoplasm of macrophages, but not in MAMIYA cells (Fig. 3e). Soluble TNF‐α was detected in the supernatant of direct co‐culture cells, however, was not detectable in the supernatants of macrophages and MAMIYA cells in unstimulated condition (Fig. 3f). The molecular size of the TNF‐α expressed on unstimulated macrophages and lipopolysaccharide‐stimulated macrophages was approximately 25 and 17 kDa, respectively (Fig. 3g), indicating that the TNF‐α on unstimulated macrophages were in a membrane‐bound form. A blocking antibody against TNF‐α, used to evaluate the involvement of TNF‐α in the upregulation of CD44, abrogated the upregulation of CD44 in MAMIYA cells co‐cultured with macrophages (Fig. 4a). Silencing of TNF‐α on the macrophages also suppressed the upregulation of CD44 in the co‐cultured MAMIYA cells (Fig. 4b,c).
Figure 3.
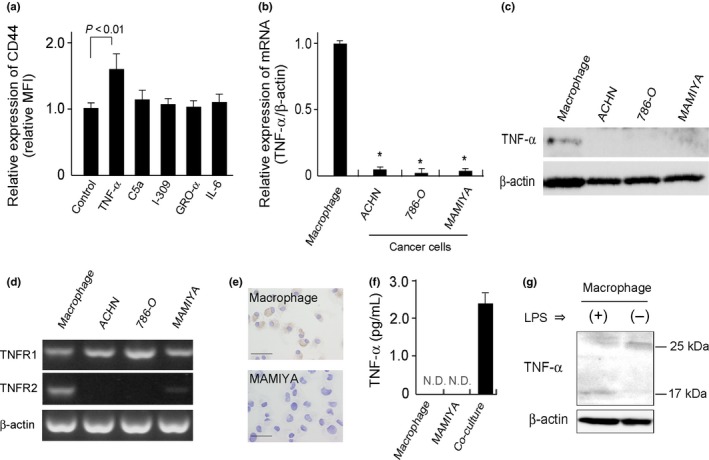
Evaluation of TNF‐α expression. (a) After stimulation of cells with C5a (1 pg/mL), TNF‐α (10 ng/mL), growth‐related oncogene (GRO)‐α (10 ng/mL), CCL1/I‐309 (10 ng/mL), or IL‐6 (10 ng/mL) for 2 days, the CD44 expression on MAMIYA cells was analyzed by flow cytometry. (b) mRNA expression of TNF‐α on macrophages, ACHN cells, 786‐O cell and MAMIYA cells were evaluated by quantitative real‐time PCR. *: P‐value < 0.05. (c) TNF‐α protein expression was detected by western blot analysis. (d) The expressions of TNF receptor superfamily 1A (TNFR1) and TNF receptor superfamily 1B (TNFR2) were tested by RT‐PCR in MAMIYA cells. (e) Immunocytostaining of TNF‐α on macrophages and MAMIYA cells. (f) High sensitive ELISA assay of TNF‐α in the cell culture supernatant. (g) Western blot analysis of TNF‐α using lysates of macrophages with or without LPS stimulation.
Figure 4.
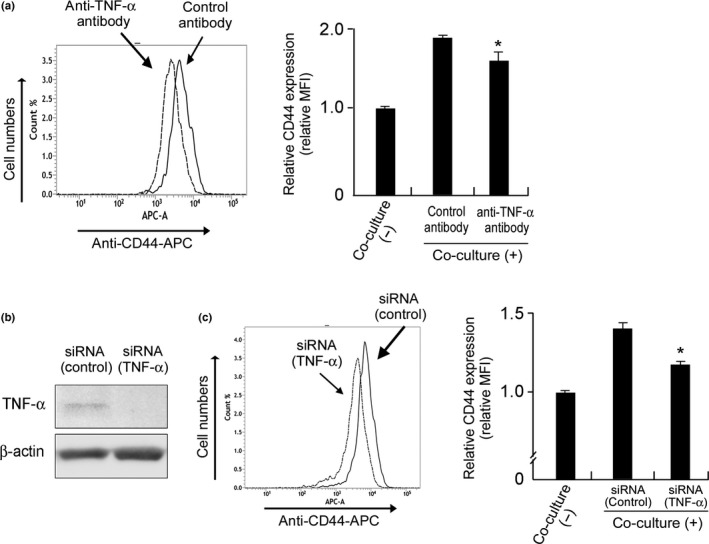
The involvement of TNF‐α in CD44 overexpression. (a) MAMIYA‐GFP cells and macrophages were co‐cultured with neutralizing antibody for TNF‐α (10 μg/mL) or control antibody for 3 days, then the CD44 expression on the MAMIYA‐GFP cells was evaluated by flow cytometry. (b) The downregulation of TNF‐α induced by siRNA on macrophages was confirmed by Western blot analysis. (c) MAMIYA‐GFP cells were co‐cultured with macrophages pre‐treated with siRNA, then the CD44 expression on the MAMIYA‐GFP cells was evaluated by flow cytometry.
