Abstract
Transmission of active transcriptional states from mother to daughter cells has the potential to foster precision in the gene expression programs underlying development. Such transcriptional memory has been specifically proposed to promote rapid reactivation of complex gene expression profiles following successive mitoses in Drosophila development [1]. By monitoring transcription in living Drosophila embryos, we provide the first evidence for transcriptional memory in animal development. We specifically monitored the activities of stochastically expressed transgenes in order to distinguish active and inactive mother cells and the behaviors of their daughter nuclei following mitosis. Quantitative analyses reveal that there is a 4-fold higher probability for rapid reactivation following mitosis when the mother experienced transcription. Moreover, memory nuclei activate transcription twice as fast as neighboring inactive mothers, thus leading to augmented levels of gene expression. We propose that transcriptional memory is a mechanism of precision, which helps coordinate gene activity during embryogenesis.
Results and Discussion
New methods for visualizing gene activity in living Drosophila embryos provide a unique opportunity to investigate the kinetics of post-mitotic reactivation during development. Is the transcriptional status of a gene (active or inactive) inherited across cell generations during embryogenesis?
To date, the extent to which transcriptional activity is inherited from mother to daughter cells, referred to as transcriptional memory, has been directly recorded only in Dictyostelium and cultured mammalian cells [2 and 3]. In the latter, live imaging of inducible fluorescent Pol II and MS2-reporter transgenes reveals that the kinetics of Pol II recruitment and production of transcripts after mitosis is accelerated 13-fold and 5-fold, respectively[3]. Thus, inheritance of transcriptional activity across a cell lineage increases the rate of transcription and may impact various developmental processes. Persistent memory was also observed at the protein level [4]. Transcriptional inheritance is lost in chromatin mutants (for example Ash2 mutants[2]) and is therefore attributed to an epigenetic bookmarking process during mitosis.
Here, we visualize the activities of stochastically-expressed transgenes in order to distinguish active and inactive mother cells and the behaviors of their daughter nuclei following mitosis. We reasoned that a variety of transcriptional mechanisms–paused RNA Polymerase II (Pol II), enhancer priming and shadow enhancers–ensure precise timing of transcription reactivation following mitosis [5]. In previous experiments, the transgenes analyzed were activated in a rapid and synchronous manner and this precluded a rigorous comparison of expressing and non-expressing nuclei through mitosis, obscuring the detection of potential transcriptional memory processes during early mitotic cycles in the Drosophila embryo [6 and 7].
We employed sensitized transgenes that produce patterns of sporadic expression in order to trace the behaviors of individual cell lineages. By comparing the timing of gene reactivation of daughters originating from expressing (called memory nuclei) and neighboring non-expressing nuclei (non memory ones), we obtained evidence for transcriptional memory.
We focus our analysis on the regulation of snail (sna), which encodes a zinc finger transcription factor that is essential for epithelial-mesenchyme transitions (EMT) in most animal systems [8]. It has been implicated in a variety of developmental and disease processes, including mesoderm invagination in Drosophila [9] and metastases of cancerous tissues [8]. In Drosophila, sna is expressed in a thousand cells comprising the embryonic mesoderm [10], where it is important for coordinating their invagination during gastrulation [9]. Paused Pol II helps to coordinate the activation of snail transcription in the different cells of the mesoderm [11], while “redundant” mesodermal enhancers provide robustness in the activation of sna expression even under stressful conditions such as elevated temperatures [12 and 13].
A 500 bp region of the distal sna (shadow) enhancer (snaE) was attached to a yellow reporter gene containing 24 MS2 stem loops (Figure 1 A). This minimal enhancer lacks 3 Zelda binding sites [14 and 15], which are likely to be important for the timely activation of the endogenous sna gene during early stages of development [16]. This truncated enhancer also lacks two twist binding sites, which are known to act synergistically with Dorsal for the activation of sna expression [17]. The minimal enhancer was used in combination with several different core promoters and each construct was integrated, as a single copy, in the same landing site using the PhiC31 integrase-mediated site-specific transgenesis method [18]. These minimal promoters include e.g. sna and brinker (brk) that contain paused Pol II and others, e.g. wntD, that lack it [11, 19]. All of the resulting snaE<MS2-yellow heterozygous transgenes exhibit slow expression dynamics (see Supplemental movies 1–2). Activation is not clearly observed until nuclear cleavage cycle (nc) 13, and only in a fraction of the nuclei (~10–20%), whereas the endogenous locus is normally expressed in all nuclei of the presumptive mesoderm during this cycle (Figure 1 C; Figure S1).
