Abstract
Since 2013, a new drugs-of-abuse trend attempts to bypass drug legislation by marketing isomers of scheduled synthetic cannabinoids (SCs), e.g., FUBIMINA (BIM-2201) and THJ-2201. It is much more challenging to confirm a specific isomer’s intake and distinguish it from its structural analog because the isomers and their major metabolites usually have identical molecular weights and display the same product ions. Here, we investigated isomers FUBIMINA and THJ-2201 and propose strategies to distinguish their consumption. THJ-2201 was scheduled in the US, Japan, and Europe; however, FUBIMINA is easily available on the Internet. We previously investigated THJ-2201 metabolism in human hepatocytes, but human FUBIMINA metabolism is unknown. We aim to characterize FUBIMINA metabolism in human hepatocytes, recommend optimal metabolites to confirm its consumption, and propose strategies to distinguish between intakes of FUBIMINA and THJ-2201. FUBIMINA (10 μM) was incubated in human hepatocytes for 3 h, and metabolites were characterized with high-resolution mass spectrometry (HR-MS). We identified 35 metabolites generated by oxidative defluorination, further carboxylation, hydroxylation, dihydrodiol formation, glucuronidation, and their combinations. We recommend 5′-OH-BIM-018 (M34), BIM-018 pentanoic acid (M33), and BIM-018 pentanoic acid dihydrodiol (M7) as FUBIMINA specific metabolites. THJ-2201 produced specific metabolite markers 5′-OH-THJ-018 (F26), THJ-018 pentanoic acid (F25), and hydroxylated THJ-2201 (F13). Optimized chromatographic conditions to achieve different retention times and careful selection of specific product ion spectra enabled differentiation of isomeric metabolites, in this case FUBIMINA from THJ-2201. Our HR-MS approach should be applicable for differentiating future isomeric SCs, which is especially important when different isomers have different legal status.
Keywords: FUBIMINA, BIM-2201, THJ-2201, Synthetic cannabinoid, Hepatocyte metabolism, Isomer
Introduction
Synthetic cannabinoids (SCs) were originally developed as tools to study the endogenous endocannabinoid system [1, 2]. Many SCs are CB1 and CB2 receptors agonists, eliciting greater cannabimimetic effects than Δ9-tetrahydrocannabinol [3, 4]. However, SCs marketed as drugs-of-abuse since the 2000s are an important global public health and safety issue [5]. SC drug abuse can produce significant human toxicity including agitation, seizures, hypertension, emesis, myocardial infarction, and even death [6–9]. For these reasons, many SCs were scheduled across the globe, and many more structural analogs emerged. However, little is known about metabolism of these new SCs, which makes it challenging to document their intake in clinical screenings, complicated by the fact that the parent SCs rarely are found in urine, the most common analytical matrix.
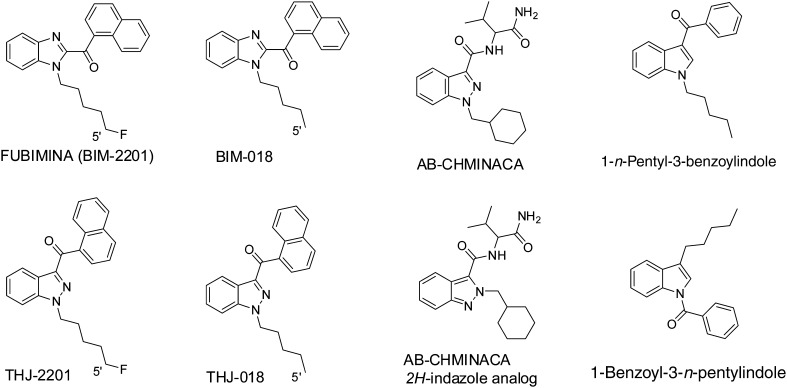
Compared to structural analogs, it is much more challenging to confirm the intake of isomers of scheduled SCs by mass spectrometry (MS) because the SCs and their major metabolites display identical molecular ions and often identical product ions on a mass spectrometer. FUBIMINA (BIM-2201), BIM-018, AB-CHMINACA, and 1-n-pentyl-3-benzoylindole are isomers of THJ-2201, THJ-018, AB-CHMINACA 2H-indazole analog, and 1-benzoyl-3-n-pentylindole, respectively (Fig. 1) [10–12]. In many cases, the isomers endow different scheduling status. For instance, THJ-2201 is scheduled in the US, Japan, and some European countries; however, FUBIMINA, while being scheduled in Japan since August 2014, is currently available over the Internet in the US. Therefore, it is essential to confirm which SC is consumed for the purpose of legal prosecution. Previously, DeRuiter et al. [12] described the differentiation of 1-n-pentyl-3-benzoylindole and 1-benzoyl-3-n-pentylindole. However, the methods are limited to the parent drugs without considering metabolites. Because all previously studied SCs are extensively metabolized, major metabolites information is more important than parents for confirmation of intake.
Fig. 1.
Structures of pairs of isomeric synthetic cannabinoids
FUBIMINA [(1-(5-fluoropentyl)-1H-benzo[d]imidazol-2-yl)(naphthalen-1-yl)methanone] was first detected in 2013 by Uchiyama et al. [11] in herbal-type products purchased on the Internet. FUBIMINA, a potent agonist for the CB2 receptor (Ki = 23.45 nM) [3], and THJ-2201 are fluoro derivatives of BIM-018 and THJ-018, respectively (Fig. 1). FUBIMINA was detected in seized samples since February 2014, according to the United States Army Criminal Investigation Laboratory (unpublished data).
