Abstract
Acute myeloid leukemia (AML) is a common and fatal form of hematopoietic malignancy. Overexpression and/or mutations of FLT3 have been shown to occur in the majority cases of AML. Our analysis of a large-scale AML patient cohort (n=562) indicates that FLT3 is particularly highly expressed in some subtypes of AML such as AML with t(11q23)/MLL-rearrangements or FLT3-ITD. Such AML subtypes are known to be associated with unfavorable prognosis. To treat FLT3-overexpressing AML, we developed a novel targeted nanoparticle system: FLT3 ligand (FLT3L)-conjugated G7 poly(amidoamine) (PAMAM) nanosized dendriplex encapsulating miR-150, a pivotal tumor-suppressor and negative regulator of FLT3. We show that the FLT3L-guided miR-150 nanoparticles selectively and efficiently target FLT3-overexpressing AML cells, and significantly inhibit viability/growth and promote apoptosis of the AML cells. Our proof-of-concept animal model studies demonstrate that the FLT3L-guided miR-150 nanoparticles tend to concentrate in bone marrow, and significantly inhibit progression of FLT3-overexpressing AML in vivo, while exhibiting no obvious side effects on normal hematopoiesis. Collectively, we have developed a novel targeted therapeutic strategy, using FLT3L-guided miR-150-based nanoparticles, to treat FLT3-overexpressing AML with high efficacy and minimal side effects.
Keywords: miR-150, AML, nanoparticles, PAMAM dendrimer, FLT3, FLT3L, MLL-rearranged, MYB, HOXA9, MEIS1, expression, targeted therapy, JQ1
INTRODUCTION
Acute myeloid leukemia (AML) is one of the most common and fatal forms of hematopoietic malignancies (1). With standard chemotherapies, only 30–40% of younger (aged <60) and 5–15% of older patients with AML survive more than 5 years (1). Thus, it is urgently needed to develop more effective therapies. FMS-like tyrosine kinase 3 (FLT3) encodes a conserved membrane-bound receptor that belongs to the class III receptor tyrosine kinase family (2). Specific binding of its ligand, FLT3-ligand (FLT3L), leads to the activation of multiple downstream signaling pathways (2–5). FLT3 is a critical oncogene in hematopoietic malignancies (5, 6). Internal-tandem duplications (ITDs) and tyrosine kinase domain (TKD) point mutations of FLT3 are found in over 30% of AML cases (5–7). FLT3-ITD-carrying AMLs are usually associated with poor prognosis (4, 8–10). Previous studies reported that FLT3 was overexpressed in more than 60% of AML cases tested, relative to normal control hematopoietic cells (8, 11–13). Although a number of FLT3 tyrosine kinase inhibitors (TKIs) have been investigated in clinical trials, the long-term therapeutic effects are disappointing, likely due to secondary mutations in FLT3, up-regulation of FLT3 expression, and/or activation of FLT3 downstream signaling pathways (5, 7, 14). Thus, decreasing the abundance of FLT3 at the RNA and protein levels would be an attractive strategy to treat AMLs with FLT3 overexpression and/or FLT3-ITD/TKD mutations.
MicroRNAs (miRNA) are a class of small, non-coding RNAs that play important roles in post-transcriptional gene regulation (15). We recently reported that miR-150 functions as a pivotal tumor-suppressor gatekeeper in MLL-rearranged and other subtypes of AML, through targeting FLT3, MYB, HOXA9 and MEIS1 directly or indirectly (16). We showed that MLL-fusion proteins and MYC/LIN28 up-regulated FLT3 level through inhibiting the maturation of miR-150 (16). These findings strongly suggest a significant clinical potential of restoration of miR-150 expression and function in treating FLT3-overexpressing AML.
However, successful clinical implementation of miRNA-based therapy requires a highly specific and efficient delivery system (17, 18). Viral delivery systems or conventional non-viral systems such as cholesterol conjugates and cationic liposome packaging have often shown severe toxicity and induced hypersensitive reactions in vivo (17). Poly(amidoamine) (PAMAM) dendrimers have been shown to be effective non-viral gene delivery vectors (19, 20). Their spherical, chemically well-defined structure, precisely controlled size, multivalency, and capability of forming complexes (dendriplex) with nucleotide oligos make them as a suitable platform for miRNA delivery (21–27).
In the present study, we first showed the particular high expression of FLT3 in some subtypes of AML through analyzing a large cohort of AML patients (n=562). We then developed targeted nanoparticles based on FLT3L-guided dendrimers complexed with miR-150 oligos, which exhibited high selectivity and efficiency in targeting leukemic cells with overexpression of FLT3. Our proof-of-principle preclinical animal model studies demonstrate the potent in vivo therapeutic effect of the FLT3L-guided miR-150 nanoparticles in treating FLT3-overexpressing AML, alone or in combination with other therapeutic agents (e.g., JQ1 (28)).
METHODS
Microarrays and data analysis of GSE37642 cohort
The patient samples (n=562) samples were analyzed by use of Affymetrix Human Genome U133Plus2.0 GeneChips or Affymetrix Human Genome U133A and B (U133A+B) GeneChips. The RMA5 method (29) was used for data normalization and the expression values were log(2) transformed. Microarrays and data analyses were conducted as described previously (30, 31). The GEO ID of the entire data set is GSE37642.
Nano-materials
G7 PAMAM dendrimer were obtained from Sigma-Aldrich (St. Louis, MO). N-hydroxysuccinimide ester of cyanine5.5 (NHS-Cy5.5) was purchased from Lumiprobe Corporation (Hallandale Beach, FL). Sulfo-SMCC was obtained from Thermo Fisher Scientific (Rockford, IL). miR-150 and miR-150 mutant RNA oligos (with or without 2’-OMe modification) were purchased from Exiqon Inc. (Woburn, MA). The RNA sequences are: miR-150: 5’-UCU CCC AAC CCU UGU ACC AGU G-3’; miR-150 mutant: 5’-UUA UUU UAC CCU UGU ACC AGU G-3’.
Human recombinant FLT3L proteins were ordered from ProSpec-Tany Technogene Ltd. (East Brunswick, NJ), which contain 155 amino acids of the extracellular domain of FLT3L (i.e., the soluble FLT3L form). The synthetic Flt3L peptide containing 74 amino acids with the sequence of SSNFKVKFRELTDHLLKDYPVTVAVNLQDEKHCKALWSLFLAQRWIEQLKTVAGSK MQTLLEDVNTEIHFVTSC was purchased from Pierce Biotechnology, Inc. (Rockford, IL).
Preparation of G7-FLT3L or G7-Flt3L Dendrimers
G7 PAMAM dendrimers obtained from Sigma-Aldrich were purified and fluorescently labelled using an N-hydroxysuccinimide ester of cyanine5.5 (NHS-Cy5.5) (Lumiprobe Corporation, Hallandale Beach, FL), as has been previously reported (32). Then, human soluble FLT3 protein- or mouse Flt3L peptide-conjugated G7 dendrimers were prepared similarly as previously described (32).
AML samples and cell lines
The patient samples were obtained at the time of diagnosis with informed consent at the University of Chicago Hospital (UCH) or other collaborative hospitals, and were approved by the institutional review board of the institutes/hospitals. All patients were treated according to the protocols of the corresponding institutes/hospitals. All the AML cell lines used herein were originally obtained from ATCC (Manassas, VA) and maintained at the Chen laboratory.
RNA extraction and quantitative RT-PCR
Total RNA was extracted with the miRNeasy extraction kit (Qiagen, Valencia, CA) and was used as template for quantitative RT-PCR (qPCR) analysis as described previously (16).
Cell culture and drug treatment
MONOMAC-6 and U937 cells were cultured as described previously (16, 33). Continued testing to authenticate these cell lines was done using qPCR and Western blot to validate the existence or absence of MLL-AF9 when the cell lines were used in this project. Both cell lines were tested for mycoplasma contamination yearly using a PCR Mycoplasma Test Kit (PromoKine) and were proven to be mycoplasma negative.