Figure 1. Live imaging of a sensitized snail transgene reveals transcriptional memory.
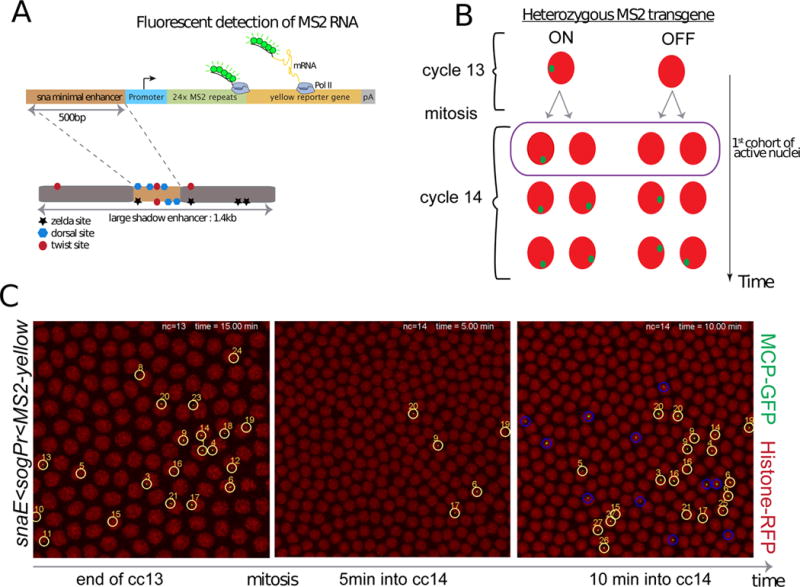
(A) Schematic view of a sensitized snail transgene. A minimal 500bp snail shadow enhancer was cloned upstream of various minimal promoters and 24X MS2 repeats, followed by a yellow reporter gene. Upon activation, the MS2 stem loops in nascent transcripts are bound by a MCP-GFP fusion protein. (B) Schematic representation of transcriptional memory. Descendants from transcriptionally active mother nuclei in nuclear cycle (cc) 13 tend to activate transcription very early during interphase 14, circled and referred to as ‘first cohort of active nuclei’. This particular population will be referred to as ‘memory’ nuclei, whereas active descendants from inactive mother nuclei at cc13 will be referred to as ‘non memory’ nuclei. (C) Live imaging of the transcriptional activity of a sensitized snail transgene (snail-enhancer<sogPr<MS2-yellow). Nascent mRNA ‘spots’ are shown in green (MS2-MCP-GFP) and nuclei are labeled in red (Histone-RFP transgene). Only selected time frames are shown, the entire movie can be obtained in Movie S1. All active nuclei at cc13 are tracked with a unique number. After mitosis their descendants are tracked, circled in yellow, while active cc14 nuclei derived from inactive mothers are circled in blue. Ventral views of a 2.1 zoomed central region of an embryo, where anterior is to the left. See also Figure S1 and Movie S1.
Transcription is not detected during mitosis 13, but is reactivated during nc14. The kinetics of this activation is slow and stochastic among nuclei and does not encompass all of the nuclei comprising the mesoderm until after 30–40 min following mitosis (Figure1, Movie S1, Figure S1). In contrast, the endogenous gene exhibits rapid and synchronous re-activation of transcription during nc14 and is expressed throughout the mesoderm in only ~10–12min following mitosis (Figure S1, [20]). The slow reactivation of snaE<MS2/yellow transgenes allowed us to trace the origins of the first nuclei to display post-mitotic reactivation following entry into nc14 (Figure 1 B).
10 to 20% of the pre-mesoderm nuclei (e.g., 26/175) exhibit transcription of the sna transgenes by the completion of nc13 (Figure 1C). After mitosis, five nuclei (~2% of the pattern) exhibit reactivation within 5 min after entry into nc14. Each of these is a descendant of a previously activated mother (“memory mothers”) (see Movie S1, Figure S2). During the next several minutes another 27 nuclei display transcription (~9% of the pattern), and over half (15/27) are descendants of memory mothers (Figure 1C, circled in yellow, false-colored in Movie S1).
