Abstract
Development of drug resistance is a major factor limiting the continued success of cancer chemotherapy. To overcome drug resistance, understanding the underlying mechanism(s) is essential. We found that HOXC10 is over-expressed in primary carcinomas of the breast, and even more significantly in distant metastasis arising after failed chemotherapy. High HOXC10 expression correlates with shorter recurrence-free and overall survival in patients with estrogen receptor (ER)-negative breast cancer undergoing chemotherapy. We found that HOXC10 promotes survival in cells treated with doxorubicin, paclitaxel or carboplatin by suppressing apoptosis and upregulating NF-κB. Overexpressed HOXC10 increases S-phase specific DNA damage repair by homologous recombination (HR) and checkpoint recovery in cells at three important phases. For double strand break repair, HOXC10 recruits HR proteins at sites of DNA damage. It enhances resection and lastly, it resolves stalled replication forks, leading to initiation of DNA replication following DNA damage. We show that HOXC10 facilitates, but is not directly involved in DNA damage repair mediated by HR. HOXC10 achieves integration of these functions by binding to, and activating cyclin dependent kinase, CDK7, which regulates transcription by phosphorylating the carboxy-terminal domain of RNA polymerase II. Consistent with these findings, inhibitors of CDK7 reverse HOXC10-mediated drug resistance in cultured cells. Blocking HOXC10 function, therefore, presents a promising new strategy to overcome chemotherapy resistance in breast cancer.
Keywords: HOXC10, DNA damage, breast, chemotherapy, CDK7
Introduction
Many chemotherapeutic agents induce DNA damage, triggering cells to activate their DNA damage repair machinery. Nucleotide excision repair (NER) is a major DNA repair pathway that mediates repair of DNA adducts that cause inter- or intra-strand cross links (ICLs) and the formation of pyrimidine dimers. Alkylating and platinum based agents often induce bulky DNA damage that is repaired via NER.
Large scale interaction screens as well as reciprocal affinity purifications have been instrumental in identifying several HOX proteins that interact with proteins critical for DNA damage repair processes [reviewed in (1)]. HOXB7 associates with Ku70, Ku80 and DNA-Pkcs (2), and with PARP1 (3), whose polyADP-ribosylation activity enhances the kinase activity of DNA-PK at the initiation of non-homologous end joining (NHEJ). Recent evidence supports HOX protein participation in the cell replication machinery by associations at DNA replication origins (4). HOXC10 is one such example (5, 6); HOXC10 also has important functions in tissue regeneration (7). Recently, loss of HOXC10 expression was implicated in the development of resistance to estrogen response modulators in estrogen receptor (ER)-positive breast cancer (8). At the present time, the function of HOXC10 in breast cancer remains poorly understood.
Here, we report that HOXC10 expression is frequently higher in ER-negative breast carcinomas that are also chemotherapy-resistant. We determined that HOXC10 contributes to chemoresistance through suppression of apoptosis and enhanced DNA damage repair, mediated through direct interaction with, and activation of CDK7. We also show that CDK7 inhibition can reverse chemotherapy resistance.
Materials and Methods
Cell Lines, Constructs, and Reagents
All cell lines were purchased from ATCC and passages used were within 6 months of purchase. Stable MCF10A-Ras cells were established by transfection of LXSN-K-Ras vector into MCF10A cells; MCF7 wt and resistant sublines were from Dr. Parissenti (9). DR-95 is a human fibroblast cell line stably expressing a pDR-GFP plasmid containing a mutated GFP gene with an 18 bp I-SceI endonuclease cleavage site and in-frame termination codon (10). Retrovirus and lentivirus were produced in HEK 293T cells. Human HOXC10 cDNA (Thermo Scientific) was cloned into the EcoRI and ClaI sites of pLPCX for retroviral production for creating HOXC10 expressing cell lines. Mutated HOXC10 constructs were generated with full length myc-tagged HOXC10 cloned in pCDNA3.1-Neo at EcoRI and XbaI sites. Reagent sources: Doxorubicin (Sigma), Paclitaxel and Gemcitabine (Tocris Bioscience) and Carboplatin, BS-181 and SNS-032 (11) (gifted by Selleckchem), TRC lentiviral Human HOXC10 shRNA (set of 4) (Thermo Scientific); FlexiTube siRNA for HOXC10 (SI04296621), E2F1 (SI00300083) or CDK7 (SI02664795) (Qiagen). All other reagents were from Sigma.
Human Tissue Samples
Fresh frozen primary human tissues were used following approval of the Johns Hopkins Institutional Review Board (IRB).
Survival Analysis in Patients with Breast Cancer
Kaplan-Meier analyses were performed using 4117 breast cancer patients. http://kmplot.com/analysis (12).
Real-time Quantitative PCR (RT-qPCR) Analysis
qRT-PCR was conducted using the MaximaSYBR Green/ROX Master Mix (Fermentas) per manufacturer protocol. The ΔΔCt method was used, with GAPDH expression for normalization (13).
Soft Agar Colony Formation Assay and Matrigel Invasion Assay
Six-well plates containing a 0.6% agar layer were overlaid with 3×103 cells i n 0.3% agar layer. Colonies were counted after 7 days. Cell invasion assay was performed using The BD BioCoat Matrigel Invasion Chamber assay system (13).
Tumor Xenograft Studies
Approved by Johns Hopkins Institutional Animal Care and Use Committee (IACUC), BALB/c nu/nu athymic mice (Sprague–Dawley-Harlan, Madison, WI) received subcutaneous (s.c) injections of 3 million cells/100 μL PBS/Matrigel (1:1), and were treated with doxorubicin (4 mg/kg BW/iv/ weekly) or paclitaxel (10 mg/kg BW/intraperitoneal /weekly).
Growth Assay
Cells were grown in 12-well plates (2000 cells/well), fixed with formalin and stained with crystal violet. To quantitate growth, the dye was solubilized by 10% acetic acid, and absorbance was measured at 560 nM.
Flow Cytometry Analysis
Cells at 70-80% confluence or after double thymidine synchronization were collected, permeabilized, pelleted, and resuspended in an isotonic buffered propidium iodide (PI)-staining solution containing RNase A (0.1 mg/ml) and PI (20 μg/mL). Samples were run on the BD FACSCalibur system (Becton Dickinson), and data analyzed using WinMDI 2.9 software.
Luciferase Assay
1×105 cells were seeded in 12-well cell culture plates, and transfected with a total of 1.6 μg of plasmids including reporter, expression and pCMV-β-galactosidase plasmids using Lipofectamine 2000 (14). NF-κB-luc (Igκ2-IFN-luc wt or mut) was a kind gift of Dr. Joel L. Pomerantz (15). The TOPFlash plasmid (16) was purchased from Addgene.
Clonogenic Cell Survival Assay
Exponentially growing cells were exposed to drugs or ultraviolet light (9). 24h later, cells were reseeded (1-2×103/well) in triplicate in 6- well plates. Viable colonies were fixed, stained with crystal violet and counted 1-2 weeks later.
