SUMMARY
Photosynthesis in C3 plants is limited by features of the carbon-fixing enzyme Rubisco, which exhibits a low turnover rate and can react with O2 instead of CO2, leading to photorespiration. In cyanobacteria, bacterial microcompartments known as carboxysomes improve the efficiency of photosynthesis by concentrating CO2 near the enzyme Rubisco. Cyanobacterial Rubisco enzymes are faster than those of C3 plants, though have lower specificity toward CO2 than the land plant enzyme. Replacement of land plant Rubisco by faster bacterial variants with lower CO2 specificity will improve photosynthesis only if a microcompartment capable of concentrating CO2 can also be installed into the chloroplast. We review current information about cyanobacterial microcompartments and carbon-concentrating mechanisms, plant transformation strategies, replacement of Rubisco in a model C3 plant with cyanobacterial Rubisco, and progress toward synthesizing a carboxysome in chloroplasts.
Keywords: carbon-concentrating mechanism, chloroplast, Rubisco, photosynthesis, carboxysome, chloroplast transformation, Nicotiana, Synechococcus elongatus, transgenic, transplastomic
INTRODUCTION
Improving the efficiency of photosynthesis has been progressively more recognized as a strategy for increasing yield of biomass and food crops (Long et al. 2015, Ort et al. 2015). While plant breeding has been enormously successful in increasing the percentage of a plant’s resources that comprise the desired product, many crop species have reached a plateau from which further improvement by traditional means is unlikely (Long et al. 2006, Long, et al. 2015). Synthetic biology offers the opportunity to increase the conversion of solar energy through such strategies as altering light capture and conversion and carbon uptake and conversion (Ort et al. 2015). This review will focus on a possible tactic to improve carbon fixation: installation of a carbon concentrating mechanism (CCM) presently utilized by cyanobacteria but not by vascular plants (Price et al. 2008, Price et al. 2013, Zarzycki et al. 2013). The cyanobacterial CCM requires the presence of a bacterial microcompartment known as the carboxysome, as well as bicarbonate transporters (Rae et al. 2013). We will discuss current progress and prospects for engineering functional carboxysomes into chloroplasts.
CARBON-CONCENTRATION MECHANISMS: COMPENSATION FOR THE UNFAVOURABLE TRAITS OF THE CARBON- FIXING ENZYME RUBISCO
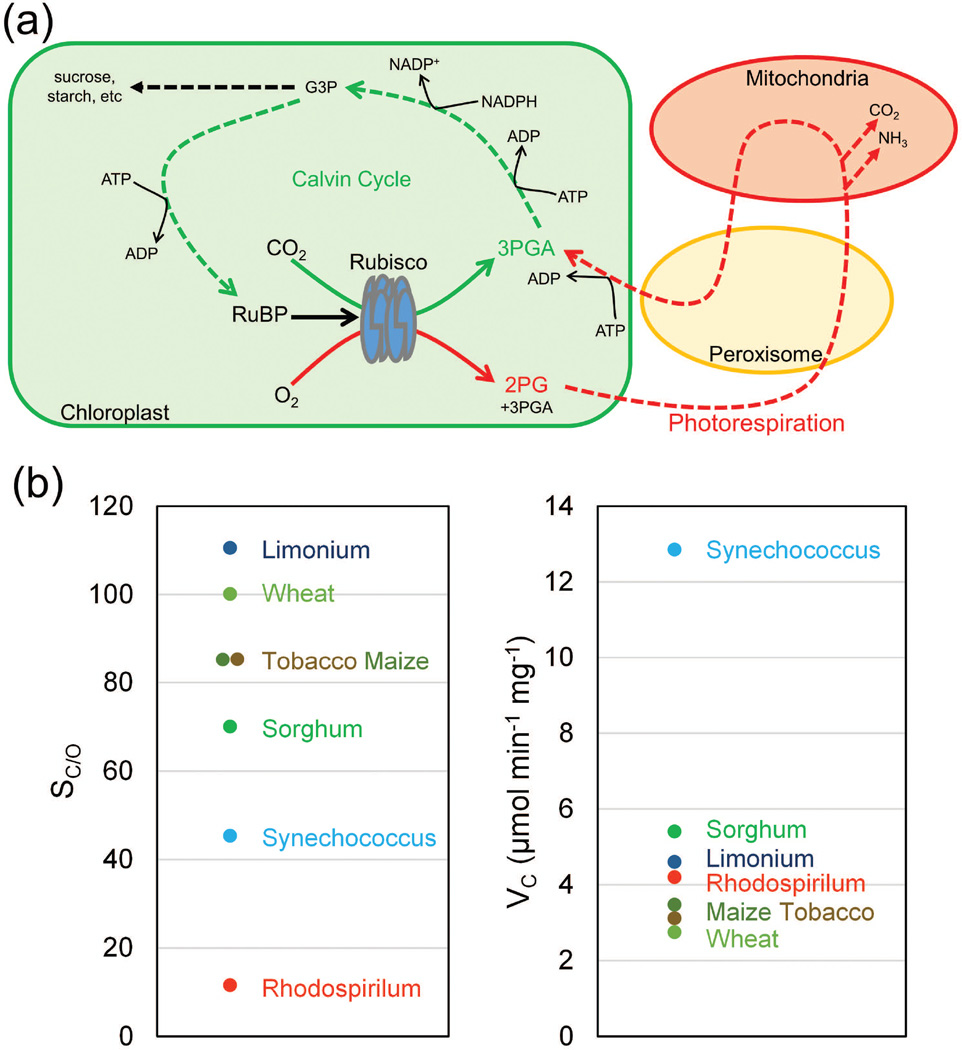
Catalysis of the reaction of ribulose-1,5-bisphosphate (RuBP) with atmospheric CO2 by the enzyme Rubisco results in fixation of carbon into organic molecules. However, as the name indicates, “Ribulose-1,5-bisphosphate carboxylase/oxygenase” can also catalyze a reaction with oxygen (O2), leading to the process known as photorespiration (Figure 1a). Reaction with O2 rather than CO2 results in loss of some fixed carbon as CO2, loss of NH3, as well as consumption of ATP, thus decreasing the efficiency of photosynthesis (reviewed by Parry et al. 2013).
Figure 1.
The carboxylation and oxygenation reactions of Rubisco. (a) Diagram of Rubisco-catalyzed reactions in C3 plants. Carboxylation of RuBP initiates the Calvin Cycle and leads to production of carbohydrates. Oxygenation of RuBP initiates photorespiration and results in the net loss of fixed CO2 and NH3, and consumption of energy. The carboxylation of RuBP by Rubisco and subsequent reactions in the Calvin Cycle are shown in green arrows. The oxygenation of RuBP by Rubisco and the subsequent photorespiration process are shown in red arrows. RuBP = Ribulose-1,5-bisphosphate; 3PGA = 3-phosphoglycerate; 2PG = 2-phosphoglycolate; G3P = glyceraldehyde 3-phosphate. (b) Rubisco specificity (SC/O) and maximum carboxylation rate (VC) in tobacco (C3 model species), wheat (C3 crop), limonium (L. gibertii, C3 with high specificity), maize and sorghum (C4 crops), S. elongatus (cyanobacteria, β-carboxysome species) and Rhodospirilum rubrum (purple bacteria, α-carboxysome species). Data are averages of values reported by: (Jordan and Ogren 1981, Jordan and Ogren 1983, Jordan and Ogren 1984, Makino et al. 1985, Parry et al. 1987, Parry et al. 1989, Sage and Seemann 1993, Kane et al. 1994, Delgado, et al. 1995, Uemura et al. 1996, Whitney et al. 1999, Whitney et al. 2001, Pearce 2006, Mueller-Cajar et al. 2007, Parry et al. 2007, Kubien et al. 2008, Mueller-Cajar and Whitney 2008, Sharwood et al. 2008, Carmo-Silva et al. 2010, Genkov et al. 2010, Whitney et al. 2011b, Occhialini et al. 2016, Prins et al. 2016).
The specificity factor (SC/O) of Rubisco defines the relative reactivity of the enzyme towards the two gaseous substrates, CO2 and O2. SC/O = Vc×Ko/Vo×Kc, where Vc and Vo represent the maximum velocities of carboxylation and oxygenation, and Kc and Ko represent the Michaelis-Menten constants for CO2 and O2. Environmental changes through geological time have shaped evolution and diversification of Rubisco by providing selective pressures that favour changes in Rubisco structure resulting in improved performance (Tcherkez et al. 2006, Christin et al. 2008b). Rubisco in higher plants is a rather complex enzyme, composed of nuclear-encoded small subunits and chloroplast-encoded large subunits (Spreitzer and Salvucci 2002), and dependent on interaction with diverse molecular chaperones essential for biogenesis and catalysis (Hauser et al. 2015b). Even a single point mutation in the Rubisco sequence can disrupt the assembly of Rubisco in tobacco (Foyer et al. 1993). As a consequence, Rubisco evolution and diversification is relatively slow. Yet, significant variation is known to exist in nature and Rubisco enzymes with higher turnover or superior catalytic efficiency and/or specificity towards CO2 have been identified. Even among the same Form 1B type Rubisco enzymes shared by higher plants including both C3 and C4 species and some cyanobacteria, remarkably diverse kinetic properties can be found (Figure 1b). Models suggest that some of these have the potential to improve photosynthetic CO2 assimilation in major C3 crops such as wheat (Delgado et al. 1995, Galmes et al. 2005, Carmo-Silva et al. 2015, Prins et al. 2016).