During cc13, a small fraction fr13 of nuclei are active. By the end of cc14, the majority of mesodermal nuclei are active. The activation of the population of nuclei shows sigmoidal kinetics (see Figure 2 A, B). Although a significant fraction of these nuclei are derived from mothers that did not transcribe the transgene during nc13 (Figure 1 C, blue circles) (referred to as “non memory mothers”), quantitative analysis provides clear evidence for memory (Figure 2). The daughters of memory mothers are ~4-times more likely to be among the first nuclei to display reactivation upon entry into nc14 as compared to a random reactivation (Movies S1, S2, Figure 2 E). To estimate the temporal dependence of the number of activated nuclei coming from active mothers upon random memoryless activation, we use as a reference the activation kinetics of the nuclei coming from inactive mothers Nin14(t). In the absence of memory, the temporal behavior of active nuclei coming from active mothers Nact14(t) will be the same as that of nuclei coming from inactive mothers Nin14(t) multiplied by the probability that they do activate fr13 : Nact14(t)~ Nin14 (t)• fr13, shown as dotted lines in Figure 2 A-A′, B-B′.
Figure 2. Quantitative analysis of transcriptional memory.
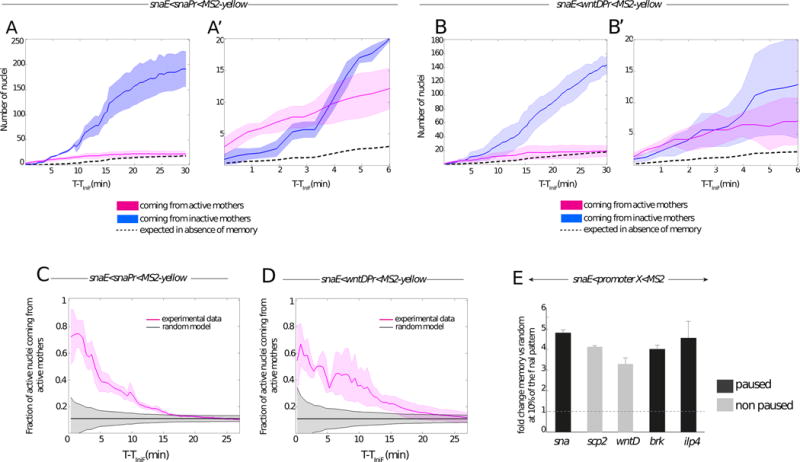
(A–B) Memory bias in transcriptional activity during cc14. Kinetics are extracted from 3 independent movies of snaE<snaPr<MS2 transgenic embryos (A) and from 3 independent movies of snaE<wntDPr<MS2 transgenic embryos (B). The distribution of active nuclei derived from transcriptionally active mother nuclei is shown in pink. The descendants of inactive mother nuclei are depicted in blue. Light colors indicate the standard deviation of these quantified kinetics. Dashed curves represent the expected average time behavior in the absence of a memory bias, for mathematical formulations please refer to the main text. (A′, B′) Zoomed view of the 6 first minutes of cc14, corresponding to the plots shown in (A) and (B) respectively.
(C, D) Temporal behavior of the fraction of nuclei derived from memory mothers during cc14 (pink curves). Two different genotypes are shown, with a paused promoter (snaE<snaPr<MS2, n=3) (C) or a non-paused promoter (snaE<wntDPr<MS2, n=3) (D). Experimental data are compared to the expected behavior of a random activation distribution, indicated in grey, assuming a binomial sampling of a constant fraction of active nuclei in each cell cycle (see Supplements for details). For all the plots (A to D) the x axes corresponds to the time (in minutes), starting at the frame where the first spot is detected (TiniF). (E) Increase in the probability that a memory daughter nucleus is among the first cohort (first 10%) of activated nuclei with respect to the random model expectations during the onset of cc14 for the following transgenes: snaE<snaPr<MS2 (n=3 movies); snaE<wntDPr<MS2 (n=3 movies), snaE<scpPr<MS2 (n=2movies), snaE<brkPr<MS2 (n=2movies), snaE<ilp4Pr<MS2 (n=2 movies). The fold change is obtained by dividing the fraction of active nuclei coming from active mothers by the fraction expected from the random model. The fold change is evaluated at the time corresponding to 10% of the total active pattern. Error bars represent standard errors. See also Figure S2 and Movie S2.