MTT assay
2.5×103 cells/well were plated in 96-well plates in triplicate and treated with drugs alone or in combination, and MTT assay was performed (17). Values are expressed as percent survival of the vehicle-treated control.
Caspase 3/7 Activity
Caspase-3 and -7 activities were measured with the Caspase-Glo®assay kit (Promega) according to the manufacturer’s instructions. Values were expressed as percent activity of the vehicle-treated control.
Western Blot Analysis
Cells were lysed with RIPA buffer and processed as described (17). Sources: Anti-CDK7 and anti-XPD (Santa Cruz); BCL-xL and BIRC3 (Abcam); all other antibodies (Cell Signaling).
Host-Cell Reactivation Assay
The assay was performed (18) with some modifications. pGL3- basic (Promega) was exposed to UV light [Stratalinker® UV-Crosslinker 1800 (Stratagene)] or treated with 100-1000 nM cisplatin. To measure HR-mediated repair, 1 μg of vector digested with Hind III was transfected into cells with pCMV-β-galactosidase. Luciferase and β-galactosidase activities were measured after 48h.
Comet Assay
The alkaline Comet assay to measure DNA strand breaks in single cells was performed according to the manufacturer’s protocol [CometAssay Kit (Trevigen)]. Comet tail moments of ≥100 cells were measured and quantified using the CometScore software.
Chromosomal aberration analysis at metaphase
Chromosomal aberration analysis was performed as described (18, 19). Exponentially growing cells were gamma irradiated (3 G) and analyzed for metaphase aberrations after 12 hr (20). Cisplatin-induced chromosome aberrations were analyzed as described (19).
Detection of γ-H2AX Foci
Cells growing in chamber slides (Nunc® Lab-Tek® II) were treated with Dox (200 nM) or Gemc (50 nM) for 24h. Following fixation and permeabilization, cells were probed with antibody against phosphorylated H2AX-Ser139 (Upstate Biotechnology); H2AX foci were visualized using a Zeiss Axio Scope fluorescent microscope and scored with the ImageJ software (v1.47, NIH). 100 or more cells were evaluated.
I-Sce1 assay for homologous recombination repair activity
DR-95 cells were transfected with pI-Sce1, pEGFP or pCMV (21,22), harvested after 72 h, and %GFP-expressing cells was measured by flow cytometry. Frequency of recombination events = Mean %GFP positive cells transfected with pI-Sce1/Mean %GFP-positive cells transfected with pEGFP.
Recruitment of HR protein at DSB sites
DR95 cells were electroporated with I-pCBASce. ChIP was done using antibodies to Rad51 (ABCAM); BRCA1 (Cell Signaling); KU80 (Cell Signaling) (20). Quantification of ChIP DNA was carried out by real-time PCR in triplicate using the LightCycler Fast Start DNA Master SYBR Green I (Roche Applied Sciences).
DNA Fiber Assay
DNA fiber spreads were prepared (19) with minor modifications. Cells in exponential phase were labeled for sites of ongoing replication with IdU (50 μM) followed by exposure to hydroxyurea (4 mM), washed and labeled with CIdU (50 μM). Fibers were analyzed using ImageJ software.
Co-immunoprecipitation to detect HOXC10/CDK7
1 mg of protein lysate was subjected to immunoprecipitation overnight at 4°C with CDK7 antibody (C-19, Santa Cruz) or normal rabbit IgG control (sc-2027, Santa Cruz); immunoblotting was performed using: Myc-Tag (9B11, Cell Signaling), RNA Polymerase II (CTD4H8, Millipore), and XPD [sc-101174, Santa Cruz) (14).
CDK7 Kinase Activity
300 μg of protein extract from drug treated cells was used for immunoprecipitation with CDK7 antibody. The immuno-precipitate was resuspended in 40 μl of kinase buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 250 μM EGTA, 10 mM β-glycerophosphate, 1 mM DTT), 10 μl of 50 nM ATP, and 20 ng GST-CDT peptide (P4016, Proteinone) as substrate, incubated 1h at 30°C; added to an equal volume of kinase-GLO™ reagent (Promega) and incubated for 15 min a t room temperature. Control reactions lacked the CTD peptide substrate. Luminescence was recorded and expressed as relative RLU to the untreated control cells.
Statistics
Results were expressed as mean ± SEM of at least 3 independent experiments. Paired Student t-test or ANOVA tests were performed for data analysis. All statistical analyses were performed using GraphPad Prism version 4.03 (GraphPad Software, Inc.). In all figures, *p< 0.05, **p< 0.01 and ***p< 0.001.
Details of constructs, materials and experimental procedures are provided in “Supplemental Materials and Methods” online.
Results
HOXC10 overexpression is linked to chemotherapy resistance
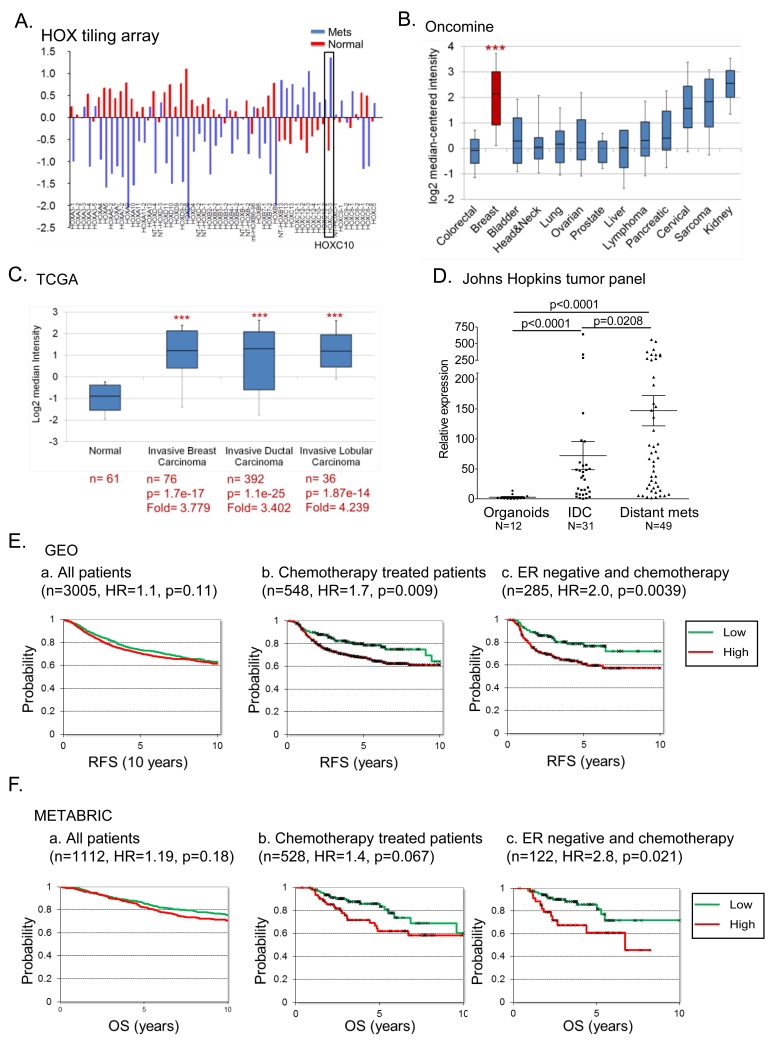
By HOX tiling mRNA expression array, compared to normal breast, HOXC10 was among the top five overexpressed genes in breast cancer (23), and higher than all 39 HOX genes in distant metastasis (Figure 1A). In Oncomine (24), HOXC10 was among the top 1% transcripts in breast compared to other carcinomas (Figure 1B), and in TCGA (https://tcga-data.nci.nih.gov/), was equally prevalent in all breast carcinomas (Figure 1C). qRT-PCR analysis of our primary tissue panel validated this finding; median HOXC10 expression was 10-fold higher in primary invasive carcinomas and 30-fold higher in distant metastatic tissues (Figure 1D). No correlation of HOXC10 expression with relapse-free (RFS) or overall survival (OS) was observed in breast cancer patients in the GEO (http://www.ncbi.nlm.nih.gov/geo) and the METABRIC cohorts (25) (Figure 1Ea, 1Fa). In chemotherapy treated patients, high HOXC10 expression correlated with short RFS (Figure 1Eb, S1A, S1B) and short OS (Figure 1Fb, S1C), and even more consistently in the subset of ER-negative patients treated with chemotherapy (Figure 1Ec, 1Fc). Cox multivariate regression analysis, taking all clinical parameters into account, revealed a highly significant (p=0.00013) inverse correlation between HOXC10 expression and RFS.