NF‐κB signaling is involved in CD44 overexpression on MAMIYA cells
It is well known that TNF‐α induces CD44 overexpression via the NF‐κB signaling pathway.20 In the present study, we used NF‐κB inhibitors (BAY‐11‐7082 and parthenolide) to evaluate the involvement of NF‐κB activation in CD44 upregulation on MAMIYA cells co‐cultured with TNF‐α or macrophages. The NF‐κB inhibitors showed cytotoxicity against MAMIYA cells at the concentrations of 2.5 nmol/L (BAY‐11‐7082) and 5 nmol/L (parthenolide) (Fig. 5a). As such, the NF‐κB inhibitors were used at the lower concentrations of 1.25 nmol/L (BAY‐11‐7082) and 2.5 nmol/L (parthenolide) in the subsequent experiments (Fig. 5a,b). As we expected, the NF‐κB inhibitors abrogated the TNF‐α‐induced CD44 overexpression on MAMIYA cells (Fig. 5c), and notably inhibited the CD44 upregulation induced by co‐culture with macrophages (Fig. 5d).
Figure 5.
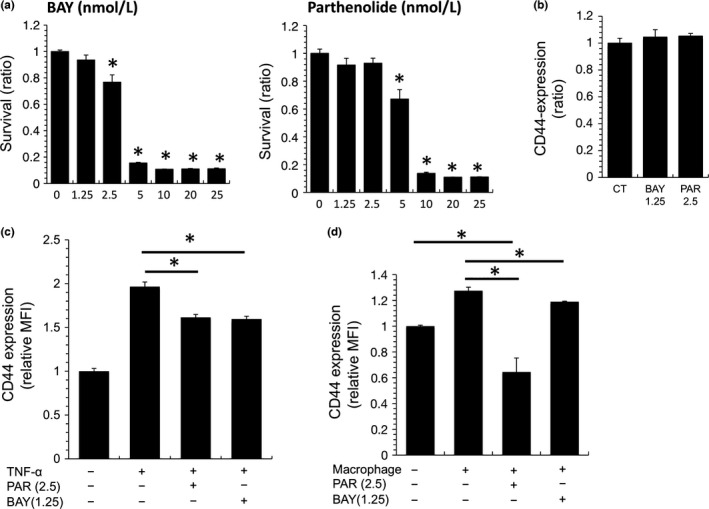
The involvement of NF‐κB in CD44 overexpression on MAMIYA cells. (a) MAMIYA cells were treated with one of two NF‐κB inhibitors (BAY‐11‐7082 or parthenolide) for 2 days, then the numbers of cancer cells were evaluated by WST assay. (b) CD44 expression on MAMIYA cells treated with NF‐κB inhibitors was evaluated by flow cytometry. (c) After stimulation of MAMIYA cells with TNF‐α in the presence or absence of NF‐κB inhibitors, CD44 expression was evaluated by flow cytometry. (d) After co‐culture of MAMIYA‐GFP cells with macrophages in the presence or absence of NF‐κB inhibitors, CD44 expression was evaluated by flow cytometry. *P value < 0.05.