All SCs investigated so far were highly metabolized, with rare detection of the parent SCs in urine, making identification of urinary metabolites essential to document SC intake in this matrix [13, 14]. However, for newly emerging SCs, metabolism data are generally unavailable. We previously determined THJ-2201 metabolism with human hepatocytes and high resolution mass spectrometry (HR-MS) [15]. Currently, the only available FUBIMINA metabolism data were reported by Wiley et al. [3], who focused on FUBIMINA affinity and potency, only mentioning detection of some metabolites. In rat urine, hydroxylation, oxidative defluorination, and carboxylation metabolites were observed. However, these helpful preliminary data also have limitations. Metabolism was in rats, rather than humans; metabolites were mentioned, but no structures were characterized; the relative abundance of metabolites was not provided; and no marker metabolites were recommended for future identification of FUBIMINA intake.
Since 2005, HR-MS has become more widely available for characterizing new drug metabolism due to its many advantages over traditional unit-resolution MS [16], including metabolite identification and bioactivation mechanism elucidation [17–19]. Accurate mass measurement efficiently confirms metabolite parent ion and fragment ion formulas and achieves metabolite identification with improved productivity and quality. Traditional unit-resolution MS requires multiple injections with different acquisition methods by experienced staff [16]; data analysis is time-consuming and less reliable due to the uncertainty of fragment elemental composition.
Our aims were to characterize human FUBIMINA metabolism with human hepatocytes, recommend appropriate metabolites for clinical screening, and propose strategies to distinguish FUBIMINA from THJ-2201 intake. Human hepatocytes contain all required cofactors, enzymes, and an intact membrane, producing ratios of metabolites more closely matched to those in authentic human urine than those produced from human liver microsomes (HLM) [20–22]. Our approach using HLM to determine parent compound half-life and properly design human hepatocyte experiments, human hepatocyte incubation to produce phase I and phase II metabolites followed by HR-MS analysis, and sophisticated data analysis have proved successful in predicting major human urine metabolites for other SCs including AB-PINACA [23], AB-FUBINACA [24], FDU-PB-22, and FUB-PB-22 [14]. The spectral data for major metabolites will be highly useful for clinical scientists to incorporate into their urine screening methods to identify FUBIMINA intake.
Materials and methods
Chemicals and reagents
FUBIMINA (purity 98.42 %) was verified and kindly provided by the Drug Enforcement Administration, Department of Justice, USA. THJ-2201 and NADPH were purchased from Cayman Chemical (Ann Arbor, MI, USA). Cryopreserved human hepatocytes (10-donor pool), GRO CP and GRO KHB (Krebs-Henseleit Buffer) buffer for hepatocytes, and HLM (50-donor pool) were acquired from BioreclamationIVT (Baltimore, MD, USA). Acetonitrile was purchased from Sigma-Aldrich (St. Louis, MO, USA). LC–MS grade water and formic acid were purchased from Fisher Scientific (Waltham, MA, USA).
Metabolic stability of FUBIMINA in human liver microsomes
The HLM metabolic stability assays were performed in the same manner as in our previous manuscripts [25, 26]. Final concentration of FUBIMINA in HLM incubation system was 1 μM. Samples were centrifuged at 15,000g (4 °C, 5 min); supernatant was stored at −80 °C until analysis. When analyzing, the samples were thawed, vortexed, and centrifuged again. Then, 10 μL supernatant was diluted in 990 μL mobile phase A/B (90: 10, v/v). Ten microliters was injected to determine remaining FUBIMINA.
The high-performance liquid chromatography (HPLC) system consisted of two LC-20ADxr pumps, a DGU-20A3R degasser, a SIL-20ACxr autosampler, and a CTO-20A column oven (Shimadzu, Columbia, MD, USA). The Kinetex C18 column (100 mm × 2.1 mm, 2.6 μm; Phenomenex, Torrance, CA, USA) was fitted with a KrudKatcher Ultra HPLC in-line filter (0.5 μm × 0.1 mm, Phenomenex). Mobile phases were 0.1 % formic acid in water (A) and 0.1 % formic acid in acetonitrile (B). The gradient elution started at 10 % B for 0.5 min, ramped to 95 % B at 10 min, then held until 12.5 min before re-equilibrating at 10 % B for 2.5 min. Total run time was 15 min with a flow rate of 0.3 mL/min. Column and autosampler temperatures were 40 and 4 °C, respectively.
Data acquisition was performed on 3200 QTRAP mass spectrometer (Sciex, Redwood City, CA, USA) with Analyst software (version 1.6) in positive electrospray ionization (+ESI) mode. Ion source parameters were as follows: source temperature 500 °C; ion spray voltage, 4000 V; curtain gas, 30 psi; gas 1, 45 psi; gas 2, 70 psi. Two ion transitions were monitored for FUBIMINA (m/z 361 → 155; m/z 361 → 177). Mass parameters were as follows: declustering potential, 61 V; collision energy, 43 eV (m/z 361 → 155) and 35 eV (m/z 361 → 177).
FUBIMINA peak areas were plotted against time, and in vitro microsomal half-life (T1/2) and intrinsic clearance (CLint, micr) were calculated. Microsomal intrinsic clearance was then scaled to whole-liver dimensions, yielding intrinsic clearance (CLint). Human hepatic clearance (CLH) and extraction ratio (ER) were predicated based on CLint without considering plasma protein binding.