Figure 1.
HOXC10 overexpression has prognostic significance in breast cancer.
(A) HOX-tiling array analysis of mRNA expression in normal breast and distant metastasis. HOXC10 expression in (B) cancers in the Bittner multi-cancer dataset ***p= 3.18 e-40. (C) invasive ductal and lobular breast carcinoma. (D) qRT-PCR of HOXC10 expression in normal breast organoids (n=12), invasive ductal carcinoma (n=31) and distant metastasis (n=49). K-M plots of correlation of HOXC10 expression with (E) recurrence free survival (RFS), and (F) overall survival (OS) in (a) all patients with breast cancer (n=4117), (b) chemotherapy treated-, (c) chemotherapy treated-ER-negative breast cancer.
HOXC10 is induced during development of chemotherapy resistance
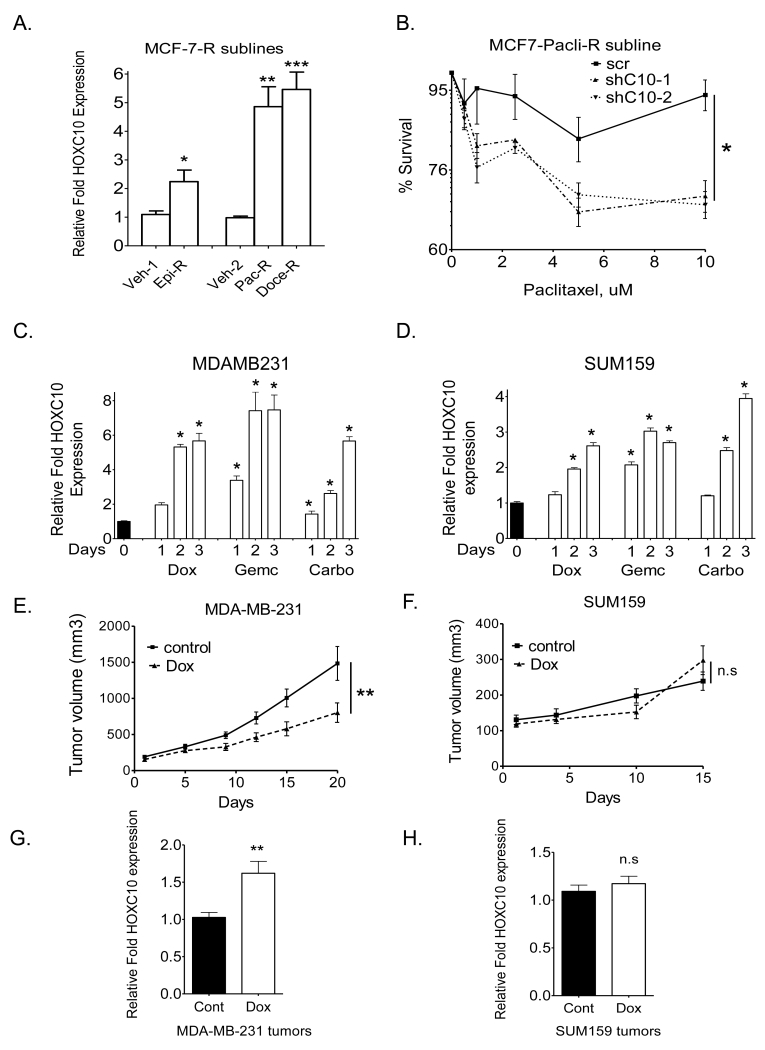
As a first step to investigating the contribution of HOXC10 to drug resistance, we analyzed HOXC10 expression in chemotherapy-resistant MCF7-sublines (9) which displayed resistance (>250-fold) to epirubicin, paclitaxel and docetaxel. HOXC10 was expressed at 2-8 fold higher levels in MCF-7- R sublines (Figure 2A) and in newly developed MDA-MB-231-R sublines that emerged (>30 days) following exposure to drugs (Figure S2A). Depleting HOXC10 levels in MCF-7-Tax-R subline restored its response to paclitaxel (Figure 2B). Further studies were carried out in eight breast cancer cell lines with varying levels of HOXC10 mRNA (Figure S2B). On exposure of cells in culture to drugs, induction of HOXC10 expression was rapid, starting at day 1 in MDA-MB-231 (Figure 2C), SUM159 (Figure 2D), and SUM149 (Figure S2C). Basal levels of HOXC10 expression determined response to Dox. Mouse xenografts of low HOXC10 expressing MDA-MB-231 (Figure 2E) responded to Dox significantly better than high HOXC10-expressing SUM159 (Figure 2F) or HCC1954 (Figure S2D). By qRT-PCR, MDA-MB-231 (Figure 2G) and SUM149 (Figure S2E) tumors that resumed growth during treatment showed significantly higher HOXC10 expression compared to SUM159 (Figure 2H) and HCC1954 (Figure S2F) supporting the argument that upregulated HOXC10 expression correlated with resistance to chemotherapy.
Figure 2.
Chemotherapy-resistant cell lines upregulate HOXC10.
(A) qRT-PCR of HOXC10 expression in drug-resistant MCF7 sublines. Epirubicin (Epi-R), Paclitaxel (Pac-R), and docetaxel (Doc-R) resistant. (B) MTT assay in HOXC10-depleted MCF7-Pac-R clones. qRT-PCR of HOXC10 expression in (C) MDA-MB-231 cells and (D) SUM159 cells treated with Dox (200nM), Gemc (200nM), or carboplatin (50uM). Growth in vivo of xenografts of (E) MDA-MB-231 (n=12) (F) SUM159 (n=12), treated with doxorubicin. qRT-PCR of HOXC10 expression in Dox-treated xenografts of (G) MDA-MB-231 and (H) SUM159.