The CCM in C4 and CAM plants
Decreased CO2 concentration in the atmosphere millions of years ago led to the evolution of photosynthetic CCMs in plants and algal species (Christin et al. 2008a, Sage and Stata 2015). In C4 plants, such as maize and sugarcane, carbon is initially fixed by phosphoenolpyruvate carboxylase (PEPC), with production of an organic acid containing 4 carbons (hence the C4 name). Subsequent decarboxylation of the C4 acids results in accumulation of high concentrations of CO2 around Rubisco, thus favouring carboxylation over oxygenation of RuBP. In addition to the specialized photosynthetic biochemistry, the leaves of many C4 plants show anatomical features associated with the CCM. These characteristics are known as Kranz anatomy (comprehensively reviewed by (Dengler and Nelson 1999). The initial fixation of inorganic carbon occurs in the mesophyll cells, where PEPC is exclusively located. C4 acids are translocated to the bundle sheath cells, where they undergo decarboxylation. The released CO2 is subsequently assimilated by Rubisco, which is exclusively located in the chloroplasts of the bundle sheath cells (Edwards et al. 1985). Chemical modification and increased thickness of the bundle sheath cell walls, and reduction of the surface area of bundle sheath exposure to intercellular spaces decrease leakage of CO2 back to the mesophyll cells so that CO2 accumulates (Furbank et al. 1989, Brown and Byrd 1993, Evans and Von Caemmerer 1996, Jenkins 1997, Kiirats et al. 2002).
The increased CO2 concentration in the bundle sheath cells results in low RuBP oxygenation and, consequently, low rates of photorespiration and increased rates and efficiency of photosynthesis in C4 plants (Kanai and Edwards 1999). The presence of a CCM makes C4 photosynthesis especially competitive in conditions that promote carbon loss through photorespiration, such as high temperatures, high light intensities and decreased water availability causing low intercellular CO2 concentrations as a consequence of stomatal closure. As a result, C4 plants tend to have lower stomatal conductance, lower transpiration rate and higher water use efficiency than their C3 counterparts.
Like C4 photosynthesis, plants with the Crassulacean acid metabolism (CAM) also use PEPC to fix carbon into organic acids with subsequent release of high concentrations of CO2 around Rubisco. However, in CAM plants the two carboxylases are located on the same cell type and there is a temporal separation of primary (nocturnal) and secondary (diurnal) fixation of CO2. Since stomata are open at night and closed during the day, CAM plants are very water-use efficient and well adapted to warm and dry environments. The prospects for introducing the CAM CCM into C3 plants has recently been reviewed by DePaoli et al. (2014).
The CCM in cyanobacteria
Cyanobacteria have evolved a mechanism that allows them to utilize faster Rubisco enzymes that are more sensitive to oxygen. Inorganic carbon mainly in the form of bicarbonate ion is accumulated within cytosol through optimally regulated bicarbonate transporters and CO2 uptake systems (Price et al. 2008). As a result, the intracellular concentration of bicarbonate ion in cyanobacteria is typically well over 10 mM or two to three orders of magnitude higher than that observed outside the cells (Woodger et al. 2005). By encapsulating Rubisco and carbonic anhydrase in a protein microcompartment known as a carboxysome and surrounding the enzyme with higher concentrations of CO2, Rubisco is less likely to react with oxygen (Badger and Price 2003). This strategy is so powerful that it has independently evolved in two lineages of bacteria that contain two different types of Rubisco. Form 1A Rubisco is found in α-carboxysomes, while Form 1B Rubisco is located in β-carboxysomes (Badger et al. 2002).
Genes present in cyanobacteria that make α- or β-carboxysomes have been well studied through analysis of mutants and considerable information is available about the structural arrangement of proteins present in the two types of carboxysomes (Figure 2). Incorporation of either α- or β-carboxysomes into crop plants is an attractive strategy for improving photosynthesis (Price, et al. 2013, Zarzycki, et al. 2013, Whitehead et al. 2014). Each type of microcompartment has potential advantages and disadvantages as a candidate for engineering into plants. One advantage of utilizing α-carboxysomes is the fact that their components have already been assembled in a heterologous system along with functional Rubisco, following expression of 10 genes from Halothiobacillus neapolitanus in E. coli (Bonacci et al. 2012), while β-carboxysomes have not yet been synthesized in E. coli or microbes other than cyanobacteria. However, phylogenetically, Form 1B Rubisco is related to Rubisco present in land plants (Badger, et al. 2002), suggesting that land plant chaperones are more likely to be effective on enzymes normally present in β-carboxysomes. Use of either α- or β-carboxysomes in chloroplasts will require replacement of the endogenous land plant Rubisco with a cyanobacterial Rubisco in order to take advantage of better kinetic properties in a high CO2 environment. Aside from the advantage of improved turnover rates with the cyanobacterial enzyme, land plant Rubisco will probably be more difficult to engineer to pack into either type of carboxysomes than replacing the endogenous Rubisco with a cyanobacterial enzyme that normally assembles into a microcompartment. This review will focus on β-carboxysomes because at this writing, there are no reports of autotrophic plants that fix carbon with Form 1A Rubisco, while plants containing cyanobacterial Form 1B Rubisco have been reported (Lin et al. 2014b, Occhialini et al. 2016).
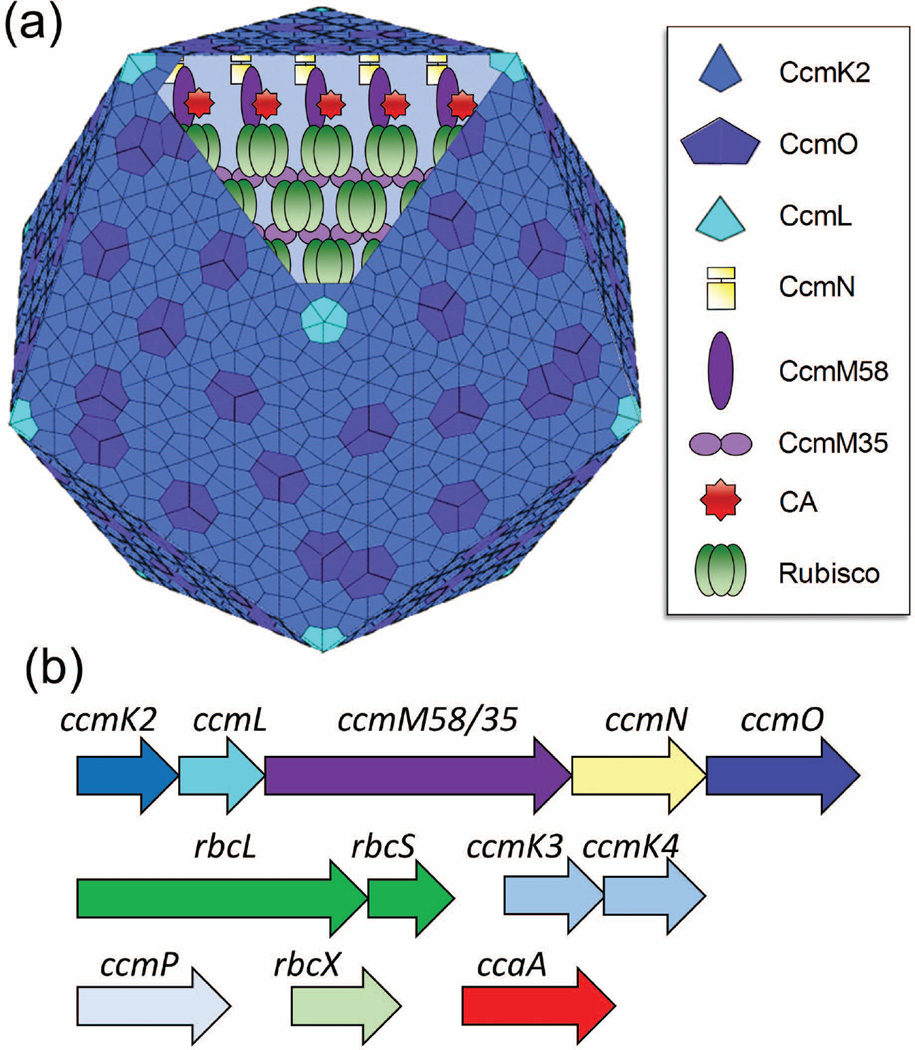
Figure 2.
The components of β-carboxysomes. (a) Structural model of β-carboxysome in S. elongatus PCC7942. Data on protein location and assembly as described by Cameron et al. (2013). Please note that the role of CcmO remains unclear although it is represented as pseudohexamers here. Carboxysome model courtesy of Kevin Hines. (b) Operons present in S. elongatus PCC7942 (Heinhorst et al. 2014).
Components Required for Introducing Microcompartments into Chloroplasts
Recently, much progress has been made in characterization and manipulation of bacterial microcompartments for applications in synthetic biology and metabolic engineering. A targeting or encapsulation peptide was first identified at the N terminus of the propionaldehyde dehydrogenase enzyme from the propanediol utilization microcompartment (Fan et al. 2010). Similar encapsulation peptides are now known to be present as N- or C-terminal extensions in core enzymes of bacterial microcompartments including CcmN protein from β-carboxysomes (Kinney et al. 2012, Aussignargues et al. 2015). These encapsulation peptides are capable of targeting foreign proteins and introducing new functions to heterologously produced bacterial microcompartments (Choudhary et al. 2012, Lassila et al. 2014, Lawrence et al. 2014, Lin et al. 2014a). A recent study demonstrated that a mixture of shell proteins from an α-carboxysome and a β-carboxysome can assemble into chimeric microcompartments (Cai et al. 2015). In addition, shell proteins with modified “pores” have been shown to be incorporated into carboxysomes (Cai, et al. 2015). Thus, it is now a real possibility to apply synthetic biology approaches to customize microcompartments for biotechnological purposes.