Metabolite identification in human hepatocytes
Hepatocyte incubation was carried out as previously described [15, 25, 26]. Cryopreserved human hepatocytes were washed twice with GRO CP medium and GRO KHB buffer. Cell viability was evaluated with Trypan blue (0.4 %, v/v) exclusion method, assuring greater than 80 % viability. FUBIMINA was incubated at a final concentration of 10 μM with human hepatocytes (1 × 106 cells/mL) in a 24-well plate (BD Biosciences, San Jose, CA, USA). Final incubation volume was 500 μL and samples were incubated at 37 °C. Chemical stability of FUBIMINA in the KHB buffer, and hepatocyte incubation without FUBIMINA also were performed (37 °C, 3 h) to determine whether metabolites are dependent on hepatocytes enzymes or are impurities from hepatocyte itself. Diclofenac was incubated as a positive control to ensure metabolic activity of the hepatocytes by monitoring formation of 4′-hydroxydiclofenac and acyl β-d-glucuronide diclofenac. The reactions were quenched by adding 500 μL ice-cold acetonitrile at 0 and 3 h incubation. Samples were stored at −80 °C until analysis. Before analysis, samples were thawed, vortexed thoroughly, and 100 µL acetonitrile added to 100 µL samples. After vortexing and centrifugation at 15,000 g (4 °C, 5 min), supernatant was transferred to a new 10-mL plastic tube, evaporated to dryness under nitrogen at 40 °C, and reconstituted in 150 µL mobile phase A/B (80:20, v/v). Fifteen microliters of the reconstituted solution was injected for metabolite identification.
The HPLC system consisted of two LC-20ADxr pumps, a DGU-20A5R degasser, a SIL-20ACxr autosampler, and a CTO-20 AC column oven (Shimadzu). Chromatographic separation was achieved on an Ultra Biphenyl column (100 × 2.1 mm, 3 μm; Restek, Bellefonte, PA, USA) with a guard column containing the same packing material. Gradient elution was performed with 0.1 % formic acid in water (A) and 0.1 % formic acid in acetonitrile (B) at a flow rate of 0.5 mL/min. Initial gradient conditions were 20 % B, held for 0.5 min; then increased to 95 % B over 10.5 min, held until 13.0 min; and returned to 20 % B at 13.1 min and held until 15.0 min. The column and autosampler were maintained at 30 and 4 °C, respectively.
Data were acquired on a 5600+ TripleTOF MS (Sciex) in +ESI mode. MS data were acquired by information-dependent acquisition (IDA) in combination with multiple mass defect filters (MDF) and dynamic background subtraction (DBS). Ion source parameters were as follows: curtain gas, 45 psi; gas 1, 60 psi; gas 2, 75 psi; source temperature 650 °C; ion spray voltage, 4000 V; declustering potential, 80 V; entrance potential, 10 V. For IDA, spectra exceeding 100 cps were selected for the metabolism-dependent tandem mass spectrometry MS/MS scan, isotopes within 1.5 Da were excluded, mass tolerance was 50 mDa, and collision energy was set to 35 ± 15 eV. The MS was calibrated automatically every three injections.
MetabolitePilot (version 1.5, Sciex) characterized metabolites with different peak-finding algorithms including MDF, predicted biotransformation, generic liquid chromatography (LC) peak finding, common product ion and neutral loss; LC peak intensity threshold was 500 cps, MS 200 cps, and MS/MS 50 cps. Special attention was given to phase II metabolites that were susceptible to in-source fragmentation.
To distinguish FUBIMINA from THJ-2201, THJ-2201 hepatocyte samples incubated in the same procedure were analyzed with the same acquisition and processing methods.
Results and discussion
Metabolic stability evaluation in human liver microsomes
After determining remaining FUBIMINA at different time points, the in vitro T1/2 was calculated to be 4.9 ± 0.03 min, and in vitro CLint, micr was 0.142 mL/min/mg, corresponding to intrinsic clearance (CLint) of 134.1 mL/min/kg after scaling to whole-liver dimensions [27]. Without considering plasma protein binding and with a simplified Rowland’s equation [21, 28], we calculated the predicted human CLH as 17.4 mL/min/kg and ER as 0.87.
Metabolic stability, expressed as in vitro T1/2 and CLint defines a drug’s susceptibility to metabolism. These values facilitate the prediction of in vivo hepatic clearance, in vivo T1/2, and bioavailability [21, 29]. According to criteria proposed by Lave et al. [30], short T1/2, large CLint, and high ER demonstrate that FUBIMINA is a high clearance drug.
Fragmentation of FUBIMINA and THJ-2201
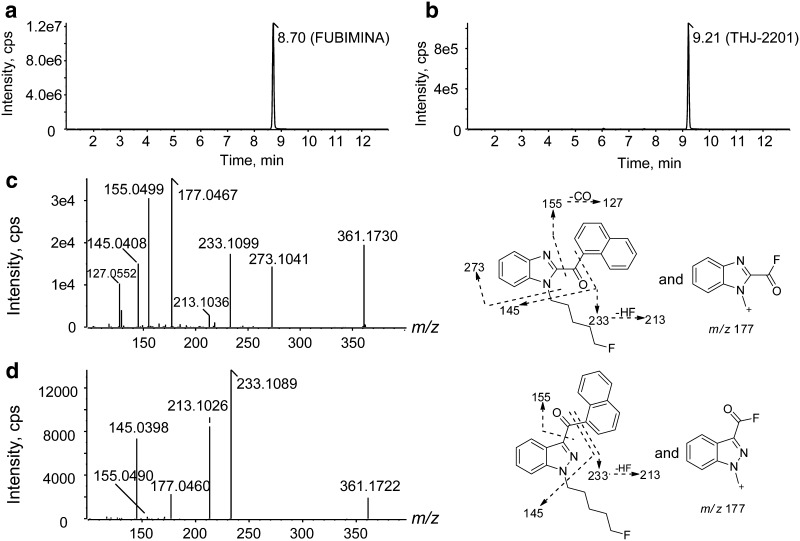
FUBIMINA ([M + H]+, m/z 361.1730) eluted at 8.70 min and displayed characteristic product ions at m/z 273.1041, 233.1099, 213.1036, 177.0467, 155.0499, 145.0408, and 127.0552 (Fig. 2a, c). Fragment m/z 273.1041 was generated via neutral loss of the fluoropentyl chain; m/z 233.1099 and 145.0408 represent the benzimidazole core with and without the fluoropentyl chain. Neutral HF loss from m/z 233.1099 yielded m/z 213.1036; m/z 177.0467 is formed by fluorine rearrangement, and m/z 155.0499 and 127.0552 correspond to the naphthalene moiety with or without the acyl group.