HOXC10 overexpression decreases susceptibility to chemotherapy
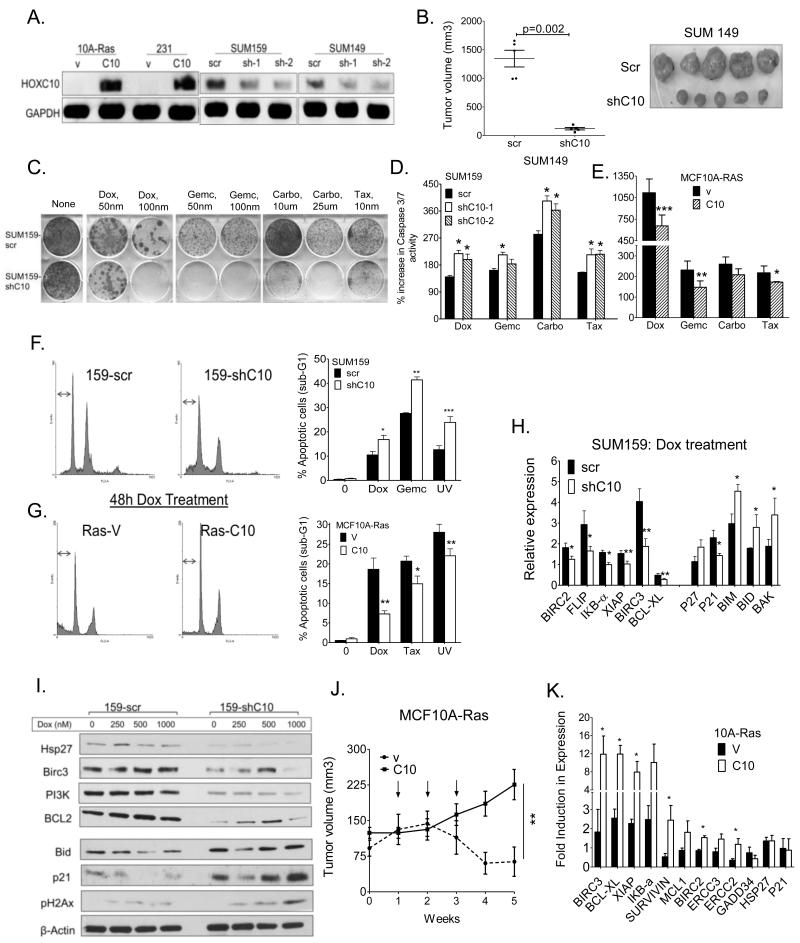
To characterize the effects of HOXC10 overexpression, we derived stable clones of MCF10A-Ras [stably expressing K-ras (Gly12-Val)] and MDA-MB-231 overexpressing myc-tagged-HOXC10, and SUM159 and SUM149 cells depleted of HOXC10 by 50-70% (immunoblot in Figure 3A) using specific shRNAs. Properties of anchorage-independent growth (Figure S3A, B), colony formation (Figure S3C), invasion through matrigel (Figure S3D), proliferation (Figure S3E), quantified in (Figure S3F), and tumor growth in mice (Figures 3B, S3G) were all significantly decreased by depleting HOXC10 and enhanced by overexpressing HOXC10 in tumor cells. By colony survival assays, Dox, Gemc, Carbo and Tax were more cytotoxic in SUM159-shC10 (Figure 3C, S3H), MCF7-shC10 and SUM149-shC10 (Figure S3H) than in MCF10A-Ras-C10 cells (Figures S3I). This body of data, supporting an increased oncogenicity and aggressiveness of HOXC10-expressing breast cancer cells, was further corroborated by MTT assays in SUM159-shC10 (Figure S3J) and in MCF10A-Ras-10 cells (Figure S3K).
Figure 3.
HOXC10 overexpression decreases susceptibility to chemotherapy treatment.
(A) Western blot analysis of MCF10A-Ras-C10 and MDA-MB-231-C10, and HOXC10 shRNAs expressing clones of SUM159 and SUM149 cells. Loading control: GAPDH (B) Growth of xenografts of SUM149–Scr and SUM149-shC10 cells in immunodeficient mice. (C) Colony survival assay of SUM159-shC10-1 cells treated with Dox, Gemc, Carbo and Tax. (D) Caspase 3/7 activity in SUM159-shC10 cells and (E) MCF10A-Ras-C10 cells following treatment with treated with Dox (1 μM), Gemc (1 μM), taxol (0.5 μM), docetaxel (0.5 μM) or carboplatin (100 μM) for 24h. (F) Flow cytometry analysis of SUM159-shC10 cells (G) MCF10A-Ras-C10 cells treated with Dox (50 nM), Gemc (50 nM), or UV; bar graphs showing quantification of cells in sub-G1. (H) qRT-PCR analysis of pro- and anti-apoptotic mRNAs with Dox (200 nM) for 72 h, (I) Western blot analysis of select proteins in SUM159-shC10 cells treated with Dox (250-1000 nM). (J) Growth of established xenografts of MCF10A-Ras-v (n=6) and MCF10A-Ras-C10 (n=9) cells in nude mice, treated 3 times at weekly intervals (arrows) with Dox (4 mg/kg). (K) qRT-PCR of anti-apoptotic genes in tumors from (I) at 5 weeks.
The activation of caspase-3/7 is a reliable marker of cells undergoing apoptosis (26). Susceptibility to the drugs was reflected by an increase in caspase 3/7 activity in SUM159-shC10 cells (Figure 3D) and a decrease in MCF10A-Ras-C10 cells (Figure 3E). Apoptotic cells were identified based on DNA content frequency histograms as cells with fractional “sub-G (1)” DNA content (27). In response to Dox, there was a decrease in sub-G1 population in SUM159shC10 cells (Figure 3F) and an increase in MCF10A-Ras-C10 cells (Figure 3G). Also, several pro- and anti-apoptotic mRNAs were modulated in SUM159-shC10 and MCF10A-Ras (Figure S3L, S3M), and further intensified by drug treatment (Figure 3H, S3N). Western blot analysis of SUM159-shC10 cells showed that anti-apoptotic genes, hsp27, Birc 3, PI3K, and BCL2 levels were decreased, while pro-apoptotic genes, p21, and BID levels were increased by depletion of HOXC10 (Figure 3I). Dox-treated MCF10A-Ras-vec xenografts regressed by week 4, while MCF10A-Ras-C10 tumors grew after week 2 (Figure 3J). Consistent with this observation, MCF10A-Ras-C10 tumors showed high mRNA expression of many anti-apoptotic genes (Figure 3K).