We chose to investigate the assembly of microcompartments by β-carboxysome shell proteins from Synechococcus elongatus PCC7942 in plant chloroplasts because they represent one of the best characterized bacterial microcompartments with regard to structural organization. Genes encoding the protein shell and internal proteins of both α- or β-carboxysomes have been identified through genetic analysis. Currently the proteins CcmK2, CcmO, and CcmL are known to be the most important for synthesis of the shell layer of the β-carboxysome. Based on its crystal structure, it is accepted that CcmK2 represents the most abundant protein of the icosahedral shell and forms its facets (Kerfeld et al. 2005). CcmL forms pentamers that are believed to cap the vertices of the icosahedral shell (Tanaka et al. 2008). Although the exact role of CcmO remains unclear, it has been proposed to occupy the edges along two adjacent facets (Rae et al. 2012). In addition, either CcmK3 or CcmK4 may be required for the fully functional β-carboxysome (Rae, et al. 2012). The hexameric shell proteins form central pores that are believed to be selectively permeable.
In β-carboxysomes from S. elongatus PCC7942, CcmM58 is an internal protein that is known to interact with Rubisco. CcmM58 carries three so-called small subunit like domains (SSLD) in its C-terminal region that are also present in CcmM35, a 35kD isoform that is translated from an internal start site (Figure 2) (Long et al. 2010). CcmM58 has also been shown to interact with the carboxysomal carbonic anhydrase (CcaA) and another internal protein, CcmN, which possesses a short encapsulation peptide that binds to the major shell protein CcmK2 (Long et al. 2007, Kinney, et al. 2012).
One rapid method to investigate interaction of proteins in chloroplasts is to express them transiently by Agroinfiltration (Fischer et al. 1999, Yang et al. 2000). The species Nicotiana benthamiana is particularly amenable to this method, which also works reasonably well for Nicotiana tabaccum, in our experience (Goodin et al. 2002). T-DNA vectors that will express one or more genes are introduced into a binary vector and incorporated into an Agrobacterium tumefaciens strain, which is then infiltrated into a leaf. More than one Agrobacterium strain, containing T-DNA vectors carrying a different set of genes, can be infiltrated simultaneously to express a variety of combinations of genes (Hanson and Sattarzadeh 2014). Following a brief period for expression (2–5 days), protein expression and interactions can be monitored.
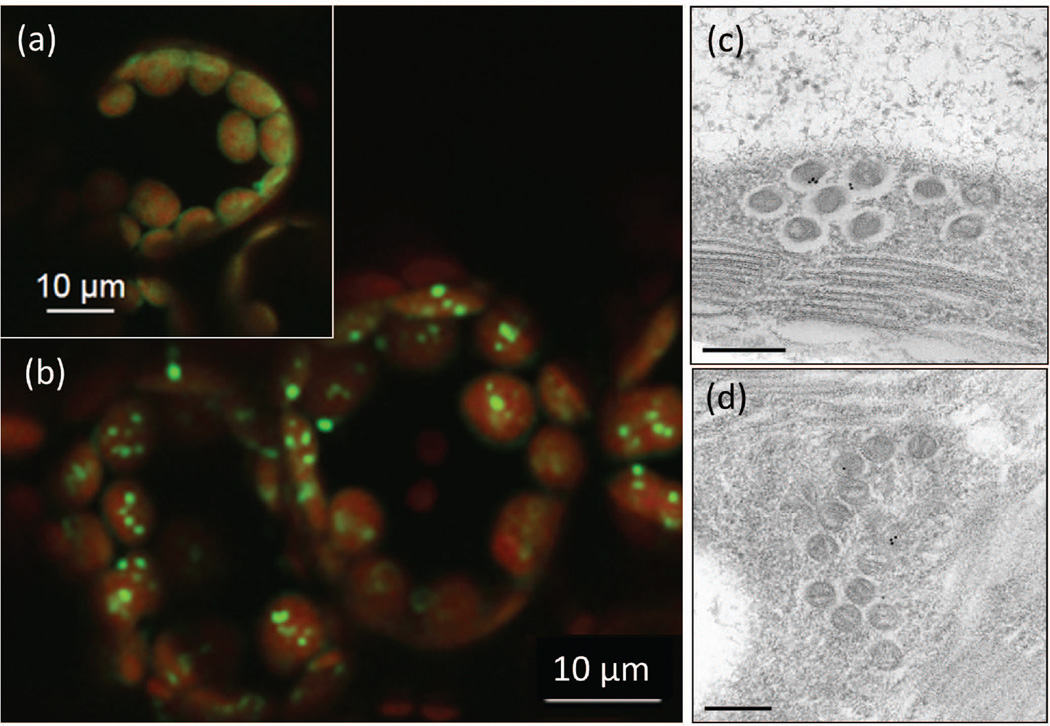
We chose to examine tissue for the localization of Yellow Fluorescent Protein (YFP) fused to CcmO. When this protein is expressed by itself, the protein remains diffuse in the stroma. However, when CcmO-YFP is co-expressed with different combinations of (1) CcmK2 and CcmL (2) CcmK2 and CcmM58, and (3) CcmK2, CcmL and CcmM58, fluorescent punctate loci are visualized in the chloroplast stroma (Figure 3). Characterization at a higher resolution by transmission electron microscopy (TEM) indicates that these punctate loci are highly organized spherical bodies about 80–110 nm in size (Lin et al. 2014a). These structures appear to have a double-layered shell with a less organized inner layer. In comparison, β-carboxysomes in native S. elongatus PCC7942 are icosahedral bodies about 175 nm in size with a single-layered shell.
Figure 3.
Transient expression of cyanobacterial shell proteins in Nicotiana benthamiana. (a) Diffuse localization of CcmO-YFP within chloroplasts when CcmK2 and CcmL are absent. (b) Formation of YFP fluorescent punctate loci (green) within the chloroplasts (red) of N. benthamiana transiently expressing CcmK2, CcmL and CcmO-YFP. (c) and (d) Spherical structures formed in plants transiently expression CcmK2-YFP, CcmL, and CcmO-YFP. Microscopic images from Lin et al. (2014a), with permission.
The smaller size and lack of angular nature of the microcompartments formed by the shell protein expressed in the chloroplasts indicate the importance of internal components in the proper assembly of β-carboxysomes. This is in agreement with recent studies illuminating the central role played by Rubisco-CcmM35 interaction during the assembly process of β-carboxysomes (Cameron et al. 2013). In our previous study, we also demonstrated that the encapsulation peptide from CcmN that is 17 amino-acid long (N17) is able to target YFP to the shell assembly formed by CcmK2 and CcmO-YFP (Lin et al. 2014a). The observation that a foreign protein can be targeted into carboxysomes by the N17 peptide opens up the possibility of engineering microcompartments with new functions in chloroplasts.
STRATEGY CONSIDERATIONS FOR REPLACEMENT OF LAND PLANT RUBISCO WITH CYANOBACTERIAL RUBISCO
The location of the gene (rbcL) encoding the large subunit (LS) of Rubisco in the chloroplast genome and genes for the Rubisco small subunit (SS) in the nuclear genome poses a special challenge for engineering cyanobacterial Rubisco into plants. Transplastomic plants (plants containing transgenes in the plastid) have been reported in which the tobacco rbcL gene has been replaced with a large subunit genes from another species, resulting in a hybrid enzyme. Unfortunately, the resultant transplastomic plants exhibited impaired photosynthesis, some requiring sucrose or high CO2 for growth (reviewed in (Whitney et al. 2011a, Hanson et al. 2013). Reduced function of hybrid enzymes is not unexpected, given that 8 large and 8 small subunits must assemble into a functional complex.
Expression of both the nuclear and chloroplast-encoding subunits must be prevented for complete replacement of a native Rubisco with a heterologous version. Most plants contain multiple nuclear genes (rbcS) encoding the small subunit of Rubisco (Spreitzer 2003), so in order to prevent their expression, either an RNA silencing or mutagenesis strategy must be carried out. Antisense tobacco lines with reduced Rubisco have been previously produced (Rodermel et al. 1988, Quick et al. 1991, Makino and Sage 2007), but any RNA silencing strategy is susceptible to possible epigenetic loss of silencing. Another strategy for removing a tobacco multigene family is to use recently developed CRISPR-Cas9 technology (Belhaj et al. 2013, Jiang et al. 2013), though at this writing the method has not yet been used to mutagenize a large plant gene family.
For those plant species in which plastid transformation is feasible, the rbcL gene can easily be targeted for deletion or replacement by the natural homologous recombination system that exists in chloroplasts (Maliga 2004; Figure 4). Often the knockout of either a nuclear or plastid component of a complex that contains subunits encoded by both genomes results in loss of all proteins in the complex due to protein instability when one partner is missing (Stern et al. 2004). Therefore, loss of the LS in a species is likely to lead to loss of the corresponding SS (Rodermel, et al. 1988). Replacement of the LS with a different species’ LS might rescue the endogenous SS, but only if the hybrid enzyme can assemble and stabilize the endogenous SS.