Fig. 2.
Extracted ion chromatograms of FUBIMINA (m/z 361.0730, a) and THJ-2201 (m/z 361.0730, b), their product ion spectra and proposed fragmentation (FUBIMINA, c; THJ-2201, d)
THJ-2201 eluted later at 9.21 min (Fig. 2b), and produced similar fragments (Fig. 2d), but with quite different relative ion abundances, as for m/z 177.0467 and 155.0499. Notably, the prominent fragment m/z 273.1041 in FUBIMINA was not observed in THJ-2201.
FUBIMINA metabolites in human hepatocytes
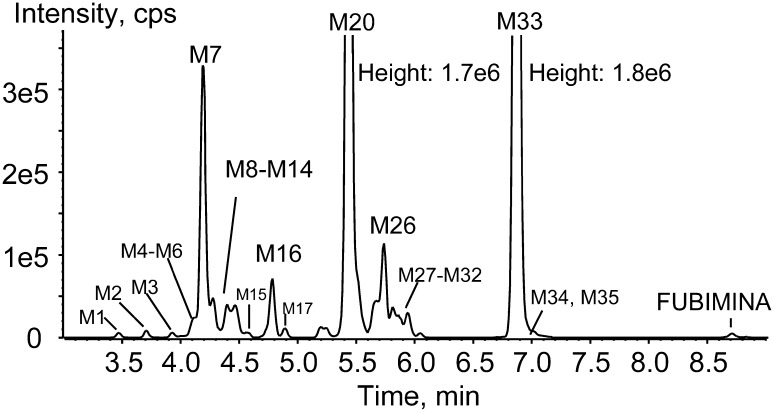
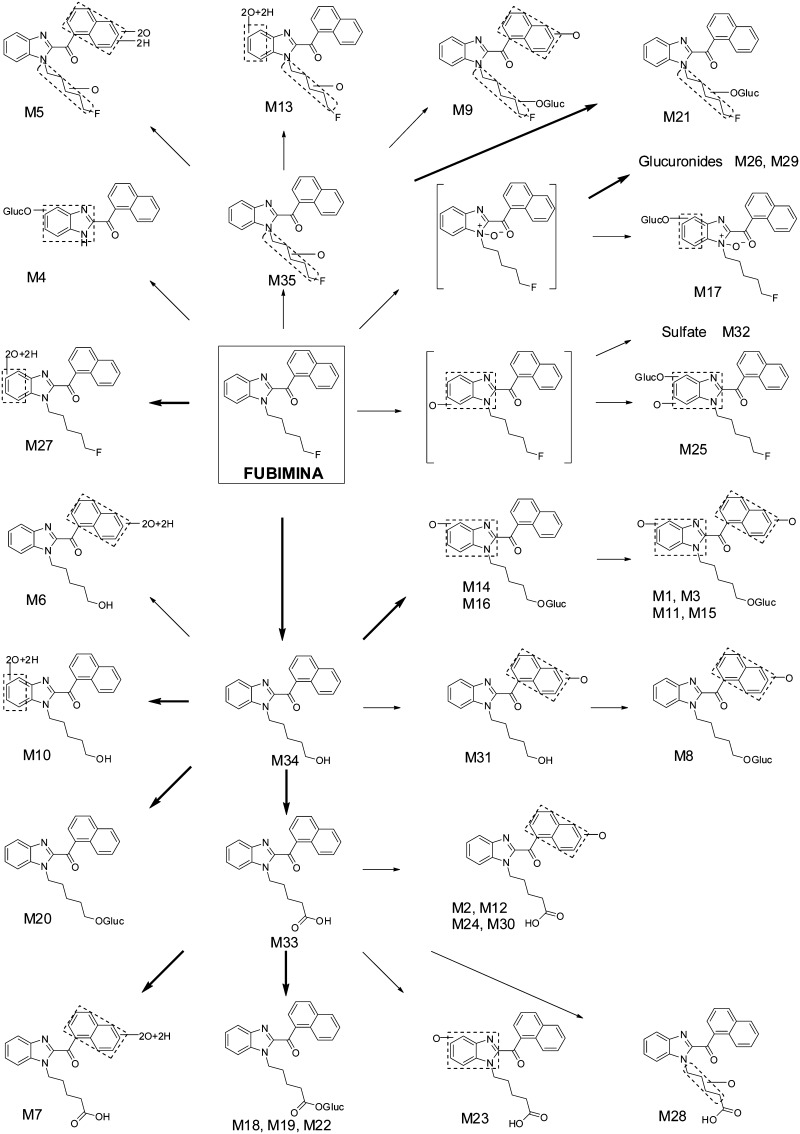
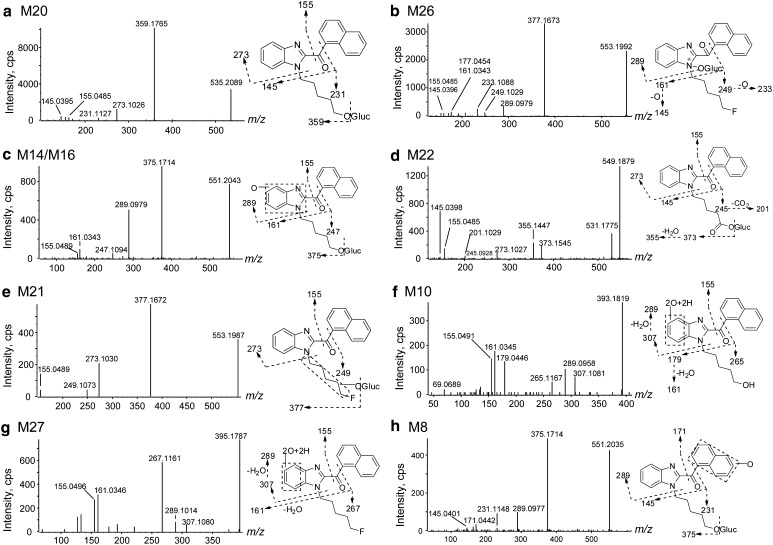
In the 3 h hepatocyte sample, 35 metabolites were detected (Fig. 3; Table 1). No metabolites were observed following FUBIMINA incubation in buffer for 3 h, indicating that metabolite formation was enzyme-dependent. An overview of the metabolic pathway is shown in Fig. 4. Metabolites were labeled “M” in sequential eluting order. Table 1 lists metabolites with pathway description, retention time, m/z, mass error, formula, fragment ions, and peak areas. Based on MS peak areas, primary metabolites were BIM-018 pentanoic acid (M33), 5′-OH-BIM-018 glucuronide (M20), and BIM-018 pentanoic acid dihydrodiol (M7). The top ten major metabolites were elucidated as described below, with product ion spectra shown in Fig. 5.