Discussion
In this paper, we demonstrated by means of in vitro and in vivo studies that CD44 expression on cancer cells is induced by cell–cell interactions with macrophages. In the in vivo study using surgically resected samples, we first found that CD44+ cancer cells were located in areas with a high infiltration of TAM. In particular, the density of CD163+ TAM appeared to be more closely related to CD44 expression in cancer cells than that of CD204+ TAM. Our previous study using lymphoma cells demonstrated the significance of CD163 in the direct cell–cell interaction between macrophages and lymphoma cells, although the detailed mechanisms were not clarified.19 In addition, our previous in vivo study using an animal model showed that tumor progression was significantly suppressed in CD163‐deficient mice (unpublished data). These data indicate that CD163, rather than CD204, is involved in the cell–cell interaction between TAM and RCC cells via unknown mechanisms.
We next performed co‐culture experiments using human RCC cell lines and monocyte‐derived macrophages to investigate the detailed mechanisms of the cell–cell interaction between cancer cells and TAM. Interestingly, our studies demonstrated that CD44 overexpression in MAMIYA cells could only be induced by direct co‐culture with macrophages, and not by indirect co‐culture in transwell culture dishes. A similar phenomenon was observed in a previous study using lymphoma cell lines in which growth factors, including C5a, TNF‐α, I‐309, GRO‐α and IL‐6, were found to be secreted by macrophages activated by direct contact with lymphoma cells.19 The results from Figure 3 indicated that TNF‐α was involved in the CD44 overexpression in MAMIYA cells. Macrophages express a high level of membrane‐bound TNF‐α in the normal condition in the cytoplasm, and TNF‐α was suggested to be secreted after direct contact with MAMIYA cells. We demonstrated that the mRNA expression of TNF‐α was significantly higher in macrophages than in cancer cells, and that blocking of TNF‐α suppressed the CD44 upregulation induced by co‐culture with MAMIYA cells; these findings indicated that TNF‐α expressed on macrophages induce CD44 overexpression in the neighboring RCC cells. It is well known that CD44 is induced by TNF‐α via NF‐κB activation.20, 21 By means of in vitro studies using NF‐κB inhibitors, NF‐κB activation was found to be important for CD44 upregulation in MAMIYA cells. These observations indicated that TNF‐α plays an important role in the cell–cell interaction between cancer cells and TAM via NF‐κB signaling. Several soluble factors are known to be secreted by macrophages that are activated by cancer cells,22 and recent studies have also identified several macrophage‐derived growth factors, such as growth differentiation factor 15.23 Thus, several unknown molecules other than TNF‐α might be involved in the cell–cell interaction between cancer cells and TAM.
In this study, macrophages differentiated with GM‐CSF and M‐CSF were used for in vitro co‐culture studies. In our previous studies, cancer cell proliferation was induced more by co‐culture with IL‐10‐stimulated M2 macrophages than by co‐culture with interferon‐γ‐stimulated M1 macrophages.16, 24 In contrast, macrophages were found to be polarized into M2 phenotype by co‐culture with RCC cells in our previous study.16 Therefore, we thought it unnecessary to use M2‐polarized macrophages in the present study.
The protumor functions of TNF‐α have been reported in several malignant tumors, including RCC.25 It is well known that TNF‐α is one of the inflammatory cytokines secreted from macrophages, and a high expression level of protumor molecules, including TNF‐α, IL‐1 and matrix metalloproteinases, was detected in isolated TAM in RCC.26 A higher level of TNF‐α in plasma was associated with poorer survival, and targeting of TNF‐α using neutralizing monoclonal antibody can prolong the survival time of patients with advanced RCC.27 Macrophage‐derived TNF‐α was found to be involved in the tumorigenesis of gastric cancer.28 TNF‐α production from macrophages was associated with mesenchymal differentiation and radiation resistance in glioblastoma.29, 30 Blocking TNF‐α might be effective in combination with other targeted agents, such as tyrosine kinase inhibitors.
Blocking effects of TNF‐α for CD44 overexpression on MAMIYA cells was limited, as shown in Figure 4, and this indicates that other unknown mechanisms are involved in cell–cell interactions. We previously demonstrated that membrane type M‐CSF expressed on cell surface membrane of cancer cells including RCC cell lines induced strong macrophage activation via M‐CSFR activation;16, 24 therefore, unknown macrophage‐derived factors are considered to be involved in CD44 overexpression in MAMIYA cells. Tunneling nanotubes are known to be the long membrane extensions connecting to other cells,31 and viruses such as HIV‐1 infected in macrophages have recently been shown to utilize tunneling nanotubes to move from cell to cell.32 Because there is no research reported related to tunneling nanotubes in cell–cell interaction between cancer cells and macrophages, we are trying to investigate the significance of tunneling nanotubes in macrophage‐induced cancer cell activation.