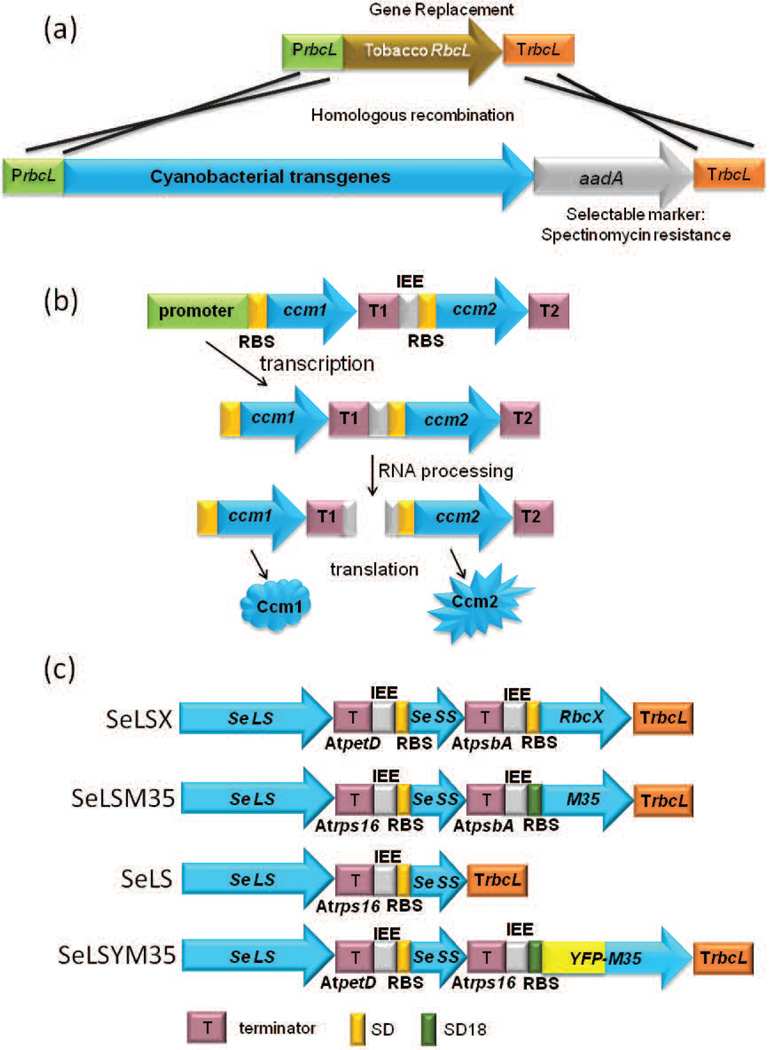
Figure 4.
Schematics of gene arrangements in synthetic operons to express cyanobacterial Rubisco from chloroplasts. (a) Replacement of the tobacco rbcL gene with cyanobacterial transgenes and a selectable marker by homologous recombination. (b) A typical gene arrangement in a synthetic operon with two generic cyanobacterial genes, ccm1 and ccm2. Each gene is followed by a different terminator sequence denoted as T1 or T2. In the intergenic region between ccm1 and ccm2, an intercistronic expression element (IEE) and a ribosome binding site (RBS) are inserted immediately upstream of ccm2 for processing of the dicistronic transcript into monocristronic ones for more efficient translation of the downstream gene, ccm2. (c) Schematics of synthetic operons in four different constructs to express cyanobacterial transgenes from the tobacco rbcL locus (Occhialini, et al. 2016). Se LS, Se SS, RbcX, and M35 represent rbcL, rbcS, rbcX, and ccmM35 genes from S. elongatus PCC7942 respectively. RBS: ribosome binding site. Single (SD) or triple (SD18) Shine-Dalgarno sequences from T7 gene 10 (Drechsel and Bock 2011).
Different strategies can be envisioned for replacement of an endogenous C3 plant Rubisco with a different species’ Rubisco. A knockout of the rbcL gene is readily accomplished in those species in which chloroplast transformation is possible. By simultaneously deleting the endogenous rbcL and replacing it with genes encoding a foreign Rubisco, it should be possible to obtain autotrophic plants, though perhaps ones requiring high CO2 or growth on sucrose unless the new Rubisco is highly functional in air and expressed at adequate levels. For those species in which no chloroplast transformation method is available, either a targeted or untargeted mutagenesis of rbcL will be required, and the mutant plants will require sucrose unless functional foreign rbcL and rbcS genes have been incorporated into the nucleus before undertaking the mutagenesis.
A barrier to the use of nuclear transgenes for introducing foreign Rubisco has been the lower expression level of nuclear-encoded transgenic proteins that is typically achieved in stably transformed plants, less than 1% total soluble protein (TSP) when a single gene including a standard 35S promoter cassette is used (Conley et al. 2011). However, new methods are promising for increasing the levels of protein from nuclear transgenes. For example, a system based on a DNA virus amplification system resulted in accumulation of foreign protein as 10% of total soluble protein (Dugdale et al. 2014). Another possibility is to use the same strategy that appears to have evolved in C3 plants for high-level nuclear expression of the SS of Rubisco: express the protein from multiple nuclear loci. The amount of Rubisco needed to support adequate rates of photosynthesis and plant growth will depend on the catalytic properties of the new Rubisco to be incorporated. Most C3 plants have leaves in which Rubisco represents 25–50% of total soluble protein. If the new Rubisco is 5 times faster, and able to carboxylate at such speed, then only 5–10% Rubisco soluble protein should be needed for growth at the same rate, though more will be needed if the goal is to enhance growth.
A FIRST STEP FOR STABLE INCORPORATION OF CARBOXYSOMES INTO CHLOROPLASTS
We decided to replace the tobacco rbcL gene with both the rbcL and rbcS genes from S. elongatus PCC7942, without altering the tobacco nuclear rbcS genes (Figure 4) (Lin et al. 2014b, Occhialini et al. 2016). For expression of a foreign gene from a chloroplast genome, a transgene construct must carry a 5’ untranslated region (UTR) that provides translation signals and a terminator region for stabilization of the RNA transcript. One of the most effective such 5’ UTRs is derived from the bacteriophage T7 gene 10 (Maliga 2004, Yang et al. 2013). Multiple tandem copies of the Shine-Dalgarno sequences from gene 10 have been shown to improve translation over single copies (Drechsel and Bock 2011). An issue regarding multigene constructs is the possibility of unwanted homologous recombination between components of the same transgene operon, or between transgenes and homologous regions of the chloroplast genome from which a gene regulatory sequence was derived (Gray et al. 2009). Thus, it can be advantageous to use a gene regulatory sequence such as a promoter or terminator from a different plant species, provided that the sequence is effective in the heterologous system. Alternatively, if the endogenous sequence that is used is not large, the probability of undesirable recombination is reduced (Mudd et al. 2014).
Two or more chloroplast genes can be transcribed in a polycistronic transcript from a single promoter. However, in chloroplasts, downstream coding regions in transcripts are sometimes not efficiently translated from polycistronic transcripts, unlike in bacteria. To enhance the possibility of effective translation, an Intercistronic Expression Element (IEE), which carries a sequence recognized by a nuclease, can be placed between different transgenes on a polycistronic transcript (Zhou et al. 2007). Presence of the IEE has been reported to result in processing of polycistronic transcripts into monocistronic transcripts, enhancing protein accumulation (Bock 2013, Bock 2015; Figure 4b).
Another enhancement of expression of foreign proteins in chloroplasts can sometimes result from changing the codon usage of the gene of interest to avoid codons that are rarely used in chloroplasts (Reed et al. 2001, Franklin et al. 2002). Nevertheless, the optimal codon usage in a transgene is not always straightforward to identify (Weiss et al. 2012, Sugiura 2014) and may need empirical optimization. A further aspect of the codon region that can be important is the N-terminal region, either because of the presence of a downstream box sequence that affects protein accumulation (Gray et al. 2011) or N-terminal features that affect protein stability (Apel et al. 2010, Elghabi et al. 2011, Gray, et al. 2011).
Chloroplast transformants (transplastomic plants) are usually made by particle bombardment with a biolistic device (Maliga and Tungsuchat-Huang 2014). Tungsten or gold microparticles can be coated with plasmid DNA and then propelled into plant cells, now usually accelerated by a blast of helium. If regenerated transgenic plants are desired, the tissue that is bombarded must be capable of regeneration from tissue cultures. While transplastomic plants can be made from a variety of crop plants in addition to tobacco (Hanson, et al. 2013, Bock 2015), Nicotiana species remain the superior model system for chloroplast transformation, due to high regenerability and rapid growth in culture. The spectinomycin/streptomycin resistance gene aadA, used in the initial report of stable transplastomic plants (Svab et al. 1990), continues to be the most effective selectable marker for chloroplast transformation. Following bombardment of tobacco leaf tissue with plasmids carrying the transgene operons and the selectable marker and incubation without selection for a few days, tissue is removed to regeneration media containing the antibiotic and induced to regenerate shoots. Usually such shoots contain a mixture of transformed and untransformed chloroplasts, a state termed “heteroplasmy”. Each cell that was bombarded contained multiple chloroplasts and each chloroplast within that cell contained multiple chloroplast genomes. In order to obtain shoots with a uniform chloroplast genome (a state termed “homoplasmy”), repeated rounds of regeneration are often needed. During dedifferentiation of leaf cells in culture and redifferentiation into shoot meristems, the number of plastids/cell is reduced, facilitating random segregation of transgenic and non-transgenic plastids. Selection ensures that only shoots resistant to the selectable marker will regenerate (Lutz et al. 2007, Maliga and Tungsuchat-Huang 2014). In our experience, homoplasmic plants can usually be obtained after a second round of regeneration and selection.