Fig. 3.
Metabolic profile of FUBIMINA after 3 h incubation in human hepatocytes
Table 1.
Identification of FUBIMINA metabolites after 3 h incubation with human hepatocytes. Fragment ions were expressed in nominal mass because of space restraint
| ID | Metabolic pathway | Time (min) | [M + H]+ (m/z) | Mass error (ppm) | Formula | Fragment ions | Area | Rank |
|---|---|---|---|---|---|---|---|---|
| Parent | 8.72 | 361.1711 | 0.1 | C23H21N2OF | 273, 233, 213, 177, 155, 145, 127 | 2.48E + 04 | NA | |
| M1 | M34 + dioxidation + glucuronidation | 3.47 | 567.1984 | 1.8 | C29H30N2O10 | 391, 305, 247, 229, 171, 161 | 1.83E + 04 | 29 |
| M2 | M33 + oxidation | 3.71 | 389.1505 | 2.3 | C23H20N2O4 | 371, 289, 201, 171, 145 | 2.59E + 04 | 27 |
| M3 | M34 + dioxidation + glucuronidation | 3.92 | 567.1973 | 0.0 | C29H30N2O10 | 391, 247, 171, 161 | 2.12E + 04 | 28 |
| M4 | N-Dealkylation + oxidation + glucuronidation | 4.09 | 465.1298 | 1.2 | C24H20N2O8 | 289, 161, 155 | 1.22E + 04 | 34 |
| M5 | Oxidation + dihydrodiol | 4.10 | 411.1720 | 1.4 | C23H23N2O4F | 393, 375, 289, 271, 189, 171, 145 | 4.27E + 04 | 20 |
| M6 | M34 + dihydrodiol | 4.14 | 393.1825 | 4.0 | C23H24N2O4 | 375, 289, 271, 189, 171, 145 | 4.20E + 04 | 21 |
| M7 | M33 + dihydrodiol | 4.19 | 407.1613 | 2.9 | C23H22N2O5 | 389, 371, 307, 289, 271, 245, 201, 189, 171, 145 | 9.27E + 05 | 3 |
| M8 | M34 + oxidation + glucuronidation | 4.21 | 551.2030 | 1.2 | C29H30N2O9 | 375, 289, 231, 171, 145 | 1.07E + 05 | 11 |
| M9 | Dioxidation + glucurnidation | 4.23 | 569.1932 | 0.4 | C29H29N2O9F | 393, 289, 249, 171, 145 | 6.40E + 04 | 17 |
| M10 | M34 + dihydrodiol | 4.28 | 393.1823 | 3.5 | C23H24N2O4 | 307, 289, 265, 179, 161, 155 | 1.10E + 05 | 9 |
| M11 | M34 + dioxidation + glucuronidation | 4.36 | 567.1978 | 0.9 | C29H30N2O10 | 391, 305, 231 | 1.40E + 04 | 32 |
| M12 | M33 + oxidation | 4.40 | 389.1506 | 2.6 | C23H20N2O4 | 289, 245, 201, 171, 145 | 9.65E + 04 | 12 |
| M13 | Oxidation + dihydrodiol | 4.40 | 411.1711 | −0.8 | C23H23N2O4F | 393, 307, 289, 161, 155 | 3.51E + 04 | 24 |
| M14 | M34 + oxidation + glucuronidation | 4.47 | 551.2033 | 1.5 | C29H30N2O9 | 375, 289, 247, 161 | 1.52E + 05 | 7 |
| M15 | M34 + dioxidation + glucuronidation | 4.57 | 567.1973 | 0.0 | C29H30N2O10 | 391, 305 | 1.75E + 04 | 30 |
| M16 | M34 + oxidation + glucuronidation | 4.78 | 551.2033 | 1.6 | C29H30N2O9 | 375, 289, 247, 161, 155 | 2.37E + 05 | 5 |
| M17 | Dioxidation + glucuronidation | 4.89 | 569.1935 | 0.9 | C29H29N2O9F | 393, 289, 249, 193, 161 | 3.58E + 04 | 23 |
| M18 | M33 + glucuronidation | 5.20 | 549.1876 | 1.5 | C29H28N2O9 | 531, 373, 355, 273, 201, 155 | 6.98E + 04 | 14 |
| M19 | M33 + glucuronidation | 5.39 | 549.1876 | 1.6 | C29H28N2O9 | 531, 373, 355, 273, 201, 155 | 6.59E + 04 | 16 |
| M20 | M34 + glucuronidation | 5.43 | 535.2084 | 1.7 | C29H30N2O8 | 359, 273, 231, 155, 145 | 4.53E + 06 | 2 |