Quantitative analysis of thousands of nuclei in living embryos unequivocally reveals the occurrence of transcriptional memory (Figure 2). Transgenes containing either the sna or wntD core promoter exhibit a memory bias (Figure 2 A-A′, B-B′). Depending on the promoter sequence, between 60–80% of the first cohort of nuclei that are reactivated during the early phases of nc14 derive from memory mothers (Figure 2 C, D), which is quite impressive considering that the vast majority of nuclei (~80–90%) do not express the transgene during nc13 (Figure 2 A, B). To test whether the enhanced number of active nuclei coming from active mothers at the beginning of cycle 14 cannot be explained by stochastic activation alone, we consider a random activation model where each nucleus makes an independent decision whether to activate or not. Since all mesodermal nuclei are eventually activated in cc14, the fraction of active nuclei at cc14 coming from active mothers is equal to the fraction fr13 of active nuclei at the end of cc13. In the absence of memory, the independent decision of each nuclei to activate or not corresponds to a binomial distribution of activated nuclei with a mean of fr13. The grey shadow in Figure 2 C, D shows the random expectation in the absence of memory calculated as the standard deviation of the fraction of activated nuclei coming from active mothers Nact14(t)/Ntot14(t) as a function of time, , where Nact14(t) is the number of active nuclei coming from active mothers and Ntot14(t) is the total number of active nuclei.
Despite the relatively small number of nuclei constituting the first cohort of activation, the observed distribution is non-random (Figure 2 C, D). The proportion of memory nuclei exhibiting reactivation progressively diminishes to 30%, 20% and 10% during the next 5, 10, and 15 min, respectively (Figure 2 C, D).
These observations leave little doubt that transcription of sna transgenes during nc13 predisposes them for rapid reactivation during nc14 as compared with inactive nuclei. In fact, transcription during nc13 results in a 3.5-fold to 5-fold increase in the likelihood of daughter nuclei exhibiting reactivation during the initial phases of nc14 (Figure 2 E, Figure S3 A–D). The efficiency of this transcriptional memory is influenced by the different promoter sequences tested in this study. The sna and ilp4 promoters yield a higher efficiency of reactivation during the initial phases of nc14 than does the wntD promoter. The basis for these differences is uncertain, but do not scale with the levels of paused Pol II identified by tissue-specific embryonic Pol II ChIP-Seq or Gro-Seq assays [11 and 21]. Moreover, memory is observed regardless of the presence (e.g sna promoter) or absence (e.g brk promoter) of the TATA box sequence, despite previous reports that TBP remains bound to mitotic chromosomes and fosters post-mitotic reactivation [22]. The brk promoter is not bound by TBP in the pre-cellular embryo [23], even though our movies clearly demonstrate a memory bias in the reactivation of the snaE<brk<MS2 transgene. Thus, other mechanisms of mitotic “bookmarking” might be at play (see below).
At least two mechanisms can be envisioned for transcriptional memory, asymmetric distribution of activators or inheritance of transcribed templates. Sna is activated by Dorsal and Twist, which are distributed in gradients. In principle, there could be slight variations in their distributions even among nuclei located in ventral regions where there are peak concentrations of both activators. Perhaps a few “jackpot” nuclei obtain higher levels of Dorsal and Twist, which are subsequently inherited by daughter nuclei. Alternatively, transcription of a DNA template might render it more susceptible to rapid reactivation following mitosis. Transcription could cause inherited changes in the distribution of DNA-bound transcription factors, nucleosomes or histone modifications favoring activation in the next cell cycle.
In an effort to distinguish between these alternative models, we quantified the inheritance patterns of neighboring active and inactive mother nuclei during the transition from nc13 to nc14 (Figure 3 A, B, F, G and Figure S3 G–J). Most active mother nuclei produce just one daughter nucleus that rapidly reactivates the sna transgene during the early phases of nc14. The activation time for memory daughters is ~2-times faster than for descendants of a neighboring non-active mother (Figure 3 B). The second “memory daughter” displays expression only after a delay (8.5min +/−6.2), however the activation of this second memory daughter is significantly faster than the second non memory daughter (Figure 3 C). To explain these observations we hypothesize that, in most cases, only one of the two sister chromatids of the homologue containing the transgene initiates transcription detected by the MS2-MCP technique (summarized in Figure S2-C).