Transplastomic tobacco plants fixing carbon with cyanobacterial Rubisco
In our initial constructs designed to replace endogenous Rubisco with S. elongatus PCC7942 Rubisco, we included either genes for the assembly factor RbcX, required for proper Rubisco folding in some cyanobacteria (Saschenbrecker et al. 2007), or for CcmM35, a protein involved in organization of Rubisco within the carboxysome (Cameron, et al. 2013) (Figure 2). We thought that one of these proteins might facilitate proper folding of the cyanobacterial Rubisco, which normally requires a GroEL chaperonin and its cofactor GroEs. Land plants contain Cpn60, a homolog of GroEL/GroEs comprised of Cpn10 and Cpn20 subunits. Both cyanobacteria and land plants require an additional Rubisco assembly factor known as Raf1 (Feiz et al. 2012, Hauser et al. 2015a). While some cyanobacterial Rubisco enzymes require RbcX for assembly, S. elongatus PCC7942 does not in its native cell (Emlyn-Jones et al. 2006); however, we considered that it might be required in a heterologous system. Recently, the inclusion of Arabidopsis Raf1 in a transplastomic plant in which the tobacco rbcL was replaced with Arabidopsis rbcL was shown to improve hybrid Rubisco assembly and photosynthesis (Whitney et al. 2015).
Our two initial lines, named SeLSX (for S. elongatus Rubisco Large and Small subunit with RbcX) and SeLSM35 (CcmM35 instead of RbcX) both were able to grow autotrophically provided that that they were given elevated CO2 in their environment. Though their growth rate was much slower than wild-type, the plants had normal morphology and could flower and set seed. Using DNA blots, we demonstrated that the tobacco rbcL coding region had been completely replaced by the cyanobacterial transgene operons. Immunoblots of total cellular protein revealed no detectable tobacco SS or LS; evidently the absence of the tobacco LS destabilized the tobacco SS even though the rbcS genes were intact (Lin et al. 2014b).
Both SeLSX and SeLSM35 were designed with rbcL and rbcS coding regions altered to be more similar to endogenous chloroplast transcripts. These same coding regions were used in two subsequent transgene operons, one in which neither RbcX nor CcmM35 was expressed (plant line designated SeLS) and a second line in which a yfp gene was fused N-terminally to ccmM35 and expressed along with the cyanobacterial rbcL and rbcS genes (plant line designated SeLSYM35). Elimination of rbcX resulted in attachment of the tobacco rbcL terminator to the rbcS gene. Each of the 4 transgene operons differed in terminator configuration; furthermore, the yfp::ccmM35 gene was synthesized with an altered codon usage, unlike the ccmM35 gene present in SeLSM35 (Figure 4).
The two new plant lines grew much faster than the original lines in 3% CO2; in fact the growth of the SeLS line was only slightly slower than wild-type tobacco (Figure 5). In order to determine whether transgene expression levels could explain the different growth rates, we examined RNA and protein-level expression. By performing RNA blots, we discovered that transcripts carrying the Arabidopsis rps16 terminators did not accumulate, thus indicating this terminator is not suitable for tobacco transformation vectors. A second important finding was that the presence of the IEE sequence did not always result in efficient cleavage between the open reading frames, resulting in the continued presence of polycistronic transcripts (Occhialini et al. 2016). Despite the absence of many monocistronic rbcS transcripts, evidently there was sufficient expression of the downstream rbcS genes for adequate protein synthesis for assembly of Rubisco. When we compared Rubisco content in the four lines in 3% CO2, we observed that the upper leaves of SeLS contained significantly more Rubisco than SeLSX, perhaps explaining the enhanced growth (Figure 5a–b). One possibility we cannot rule out is that the original transgene constructs had an unknown inhibitory effect on other aspects of chloroplast gene expression, completely unrelated to Rubisco expression. Nevertheless, the rapid growth of SeLS in 3% CO2 demonstrates that concept that nitrogen use efficiency may improve when less enzyme is needed for carbon fixation (Carmo-Silva et al. 2015).
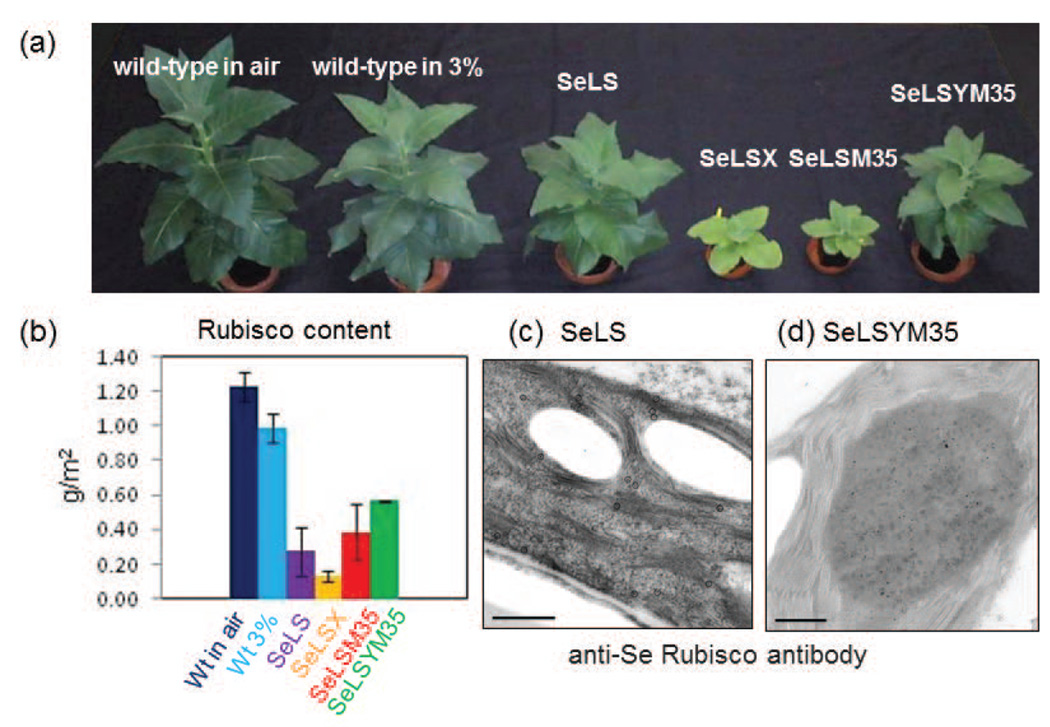
Figure 5.
Characterization of transgenic tobacco plants engineered with cyanobacterial Rubisco. (a) Relative size of wild-type (wt) and SeLS, SeLSX, SeLSM35 and SeLSYM35 tobacco plants when grown for 42 days in 3% CO2. (b) Rubisco content per leaf area in wt tobacco growing in air vs. wt and transgenic lines growing in 3% CO2. (c–d) Localization of cyanobacterial Rubisco in the chloroplast stroma of the SeLS (c) and SeLSYM35 (d) tobacco transplastomic lines probed with anti-Se Rubisco antibody and a secondary antibody conjugated with 10 nm gold particles (black circles or dots). Scale bars = 500 nm. Modified from Occhialini et al. 2016, with permission.
Despite the fact that plants SeLSM35 and SeLSYM35 had greater Rubisco content than SeLS or SeLSX, their growth was slower than SeLS in 3% CO2 (Figure 5) (Occhialini et al. 2016). The unusual organization of Rubisco in the M35-containing plants may explain the slower growth compared to SeLS. Chloroplasts from the SeLSX, SeLS, SeLSM35, and SeLSM35Y plants were imaged by electron microcopy and the location of Rubisco was determined with specific antibodies. These experiments revealed that the cyanobacterial Rubisco was dispersed within the stroma in SeLSX and SeLS, but aggregated in large bodies in the SeLSM35 and SeLSYM35 plants, suggesting that M35 in the chloroplast binds to the cyanobacterial LS and results in aggregation of Rubisco (Figure 5c–d). The Rubisco/M35 aggregates in tobacco chloroplasts appear analogous to aggregates observed when an M35::YFP fusion was expressed in S. elongatus PCC7942 mutant carrying a ccmK2-ccmO deletion (Cameron, et al. 2013).