| M21 | Oxidation + glucuronidation | 5.45 | 553.1985 | 0.7 | C29H29N2O8F | 377, 273, 249, 155 | 1.26E + 05 | 8 |
| M22 | M33 + glucuronidation | 5.51 | 549.1876 | 1.5 | C29H28N2O9 | 531, 373, 355, 273, 245, 201, 155, 145 | 2.35E + 05 | 6 |
| M23 | M33 + oxidation | 5.56 | 389.1500 | 1.1 | C23H20N2O4 | 289, 261, 217, 161, 155 | 2.88E + 04 | 25 |
| M24 | M33 + oxidation | 5.66 | 389.1507 | 2.9 | C23H20N2O4 | 289, 201, 171, 145 | 6.07E + 04 | 19 |
| M25 | Dioxidation + glucuronidation | 5.67 | 569.1935 | 0.9 | C29H29N2O9F | 393, 305, 265, 177 | 6.28E + 04 | 18 |
| M26 | N-Oxide + glucuronidation | 5.74 | 553.1992 | 2.1 | C29H29N2O8F | 377, 289, 249, 233, 177, 161 | 4.11E + 05 | 4 |
| M27 | Dihydrodiol | 5.81 | 395.1773 | 2.0 | C23H23N2O3F | 307, 289, 267, 161, 155 | 1.07E + 05 | 10 |
| M28 | M33 + oxidation | 5.84 | 389.1505 | 2.3 | C23H20N2O4 | 273, 217, 155, 145, 127 | 2.70E + 04 | 26 |
| M29 | N-Oxide + glucuronidation | 5.86 | 553.1990 | 1.6 | C29H29N2O8F | 377, 289, 249, 207, 193, 177, 161, 145 | 6.82E + 04 | 15 |
| M30 | M33 + oxidation | 5.93 | 389.1502 | 1.5 | C23H20N2O4 | 289, 201, 171, 145 | 8.81E + 04 | 13 |
| M31 | M34 + oxidation | 5.96 | 375.1712 | 2.4 | C23H22N2O3 | 289, 231, 171, 145 | 1.24E + 04 | 33 |
| M32 | Oxidation + sulfation | 6.04 | 457.1236 | 1.8 | C23H21N2O5FS | 377, 249, 193, 161, 155 | 1.52E + 04 | 31 |
| M33 | Oxidative defluorination to carboxylic acid | 6.87 | 373.1558 | 3.0 | C23H20N2O3 | 273, 245, 201, 155, 145, 127 | 5.62E + 06 | 1 |
| M34 | Oxidative defluorination | 6.92 | 359.1756 | 0.6 | C23H22N2O2 | 273, 231, 155, 145, 127 | 3.87E + 04 | 22 |
| M35 | Oxidation | 7.01 | 377.1661 | 0.4 | C23H21N2O2 | 273, 249, 231, 155, 145, 127 | 1.15E + 04 | 35 |
NA not applicable
Fig. 4.
Tentative metabolic pathways of FUBIMINA in human hepatocytes
Fig. 5.
FUBIMINA major metabolites, product ion spectra, proposed structures, and fragmentation patterns (M7 and M33 are shown in Fig. 6)
Oxidative defluorination and further oxidation plus glucuronidation
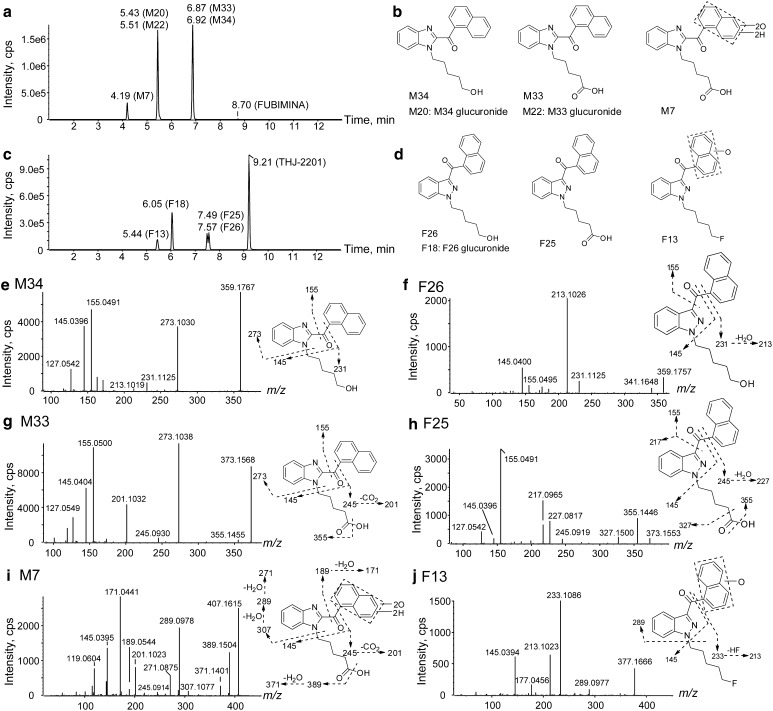
For M34, precursor m/z 359.1767 and fragment m/z 231.1125 were 1.9963 Da (–F + OH) less than the corresponding ions of FUBIMINA (Fig. 6e), indicating fluorine replacement by a hydroxyl group. Other fragments m/z 213.1019, 155.0491, 145.0396, and 127.0542 were the same as for FUBIMINA, indicating that the benzimidazole and naphthalene moieties were unmodified. Therefore, M34 was proposed as the oxidative defluorination metabolite, i.e., 5′-OH-BIM-018. Oxidative defluorination also was the primary metabolic pathway of THJ-2201 [15]. M34 was easily glucuronidated to yield 5′-OH-BIM-018 glucuronide (M20) (Fig. 5a), the second most abundant FUBIMINA metabolite. M20 fragment m/z 359.1765 was formed via neutral glucuronide loss (−176.0324 Da). Other fragments, m/z 273.1026, 231.1127, 155.0485, and 145.0395, were the same as for M34.