BM mononuclear cells from healthy donors or AML patients were grown in RPMI medium 1640 containing 10% FBS, 1% HEPES and 1% penicillin-streptomycin, supplemented with10 ng/mL SCF, TPO, Flt3L, IL-3 and IL-6. Cells were treated with PBS, G7-NH2-miR-150mut, G7-NH2-miR-150, G7-Flt3L-miR-150mut or G7-Flt3L-miR-150 at the indicated doses.
Cell apoptosis, viability and proliferation assays
These experiments were conducted as described previously (16, 33) with ApoLive-Glo Multiplex Assay Kit, or CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay Kit (Promega).
Mouse bone marrow transplantation (BMT) followed with drug treatment
Secondary mouse BMT was carried out as described previously (16). After the onset of leukemia (when mice had an engraftment (CD45.1) of over 20% and/or white blood cell counts higher than 4×109/L, usually 10 days post transplantation), the recipient mice were injected with PBS control, 0.5 mg/kg G7-NH2-miR-150mut, G7-NH2-miR-150, G7-Flt3L-miR-150mut or G7-Flt3L-miR-150, i.v., every other day, until the PBS-treated control group all died of leukemia. For the JQ1 and nanoparticle combination treatment experiment, after the onset of AML, the recipient mice were treated with PBS control, or 50 mg/kg JQ1, i.p., alone, or together with 0.5 mg/kg G7-NH2-miR-150, or G7-Flt3L-miR-150, i.v., every other day, until the PBS-treated control group all died of leukemia.
The maintenance, monitoring, and end-point treatment of mice
All laboratory mice were maintained and monitored as previously described (16).
Statistical software and statistical analyses
The microarray data analysis was conducted by use of Partek Genomics Suite (Partek Inc, St. Louis, MI). The t-test, Kaplan-Meier method, and log-rank test, etc. were performed with WinSTAT (R. Fitch Software), GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA), and/or Partek Genomics Suite (Partek Inc). The P-values less than 0.05 were considered as statistically significant.
RESULTS
Overexpression of FLT3 in AMLs
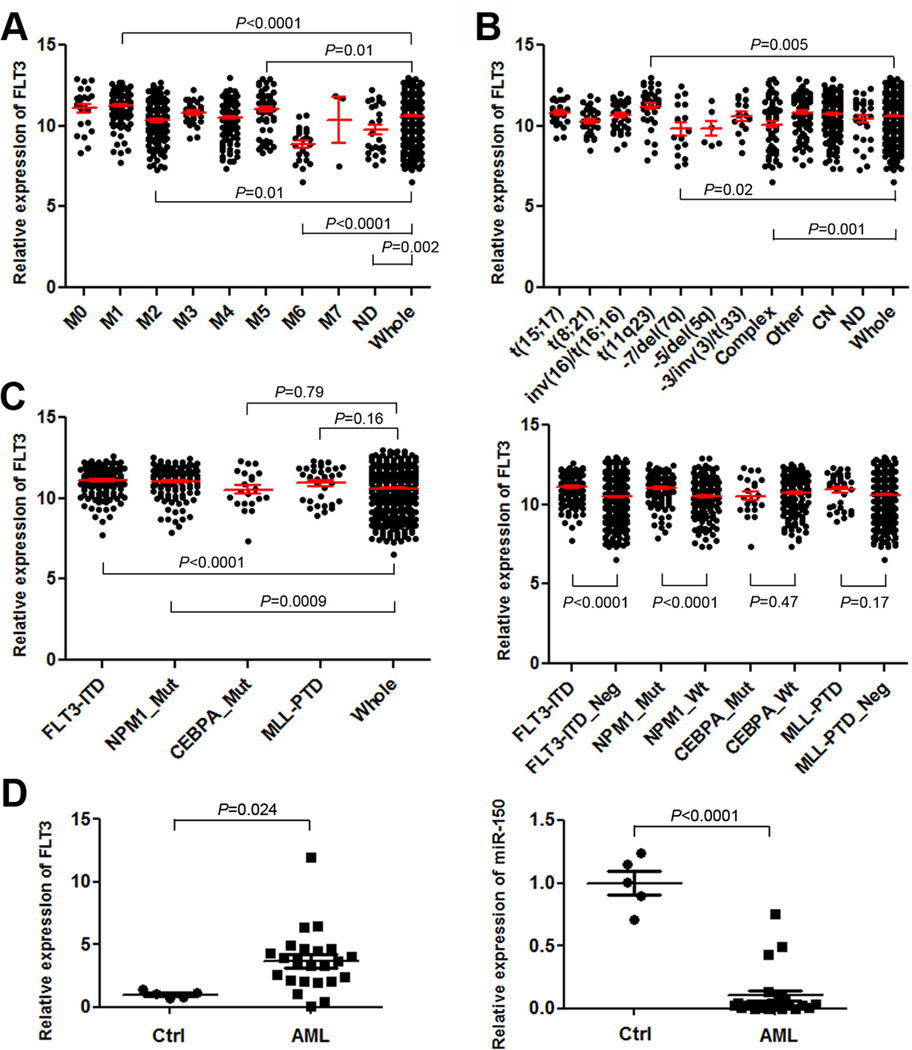
Previous studies, based on relatively small numbers of AML patients (merely 200 patients or fewer), have shown that FLT3 is overexpressed in the majority cases of primary AML relative to normal control cells, whereas controversial observations have been reported regarding the association of FLT3 overexpression with specific AML subtypes (11, 13, 34, 35). Here we analyzed FLT3 expression across various AML subtypes in a microarray-based gene-expression dataset of 562 AML patients (GSE37642) (Supplemental Table 1) (30, 31). Amongst various FAB subtypes, the expression level of FLT3 in the FAB M1, M2 and M5 AML subgroups was significantly higher than its average level in the entire set of AML samples (Figure 1A). Amongst AML subtypes with different cytogenetic abnormalities, the average expression level of FLT3 in AML with t(11q23)/MLL-rearrangements was significantly higher than that in the whole set of AML samples, whereas the opposite was true in AML with −7/del(7q) or complex karyotypes (Figure 1B). In AML with distinct molecular abnormalities, the average expression level of FLT3 in AML with FLT3-ITD and/or NPM1 mutations was significantly higher than that in the whole set of AML or AML without the relevant mutations (Figure 1C). We have also conducted qPCR analysis of expression of both FLT3 and miR-150 in 23 human primary AML samples and 5 normal control samples. As shown in Figure 1D, FLT3 is expressed at a higher level in 21 (91%) out of the 23 AML samples, whereas miR-150 is expressed at a lower level in all the 23 AML samples than its average level in the 5 normal controls.
Figure 1. FLT3 is highly expressed and miR-150 is repressed in AML.
(A–C) The expression patterns of FLT3 across different FAB subtypes (A), cytogenetic subtypes (B) or subtypes with different molecular mutations (C) in the GSE37642 AML cohort. In Figure plots A and B, P-value was shown for the particular AML subtypes in which FLT3 was expressed at a significantly higher or lower level than that in the whole AML set (n=562). In Figure plot C, the P-value was shown for comparison between each individual AML subtype (with a particular type of molecular abnormalities) and the whole AML set (left panel), or between AML cases with and without a particular type of molecular abnormalities (right panel). ND, not determined; complex, complex karyotypes; other, other cytogenetic abnormalities; CN, cytogenetically normal; _Mut, with a particular type of molecular mutations; _Neg, without a particular type of molecular mutations. Note: The expression values were log(2) transformed and normalized by RMA (29). Over 40% of the AML cases with NPM1 mutations also have FLT3-ITD. (D) qPCR analysis of expression of both FLT3 and miR-150 in 23 human primary AML mononuclear (MNC) samples including 3 t(4;11), 1 t(6;11), 9 t(9;11), 1 t(11;19), 3 t(8;21), 1 t(15;17), 3 inv(16), 1 FLT3-ITD, and 1 t(4;11)/FLT3-ITD, along with 5 normal BM cell controls (Ctrl; including 3 MNC and 2 CD34+ samples). The P-values were calculated by two-tailed t-test.