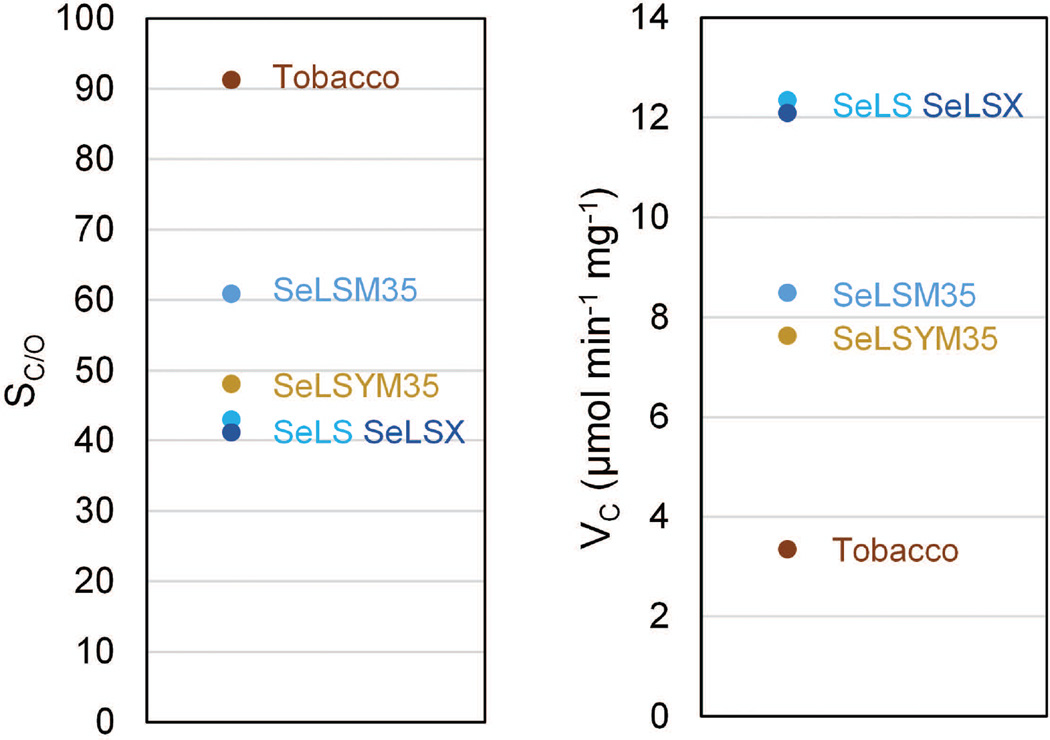
Aggregation of Rubisco in the absence of a functional carboxysome could potentially prevent supply of CO2 to the enzyme, thereby reducing plant growth rate. Purified Rubisco isolated from the transgenic lines expressing the cyanobacterial Rubisco (SeLS and SeLSX) had essentially identical kinetic constants (Figure 6) to wild type Rubisco isolated from the cyanobacteria (Figure 1b). However, where the enzyme was expressed in association with CcmM35, the Rubisco was slower and had a slightly greater affinity for CO2. This suggests that the incorporation of CcmM35 into the Rubisco complex disrupts the enzyme structure possibly by displacing the Rubisco small subunits and changes the catalytic properties; 3D structural determination of the complexes will be required to clarify the mechanism for these changes.
Figure 6.
Rubisco specificity (SC/O) and maximum carboxylation rate (VC) in wild-type and SeLS, SeLSX, SeLSM35 and SeLSYM35 transplastomic tobacco plants. Data from Occhialini et al. (2016).
While SeLS grows substantially faster than SeLSX 3% CO2, we have observed that growth rates of the two lines are quite similar in 0.9% CO2. This observation is in line with the observation that CO2 assimilation in SeLS and SeLSX is quite similar when measured on a leaf area basis. Also, Rubisco active sites/m2 is not significantly different in the two lines when grown in 0.9% CO2 (Occhialini et al. 2016). Currently we do not understand why there is relatively more Rubisco in SeLS in 3% CO2 than in 0.9% CO2.
ADDITIONAL ENGINEERING NEEDED TO INSTALL THE CYANOBACTERIAL CCM IN CHLOROPLASTS
As the next step, shell proteins such as CcmK2, CcmO and CcmL and other internal proteins of the β-carboxysome including CcmM58, CcaA and CcmN will need to be co-expressed with the core components, made up of Rubisco and CcmM35. These additional proteins may be expressed from similar synthetic operons from chloroplast genome as described above. Alternatively, proteins that are known to be less abundant such as CcmL, CcaA, and either CcmK3 or CcmK4 may be expressed through nuclear transformation and imported into chloroplast stroma with stromal transit peptides. A recent study described the surprising finding that a chimeric protein derived from fusing the three SSU domains from CcmM58, CcaA and the encapsulation peptide from CcmN is able to functionally replace four internal proteins, CcmM58, CcmM35, CcaA and CcmN, thus greatly simplifying the engineering of β-carboxysomes into chloroplasts (Gonzalez-Esquer et al. 2015).
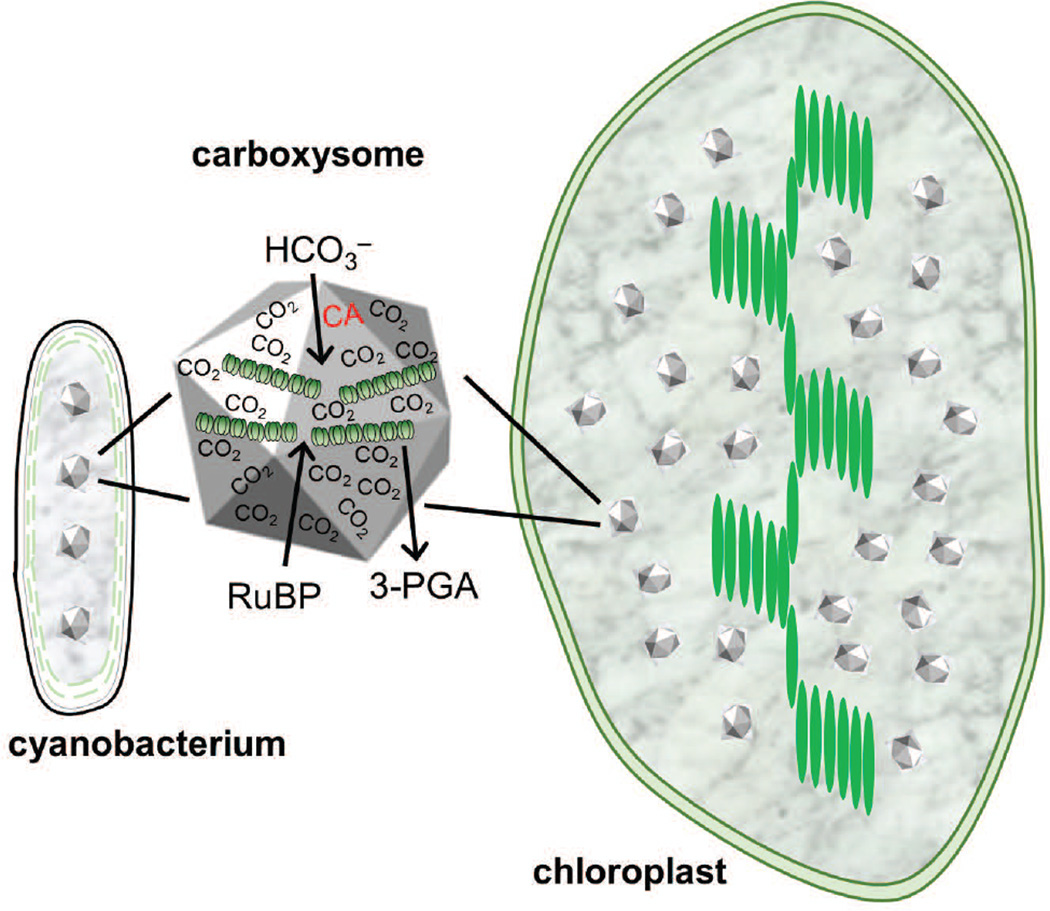
It may be necessary to use synthetic operons with different gene arrangements and regulatory elements such as the ribosome binding sites to attain the optimal expression ratios for successful assembly of these proteins into functional β-carboxysomes in chloroplasts. In addition, the number of carboxysomes per chloroplast will also need to be optimized so that the metabolic flux through these carboxysomes is efficiently integrated into the Calvin Cycle. In considering only the relative volume of a cyanobacterium vs. a chloroplast, to maintain the same number of microcompartments per unit volume, a typical tobacco chloroplast would need about 70 carboxysomes (Figure 7). However, a recent theoretical analysis indicates that approximately 2700 carboxysomes per chloroplast would be needed to achieve the same amount of Rubisco per chloroplast as in wild-type plants (McGrath and Long 2014). On the other hand, we expect the optimal number of carboxysomes per chloroplast would be much lower than 2700 due to the significantly higher catalytic rate of the cyanobacterial Rubisco and altered metabolic flux through photorespiration upon installation of the cyanobacterial CCM in the chloroplast.
Figure 7.
Diagram of carboxysomes in cyanobacteria and hypothetically in a chloroplast. Diagram visualizes approximate relative size of S. elongatus cell and typical tobacco chloroplast. A typical S. elongatus cell contains an average of 4 carboxysomes (Savage et al. 2010). However, modeling indicates more will be needed for adequate Rubisco to be present (McGrath and Long 2014).
Incorporating a functional microcompartment containing Rubisco and other internal proteins is not sufficient for creating a complete CCM in plants
Chloroplasts naturally contain carbonic anhydrase within the stroma (the soluble portion of the chloroplast). Expression of stromal carbonic anhydrase must be prevented through RNA silencing or mutagenesis in order for the carboxysome to function properly. In previous studies with transgenic plants in which stromal carbonic anhydrase was knocked down, little effect on photosynthetic efficiency in mature leaves was detected (Majeau et al. 1994, Ferreira et al. 2008). In cyanobacteria, when carbonic anhydrase was expressed in the cytoplasm (the bacterial equivalent of the chloroplast stroma), the cells required high CO2 because the carbon-concentrating mechanism did not function (Price and Badger 1989).