Figure 3. Transcriptional kinetics of memory.
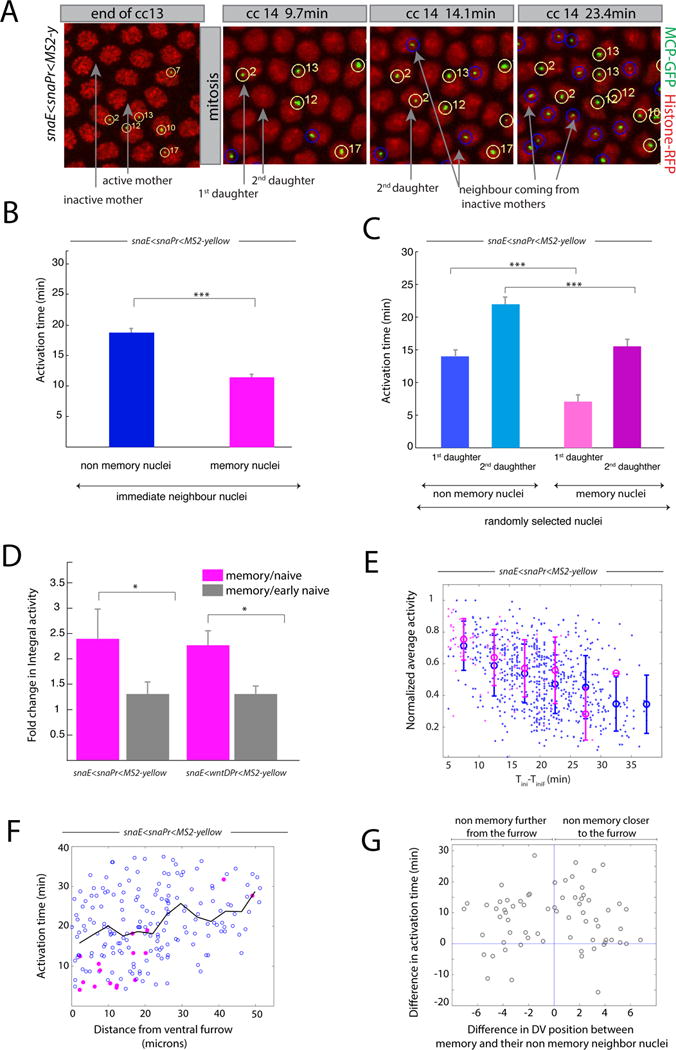
(A) Snapshots of live imaging of a snaE<snaPr<MS2 transgenic embryo. Confocal z-projected images of selected ventral regions are shown at different time points during cc13 and cc14. Tracked active nuclei at cc13 and their descendants (first and second daughter to exhibit transcription) are circled and numbered in yellow. Examples of immediate neighboring nuclei coming from inactive mother nuclei are circled in blue. (B) Mean activation time for descendants of active mother nuclei and for immediate neighboring nuclei coming from inactive mothers. Error bars represent standard errors (non-memory nuclei are shown in blue, n=68; memory nuclei are shown in pink, n=68; memory versus non-memory p-value=3*10−10). (C) Mean activation time for descendants of active mother nuclei or those from inactive mothers, tracking the first daughter and second daughter separately, randomly selected from the entire imaged region. For panels B, C, quantification was performed on 3 movies of snaE<snaPr<MS2 transgenic embryos (total of 750 nuclei). Error bars represent standard errors (first daughter non memory, n=54; second daughter non memory, n=54; first daughter memory, n=24; second daughter non memory n=24). Statistics: first daughter non memory versus first daughter memory, p-value= 3*10−8; second daughter non memory versus second daughter memory, p-value=1*10−4). (D) Fold change in integral activity, defined as the ratio between the integral activity of memory nuclei divided by that of non memory nuclei. Integral activity corresponds to the sum of activities across all time frames. Error bars represent standard errors computed on 3 different movies for each genotype. Statistics: snaE<snaPr<MS2 p-value=0.02; snaE<wntDPr<MS2 p-value=0.03). (E) This scatter plot shows the behavior of the average activity as a function of the activation time. The data points are extracted from three movies and the value of the average activity is normalized by its maximum. Nuclei coming from active mothers are depicted in pink while those coming from inactive mothers are represented in blue. Error bars are standard deviations evaluated by binning the activation time in intervals of 5 minutes each. (F) Scatter plot of the activation time as function of the distance from the ventral furrow at cc14 for a snaE<snaPr<MS2 transgenic embryo. Pink symbols represent memory nuclei and blue symbols non memory ones. The black line is the average behavior. (G) Scatter plot of the difference in activation time between a memory nucleus and its non memory closest neighbor as function of their distance. The statistical test used in Figure 3 is a two samples t-test with the following convention: * for a p-value comprised between 0.05 and 0.01; ** for a p-value comprised between 0.01 and 0.001; *** for a p-value inferior or equal to 0.001. See also Figure S3, Movie S2 and Movie S3.