Development of FLT3L-conjugated Generation 7 (G7) PAMAM (G7-FLT3L) dendrimers for selective delivery
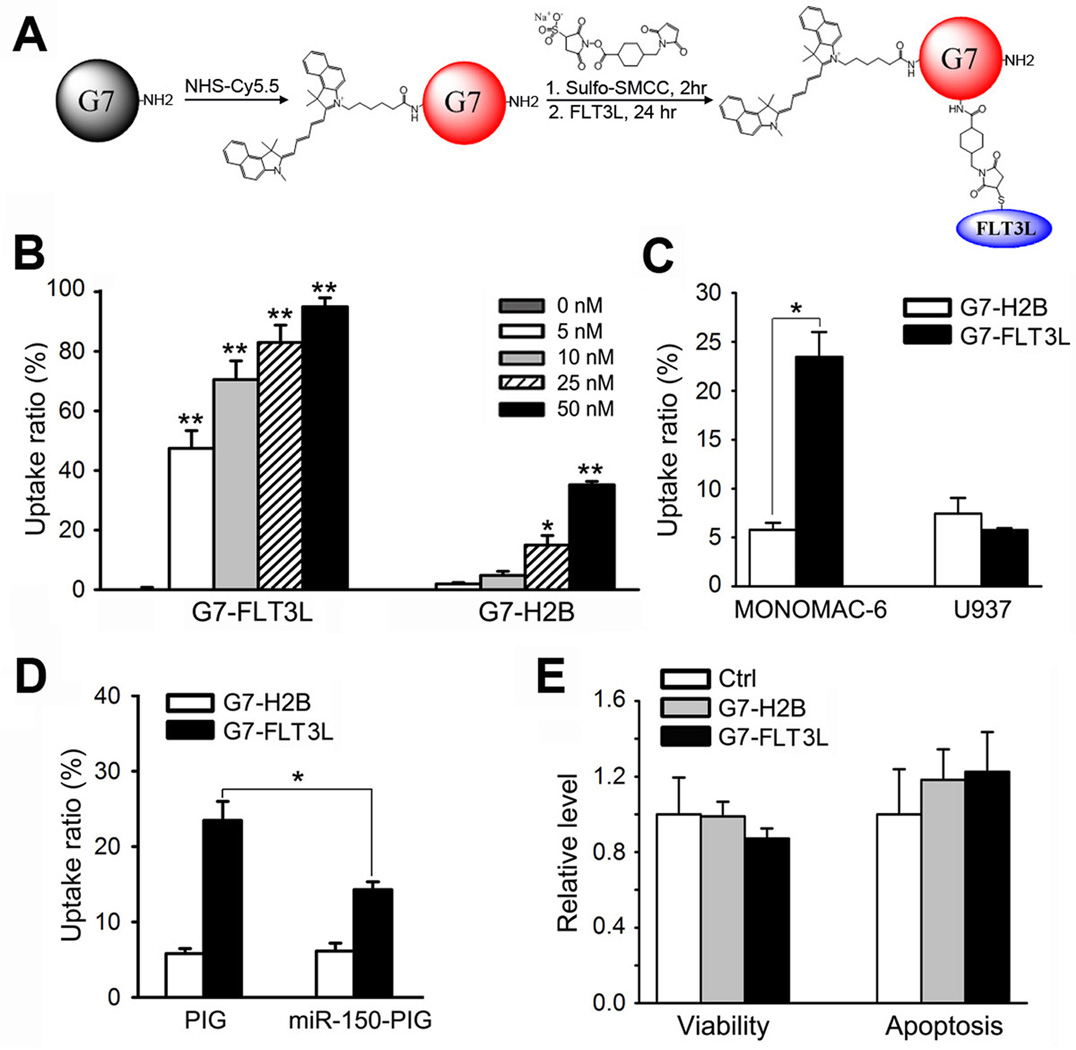
In order to selectively deliver miR-150 oligos to FLT3-overexpressing AML cells, we chose Poly(amidoamine) (PAMAM) dendrimers (21, 32, 36–38) as the basis of our nanoparticle carriers, and conjugated these nanoparticles with the near-infrared dye Cyanine 5.5 (Cy5.5) for monitoring the dynamic distributions of the nanoparticles, and FLT3 ligand (FLT3L) proteins for specific targeting FLT3 on the cell surface. H2B, a nuclear histone protein with similar molecular weight as FLT3L, was conjugated as a negative control (Figure 2A; Supplemental Figure 1A–C).
Figure 2. Development of G7-FLT3L dendrimers and their selective targeting to FLT3-overexpressing AML cells.
(A) G7-NH2 PAMAM dendrimers were first conjugated with the near-infrared dye Cy5.5 using an N-hydroxysuccinimide ester linker sulfo-SMCC. Next, the fluorescently-tagged G7-NH2 (G7-Cy5.5) was conjugated to human recombinant FLT3L (i.e., the soluble FLT3L form with 155 amino acids; ProSpec-Tany Technogene Ltd., East Brunswick, NJ) at a 1:2 ratio using the heterofunctional linker sulfosuccinimidyl 4-((N-maleimidomethyl) cyclohexane-1-carboxylate) (sulfo-SMCC), resulting in the G7-FLT3L conjugates. (B) MONOMAC-6 cells were treated with Cy5.5-conjugated G7-FLT3L or G7-H2B nanoparticles for 24 hrs at the indicated doses. The proportion of Cy5.5+ cells were detected through flow cytometry analysis. (C) MONOMAC-6 and U937 cells were treated with 50 nM Cy5.5-conjugated G7-FLT3L or G7-H2B nanoparticles for 1 hr. The proportion of Cy5.5+ cells were detected through flow cytometry analysis. (D) MONOMAC-6 cells transfected with MSCV-PIG-miR-150 (miR-150-PIG) or MSCV-PIG (PIG) were treated with 50 nM Cy5.5-conjugated G7-FLT3L or G7-H2B nanoparticles for 1 hr. The proportion of Cy5.5+ cells were detected through flow cytometry analysis. Each experiment was repeated independently for at least three times. Average levels of at least three replicates are shown. (E) Cell viability (left panel) and apoptosis (right panel) of MONOMAC-6 cells treated with PBS (Ctrl), 50 nM G7-FLT3L or G7-H2B nanoparticles for 48 hr. *, P<0.05; **, P<0.01.
We then investigated whether G7-FLT3L dendrimers can efficiently and selectively target AML cells with FLT3 overexpression. After 24 hours’ treatment of the nanoparticles, the uptake ratios of the G7-FLT3L dendrimers were significantly higher than the G7-H2B control nanoparticles in MONOMAC-6 cells, an AML cell line carrying the t(9;11)/MLL-AF9 (i.e., the most common form of MLL-rearranged AML (39, 40)). The nanoparticles were taken up in a dose dependent manner (Figure 2B; Supplemental Figure 1D). In addition, the cellular uptake of G7-FLT3L nanoparticles by FLT3-overexpressing AML cells is rapid. Just one hour post the treatment of the nanoparticles, the uptake ratio of G7-FLT3L dendrimers was already 23.6%, significantly higher than that (5.9%) of G7-H2B dendrimers (Figure 2C; Supplemental Figure 1E). In contrast, the uptake ratios between G7-FLT3L and G7-H2B dendrimers showed no significant difference in U937 cells (a cell line with very low levels of FLT3 (16, 41)) (Figure 2C; Supplemental Figure 1E).
Further, we would like to determine whether the high level of FLT3 expression of target cells is required for the high uptake ratio of G7-FLT3L nanoparticles. As reported previously (16), forced expression of miR-150 significantly reduced endogenous expression of FLT3. Suppression of endogenous FLT3 expression in MONOMAC-6 cells by overexpressing miR-150 resulted in a significant decrease of the uptake ratio of G7-FLT3L nanoparticles, but not that of G7-H2B nanoparticles (Figure 2D; Supplemental Figure 1F). Neither G7-FLT3L nor G7-H2B dendrimers showed significant effects on the viability and apoptosis of MONOMAC-6 cells as compared with PBS, indicating low, if any, cytotoxicity of both dendrimer constructs (Figure 2E).