Given that the anti-apoptotic genes examined are known direct targets of NF-κB (28), we measured NF-κB activity using the NF-κB-responsive reporter Igκ2-IFN-LUC, with and without Dox treatment. The basal activity of NF-κB was lower in SUM159-shC10 (Figure S3O) and high in MCF10A-Ras-C10 (Figure S3P) compared to their respective controls, and further enhanced by Dox treatment. This data is consistent with findings that the basal activity of NF-κB is much higher, and maintained as such, in Dox-resistant MCF7 cells (29). Thus, HOXC10 upregulates NF-κB to achieve both activation of anti-apoptotic pathways, and increases cell survival following stress.
HOXC10 enhances DNA damage repair and checkpoint recovery
Several homeodomain related proteins have been functionally related to DNA damage repair (1, 2, 30, 31). Overexpression of HOXA5 in S. cerevisiae up-regulates components of the mismatch repair (MMR) system, important for the detection and repair of DNA damage (32). Hence, we investigated if HOXC10 participates directly in DNA repair and if so, the type of damage and cell cycle stage of repair.
A host-cell reactivation assay was used to assess cellular ability to repair exogenously damaged DNA. A pGL3 (luciferase-reporter) plasmid was exposed to UV or cisplatin, or digested with Hind III (to generate double strand breaks, DSBs) followed by transfection into SUM159-shC10 cells or MCF-10A-Ras-C10 cells and their respective controls. SUM159-shC10 cells displayed a significantly decreased ability to repair DNA damage (Figure 4A), while the reverse was shown for UV-induced DNA damage in MCF-10A-Ras-C10 cells (Figure 4B). Clonogenic cell survival assays in both MCF10A-Ras-C10 and SUM159-shC10 cells exposed to UV (Figure 4C, quantified in S4A) supported these findings. However, repair of DNA DSBs caused by Hind III digestion (Figure 4A) remained unchanged in SUM159-shC10 cells, suggesting no involvement of the NHEJ pathway. Further, ionizing radiation (IR) induced G1-chromosome damage repair was not affected in SUM159-shC10 cells (Figure S4B), wherein DSB repair by NHEJ is predominant. On the other hand, SUM159-shC10 cells had a higher frequency of S-phase specific IR-induced chromosomal aberrations (Figure S4C), suggesting that HOXC10 expression enhances repair, possibly by the homologous recombination (HR) pathway. MCF10A-Ras-C10 cells treated with cisplatin showed significantly reduced number of cells with chromosomal aberrations at metaphase (Figure S4D) compared to the vector control cells. Collectively, the data strongly support the hypothesis that HOXC10 is an important participant in DNA DSB repair by HR.
Figure 4.
HOXC10 facilitates repair of DNA damage.
Host cell reactivation assay in, (A) SUM159-shC10 and (B) MCF10A-Ras-C10 cells following UV and/or cisplatin exposure. (C) Colony survival assay of SUM159-shC10 and MCF10A-Ras-C10 cells, 7 days after UV exposure. (D) Alkaline comet assay measure of double strand breaks (DSB) caused by UV or Dox in SUM159-shC10 cells. Quantification of tail moments in (E) SUM159 and (F) MCF10A-Ras treated with Dox (200nM), Gemc (200nM), and UV. (G) Quantification of phosphorylated γ-H2AX using fluorescent anti-phospho-H2AX Ser139 antibodies in SUM159-shC10 cells treated with Dox (200 nM) or Gemc (50 nM) for 24h (H) I-Sce1 assay for HR repair activity in DR95-siC10, 72 h after transfection. HR frequencies are shown with (+) or without (−) I-SceI induction. Positive control: BRCA-1 siRNA transfected cells. (I) ChIP analysis of recruitment of repair proteins to I-Sce I DSB site (*p <0.05, Chi-square test). (J) Representative images of DNA fiber assay on SUM159-shC10 and MCF10-Ras-C10 at 21 h. (K) Quantification of stalled forks (green) and new origins (red) in the DNA fiber assay showing correlation between HOXC10 expression and initiation of new origins of replication.
We had noted that HOXC10 expression had a dramatic effect on survival following exposure to UV light or the platinum drugs (Figures 4A-C and S4A). DNA damage induced by such agents is repaired by nucleotide excision repair (NER) and HR. Therefore, we measured the activity of the NER pathway by the alkaline comet assay (Figure 4D), a single-cell gel electrophoresis method in which the intensity of the comet tail of the migrating cells relative to the head reflects the number of DNA breaks (33). Cells depleted of HOXC10 had significantly more residual UV-, Dox- and Gemc-induced DNA damage after 24h after treatment (Figure 4E). Furthermore, time-course analysis after UV treatment showed that DNA damage in SUM159-scr cells had returned to baseline levels in 24h; but was still 4-fold higher in SUM159-shC10 cells (Figure 4E), revealing inefficient DNA repair in the absence of HOXC10. Improved response to DNA damage was evident in MCF10A-Ras-C10 cells compared to the vector-control cells (Figure 4B, 4C, 4F). The reduced survival of SUM159-shC10 cells in colony formation assays (Figure 4C) also correlated with higher levels of residual DSBs.
DNA damage that results in DSBs is always followed by the phosphorylation of the histone, H2AX (34). Newly formed phosphorylated protein γ-H2AX is the initial step of damage detection to recruit repair proteins (34). We quantified the number of residual γ-H2AX foci 24 h after treatment of SUM159-shC10 cells with Dox or Gemc. On average, 20-30% more γ-H2AX foci accumulated in SUM159-shC10 cells after drug treatment (Figure 4G) with almost 50-60% more cells displaying ≥100 foci/nucleus (Figure S4E). A time-course analysis of γ-H2AX protein after UV or Dox treatment showed no differences in the initial induction of γ-H2AX (10 min to 8h) (Figure S4F, S4G). Residual γ-H2AX measured 24h post treatment, and not the initial kinetics of γ-H2AX formation, is a better predictor of cell viability (35, 36). These results indicated HOXC10 might be more involved in DNA damage processing rather than in damage sensing. No major difference was observed in the phosphorylation status of known DNA damage sensors of the ATR/ATM pathways after Dox or Gemc treatment of SUM159-shC10 (Figure S4H), thus confirming that HOXC10 is not involved in DNA damage sensing.
These findings were further substantiated by the HR-DSB repair assay of lesions induced by I-Sce1 endonuclease performed in DR-95 cells engineered to stably express a pDR-GFP plasmid containing a mutated GFP gene with an 18 bp I-Sce1 endonuclease cleavage site and an in-frame termination codon (10, 20). Efficient repair of the Sce-1 cleavage site restores expression of GFP. DR-95 cells, with and without HOXC10 silencing (using siRNA) were transfected with the I-Sce1 expression plasmid. DR95-siHOXC10 showed reduced repair of I-Sce-1 induced lesions. Consequently, restoration of GFP expression occurred at a significantly lower efficiency than in control cells (Figure 4H). Reduction of GFP expression was comparable to cells with reduced expression of BRCA-1, a known DNA repair protein, used as a positive control (Figure 4H). Thus, repair of DSB by HR is impaired in cells lacking HOXC10 expression.