In the present study, CD44 expression in cancer cells was not associated with PFS or OS, but these results are not consistent with the results of previous research.10, 11 However, similar to our current results, Costa et al.33 demonstrated that CD44 expression was not associated with clinical prognosis. New anti‐cancer therapy using tyrosine kinase inhibitors, such as sorafenib and sunitinib, for advanced or recurrent cancer patients was developed around 2005, but the samples used in this study were resected between 1998 and 2008, as described in the Materials and Methods; the different types of therapies offered during this period may have caused discrepancies in the results.
In conclusion, we showed that TAM are closely associated with CD44 expression on RCC cells using an in vivo study and an in vitro study. Although CD44 expression in ccRCC cells was not associated with clinical prognosis, the results of this study suggest a significant involvement of TAM in the maintenance of CSC in ccRCC. TNF‐α expressed on TAM is suggested to induce CD44 upregulation on neighboring cancer cells via activation of the NF‐κB signaling pathway. The cell–cell interaction between RCC cells and TAM might be a target in the treatment of patients with ccRCC.
Disclosure Statement
The authors have no conflict of interest to declare.
Acknowledgments
We thank Ms Emi Kiyota, Mr Osamu Nakamura, Ms Yui Hayashida and Mr Takenobu Nakagawa for their technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 25460497 and No. 25293089).
Cancer Sci 107 (2016) 700–707
Funding Information
Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 25460497 and No. 25293089)
References
- 1. Bukowski RM. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin‐2. Cancer 1997; 80: 1198–220. [DOI] [PubMed] [Google Scholar]
- 2. Park YH, Baik KD, Lee YJ, Ku JH, Kim HH, Kwak C. Late recurrence of renal cell carcinoma >5 years after surgery: clinicopathological characteristics and prognosis. BJU Int 2012; 110: E553–8. [DOI] [PubMed] [Google Scholar]
- 3. Kim SP, Weight CJ, Leibovich BC et al Outcomes and clinicopathologic variables associated with late recurrence after nephrectomy for localized renal cell carcinoma. Urology 2011; 78: 1101–6. [DOI] [PubMed] [Google Scholar]
- 4. Motzer RJ, Hutson TE, Tomczak P et al Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol 2009; 27: 3584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie W, Ren B. Developmental biology. Enhancing pluripotency and lineage specification. Science 2013; 341: 245–7. [DOI] [PubMed] [Google Scholar]
- 6. Garg N, Vijayakumar T, Bakhshinyan D, Venugopal C, Singh SK. MicroRNA regulation of brain tumour initiating cells in central nervous system tumours. Stem Cells Int 2015; 2015: 141793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshida GJ, Saya H. Therapeutic strategies targeting cancer stem cells. Cancer Sci 2016; 107: 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagano O, Okazaki S, Saya H. Redox regulation in stem‐like cancer cells by CD44 variant isoforms. Oncogene 2013; 32: 5191–8. [DOI] [PubMed] [Google Scholar]
- 9. Shimada Y, Ishii G, Nagai K et al Expression of podoplanin, CD44, and p63 in squamous cell carcinoma of the lung. Cancer Sci 2009; 100: 2054–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y, Sun B, Zhao X et al Clinical significances and prognostic value of cancer stem‐like cells markers and vasculogenic mimicry in renal cell carcinoma. J Surg Oncol 2013; 108: 414–9. [DOI] [PubMed] [Google Scholar]
- 11. Jeong BJ, Liang ZL, Huang SM, Lim JS, Kim JM, Lee HJ. CD44 is associated with tumor recurrence and is an independent poor prognostic factor for patients with localized clear cell renal cell carcinoma after nephrectomy. Exp Ther Med 2012; 3: 811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rioux‐Leclercq N, Epstein JI, Bansard JY et al Clinical significance of cell proliferation, microvessel density, and CD44 adhesion molecule expression in renal cell carcinoma. Hum Pathol 2001; 32: 1209–15. [DOI] [PubMed] [Google Scholar]