Fig. 6.
Profiles of marker metabolites for FUBIMINA (a) and THJ-2201 (c); structures of three metabolite markers for FUBIMINA (b) and THJ-2201 (d), and their product ion spectra and proposed fragmentation (e M34; f F26; g M33; h F25; i M7; j F13)
The predominant M34 metabolic pathway was oxidation to BIM-018 pentanoic acid (M33, Fig. 6g). M33 generated fragments at m/z 273.1038, 245.0930, 201.1032, 155.0500, 145.0404, and 127.0549. Among them, m/z 273.1038, 155.0500, 145.0404, and 127.0549 also were present in FUBIMINA, indicating naphthalene and benzimidazole moieties were intact. The m/z 201.1032 was formed via neutral CO2 loss (−43.9898 Da) from m/z 245.0930. Therefore, M33 was tentatively assigned as BIM-018 pentanoic acid. Subsequent glucuronidation of M33 led to M18/M19/M22 that shared similar product ion spectra, all of them displaying neutral loss of glucuronide (Fig. 5d). M18/M19/M22 appeared in a cluster probably because of acyl glucuronide migration [31].
M34 also underwent benzimidazole hydroxylation and glucuronidation to M14/M16. Their major fragment, m/z 375.1714 was generated via loss of glucuronide (Fig. 5c). Fragments m/z 289.0979 and 161.0343 were 15.9947 Da (+O) larger than corresponding M34 ions, suggesting benzimidazole hydroxylation. Glucuronidation may also occur on the benzimidazole hydroxyl group.
M34 also underwent dihydrodiol formation yielding M10. For M10 (Fig. 5f), fragments m/z 307.1081 and 179.0446 were 34.0051 Da (+2O + 2H) larger than M34 corresponding ions, indicating dihydrodiol formation on benzimidazole. Dihydrodiol ions lost water easily within the MS collision cell, generating ions at m/z 289.0957 and 161.0345. Similarly, M33 (BIM-018 pentanoic acid) produced a dihydrodiol metabolite (M7), the 3rd largest metabolite. M7 shared fragments m/z 245.0914, 201.1023, and 145.0395 with M33 (Fig. 6i); fragments m/z 307.1077 and 189.0544 were 34.0039 Da larger than corresponding ions of M33, suggesting naphthalene dihydrodiol formation. Additionally, FUBIMINA itself underwent dihydrodiol formation yielding M27 (Fig. 5g). It shared fragment m/z 155.0496 with FUBIMINA; fragments m/z 307.1080 and 267.1161 were 34.0039 Da larger than m/z 273.1041 and 233.1099 of FUBIMINA, respectively, suggesting dihydrodiol formation on the benzimidazole.
Fluoropentyl chain hydroxylation and further conjugation
Fluoropentyl chain hydroxylation led to M35. M35 shared fragments m/z 273.1024, 155.0487, 145.0393, and 127.0543 with FUBIMINA. M35 fragment m/z 249.1038 was 15.9939 Da (+O) larger than m/z 233.1099 of FUBIMINA, indicating fluoropentyl chain hydroxylation. The exact position of hydroxylation cannot be determined by MS. Sequential glucuronidation generated M21 (Fig. 5e).
N-Oxide and glucuronidation
Similar to THJ-2201 [15], an unusual pathway for SC, i.e., N-oxide glucuronidation was observed for FUBIMINA generating M26 and M29. Their most intense fragment m/z 377.1673 was generated via glucuronide loss (Fig. 5b). Both metabolites shared product ions m/z 233.1088, 177.0454, 155.0485, and 145.0396 with FUBIMINA, suggesting naphthalene, benzimidazole, and fluoropentyl chain were unmodified. However, fragments m/z 289.0979, 249.1029, and 161.0343 indicate benzimidazole oxidation. Therefore, we propose M26/M29 as N-oxide glucuronides rather than glucuronides of hydroxylated metabolites. Generally, neutral H2O loss, instead of O, was observed between fragments of hydroxylated metabolites, whereas it is common for N-oxides to lose O or an OH radical [32, 33]. This unusual biotransformation dominated metabolism of an antidepressant vortioxetine [32], and anticonvulsants GI265080 and GW273293 [34]. FUBIMINA N-oxide itself was not observed, probably because it was easily glucuronidated.
THJ-2201 major metabolites under the same condition
THJ-2201 underwent similar metabolism in human hepatocytes as FUBIMINA, including oxidative defluorination, further carboxylation or glucuronidation, hydroxylation on the fluoropentyl chain, and dihydrodiol formation on naphthalene. THJ-2201 primary metabolites were 5′-OH-THJ-018 (F26) and its glucuronide F18, THJ-018 pentanoic acid (F25), and hydroxylated THJ-2201 (F13); their product ion spectra and fragmentation are shown in Fig. 6f, h, j. THJ-2201 metabolite names were taken from our previous manuscript characterizing its metabolic profile [15]. THJ-2201 and FUBIMINA marker metabolites are shown in Fig. 6b, d; notably, there are two isomer pairs (M34 and F26, M33 and F25).