Collectively, the above results indicate that G7-FLT3L nanoparticles can rapidly, efficiently, and selectively target FLT3-overexpressing AML cells, with minimal non-specific cellular toxicity.
G7-FLT3L-miR-150 nanoparticles significantly inhibit viability/proliferation and enhance apoptosis of FLT3-overexpressing AML cells
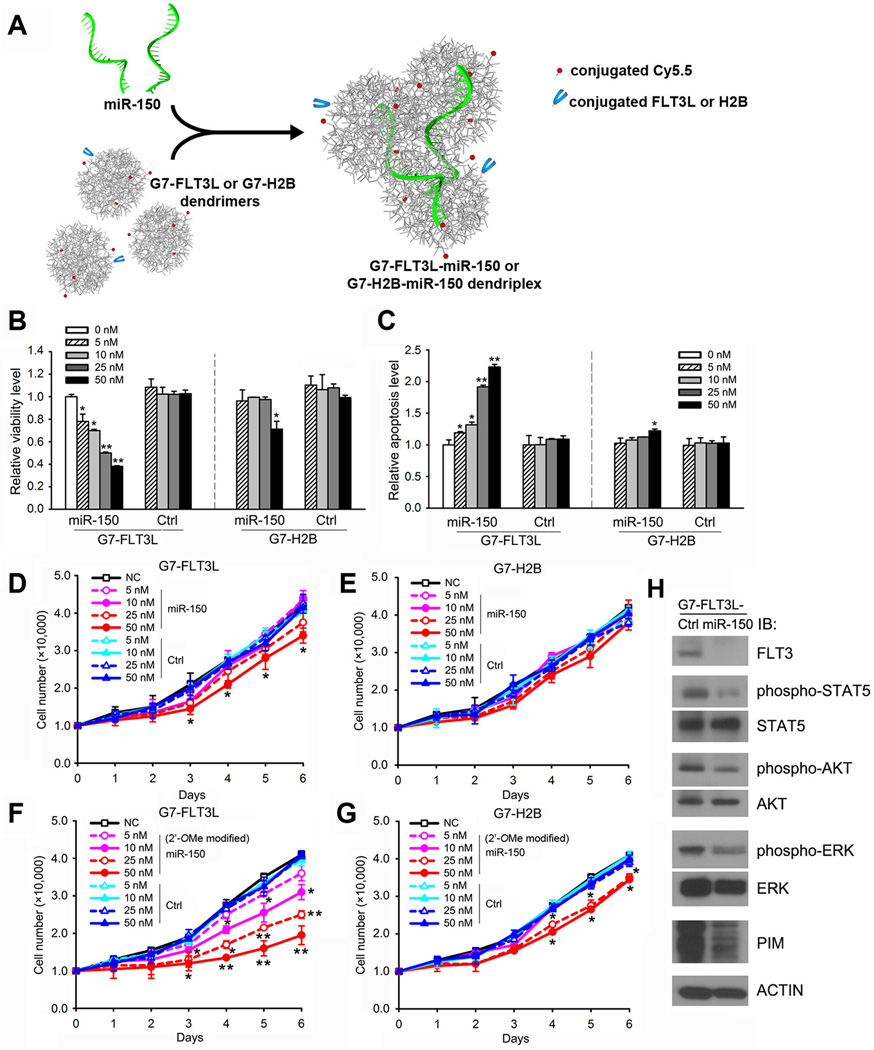
We then integrated miR-150 oligo with G7-FLT3L dendrimers to form G7-FLT3L-miR-150 dendriplexes (Figure 3A; Supplemental Figure 2). G7-FLT3L-miR-150 significantly reduced the viability and increased the apoptosis of MONOMAC-6 cells in a dose-dependent manner, while G7-H2B-miR-150 showed mild effects only at a high concentration (Figure 3B,C). Neither G7-FLT3L-control nor G7-H2B-control carrying miR-150 mutant control oligos (i.e., Control/Ctrl, which has a mutated “seed” sequence (i.e., mutations in the 2nd–6th nucleotides at the 5' end), as a specific loss-of-function control for the wild type miR-150) exhibited significant influence (Figure 3B,C). G7-FLT3L-miR-150 also remarkably inhibited AML cell proliferation even at a dose as low as 10 nM (Figure 3D), while G7-H2B-miR-150 showed no significant inhibitory effects on the cell growth (Figure 3E).
Figure 3. The formation of the G7-FLT3L-miR-150 nanoparticles and their inhibitory effect on MONOMAC-6 cell growth, as well as the FLT3 signaling pathway in vitro.
(A) G7-FLT3L dendrimers were integrated with miR-150 oligos to form stabilized G7-FLT3L-miR-150 dendriplexes. (B, C) MONOMAC-6 cells were treated with G7-FLT3L or G7-H2B nanoparticles complexed with miR-150 or miR-150 mutant RNA (i.e., Control/Ctrl) oligos at the indicated doses. Cell viability (B) or cell apoptosis (C) 48 hrs post treatment are shown. (D, E) The treated cells were counted at indicated time points post-drug treatment. Total cell numbers of G7-FLT3L (D) or G7-H2B (E) treated groups are shown. (F, G) MONOMAC-6 cells were treated with G7-FLT3L or G7-H2B nanoparticles complexed with 2’-OMe-modified miR-150 or miR-150 mutant RNA oligos at the indicated doses. Total cell numbers of G7-FLT3L (F) or G7-H2B (G) treated groups at the indicated time points are shown. *, P<0.05; **, P<0.01. (H) MONOMAC-6 cells were treated with 50 nM G7-FLT3L-miR-150 or G7-FLT3L control (Ctrl) nanoparticles for 72 hours. Levels of FLT3, phosphorylated STAT5 (Y694), STAT5, phosphorylated AKT (S473), AKT, phosphorylated ERK (T202/Y204), ERK, PIM and ACTIN were determined by Western blotting.
However, the major problem with the G7-FLT3L-miR-150 nanoparticles is that their inhibitory effect on cell growth is not sustained. Cells treated with even the highest dose of G7-FLT3L-miR-150 tend to grow at a normal speed 4 days post-treatment (Figure 3D). To increase the stability of miR-150 oligos, we incorporated a 2’-o-methyl (2’-OMe) modification (42) into our miRNA oligos. Indeed, the G7-FLT3L nanoparticles carrying modified miR-150 exhibited a more sustained inhibition on cell growth (Figure 3F). G7-H2B nanoparticles carrying 2’-OMe modified miR-150 showed a slight inhibitory effect at higher doses (Figure 3G). Because 2’-OMe modified miR-150 oligos exhibited a more stable and potent effect than unmodified oligos, we used modified oligos in all our following studies.
The increase of intracellular miR-150 level in cells treated with G7-FLT3L-miR-150 resulted in a significant down-regulation of miR-150’s direct or indirect target genes, including FLT3, MYB, HOXA9 and MEIS1 (16) (Supplemental Figure 3), as well as a significant repression on FLT3 level, which in turn, led to the suppression of the FLT3 signaling pathway (5, 7, 14, 43), featured with reduced activation of ERK, AKT and STAT5, and down-regulation of PIM (Figure 3H).
Therapeutic potential of G7-Flt3L-miR-150 nanoparticles in vivo
We then investigated the potential anti-leukemia effect of G7-Flt3L-miR-150 nanoparticles in treating AML in vivo. To reduce the cost and the size of the guiding molecule, we used synthetic peptides containing the major functional domains of soluble Flt3L protein rather than the entire soluble FLT3L (see Supplemental Figure 4A). The new G7-Flt3L dendrimers exhibited a similar high uptake ratio and similar effects on inhibiting viability and promoting apoptosis (Supplemental Figure 4B,C,D) as compared with the original G7-FLT3L dendrimers (Figure 2B; Figure 3B,C).
