To the editor
Originally developed as sentinels of transcriptional activity to map the regulatory function of genetic elements, reporter-gene assays have been extensively used in high throughput screening (HTS) to identify chemical modulators of cellular pathways.1 However, HTS chemical libraries consist of structurally diverse small molecules which frequently exhibit reporter-specific interactions, thus skewing data interpretation and complicating candidate selection. For example, a recent study indicated >80% of the actives from a 500K compound HTS were assay artifacts.2 In general, for reporter-gene assays this poses a formidable challenge to distinguish compounds targeting a biological pathway from those interfering with the assay technique the mechanistic basis of which varies widely and is often unpredictable. To avoid interferences predisposed to any given reporter protein we design a coincidence ‘biocircuit’ based on the principle that discrimination of signal from noise increases when registered simultaneously by two or more detectors. The concept is functionally embodied here using 2A-mediated co-translational expression of non-homologous reporters, for example, proteins having dissimilar catalytic, light emitting, or fluorescence mechanisms (Fig. 1a).
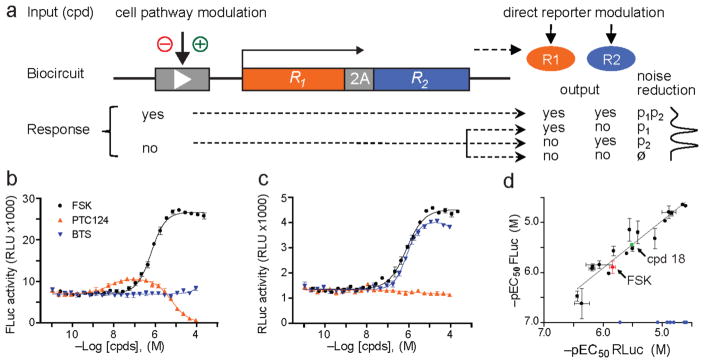
Figure 1.
Coincidence reporter biocircuit. (a) Arrangement of elements in a (b) 4XCRE-driven FLuc-P2A-RLuc dual reporter (pCI-6.24). P2A amino acid sequence is underlined; arrow indicates ribosomal ‘skipping’ site. Response output from FLuc (c) and RLuc (d) reporters to treatment with FSK, PTC124 or BTS. PTC124 and BTS are inhibitors of FLuc and RLuc, respectively, and increase reporter activity by stabilizing their cellular half-life. RLU = relative luminescence units. (e) qHTS-derived EC50 correlation plot for compounds activating the CRE element as determined by equal FLuc and RLuc expression; r2 = 0.87. Three classes of compounds identified, purinergic P1 receptor agonists, compounds 1–17 (black circles), a muscarinic receptor agonist, compound 18, and the adenylyl cyclase activator FSK. EC50 of compounds that selectively increased RLuc (blue circles) are plotted along the x-axis for comparison range in potency from 2–20 μM. Data plotted are average of replicate (n = 2) determinations; error bars represent s.d.
We confirmed the function of a preliminary biocircuit design by stoichiometric co-expression of the unrelated bioluminescent reporters, firefly (FLuc) and Renilla (RLuc) luciferase employing “ribosome skipping” facilitated by the short P2A peptide3 in a HEK293 cell type transiently transfected with the SV40-driven construct pCI-6.20 (Supplementary Fig. 1 and Supplementary Methods). Next we demonstrated the accurate discrimination of forskolin (FSK)-activated adenylyl cyclase signaling through the cAMP-response element (CRE) from reporter-dependent pharmacology mediated by the known FLuc and RLuc stabilizers, PTC124 and BTS, respectively (Figure 1b–d and Supplementary Fig. 2). Finally using the LOPAC1280 chemical library we conducted a quantitative HTS (qHTS) experiment where full titrations of each compound are tested4 to identify potentiators of the CREB pathway (Supplementary Fig. 3 and 4, and Supplementary Table 1). The screen revealed, for example, coincident FLuc and RLuc signal outputs for 17 adenosine analog agonists of endogenous purinergic P1 receptors known to signal through G-proteins in this cell type (Supplementary Table 2a).5 Excellent correlation between the EC50s calculated from the orthogonal reporter outputs was observed (Fig. 1e). Strikingly illustrating the phenomenon of reporter-dependent artifacts, we identified five aryl sulfonamides that showed selective agonist activity for the RLuc output (Supplementary Table 2b, compounds 20–24). Compounds 20–24 share a similar core scaffold with BTS and H-89, two known RLuc inhibitors and selectively inhibit the enzymatic activity of RLuc over FLuc (Supplementary Fig. 5), thus tying this particular artifact series to the phenomenon of reporter stabilization.4 Furthermore, cross-section data analysis of the screen (Supplementary Fig. 6) clearly demonstrates how coincidence detection enhances the testing of compound libraries in single concentration format.
We conclude that coincidence reporter strategies rapidly discriminate compounds of relevant biological activity from those interfering with reporter function and stability using a single assay platform. This study outlines an approach to improved use of reporter genes in HTS with numerous coincidence combination types and stoichiometries possible.
Supplementary Material
Acknowledgments
We thank P. Dranchak and R. MacArthur for assistance with supplementary data and P. Shinn for compound management. This work was supported by the NIH Roadmap for Medical Research (J.I.).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A. Anal Bioanal Chem. 2010;398(1):227–238. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- 2.Lyssiotis CA, et al. Proc Natl Acad Sci U S A. 2009;106(22):8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. Trends Biotechnol. 2006;24(2):68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Thorne N, Inglese J, Auld DS. Chem Biol. 2010;17(6):646–657. doi: 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mundell SJ, Benovic JL. J Biol Chem. 2000;275(17):12900–12908. doi: 10.1074/jbc.275.17.12900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.