Many genes are duplicated on tightly associated sister chromatids during late S phase and G2 phases of cc14 [24]. Among others, one possible interpretation of these inheritance patterns is that the sna transgene is transcribed by only one of the two sister chromatids during nc13 (Figure S2).
The inheritance patterns observed for homozygotes carrying two copies of the transgene are consistent with this interpretation (Supplemental Movie S3). Mother nuclei exhibiting transcription of both copies of the transgene during nc13 display two different inheritance patterns at roughly equal frequencies. Some produce two daughters that each contains a single transcription dot at the onset of nc14 (1,1 configuration). Others produce one daughter with 2 dots while its sister lacks early reactivation signals (2,0 configuration) (Figure S2 B, D). A simple interpretation of these observations is the independent assortment of active and inactive sister chromatids during mitosis (summarized in Figure S2 D).
These observations could be explained by an allelic specific inheritance of memory but also by an unequal distribution of transcriptional activators between two daughters. However, if one ‘jackpot’ descendant inherited higher amounts of activators, this would increase the probability of activation of the MS2 transgene and cause frequent asymmetric inheritance configurations, namely (1,0) or (2,0). Manual analysis of three independent snaE<snaPr<MS2 homozygous movies disfavor such asymmetric inheritance patterns (Movie S3, Figure S2). Moreover, unequal distribution of activators seems unlikely in syncytial embryos, where there is extensive intermingling of nuclei during mitosis (see Movies S1 to S3). Our analysis is restricted to the ventral-most regions where there are peak levels of the Dorsal and Twist activators. We have not included nuclei located in ventro-lateral regions where there are diminishing levels of activator gradients. Indeed measuring the activation time of nuclei as a function of the distance to the ventral furrow dismisses the existence of a spatial bias responsible for transcriptional memory (Figure 3 F,G and Figure S3 G–J).
The basis for transcriptional memory is uncertain. It can be as simple as the displacement of nucleosomes at the core promoter [25], thereby facilitating the recruitment of Pol II upon entry into nc14. Alternatively, active transcription during nc13 might lead to certain histone modifications at the enhancer and/or promoter (e.g., H4K5ac or H3K4me) that are retained during mitosis and foster rapid reactivation in the ensuing cell cycle, as observed in cultured cells [3], Dictyostelium [2] and yeast [26].
In the early Drosophila embryo, genome-wide H3K4methylation (H3K4me3 or H3K4me1) is detected at the maternal-zygotic transition (MZT) [23 and 27], occurring at the transition between nc13 and nc14, precisely when we observe transcriptional memory. It is therefore possible that histone methylation, established during zygotic genome activation provides the basis for transcriptional memory.
Is transcriptional memory used to help pattern the Drosophila embryo? To address this question we investigated the levels of transcription produced from nuclei derived from active and inactive mothers. The goal was to determine whether memory nuclei exhibit more efficient expression of the sna transgene than non-memory nuclei during nc14. According to such a scenario, transcription might be progressive, leading to increasing efficiencies in the production of mRNAs due to prior “priming” events. Rates of RNA synthesis can be extrapolated by measuring the average fluorescence intensities (MS2-MCP-GFP) at the transcription foci [6 and 7].
Average fluorescence intensities are inversely correlated with the delay in gene reactivation (Figure 3 D, E and Figure S3 E, F). The earlier a nucleus is activated, the higher its time-averaged rate of RNA synthesis (Figure 3 D, E). Consequently, memory nuclei produce during nc14, on average, ~2-fold more mRNAs than non-memory nuclei (Figure 3 D). Moreover, there is a reduction in the levels of RNAs produced by late-expressing nuclei (mainly non-memory daughters) (Figure 3 E and Figure S3F) due to an overall reduction in the levels of sna expression during nc14 [28].