To investigate a role of HOXC10 in the recruitment of HR repair proteins at the site of a single DNA DSB, we performed chromatin immunoprecipitation (ChIP) in DR-95 cells (10, 20). We compared the levels of BRCA1, RAD51, KU80 and HOXC10 at different distances from an I-Sce1-induced DSB site using ChIP analysis with specific primers (10) in cells with and without expression of HOXC10. Depletion of HOXC10 using siRNAs did not decrease KU80 levels in close proximity to the DSB, whereas levels of RAD51 and BRCA1 at the break site were significantly reduced (Figure 4I). This finding in HOXC10-depleted cells strongly supported the role of HOXC10 in facilitating the recruitment of proteins involved in HR. However, HOXC10 itself was undetectable at the I-Sce1 cleavage site, ruling out a direct role for HOXC10 in DSB repair.
We therefore considered the possibility that HOXC10 has a role in the initiation of new origins of DNA replication. Single molecule examination of replication dynamics by DNA fiber analysis (19) after hydroxyurea (HU) treatment in SUM159-shC10 and MCF10A-Ras-C10 cells showed that HOXC10 is required for the initiation of new origins, but not for the accumulation of stalled forks (Figures 4J, 4K).
Collectively, the data suggest that HOXC10, as a transcriptional factor, impacts the chromatin status and sets the stage for HR by, 1) decreasing S-phase chromosome aberrations, 2) enhancing resection as shown by gamma-H2AX foci, 3) increasing homologous recombination based on l-Scel endonuclease-induced DSB repair, and 4) resolving stalled replication forks as shown by DNA fiber analysis, possibly leading to initiation of DNA replication following DNA damage.
HOXC10 binds to CDK7 in vivo after chemotherapy treatment
We had thus far observed that HOXC10 enhances cell survival by affecting apoptosis, NF-kB activity and damage repair, mainly by the HR pathways. One key protein that links all three pathways is cyclin-dependent kinase 7 (CDK7). In metazoans, CDK7 has essential roles in transcription as a component of the general transcription factor, TFIIH. CDK7 is an effector kinase which also phosphorylates RNA Pol II and other proteins during transcription after the pausing of the TFIIH complex following DNA damage (37). Highly relevant to this study, HOXC10 was found to be the tightest binding protein linking the CAK (Cdk-activating kinase, CDK7) complex to TFIIH in a yeast 4-hybrid system (38). We therefore tested for an association of HOXC10 with CDK7 by co-immunoprecipitation. A weak interaction between the two proteins increased significantly upon treatment with Dox and Gemc (Figure 5A). This interaction was reduced significantly upon addition of a pharmacologic inhibitor of CDK7 activity, SNS-032 (Figure 5A), or by expressing the dominant negative, kinase-dead mutant CDK7 (D155A) in the cells (Figure 5B). We concluded that CDK7 and HOXC10 formed a complex in breast cancer cells, which became more abundant upon exposure to drugs.
Figure 5.
HOXC10 binds to CDK7 during DNA damage response.
293T cells were transfected with HOXC10, and treated with Dox (200 nM), Gemc (200 nM), taxol (50 nM) or carboplatin (50 uM) for 24 h. Cell lysates were co-immunoprecipitated (co-IP) with CDK7 (D: doxorubicin; SNS-032: CDK7-inhibitor). (B) Co-IP of HOXC10 with CDK7 (wt) and its kinase mutant (D155A). Co-IP of XPD by CDK7 in (C) SUM159 cells (-scr and -shC10) and (D) MCF10A-Ras (-vec and -C10) after treatment with Dox and Gemc for 24 h. Co-IP of RNA Pol II by CDK7 in (E) SUM159 cells (-scr and -shC10) and (F) MCF10A-Ras (-vec and -C10) after treatment with Dox and Gemc for 24 h. (G) Kinase activity of CDK7 on recombinant GST-CTD substrate in SUM159-shC10 cells treated with Dox (100 nM) or Gemc (100 nM) for 24 h. qRT-PCR analysis of MCL1 (as marker of CDK7 activity) in (H) SUM159-shC10 and (I) MCF10A-Ras-C10 cells treated with Dox (100 nM) or Gemc (100 nM) for 24 h.
Different sets of proteins are phosphorylated by CDK7 depending on its function in cell division (i.e. CDK1, 2, 4, 6) or in transcription as a complex with TFIIH (37). We therefore investigated if binding of HOXC10 to CDK7 altered its substrate preference. Also, elongating RNA Pol II can be arrested by endogenous and exogenous DNA lesions such as UV-induced pyrimidine dimers, adducts induced by anticancer drugs, and DSBs. These transcription-blocking lesions located on the transcribed strand are primarily repaired by transcription-coupled repair, the NER pathway (39). XPD helicase is a key member of the human TFIIH complex that anchors CAK kinase (cyclin H, MAT1, and CDK7) to TFIIH and unwinds DNA for transcription and for repair of duplex warping damage by nucleotide excision repair (NER), thereby maintaining genomic integrity (40). We therefore examined if HOXC10 affects the composition of the TFIIH-XPD-CAK complex through its protein/protein interaction with CDK7. The binding of XPD to CDK7 was reduced in SUM159-shC10 cells (Figure 5C) and was increased in MCF10A-Ras-C10 cells, compared to their respective control cells (Figure 5D). This finding suggested that HOXC10 may have a role in anchoring CAK to XPD, thus maintaining the integrity of the holoenzyme TFIIH during response to DNA damage. This finding is also consistent with reports that in repair-deficient cells, the association of CAK kinase, but not of XPD, to damaged DNA was reduced (41).
Next, we investigated whether the association of CDK7 with the RNA Pol II is modulated by HOXC10. We found that the association between CDK7 and RNA Pol II in drug treated cells was significantly stronger in the presence of HOXC10 in MCF10A-Ras-C10 (Figure 5E) and weaker in the absence of HOXC10 in SUM150-shC10 (Figure 5F). Further, the kinase activity of CDK7 on the C-terminal domain (CTD) of RNA Pol II was significantly reduced in SUM159-shC10 cells after DNA damage (Figure 5G). MCL1 is an anti-apoptotic protein that is rapidly depleted upon inhibition of RNA Pol II, permitting measures of its mRNA to serve as a surrogate for CDK7 activity (42). The lack of CDK7 activity is likely to impede recovery of the transcriptional machinery, thereby promoting cell death after DNA damage as was reflected by MCL1 expression (Figure 5H). Conversely, the enhanced interaction between Pol II and CDK7 in MCF10A-Ras-C10 cells (Figure 5E) promotes cell survival following DNA damage (Figure 5I). Consistent with this data, newly emerging drug-resistant-MCF7 sublines in cell culture showed increases in both MCL1 and HOXC10 mRNA expression (Figure S5).