In addition to the carboxysome, transporters will be needed to supply the chloroplast with bicarbonate, which can then be converted to CO2 by carbonic anhydrase within the carboxysome. Heterologous expression of such transporters has already begun (Pengelly et al. 2014, Atkinson et al. 2015). Transporters alone, without a microcompartment, are predicted to be able to enhance photosynthesis (McGrath and Long 2014), but additional transporter types and further engineering will be needed to realize potential gains.
Acknowledgments
Research on engineering carboxysomes into chloroplasts was supported by Biotechnology and Biological Sciences Research Council under grant number BB/I024488/1 to M.A.J.P., the National Science Foundation under grant number EF-1105584 to M.R.H. and the National Institute of General Medical Sciences of the National Institutes of Health under award number F32GM103019 to M.T.L. M.R.H. and M.T.L. also acknowledge support from the Cornell University Biotechnology Resource Center (NIH S10RR025502) for the shared Zeiss LSM 710 Confocal Microscope. E.C.S. and M.A.J.P. also acknowledge support from the 20:20 Wheat Institute Strategic Program (BBSRC BB/J/00426X/1). We thank Kevin Hines for the carboxysome model and Alessandro Occhialini for transmission electron microscopy and immunolocalization.
Contributor Information
Myat T. Lin, Email: mtl84@cornell.edu.
A. Elizabete Carmo-Silva, Email: e.carmosilva@lancaster.ac.uk.
Martin A.J. Parry, Email: m.parry@lancaster.ac.uk.
REFERENCES
- Apel W, Schulze WX, Bock R. Identification of protein stability determinants in chloroplasts. Plant J. 2010;63:636–650. doi: 10.1111/j.1365-313X.2010.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotech. J. 2015 doi: 10.1111/pbi.12497. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussignargues C, Paasch BC, Gonzalez-Esquer R, Erbilgin O, Kerfeld CA. Bacterial microcompartment assembly: The key role of encapsulation peptides. Commun. Integr. Biol. 2015;8:e1039755. doi: 10.1080/19420889.2015.1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Hanson D, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 2002;29:161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Meth. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Strategies for metabolic pathway engineering with multiple transgenes. Plant Mol. Biol. 2013;83:21–31. doi: 10.1007/s11103-013-0045-0. [DOI] [PubMed] [Google Scholar]
- Bock R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015;66:211–241. doi: 10.1146/annurev-arplant-050213-040212. [DOI] [PubMed] [Google Scholar]
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. USA. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RH, Byrd GT. Estimation of bundle sheath cell conductance in C4 species and O2 insensitivity of photosynthesis. Plant Physiol. 1993;103:1183–1188. doi: 10.1104/pp.103.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth. Biol. 2015;4:444–453. doi: 10.1021/sb500226j. [DOI] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Keys AJ, Andralojc PJ, Powers SJ, Arrabaca MC, Parry MA. Rubisco activities, properties, and regulation in three different C4 grasses under drought. J. Exp. Bot. 2010;61:2355–2366. doi: 10.1093/jxb/erq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva E, Scales JC, Madgwick PJ, Parry MA. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Env. 2015;38:1817–1832. doi: 10.1111/pce.12425. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Samaritani E, Duvall MR, Hodkinson TR, Savolainen V, Salamin N. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 2008a;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Muasya AM, Roalson EH, Russier F, Besnard G. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Mol. Biol. Evol. 2008b;25:2361–2368. doi: 10.1093/molbev/msn178. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Zhu H, Le LC, Jevnikar AM, Lee BH, Brandle JE, Menassa R. Recombinant protein production in a variety of Nicotiana hosts: a comparative analysis. Plant Biotech. J. 2011;9:434–444. doi: 10.1111/j.1467-7652.2010.00563.x. [DOI] [PubMed] [Google Scholar]
- Delgado E, Medrano H, Keys AJ, Parry MAJ. Species variation in Rubisco specificity factor. J. Exp. Bot. 1995;46:1775–1777. [Google Scholar]
- Dengler NG, Nelson T. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, editors. C4 Plant Biology. New York: Academic Press; 1999. pp. 133–172. [Google Scholar]
- DePaoli HC, Borland AM, Tuskan GA, Cushman JC, Yang XH. Synthetic biology as it relates to CAM photosynthesis: challenges and opportunities. J. Exp. Bot. 2014;65:3381–3393. doi: 10.1093/jxb/eru038. [DOI] [PubMed] [Google Scholar]
- Drechsel O, Bock R. Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res. 2011;39:1427–1438. doi: 10.1093/nar/gkq978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugdale B, Mortimer CL, Kato M, James TA, Harding RM, Dale JL. Design and construction of an in-plant activation cassette for transgene expression and recombinant protein production in plants. Nat. Protocols. 2014;9:1010–1027. doi: 10.1038/nprot.2014.068. [DOI] [PubMed] [Google Scholar]
- Edwards GE, Ku MSB, Monson RK. C4 photosynthesis and its regulation. In: Barber J, Baker NR, editors. Photosynthetic Mechanisms and the Environment. Amsterdam: Elsevier Science Publishers B.V. (Biomedical Division); 1985. pp. 287–327. [Google Scholar]
- Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R. Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotech. J. 2011;9:599–608. doi: 10.1111/j.1467-7652.2011.00598.x. [DOI] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol. 2006;47:1630–1640. doi: 10.1093/pcp/pcl028. [DOI] [PubMed] [Google Scholar]
- Evans JR, Von Caemmerer S. Carbon dioxide diffusion inside leaves. Plant Physiol. 1996;110:339–346. doi: 10.1104/pp.110.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CG, Cheng SQ, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. USA. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Williams-Carrier R, Wostrikoff K, Belcher S, Barkan A, Stern DB. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell. 2012;24:3435–3446. doi: 10.1105/tpc.112.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira FJ, Guo C, Coleman JR. Reduction of plastid-localized carbonic anhydrase activity results in reduced Arabidopsis seedling survivorship. Plant Physiol. 2008;147:585–594. doi: 10.1104/pp.108.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Vaquero-Martin C, Sack M, Drossard J, Emans N, Commandeur U. Towards molecular farming in the future: transient protein expression in plants. Biotech. Appl. Biochem. 1999;30(Pt 2):113–116. [PubMed] [Google Scholar]
- Foyer CH, Nurmi A, Dulieu H, Parry MAJ. Analysis of 2 Rubisco-deficient tobacco mutants, H7 and Sp25 - Evidence for the production of Rubisco large subunits in the Sp25 mutant that form clusters and are inactive. J. Exp. Bot. 1993;44:1445–1452. [Google Scholar]
- Franklin S, Ngo B, Efuet E, Mayfield SP. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 2002;30:733–744. doi: 10.1046/j.1365-313x.2002.01319.x. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Jenkins CLD, Hatch MD. CO2 concentrating mechanism of C4 photosynthesis - Permeability of isolated bundle sheath-cells to inorganic carbon. Plant Physiol. 1989;91:1364–1371. doi: 10.1104/pp.91.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmes J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, Madgwick PJ, Haslam RP, Medrano H, Parry MAJ. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ. 2005;28:571–579. [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. J. Biol. Chem. 2010;285:19833–19841. doi: 10.1074/jbc.M110.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Esquer CR, Shubitowski TB, Kerfeld CA. Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell. 2015;27:2637–2644. doi: 10.1105/tpc.15.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Gray BN, Ahner BA, Hanson MR. Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res. 2009;18:559–572. doi: 10.1007/s11248-009-9246-3. [DOI] [PubMed] [Google Scholar]
- Gray BN, Yang H, Ahner BA, Hanson MR. An efficient downstream box fusion allows high-level accumulation of active bacterial β-glucosidase in tobacco chloroplasts. Plant Mol. Biol. 2011;76:345–355. doi: 10.1007/s11103-011-9743-7. [DOI] [PubMed] [Google Scholar]
- Hanson MR, Gray BN, Ahner BA. Chloroplast transformation for engineering of photosynthesis. J. Exp. Bot. 2013;64:731–742. doi: 10.1093/jxb/ers325. [DOI] [PubMed] [Google Scholar]
- Hanson MR, Sattarzadeh A. Fluorescent labeling and confocal microscopic imaging of chloroplasts and non-green plastids. Methods Mol Biol. 2014;1132:125–143. doi: 10.1007/978-1-62703-995-6_7. [DOI] [PubMed] [Google Scholar]
- Hauser T, Bhat JY, Milicic G, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. Structure and mechanism of the Rubisco-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 2015a;22:720–728. doi: 10.1038/nsmb.3062. [DOI] [PubMed] [Google Scholar]
- Hauser T, Popilka L, Hartl FU, Hayer-Hartl M. Role of auxiliary proteins in Rubisco biogenesis and function. Nature Plants. 2015b;1:1–11. doi: 10.1038/nplants.2015.65. [DOI] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC, Shively JM. Carboxysomes and Their Structural Organization in Prokaryotes. In: Barton LL, Bazylinski DA, Xu H, editors. Nanomicrobiology. New York: Springer; 2014. pp. 75–101. [Google Scholar]
- Jenkins CLD. The CO2 concentrating mechanism of C-4 photosynthesis: Bundle sheath cell CO2 concentration and leakage. Aust. J. Plant Physiol. 1997;24:543–547. [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. Species variation in the specificity of ribulose-biphosphate carboxylase-oxygenase. Nature. 1981;291:513–515. [Google Scholar]
- Jordan DB, Ogren WL. Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 1983;227:425–433. doi: 10.1016/0003-9861(83)90472-1. [DOI] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. The CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase oxygenase : Dependence on ribulosebisphosphate concentration, pH and temperature. Planta. 1984;161:308–313. doi: 10.1007/BF00398720. [DOI] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, editors. C4 Plant Biology. New York: Academic Press; 1999. pp. 49–87. [Google Scholar]