In summary, we propose that transcriptional memory contributes to the rapid and synchronous activation of gene expression seen in the early Drosophila embryo. Memory might also be important for the sustained expression of constitutive “house-keeping” genes during successive cell cycles. Indeed, transcriptional memory has been documented for the actin5 gene in Dictyostelium [2]. Finally, we suggest that transcriptional memory is a mechanism of developmental homeostasis, which helps ensure, when needed, that cells will tend to retain the properties of their progenitors. However this transcriptional persistence could in principle impede the plasticity required during cell specification.
Supplementary Material
Acknowledgments
We are grateful to Thomas Gregor and Hernan Garcia for sharing the MCP-GFP fly stock. We would like to thank Wei Zhang for his help with genomic data analysis. M.L was a recipient of a HFSP long-term fellowship and is currently sponsored by the CNRS. T.F, N.D, A.M.W and M.C are supported by the grants: ANR AXOMORPH, ANR-11-LABX-0044 and ANR-10-IDEX-0001-02PSL. T.F and A.M.W. are supported by MCCIG grant no. 303561. This research was initiated in the Levine lab with a support from a NIH Grant RO1 GM34431. The Lagha lab is supported by an ATIPE-Avenir CNRS grant and an HSFP CDA grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Experimental Procedures
All MS2 containing transgenic embryos were generated by the targeted integration (VK33) of snaEnhancer<minimal promoter<24X-MS2 repeats-yellow plasmids [10]. Histone-RFP and MCP-GFP are maternally provided by a stock described in [6]. All movies have been acquired using a 780lsm Zeiss confocal microscope with the following settings: 40× oil objective, a 2.1 zoomed 512×512 16bit/pixel image, with bidirectional scanning, 21 z-stacks, 0.5μm apart. Under these conditions, the time resolution is in the range of 15–20 seconds per frame. Image processing of the MS2-MCP-GFP signal is performed in a semi-automatic way using custom Matlab algorithms, based on our previously published work [5]. See supplementary information for details on transgenic fly stocks, molecular biology, imaging and quantification.
References
- 1.Porcher A, Abu-Arish A, Huart S, Roelens B, Fradin C, Dostatni N. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development. 2010;137:2795–2804. doi: 10.1242/dev.051300. [DOI] [PubMed] [Google Scholar]
- 2.Muramoto T, Müller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is Required for Inheritance of Active Transcriptional States. Current Biology. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Cell Biology. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 5.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas T, Ferraro T, Roelens B, De Las Heras Chanes J, Walczak AM, Coppey M, Dostatni N. Live Imaging of Bicoid-Dependent Transcription in Drosophila Embryos. Current Biology. 2013;23:2135–2139. doi: 10.1016/j.cub.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Current Biology. 2013;23:2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014 doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemavathy K, Meng X, Ip YT. Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development. 1997;124:3683–3691. doi: 10.1242/dev.124.19.3683. [DOI] [PubMed] [Google Scholar]
- 10.Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes & Development. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 11.Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow Enhancers Foster Robustness of Drosophila Gastrulation. Current Biology. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4566–4566. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB. Zelda Binding in the Early Drosophila melanogaster Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal Coordination of Gene Networks by Zelda in the Early Drosophila Embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien CY, Kirov N, Shvartsman SY, Rushlow CA. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol. 2014;24:1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. The EMBO Journal. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature Genetics. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife. 2015;4 doi: 10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes & Development. 2013;27:1146–1158. doi: 10.1101/gad.215459.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nature Cell Biology. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. Author response. eLife. 2013;2 doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKnight SL, Miller OL. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell. 1979;17:551–563. doi: 10.1016/0092-8674(79)90263-0. [DOI] [PubMed] [Google Scholar]
- 25.Lenhard B, Sandelin A, Carninci P. Regulatory elements: Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nature Publishing Group; 2012. pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 26.Brickner JH. Transcriptional memory at the nuclear periphery. Current Opinion in Cell Biology. 2009;21:127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X-Y, Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife. 2014;3 doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boettiger AN, Levine M. Rapid transcription fosters coordinate snail expression in the Drosophila embryo. CellReports. 2013;3:8–15. doi: 10.1016/j.celrep.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