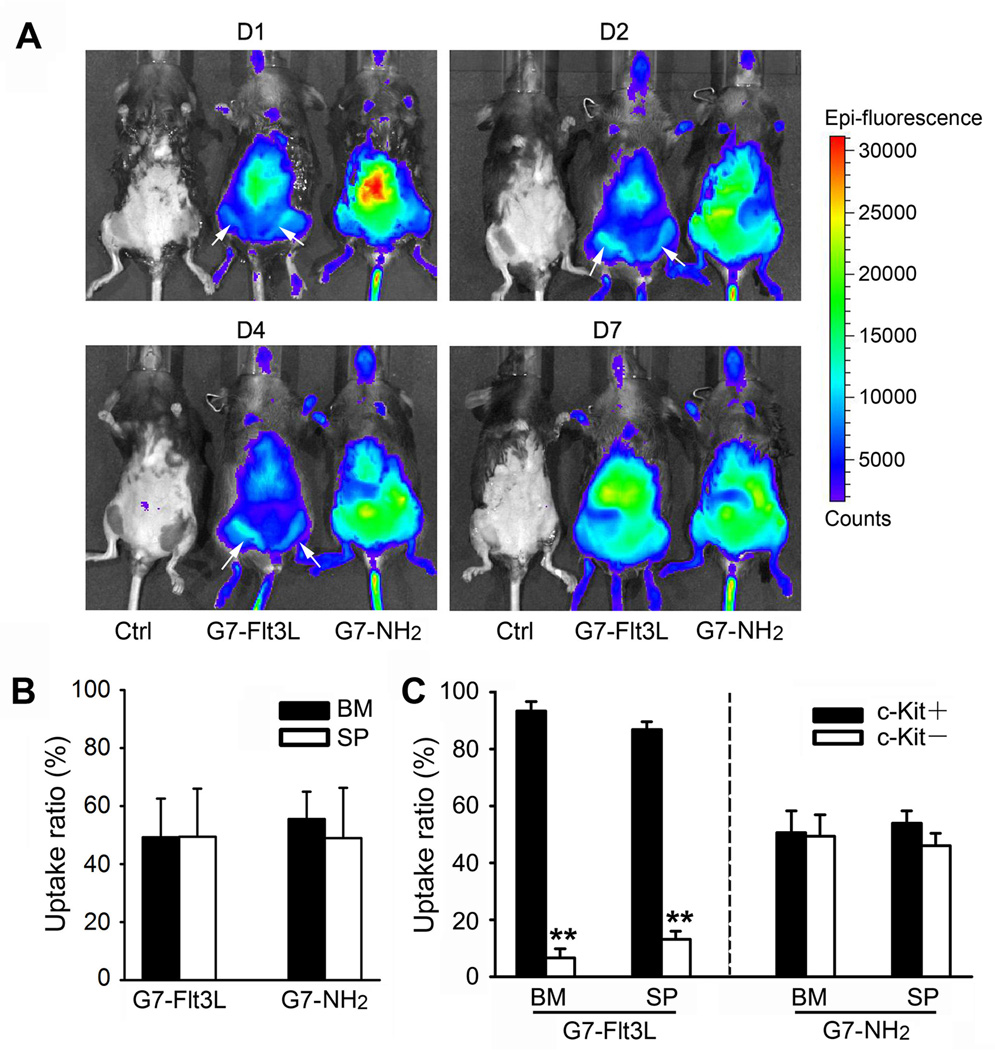
To show the in vivo distribution of the nanoparticles, we injected the G7-Flt3L or control nanoparticles into normal mice. Whole animal imaging shows different distributions of the two nanoparticles: G7-Flt3L was mainly enriched in the bone marrow and the liver, while G7-NH2 showed a more diffuse distribution all over the abdomen (Figure 4A). Both nanoparticles were taken up at a similar ratio in the bone marrow and the spleen of the injected mice, but more G7-Flt3L+ cells are c-Kit+ progenitor cells, indicating a preferential recruitment of the G7-Flt3L nanoparticles to leukemic stem cells or progenitors with overexpressed Flt3 (Figure 4B,C). The level of miR-150 was detected in mouse BM c-Kit+ cells from 30 minutes to 48 hours after a single dosage of G7-Flt3L-miR-150. Results showed that the cellular level of miR-150 was maintained at 2.01 folds of the control group 24 hours post-treatment, and at 1.32 folds 48 hours post-treatment (Supplemental Figure 5A).
Figure 4. In vivo distribution of the G7-Flt3L or G7-NH2.
(A) C57BL/6 wild type mice were treated with 0.5 mg/kg Cy5.5-conjugated G7-Flt3L or G7-NH2, i.v., once. Whole animal images at the indicated time points are shown (D1 = day 1, etc.). The white arrows indicate the location of femur bone marrow (BM). (B) Uptake ratio of Cy5.5-conjugated G7-Flt3L or G7- NH2 in mouse BM and spleen (SP) cells. Animals were sacrificed 7 days post treatment, and the Cy5.5+ ratio was determined through flow cytometry analysis. (C) Uptake ratio of Cy5.5-conjugated G7-Flt3L or G7-NH2 in mouse BM and SP c-Kit+ or c-Kit− cells. Cy5.5+ ratio in c-Kit+ or c-Kit− cell populations is shown. Each experiment was repeated independently for at least three times. Average levels of at least three replicates are shown. *, P<0.05.
We further investigated the in vivo therapeutic effects of the miR-150 nanoparticles in a MLL-rearranged leukemic mouse model. G7-Flt3L-miR-150 nanoparticles showed the best therapeutic effect as compared with the control group or the G7-NH2-miR-150 treated group, while miR-150-mutant nanoparticles showed no anti-leukemia effect (Figure 5A). Notably, G7-Flt3L-miR-150 treatment almost completely blocked MLL-AF9-induced leukemia in 20% of the mice. The therapeutic effect of G7-Flt3L-miR-150 was also evidenced by the morphological changes in peripheral blood (PB), bone marrow (BM), spleen and liver between the treated mice and control AML mice (Figure 5C). The anti-leukemia effect of G7-Flt3L-miR-150 nanoparticles is associated with a significant increase in intracellular miR-150 level and decrease in the expression of miR-150’s critical direct targets (e.g., Flt3 and Myb) and indirect targets (e.g., Hoxa9 and Meis1) (Supplemental Figure 5B). Furthermore, neither G7-Flt3L-miR-150 nor G7-NH2-miR-150 nanoparticles exhibited noticeable effects on normal hematopoiesis (Supplemental Table 2; Supplemental Figure 6).