- 13. Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci 2004; 95: 930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu G, Li H, Wang J et al miRNA‐34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J Urol 2014; 192: 1229–37. [DOI] [PubMed] [Google Scholar]
- 15. Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009; 9: 239–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Komohara Y, Hasita H, Ohnishi K et al Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci 2011; 102: 1424–31. [DOI] [PubMed] [Google Scholar]
- 17. Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci 2014; 105: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitada S, Yamada S, Kuma A et al Polypeptide N‐acetylgalactosaminyl transferase 3 independently predicts high‐grade tumours and poor prognosis in patients with renal cell carcinomas. Br J Cancer 2013; 109: 472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komohara Y, Niino D, Saito Y et al Clinical significance of CD163⁺ tumor‐associated macrophages in patients with adult T‐cell leukemia/lymphoma. Cancer Sci 2013; 104: 945–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pasparakis M. Regulation of tissue homeostasis by NF‐κB signalling: implications for inflammatory diseases. Nat Rev Immunol 2009; 9: 778–88. [DOI] [PubMed] [Google Scholar]
- 21. Mikami S, Mizuno R, Kosaka T, Saya H, Oya M, Okada Y. Expression of TNF‐α and CD44 is implicated in poor prognosis, cancer cell invasion, metastasis and resistance to the sunitinib treatment in clear cell renal cell carcinomas. Int J Cancer 2015; 136: 1504–14. [DOI] [PubMed] [Google Scholar]
- 22. Hagemann T, Wilson J, Burke F et al Ovarian cancer cells polarize macrophages toward a tumor‐associated phenotype. J Immunol 2006; 176: 5023–32. [DOI] [PubMed] [Google Scholar]
- 23. Urakawa N, Utsunomiya S, Nishio M et al GDF15 derived from both tumor‐associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Lab Invest 2015; 95: 491–503. [DOI] [PubMed] [Google Scholar]
- 24. Komohara Y, Horlad H, Ohnishi K et al Importance of direct macrophage‐tumor cell interaction on progression of human glioma. Cancer Sci 2012; 103: 2165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009; 9: 361–71. [DOI] [PubMed] [Google Scholar]
- 26. Chittezhath M, Dhillon MK, Lim JY et al Molecular profiling reveals a tumor‐promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014; 41: 815–29. [DOI] [PubMed] [Google Scholar]
- 27. Harrison ML, Obermueller E, Maisey NR et al Tumor necrosis factor alpha as a new target for renal cell carcinoma: two sequential phase II trials of infliximab at standard and high dose. J Clin Oncol 2007; 29: 4542–9. [DOI] [PubMed] [Google Scholar]
- 28. Oshima H, Ishikawa T, Yoshida GJ et al TNF‐α/TNFR1 signaling promotes gastric tumorigenesis through induction of Noxo1 and Gna14 in tumor cells. Oncogene 2014; 33: 3820–9. [DOI] [PubMed] [Google Scholar]
- 29. Roessler K, Suchanek G, Breitschopf H et al Detection of tumor necrosis factor‐alpha protein and messenger RNA in human glial brain tumors: comparison of immunohistochemistry with in situ hybridization using molecular probes. J Neurosurg 1995; 83: 291–7. [DOI] [PubMed] [Google Scholar]
- 30. Bhat KP, Balasubramaniyan V, Vaillant B et al Mesenchymal differentiation mediated by NF‐κB promotes radiation resistance in glioblastoma. Cancer Cell 2013; 24: 331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science 2004; 303: 1007–10. [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto M, Bhuyan F, Hiyoshi M et al HIV spreads in macrophages by promoting tunneling nanotube formation. J Immunol 2016; 196: 1832–41. [DOI] [PubMed] [Google Scholar]
- 33. Costa WH, Rocha RM, Cunha IW, Guimaraes GC, Zequi Sde C. Imunohistochemical expression of CD44s in renal cell carcinoma lacks independent prognostic significance. Int Braz J Urol 2012; 38: 456–65. [DOI] [PubMed] [Google Scholar]