- Kane HJ, Viil J, Entsch B, Paul K, Morell MK, Andrews TJ. An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust. J. Plant Physiol. 1994;21:449–461. [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kiirats O, Lea PJ, Franceschi VR, Edwards GE. Bundle sheath diffusive resistance to CO2 and effectiveness of C4 photosynthesis and refixation of photorespired CO2 in a C4 cycle mutant and wild-type Amaranthus edulis. Plant Physiol. 2002;130:964–976. doi: 10.1104/pp.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 2012;287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. The biochemistry of Rubisco in Flaveria. J. Exp. Bot. 2008;59:1767–1777. doi: 10.1093/jxb/erm283. [DOI] [PubMed] [Google Scholar]
- Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J. Mol. Biol. 2014;426:2217–2228. doi: 10.1016/j.jmb.2014.02.025. [DOI] [PubMed] [Google Scholar]
- Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 2014;3:454–465. doi: 10.1021/sb4001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR. b-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014a;79:1–12. doi: 10.1111/tpj.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014b;513:547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD. Functional cyanobacterial b-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 2010;153:285–293. doi: 10.1104/pp.110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell. 2015;161:56–66. doi: 10.1016/j.cell.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR. Can improvement in photosynthesis increase crop yields? Plant Cell Enev. 2006;29:315–330. doi: 10.1111/j.1365-3040.2005.01493.x. [DOI] [PubMed] [Google Scholar]
- Lutz KA, Azhagiri AK, Tungsuchat-Huang T, Maliga P. A guide to choosing vectors for transformation of the plastid genome of higher plants. Plant Physiol. 2007;145:1201–1210. doi: 10.1104/pp.107.106963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau N, Arnoldo MA, Coleman JR. Modification of carbonic anhydrase activity by antisense and over-expression constructs in transgenic tobacco. Plant Mol. Biol. 1994;25:377–385. doi: 10.1007/BF00043867. [DOI] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K. Enzymic properties of ribulose-1,5-bisphosphate carboxylase/oxygenase purified from rice leaves. Plant Physiol. 1985;79:57–61. doi: 10.1104/pp.79.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Sage RF. Temperature response of photosynthesis in transgenic rice transformed with 'sense' or 'antisense' rbcS. Plant Cell Physiol. 2007;48:1472–1483. doi: 10.1093/pcp/pcm118. [DOI] [PubMed] [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Ann. Rev. Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- Maliga P, Tungsuchat-Huang T. Plastid transformation in Nicotiana tabacum and Nicotiana sylvestris by biolistic DNA delivery to leaves. Methods Mol. Biol. 2014;1132:147–163. doi: 10.1007/978-1-62703-995-6_8. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 2014;164:2247–2261. doi: 10.1104/pp.113.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd EA, Madesis P, Avila EM, Day A. Excision of plastid marker genes using directly repeated DNA sequences. Methods Mol Biol. 2014;1132:107–123. doi: 10.1007/978-1-62703-995-6_6. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Morell M, Whitney SM. Directed evolution of rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry. 2007;46:14067–14074. doi: 10.1021/bi700820a. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Whitney SM. Directing the evolution of Rubisco and Rubisco activase: first impressions of a new tool for photosynthesis research. Photosynth. Res. 2008;98:667–675. doi: 10.1007/s11120-008-9324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A, Lin MT, Andralojc PJ, Hanson MR, Parry MA. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J. 2016;85:148–160. doi: 10.1111/tpj.13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, Moore TA, Moroney J, Niyogi KK, Parry MA, Peralta-Yahya PP, Prince RC, Redding KE, Spalding MH, van Wijk KJ, Vermaas WF, von Caemmerer S, Weber AP, Yeates TO, Yuan JS, Zhu XG. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. USA. 2015;112:8529–8536. doi: 10.1073/pnas.1424031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013;64:717–730. doi: 10.1093/jxb/ers336. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Gutteridge S. Variation in the specificity factor of C3 higher plant Rubiscos determined by the total consumption of ribulose-P2. J. Exp. Bot. 1989;40:317–320. [Google Scholar]
- Parry MAJ, Madgwick PJ, Carvalho JFC, Andralojc PJ. Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J. Agr. Sci. 2007;145:31–43. [Google Scholar]
- Parry MAJ, Schmidt CNG, Cornelius MJ, Millard BN, Burton S, Gutteridge S, Dyer TA, Keys AJ. Variations in properties of ribulose-1,5-bisphosphate carboxylase from various species related to differences in amino-acid-sequences. J. Exp. Bot. 1987;38:1260–1271. [Google Scholar]
- Pearce FG. Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem. J. 2006;399:525–534. doi: 10.1042/BJ20060430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly JJ, Forster B, von Caemmerer S, Badger MR, Price GD, Whitney SM. Transplastomic integration of a cyanobacterial bicarbonate transporter into tobacco chloroplasts. J. Exp. Bot. 2014;65:3071–3080. doi: 10.1093/jxb/eru156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO(2)-requiring phenotype : evidence for a central role for carboxysomes in the CO(2) concentrating mechanism. Plant Physiol. 1989;91:505–513. doi: 10.1104/pp.91.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 2013;64:753–768. doi: 10.1093/jxb/ers257. [DOI] [PubMed] [Google Scholar]
- Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo-Silva E, Parry MAJ. Rubisco catalytic properties of wheat relatives provide scope for improving photosynthesis. J. Exp. Bot. 2016 doi: 10.1093/jxb/erv574. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick WP, Schurr U, Scheibe R, Schulze ED, Rodermel SR, Bogorad L, Stitt M. Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with "antisense" rbcS : I. Impact on photosynthesis in ambient growth conditions. Planta. 1991;183:542–554. doi: 10.1007/BF00194276. [DOI] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD. Structural determinants of the outer shell of b-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One. 2012;7:e43871. doi: 10.1371/journal.pone.0043871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Whitehead LF, Forster B, Badger MR, Price GD. Cyanobacterial carboxysomes: microcompartments that facilitate CO2 fixation. J. Mol. Microbiol. Biotechnol. 2013;23:300–307. doi: 10.1159/000351342. [DOI] [PubMed] [Google Scholar]
- Reed ML, Wilson SK, Sutton CA, Hanson MR. High-level expression of a synthetic red-shifted GFP coding region incorporated into transgenic chloroplasts. The Plant J. 2001;27:257–265. doi: 10.1046/j.1365-313x.2001.01088.x. [DOI] [PubMed] [Google Scholar]
- Rodermel SR, Abbott MS, Bogorad L. Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell. 1988;55:673–681. doi: 10.1016/0092-8674(88)90226-7. [DOI] [PubMed] [Google Scholar]
- Sage RF, Seemann JR. Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase activity in response to reduced light intensity in C4 plants. Plant Physiol. 1993;102:21–28. doi: 10.1104/pp.102.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Stata M. Photosynthetic diversity meets biodiversity: the C4 plant example. J. Plant Physiol. 2015;172:104–119. doi: 10.1016/j.jplph.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Saschenbrecker S, Bracher A, Rao KV, Rao BV, Hartl FU, Hayer-Hartl M. Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell. 2007;129:1189–1200. doi: 10.1016/j.cell.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM. The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large Subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol. 2008;146:83–96. doi: 10.1104/pp.107.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 2003;414:141–149. doi: 10.1016/s0003-9861(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 2004;9:293–301. doi: 10.1016/j.tplants.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Sugiura M. Plastid mRNA translation. Methods Mol Biol. 2014;1132:73–91. doi: 10.1007/978-1-62703-995-6_4. [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tcherkez GGB, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K, Suzuki Y, Shikanai T, Wadano A, Jensen RG, Chmara W, Yokota A. A rapid and sensitive method for determination of relative specificity of RuBisCO from various species by anion-exchange chromatography. Plant Cell Physiol. 1996;37:325–331. [Google Scholar]
- Weiss C, Bertalan I, Johanningmeier U. Effects of rare codon clusters on the expression of a high-turnover chloroplast protein in Chlamydomonas reinhardtii. J. Biotech. 2012;160:105–111. doi: 10.1016/j.jbiotec.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Whitehead L, Long BM, Price GD, Badger MR. Comparing the in vivo function of alpha-carboxysomes and beta-carboxysomes in two model cyanobacteria. Plant Physiol. 2014;165:398–411. doi: 10.1104/pp.114.237941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 2001;26:535–547. doi: 10.1046/j.1365-313x.2001.01056.x. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Birch R, Kelso C, Beck JL, Kapralov MV. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl. Acad. Sci. USA. 2015;112:3564–3569. doi: 10.1073/pnas.1420536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011a;155:27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmes J. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proc. Natl. Acad. Sci. USA. 2011b;108:14688–14693. doi: 10.1073/pnas.1109503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ. Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiol. 1999;121:579–588. doi: 10.1104/pp.121.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger FJ, Badger MR, Price GD. Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol. 2005;139:1959–1969. doi: 10.1104/pp.105.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gray BN, Ahner BA, Hanson MR. Bacteriophage 5' untranslated regions for control of plastid transgene expression. Planta. 2013;237:517–527. doi: 10.1007/s00425-012-1770-3. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 2013;64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]