Inhibiting CDK7 reverses HOXC10-dependent chemoresistance
Inhibiting CDK7 is of special interest in cancer therapeutics since it affects multiple signaling pathways (43). Two known selective CDK7 inhibitors are BS-181 (44) and SNS-032 (11); the latter is in Phase 1 studies (http://clinicaltrials.gov/). Combining SNS-032 or BS-181 at minimally cytotoxic doses (~20%) with chemotherapy significantly improved the response of both MCF10-Kras-C10 cells (Figure 6A, 6B) and SUM159-shC10cells (Figure S6A, S6B). In taxol and epirubicin-resistant MCF7 sublines, western blot analysis showed that CDK7 (pThr170) and CTD (pSer5) were activated (Figure 6C) with no change in cell cycle kinases, CDK1 and CDK2. qRT-PCR analysis showed an increase in both HOXC10 and MCL1 mRNAs (Figure 6D). Treatment with taxol combined with BS-181 restored drug susceptibility of Tax-R-MCF-7 cells, as seen in colony formation assays (Figure 6E), and caused cell kill in parental MCF-7 cells as shown by MTT assays (Figure 6F). Similarly, BS-181 restored sensitivity of two MDA-MB-231-Tax-R sublines to taxol (Figure S6C, S6D), an effect not achieved using inhibitors to CDK1/2/9, AZD5438 (45) and CDK4/6, PD0332991 (46) (Figure S6E, S6F). These data suggest a specific role for CDK7 in resistance to chemotherapeutic drugs.
Figure 6.
Inhibiting CDK7 restores chemo-sensitivity to breast cancer cells.
MTT assay of MCF10A-Ras-C10 cells treated with CDK7 inhibitors, (A) SNS-032 (40 nM) or (B) BS-181 (5 uM) in combination with Dox, Gemc and/or carboplatin for 48 h (C) Western blot analysis of p-CDK7 and p-CTD, p-CDK1, p-CDK2 in parental MCF (WT) and drug-resistant sublines. (D) qRT-PCR analysis of MCL1 and HOXC10 in MCF7-drug-resistant sublines, Tax-R and Epi-R. (E) Colony survival assay after treatment of MCF-7-Tax-R with taxol, 100 nM +/− BS-181 (20uM) for 7 days. (F) MTT assay of MCF7-Tax-R treated with taxol, (0-250 nM) +/− BS-181 (10 um) for 48 h. Parental MCF-7 cells serve as a control for taxol susceptibility.
In summary, our data supports a model of HOXC10 action wherein it is intimately involved in multiple steps in the process of HR-mediated DNA repair following chemotherapy exposure of breast cancer cells and in the development of drug resistance in the long term. HOXC10 is an integral component of the CAK complex, which allows restart of transcription following DNA damage (Figure 7).
Figure 7.
A model for HOXC10 action in breast cancer cells.
Upon exposure to chemotherapy- or ionizing radiation (IR), DNA damage mediator genes such as ɣ-H2AX/MDC1/53BP1/BRCA1 are activated, triggering DNA double strand break repair (DSB). HOXC10 is upregulated following chemotherapy or IR in ER-negative breast cancer. HOXC10, as part of the CAK complex, participates in the late stages of DNA repair that involves restart of transcription for recovery; and acts as an inhibitor of apoptosis.
Discussion
Chemotherapy failure in breast cancer claims at least 80,000 lives each year worldwide. Here we present evidence that HOXC10 overexpression is common and functionally important for onset of resistance to chemotherapy in ER-negative breast cancer and is also prognostic of poor outcome primarily in patients treated with chemotherapy.
Upregulation of HOXC10 is likely to be an important cancer adaptation mechanism. Ectopic expression of HOXC10 leads to decreased drug susceptibility, while decreasing its levels enhances the cytotoxic effects of chemotherapy in both in vitro and in vivo models of breast cancer. Mechanistically, HOXC10 is involved in activating different survival pathways, including driving checkpoint recovery after DNA damage, a key pathway that allows cancer cells to overcome damage response arrest (Figure 4, S4) (47). HOXC10 does not affect the sensing of DDR but is required at later stages of the DNA damage repair by NER and HR (Figure 4, S4). Cells expressing high levels of HOXC10 repaired DNA damage more efficiently and resumed transcription and growth; while their low-HOXC10 expressing counterparts eventually committed to apoptosis. Part of this response, we propose, may be attributed to the observed association of HOXC10 with the CAK complex.
The CAK complex along with TFIIH can participate in diverse functions, including transcription, DNA repair (NER) and cell cycle regulation. We confirmed that HOXC10 binding to CDK7 enhances its kinase activity towards the CTD domain of RNAPII after DNA damage. We surmise that HOXC10 plays a role in bridging the gap between these 2 complexes, enhancing the recovery process after DNA damage. Indeed, treatment of drug-resistant MCF-7 cells with CDK7 inhibitors restored their susceptibility to chemotherapy (Figure 6). The importance of current findings to breast cancer therapy stems from the recent attention given to CDK inhibition, including CDK7, in clinical trials either as single agents or in combination with chemo- or targeted therapies to overcome resistance (43, 48).
The second survival pathway that is activated by HOXC10 is shifting the balance between growth and apoptosis to allow continuous proliferation and survival under adverse conditions. Our data showed that HOXC10 facilitates the transition from G1 to S phase and progression through the S phase during the cell cycle (Figure 3). As a consequence, cells with high HOXC10 levels have a growth advantage, especially under non-ideal conditions, and restarted their replication to overcome stressful conditions. At the same time, the increase in NF-κβ activity and the consequent increase in the levels of many anti-apoptotic proteins likely decreased cell sensitivity to many stressors.
Because of HOXC10 involvement in survival and proliferation of cancer cells despite exposure to chemotherapy, HOXC10 is an attractive target to reverse chemotherapeutic resistance in breast cancer. Future studies could focus on developing direct inhibitors of HOXC10 or indirect modulators of its function by targeting its downstream effectors, such as CDK7.
Supplementary Material
Acknowledgements
We thank Dr Joel L. Pomerantz for providing us their responsive reporter plasmids NF-κB-luc, Dr Amadeo M Parissenti for the drug-resistant MCF7 cell lines, and Dr. Alan Rein for reviewing the manuscript.
Financial Support: This work was supported by funding from the DOD-BCRP BC093970 to H.S, NCI-P50-CA88843 and P30-CA006973 (S.S.) and NIH grants CA129537, CA154320 and GM109768 (T.K.P).
Footnotes
Conflict of Interest: All the authors declare no conflict of interest pertaining to the contents of this paper.