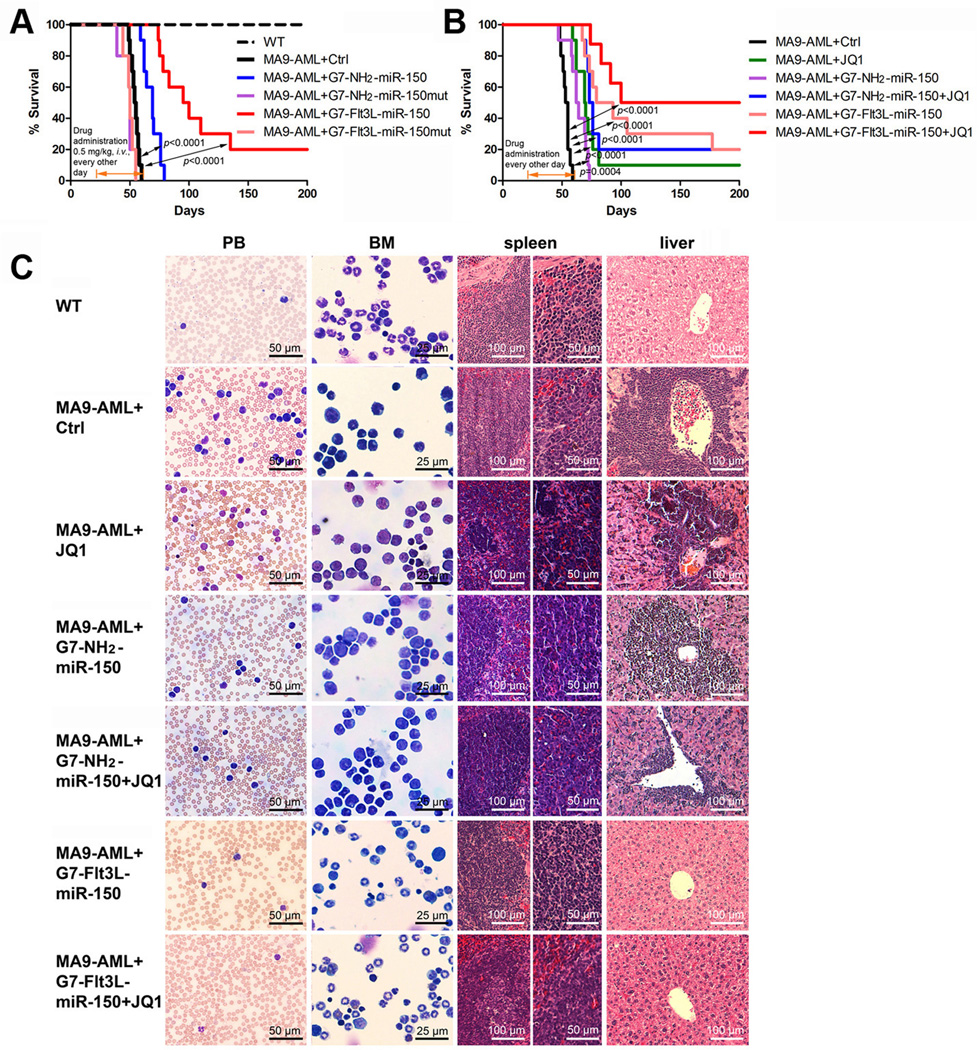
Figure 5. Therapeutic effect of G7-Flt3L-miR-150 nanoparticles in treating AML.
(A) Secondary BMT recipient mice transplanted with primary MLL-AF9 AML cells were treated with nanoparticles (G7-NH2-miR-150mut, G7-NH2-miR-150, G7-Flt3L-miR-150mut or G7-Flt3L-miR-150) or PBS control after the onset of AML. The medium survivals of G7-Flt3L-miR-150 group, the control group and the G7-NH2-miR-150 treated group are 86 days, 54 days, and 63 days, respectively. (B) Synergistic therapeutic effect of G7-Flt3L-miR-150 and JQ1. The same MLL-AF9 AML mouse model was employed. Kaplan-Meier curves are shown. G7-NH2-miR-150+JQ1 v.s. JQ1 alone: P=0.2062; G7-Flt3L-miR-150 v.s. JQ1 alone: P=0.0051. All the P values were detected by log-rank test. (C) Wright-Giemsa stained PB and BM, and hematoxylin and eosin (H&E) stained spleen and liver of the MLL-AF9-secondary leukemic mice treated with PBS control, JQ1 and/or miR-150-formulated nanoparticles. Samples were taken at the endpoints or 200 days post-BM transplantation. Representative images are shown.
JQ1 is a small-molecule inhibitor of BET bromodomains and has been proven to be effective in repressing MLL-AF9/NrasG12D-induced AML progression in vivo (28, 44). Because MYC is also a downstream target of FLT3 (6, 16), we sought to investigate whether the combination of miR-150 nanoparticles and JQ1 exhibits a stronger therapeutic effect than each alone. Our animal leukemia model studies showed that JQ1 alone and G7-Flt3L-miR-150 nanoparticles alone significantly (P<0.0001) extended the median survival of MLL-AF9 AML mice from 54 days (Ctrl; PBS treated) to 69.5 and 86 days, respectively, while their combination substantially extended the median survival to >150 days, suggesting a synergistic effect, or at least an additive effect, between JQ1 and miR-150 nanoparticles in treating FLT3-overexpressing AML (Figure 5B). Notably, 50% of the leukemic mice treated with this combination survived for more than 200 days (Figure 5B) and their pathological morphologies in PB, BM, spleen and liver tissues became normal (Figure 5C). JQ1 and G7-Flt3L-miR-150, each alone or in combination, did not cause any noticeable side effects on normal hematopoiesis in vivo (Supplemental Table 3).
The broad anti-leukemic effect of G7-Flt3L-miR-150 nanoparticles on various subtypes of human AMLs
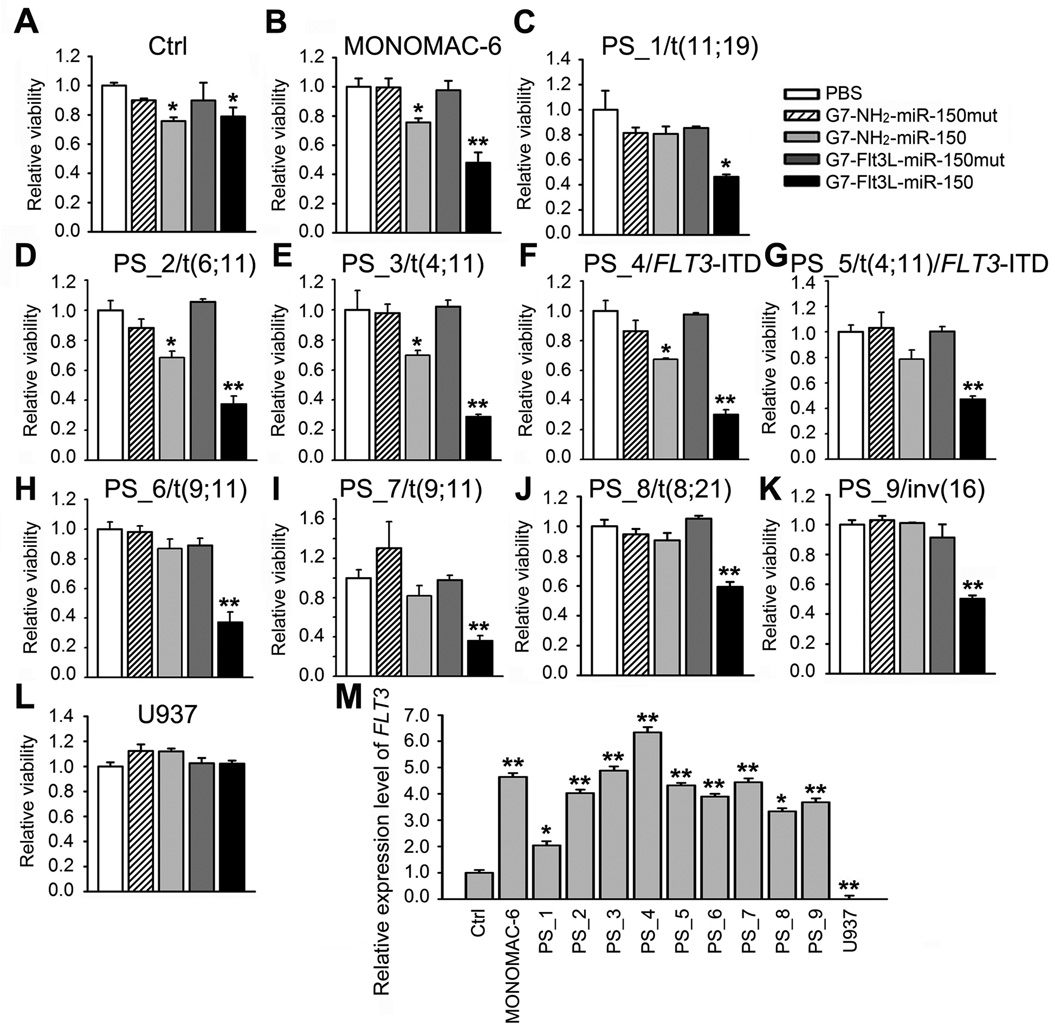
AML cell lines and primary patient samples carrying various cytogenetic or molecular abnormalities were treated with the nanoparticles. While exhibiting very minor inhibitory effect on the viability of normal control mononuclear cells from healthy donors (Control/Ctrl), G7-Flt3L-miR-150 nanoparticles exhibited a significant inhibitory effect on the viability of leukemic mononuclear BM blast cells collected from 9 primary AML patients carrying t(11;19)/MLL-ENL, t(6;11)/MLL-AF6, t(4;11)/MLL-AF4, FLT3-ITD, t(4;11)/FLT3-ITD, t(9;11)/MLL-AF9, t(8;21)/AML1-ETO or inv(16)/CBFB-MYH11, to a degree similar to that of MONOMAC-6 cells, or even better (Figure 6A–K). In contrast, no inhibitory effect was observed on the viability of U937 cells (Figure 6L). The sensitivity of the AML cells to G7-Flt3L-miR-150 nanoparticle treatment seems to be related to the expression level of endogenous FLT3 in the cells (Figure 6M). As expected, only the nanoparticles with both Flt3L guiding peptides and functional miR-150 oligos can exhibit a potent and selective inhibitory effect on FLT3-overexpressing AML cells. Taken together, these results indicate that G7-Flt3L-miR-150 nanoparticles exhibit a broad anti-leukemic effect by selectively targeting a variety of AML subtypes with overexpression of FLT3.