References
- 1.Rezsohazy R. Non-transcriptional interactions of Hox proteins: inventory, facts, and future directions. Dev Dyn. 2014;243(1):117–31. doi: 10.1002/dvdy.24060. [DOI] [PubMed] [Google Scholar]
- 2.Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, et al. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007;67(4):1527–35. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Ellmann S, Rubin E, Gil M, Jin K, Han L, et al. ADP ribosylation by PARP-1 suppresses HOXB7 transcriptional activity. PLoS One. 2012;7(7):e40644. doi: 10.1371/journal.pone.0040644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miotto B, Graba Y. Control of DNA replication: a new facet of Hox proteins? Bioessays. 2010;32(9):800–7. doi: 10.1002/bies.201000048. [DOI] [PubMed] [Google Scholar]
- 5.Marchetti L, Comelli L, D’Innocenzo B, Puzzi L, Luin S, Arosio D, et al. Homeotic proteins participate in the function of human-DNA replication origins. Nucleic acids research. 2010;38(22):8105–19. doi: 10.1093/nar/gkq688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Stanchina E, Gabellini D, Norio P, Giacca M, Peverali FA, Riva S, et al. Selection of homeotic proteins for binding to a human DNA replication origin. J Mol Biol. 2000;299(3):667–80. doi: 10.1006/jmbi.2000.3782. [DOI] [PubMed] [Google Scholar]
- 7.Christen B, Beck CW, Lombardo A, Slack JM. Regeneration-specific expression pattern of three posterior Hox genes. Dev Dyn. 2003;226(2):349–55. doi: 10.1002/dvdy.10231. [DOI] [PubMed] [Google Scholar]
- 8.Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, et al. Epigenetic Reprogramming of HOXC10 in Endocrine-Resistant Breast Cancer. Sci Transl Med. 2014;6(229):229ra41. doi: 10.1126/scitranslmed.3008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hembruff SL, Laberge ML, Villeneuve DJ, Guo B, Veitch Z, Cecchetto M, et al. Role of drug transporters and drug accumulation in the temporal acquisition of drug resistance. BMC Cancer. 2008;8:318. doi: 10.1186/1471-2407-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, et al. Interplay between human DNA repair proteins at a unique double-strand break in vivo. Embo J. 2006;25(1):222–31. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ, et al. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009;113(19):4637–45. doi: 10.1182/blood-2008-12-190256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 13.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer research. 2010;70(14):6047–58. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah N, Jin K, Cruz LA, Park S, Sadik H, Cho S, et al. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERalpha and inducing IL-6 expression. Cancer research. 2013;73(17):5449–58. doi: 10.1158/0008-5472.CAN-13-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. Embo J. 2002;21(19):5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–5. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 17.Alanee S, Shah S, Vijai J, Schrader K, Hamilton R, Rau-Murthy R, et al. Prevalence of HOXB13 mutation in a population of Ashkenazi Jewish men treated for prostate cancer. Fam Cancer. 2013;12(4):597–600. doi: 10.1007/s10689-013-9618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26(5):1850–64. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol. 2013;33(6):1210–22. doi: 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Hunt CR, Hegde ML, Chakraborty S, Udayakumar D, Horikoshi N, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014;8(1):177–89. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Molecular cancer therapeutics. 2009;8(8):2243–54. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008;9(5):378–90. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- 27.Kajstura M, Halicka HD, Pryjma J, Darzynkiewicz Z. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-G1” peaks on DNA content histograms. Cytometry A. 2007;71(3):125–31. doi: 10.1002/cyto.a.20357. [DOI] [PubMed] [Google Scholar]
- 28.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangadharan C, Thoh M, Manna SK. Inhibition of constitutive activity of nuclear transcription factor kappaB sensitizes doxorubicin-resistant cells to apoptosis. J Cell Biochem. 2009;107(2):203–13. doi: 10.1002/jcb.22115. [DOI] [PubMed] [Google Scholar]
- 30.Ramdzan ZM, Pal R, Kaur S, Leduy L, Berube G, Davoudi S, et al. The function of CUX1 in oxidative DNA damage repair is needed to prevent premature senescence of mouse embryo fibroblasts. Oncotarget. 2015;6(6):3613–26. doi: 10.18632/oncotarget.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer research. 2010;70(8):3089–97. doi: 10.1158/0008-5472.CAN-09-3138. [DOI] [PubMed] [Google Scholar]
- 32.Duriseti S, Winnard PT, Jr., Mironchik Y, Vesuna F, Raman A, Raman V. HOXA5 regulates hMLH1 expression in breast cancer cells. Neoplasia. 2006;8(4):250–8. doi: 10.1593/neo.05766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nandhakumar S, Parasuraman S, Shanmugam MM, Rao KR, Chand P, Bhat BV. Evaluation of DNA damage using single-cell gel electrophoresis (Comet Assay) J Pharmacol Pharmacother. 2011;2(2):107–11. doi: 10.4103/0976-500X.81903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–86. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 35.Olive PL, Banath JP. Kinetics of H2AX phosphorylation after exposure to cisplatin. Cytometry B Clin Cytom. 2009;76(2):79–90. doi: 10.1002/cyto.b.20450. [DOI] [PubMed] [Google Scholar]
- 36.Banath JP, Klokov D, MacPhail SH, Banuelos CA, Olive PL. Residual gammaH2AX foci as an indication of lethal DNA lesions. BMC Cancer. 2010;10:4. doi: 10.1186/1471-2407-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher RP. The CDK Network: Linking Cycles of Cell Division and Gene Expression. Genes Cancer. 2012;3(11-12):731–8. doi: 10.1177/1947601912473308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandrock B, Egly JM. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. The Journal of biological chemistry. 2001;276(38):35328–33. doi: 10.1074/jbc.M105570200. [DOI] [PubMed] [Google Scholar]
- 39.Tornaletti S. Transcription arrest at DNA damage sites. Mutat Res. 2005;577(1-2):131–45. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10(7):697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arab HH, Wani G, Ray A, Shah ZI, Zhu Q, Wani AA. Dissociation of CAK from core TFIIH reveals a functional link between XP-G/CS and the TFIIH disassembly state. PLoS One. 2010;5(6):e11007. doi: 10.1371/journal.pone.0011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conroy A, Stockett DE, Walker D, Arkin MR, Hoch U, Fox JA, et al. SNS-032 is a potent and selective CDK 2, 7 and 9 inhibitor that drives target modulation in patient samples. Cancer Chemother Pharmacol. 2009;64(4):723–32. doi: 10.1007/s00280-008-0921-5. [DOI] [PubMed] [Google Scholar]
- 43.Wesierska-Gadek J, Kramer MP. The impact of multi-targeted cyclin-dependent kinase inhibition in breast cancer cells: clinical implications. Expert Opin Investig Drugs. 2011;20(12):1611–28. doi: 10.1517/13543784.2011.628985. [DOI] [PubMed] [Google Scholar]
- 44.Ali S, Heathcote DA, Kroll SH, Jogalekar AS, Scheiper B, Patel H, et al. The development of a selective cyclin-dependent kinase inhibitor that shows antitumor activity. Cancer research. 2009;69(15):6208–15. doi: 10.1158/0008-5472.CAN-09-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byth KF, Thomas A, Hughes G, Forder C, McGregor A, Geh C, et al. AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther. 2009;8(7):1856–66. doi: 10.1158/1535-7163.MCT-08-0836. [DOI] [PubMed] [Google Scholar]
- 46.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3(11):1427–38. [PubMed] [Google Scholar]
- 47.Peng A. Working hard for recovery: mitotic kinases in the DNA damage checkpoint. Cell Biosci. 2013;3(1):20. doi: 10.1186/2045-3701-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallorini M, Cataldi A, di Giacomo V. Cyclin-dependent kinase modulators and cancer therapy. BioDrugs. 2012;26(6):377–91. doi: 10.1007/BF03261895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.