Figure 6. Broad inhibitory effects of G7-Flt3L-miR-150 nanoparticles on AML cell viability.
(A–L) Cell viability of BM mononuclear cells from a healthy donor (A), MONOMAC-6 cells (B), BM mononuclear cells from AML patients bearing t(11;19) (C), t(6;11) (D), t(4;11) (E), FLT3-ITD (F), t(4;11)/FLT3-ITD (G), t(9;11) (H and I), t(8;21) (J), inv(16) (K) and U937 cells (L). Cells were treated with PBS, 50 nM G7-NH2-miR-150mut, G7-NH2-miR-150, G7-Flt3L-miR-150mut or G7-Flt3L-miR-150. Cell viability was tested through MTS assays 48 hrs post treatment. (M) Relative gene expression level of FLT3 in the above samples. *, P<0.05; **, P<0.01.
DISCUSSION
Through analysis of a large cohort of AML patients (n=562), here we provide a more precise understanding of the patterns of FLT3 expression in AML. While FLT3 is likely overexpressed in the vast majority of primary AML cases, it is particularly highly expressed in a set of AML subtypes including FAB M1/M2/M5 AML and AML with t(11q23)/MLL-rearrangements, or FLT3-ITD and/or NPM1 mutations. As AMLs with t(11q23) or FLT3-ITD, accounting for over 40% of total AML cases, are often associated with poor outcome (4, 8–10, 45, 46), it is urgent to develop targeted therapeutic strategies to treat such FLT3-overexpressing, poor-prognosis AMLs.
To selectively target FLT3-overexpressing AMLs, we developed a novel, FLT3L-guided, dendrimer-based nanoparticles for the targeted delivery of miR-150 oligos to FLT3-overexpressing AML cells. Theoretically, our nanoparticle package has the following advantages: (i) It inhibits the growth and proliferation of FLT3-overexpressing AML cells by restoration of the expression and function of a pivotal tumor-suppressor miRNA (i.e., miR-150) that post-transcriptionally inhibits the expression of FLT3 (at the RNA level). The direct repression of FLT3 expression will avoid the potential rebound of FLT3 expression induced by other treatments, e.g., FLT3 TKI treatment (5, 7, 14, 43); (ii) It employs FLT3L as the guiding molecule to specifically deliver miR-150 oligos to FLT3-overexpressing AML cells and thus achieves selective targeting; and (iii) It uses G7 PAMAM dendrimers as the carriers for miRNA delivery. G7 PAMAM dendrimers have a big surface area, flexible architecture, and multivalent binding capability that facilitate tight cell surface binding, and the high density of positive charges on their surface also facilitate interaction with the cell membrane and subsequent cellular uptake (21, 36, 37). Indeed, we have shown that our G7-FLT3L-miR-150 nanoparticles have the above unique advantages and exhibit specific targeting and potent anti-leukemia effects on FLT3-overexpressing AML cells both in vitro and in vivo.
Therefore, our work has made the following conceptual advances and innovative discoveries: (i) Our data demonstrate that restoration of miR-150 expression/function holds a great therapeutic potential in treating AML with FLT3 overexpression. The 2’-OMe-modified miR-150 oligos delivered by targeted nanoparticles show a high stability and exhibit long-term and potent inhibitory effects on the proliferation and growth of FLT3-overexpressing AML cells in vitro and on the progression of the leukemia in vivo; (ii) We provide the first successful model, to our knowledge, showing that an engineered ligand protein/peptide can be used as a guiding molecule when conjugated to a G7 PAMAM-miRNA dendriplex for specific targeting of leukemic cells with an abnormally high level of the corresponding cell surface receptors. We show that both the entire soluble FLT3L proteins (with 155 amino acids) and synthetic Flt3L peptides (with 74 amino acids) work very well as the guiding molecules for specific targeting of FLT3-overexpressing AML cells. Compared to nanoparticles conjugated with control proteins, FLT3L-conjugated nanoparticles showed more than 4 times higher specificity and efficiency in targeting and uptake by FLT3-overexpressing AML cells. Compared to control nanoparticles, Flt3L-conjugated nanoparticles tend to concentrate in the BM rather than distribute pervasively throughout the abdomen following injection, and their accumulation is significantly enhanced in c-Kit+ leukemic progenitor cells in both BM and spleen tissues. This suggests that we might be able to extend the application of the nanoparticle system to other AMLs (e.g., AMLs with abnormally high level of c-KIT) by changing the targeting molecule from FLT3L to other molecules (e.g., c-KIT ligand); (iii) We show that G7 PAMAM dendrimer-based conjugates are ideal carriers for miRNA delivery to treat AML. Such carriers do not trigger any noticeable side effects on normal hematopoiesis in vivo; (iv) Our proof-of-concept leukemia animal model study demonstrated that the G7-Flt3L-miR-150 nanoparticles was able to substantially inhibit leukemia progression and prolong survival of the treated mice that carried FLT3-overexpressing AML, while exhibiting no obvious side effects on normal hematopoietic system. Furthermore, the combination of G7-Flt3L-miR-150 nanoparticles and JQ1 (a bromodomain inhibitor) exhibited a synergistic or at least an additive effect on inhibition of FLT3-overexpressing AML in vivo compared to administration of each agent alone. Notably, the dosages of both G7-Flt3L-miR-150 nanoparticles and JQ1 used in our animal model studies were at a low level (in order to minimize the costs of experiments), and thus a more profound anti-leukemia effect might be observed if higher dosages are used; and (v) Our data demonstrate that specific inhibition of FLT3 expression at the transcript/RNA level by miRNA-based targeted nanoparticles (or other strategies) is a feasible and effective approach to treat FLT3-overexpressing AML.
G7-Flt3L-miR-150 nanoparticles could also be combined with FLT3 TKIs to treat AML carrying FLT3-ITD or TKD mutations, which will inhibit both the expression and enzymatic activity of FLT3 and thereby will likely result in a potent long-term therapeutic effect, as FLT3-TKI-induced rebound of FLT3 level and secondary mutations in FLT3 TKD (5, 7, 14, 43) can be inhibited by G7-Flt3L-miR-150 nanoparticles at the transcript level. With no doubt, our G7-Flt3L-miR-150 nanoparticles can also be combined with other newly developed targeted therapeutic agents (e.g., JQ1), and such combinations may represent promising novel therapeutic strategies with high efficacy and minimal side effects.
In conclusion, our results show that FLT3L-guided nanoparticles complexed with miR-150, a potent tumor suppressing miRNA, exhibit a high specificity and efficiency in targeting FLT3-overexpressing AML. Our proof-of-principle leukemia animal model studies demonstrate the feasibility and effectiveness of our approach using the targeted miR-150 nanoparticles in treating FLT3-overexpressing AML in vivo. Our studies have made a number of conceptual advances and novel discoveries in the fields of nanoparticle design/application and targeted leukemia/cancer therapy as discussed above, and also provided solid basis for the development of new therapeutic strategies for clinical applications in treating FLT3-overexpressing AMLs, the majority of AML cases that are resistant to presently available standard chemotherapies.
Supplementary Material
Acknowledgments
Special thanks to our late mentor and colleague, Dr. Janet Rowley for her long-term support. We thank Drs. Scott Armstrong, Gregory Hannon, and Lin He for providing retroviral constructs. The authors thank all participants and recruiting centers of the Acute Myeloid Leukemia Cooperative Group (AMLCG) trials.
Financial Support: This work was supported by the National Institutes of Health (NIH) R01 Grants CA182528, CA178454, and CA127277 (J. Chen), Alex's Lemonade Stand Foundation for Childhood Cancer (J. Chen); Leukemia & Lymphoma Society (LLS) Translational Research Grant (J. Chen), American Cancer Society (ACS) Research Scholar grant (J. Chen), The University of Chicago Committee on Cancer Biology (CCB) Fellowship Program (X. Jiang), LLS Special Fellowship (Z. Li), Gabrielle’s Angel Foundation for Cancer Research (J. Chen, Z. Li, X. Jiang and H. Huang), the family of Marijanna Kumerich and Leukemia & Blood Cancer New Zealand (S. K. Bohlander).
Abbreviations
- AML
acute myeloid leukemia
- FLT3
FMS-like tyrosine kinase 3
- FLT3L (or FLT3LG)
FLT3 ligand
- FLT3-ITD
internal tandem duplications of FLT3
- TKD
tyrosine kinase domain
- TKI
tyrosine kinase inhibitor
- WBC
white blood cell
- miRNA
microRNA
- G7 PAMAM
generation 7 Poly(amidonamine)
- BM
bone marrow
- BMT
bone marrow transplantation
- PB
peripheral blood
- SP
spleen
- MLL
mixed lineage leukemia
Footnotes
Confliction of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Lyman SD, James L, Vanden Bos T, de Vries P, Brasel K, Gliniak B, et al. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: a proliferative factor for primitive hematopoietic cells. Cell. 1993;75:1157–1167. doi: 10.1016/0092-8674(93)90325-k. [DOI] [PubMed] [Google Scholar]
- 3.Bullinger L, Dohner K, Kranz R, Stirner C, Frohling S, Scholl C, et al. An FLT3 gene-expression signature predicts clinical outcome in normal karyotype AML. Blood. 2008;111:4490–4495. doi: 10.1182/blood-2007-09-115055. [DOI] [PubMed] [Google Scholar]
- 4.Whitman SP, Maharry K, Radmacher MD, Becker H, Mrozek K, Margeson D, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116:3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayser S, Levis MJ. FLT3 tyrosine kinase inhibitors in acute myeloid leukemia: clinical implications and limitations. Leuk Lymphoma. 2014;55:243–255. doi: 10.3109/10428194.2013.800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J Hematol Oncol. 2011;4:13. doi: 10.1186/1756-8722-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung AY, Man CH, Kwong YL. FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia. 2013;27:260–268. doi: 10.1038/leu.2012.195. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 9.Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 10.Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 11.Kuchenbauer F, Kern W, Schoch C, Kohlmann A, Hiddemann W, Haferlach T, et al. Detailed analysis of FLT3 expression levels in acute myeloid leukemia. Haematologica. 2005;90:1617–1625. [PubMed] [Google Scholar]
- 12.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 13.Ozeki K, Kiyoi H, Hirose Y, Iwai M, Ninomiya M, Kodera Y, et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood. 2004;103:1901–1908. doi: 10.1182/blood-2003-06-1845. [DOI] [PubMed] [Google Scholar]
- 14.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 15.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 16.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, et al. Blockade of miR-150 Maturation by MLL-Fusion/MYC/LIN-28 Is Required for MLL-Associated Leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 20.Peer D, Karp JM, Hong S, FaroKHzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 21.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, et al. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport. Bioconjug Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 22.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew Chem. 2011;50:11769–11772. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Sunoqrot S, Stowell C, Ji J, Lee CW, Kim JW, et al. Effect of size, surface charge, and hydrophobicity of poly(amidoamine) dendrimers on their skin penetration. Biomacromolecules. 2012;13:2154–2162. doi: 10.1021/bm300545b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugno J, Hsu HJ, Hong S. Recent advances in targeted drug delivery approaches using dendritic polymers. Biomater Sci-Uk. 2015;3:1025–1034. doi: 10.1039/C4BM00351A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XX, Rocchi P, Qu FQ, Zheng SQ, Liang ZC, Gleave M, et al. PAMAM dendrimers mediate siRNA delivery to target Hsp27 and produce potent antiproliferative effects on prostate cancer cells. ChemMedChem. 2009;4:1302–1310. doi: 10.1002/cmdc.200900076. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Camb) 2006:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 28.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold T, Metzeler KH, Vosberg S, Hartmann L, Rollig C, Stolzel F, et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124:1304–1311. doi: 10.1182/blood-2013-12-540716. [DOI] [PubMed] [Google Scholar]
- 32.Modi DA, Sunoqrot S, Bugno J, Lantvit DD, Hong S, Burdette JE. Targeting of follicle stimulating hormone peptide-conjugated dendrimers to ovarian cancer cells. Nanoscale. 2014;6:2812–2820. doi: 10.1039/c3nr05042d. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;2:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang YG, Liu XH, Liang Y, Jin J. Correlation of FLT3 gene expression level and internal tandem duplication mutation in acute myeloid leukemia and its clinical significance. Zhonghua xue ye xue za zhi. 2008;29:741–745. [PubMed] [Google Scholar]
- 35.Riccioni R, Pelosi E, Riti V, Castelli G, Lo-Coco F, Testa U. Immunophenotypic features of acute myeloid leukaemia patients exhibiting high FLT3 expression not associated with mutations. Br J Haematol. 2011;153:33–42. doi: 10.1111/j.1365-2141.2011.08577.x. [DOI] [PubMed] [Google Scholar]
- 36.Hong S, Leroueil PR, Janus EK, Peters JL, Kober M-M, Islam MT, et al. Interaction of Polycationic Polymers with Supported Lipid Bilayers and Cells: Nanoscale Hole Formation and Enhanced Membrane Permeability. Bioconjugate Chemistry. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 37.Hong S, Rattan R, Majoros IJ, Mullen DG, Peters JL, Shi X, et al. The Role of Ganglioside GM1 in Cellular Internalization Mechanisms of Poly(amidoamine) Dendrimers. Bioconjugate Chemistry. 2009;20:1503–1513. doi: 10.1021/bc900029k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunoqrot S, Bugno J, Lantvit D, Burdette JE, Hong S. Prolonged blood circulation and enhanced tumor accumulation of folate-targeted dendrimer-polymer hybrid nanoparticles. J Control Release. 2014;191:115–122. doi: 10.1016/j.jconrel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 40.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Q, Nishiuchi R, Li Q, Kumar AR, Hudson WA, Kersey JH. FLT3 expressing leukemias are selectively sensitive to inhibitors of the molecular chaperone heat shock protein 90 through destabilization of signal transduction-associated kinases. Clin Cancer Res. 2003;9:4483–4493. [PubMed] [Google Scholar]
- 42.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 43.Daver N, Cortes J, Ravandi F, Patel KP, Burger JA, Konopleva M, et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015;125:3236–3245. doi: 10.1182/blood-2014-10-605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 46.Marcucci G, Mrozek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005;12:68–75. doi: 10.1097/01.moh.0000149608.29685.d1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.