Specific metabolites to distinguish FUBIMINA intake from THJ-2201
Because FUBIMINA and THJ-2201 possess different legal status in different countries, unequivocal confirmation of which SC was taken is essential in forensic cases. In clinical and forensic urine screening tests, urine samples are usually hydrolyzed by β-glucuronidase [13, 35], and major phase I metabolites monitored. Additionally, glucuronide conjugates may degrade spontaneously at room temperature [36, 37]. Therefore, we only propose phase I metabolites or aglycones of dominant phase II metabolites as urinary marker metabolites. This proved successful in documenting intake of AB-FUBINACA [24], AB-PINACA [23], FDU-PB-22 and FUB-PB-22 [14].
Based on our results, the most appropriate marker metabolites to document FUBIMINA intake (Fig. 6a, b) are 5′-OH-BIM-018 (M34), which is derived from the intense M20, BIM-018 pentanoic acid (M33), and BIM-018 pentanoic acid dihydrodiol (M7). For THJ-2201 (Fig. 6c, d), optimal marker metabolites include 5′-OH-THJ-018 (F26), THJ-018 pentanoic acid (F25), and hydroxylated THJ-2201 (F13). The isomers M34 and F26 as well as M33 and F25 elute at different retention times, i.e., 6.92 (M34) versus 7.57 min (F26) and 6.87 (M33) versus 7.49 min (F25) (Fig. 6a, c), and are distinguishable by different product ion spectra. A unique fragment, m/z 273.1030 was always a major fragment in FUBIMINA and its metabolites, whereas it was only a negligible ion in THJ-2201 and its metabolites. For instance, m/z 273.1030 and 155.0491 were the dominant M34 fragments (Fig. 6e) whereas m/z 213.1026 and 145.0400 were the top F26 fragments and m/z 273.1030 was not observed at all (Fig. 6f). In M33, m/z 273.1038 was the most intense fragment; but it is absent in F25. Additionally, presence of both m/z 245.0930 and 201.1032 are characteristic for M33, versus m/z 245.0930, 227.0817, and 217.0965 for F25 (Fig. 6g, h).
Urine is the most common routine matrix for drug screening because of easy accessibility and high drug concentration compared to blood or oral fluid. There are limitations to extrapolating human hepatocyte data to human urine. Extrahepatic metabolism [38], transporter involvement [39], and metabolite enrichment in urine [40] might change relative abundance of urinary metabolites, and thus selection of marker metabolites. Therefore, it is preferable to obtain FUBIMINA and THJ-2201 positive urine case samples, and confirm whether recommended hepatocyte marker metabolites match those present in authentic urine after SC intake. Unfortunately, such samples are currently unavailable despite our efforts to obtain them.
Conclusions
In summary, we characterized FUBIMINA metabolism in human hepatocytes with HR-MS. We recommend 5′-OH-BIM-018 (M34), BIM-018 pentanoic acid (M33), and BIM-018 pentanoic acid dihydrodiol (M7) as the most promising urinary marker metabolites for documenting FUBIMINA intake. Their retention times and unique product ion spectra facilitate differentiation of FUBIMINA from THJ-2201 for clinical and forensic scientists. The data will enable linkage of potential adverse toxicological events to FUBIMINA or THJ-2201, in order that the public can be informed of their toxicity. These data also empower manufacturers to focus their synthesis efforts on optimal metabolites. We suggest workflow to differentiate isomeric SCs as follows: incubate SC isomers in human hepatocytes, analyze incubation samples by HPLC-HR-MS, characterize unique major metabolites’ retention times and fragmentation profile, and confirm with case urine samples. Our strategies are applicable for distinguishing other SC isomers with different legal status.
Acknowledgments
This research is supported by the Intramural Research Program of the National Institute on Drug Abuse, National Institutes of Health. FUBIMINA was generously donated by the Drug Enforcement Administration, Department of Justice, USA.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Castaneto MS, Gorelick DA, Desrosiers NA, Hartman RL, Pirard S, Huestis MA. Synthetic cannabinoids: epidemiology, pharmacodynamics, and clinical implications. Drug Alcohol Depend. 2014;144:12–41. doi: 10.1016/j.drugalcdep.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patton AL, Seely KA, Chimalakonda KC, Tran JP, Trass M, Miranda A, Fantegrossi WE, Kennedy PD, Dobrowolski P, Radominska-Pandya A, McCain KR, James LP, Endres GW, Moran JH. Targeted metabolomic approach for assessing human synthetic cannabinoid exposure and pharmacology. Anal Chem. 2013;85:9390–9399. doi: 10.1021/ac4024704. [DOI] [PubMed] [Google Scholar]
- 3.Wiley JL, Marusich JA, Lefever TW, Antonazzo KR, Wallgren MT, Cortes RA, Patel PR, Grabenauer M, Moore KN, Thomas BF. AB-CHMINACA, AB-PINACA, and FUBIMINA: affinity and potency of novel synthetic cannabinoids in producing Δ9-tetrahydrocannabinol-like effects in mice. J Pharmacol Exp Ther. 2015;354:328–339. doi: 10.1124/jpet.115.225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tait RJ, Caldicott D, Mountain D, Hill SL, Lenton S. A systematic review of adverse events arising from the use of synthetic cannabinoids and their associated treatment. Clin Toxicol. 2016;54:1–13. doi: 10.3109/15563650.2015.1110590. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson SM, Banister SD, Kassiou M. Bioisosteric fluorine in the clandestine design of synthetic cannabinoids. Aust J Chem. 2015;68:4–8. doi: 10.1071/CH14198. [DOI] [Google Scholar]
- 6.Hermanns-Clausen M, Kneisel S, Szabo B, Auwärter V. Acute toxicity due to the confirmed consumption of synthetic cannabinoids: clinical and laboratory findings. Addiction. 2013;108:534–544. doi: 10.1111/j.1360-0443.2012.04078.x. [DOI] [PubMed] [Google Scholar]
- 7.Seely KA, Lapoint J, Moran JH, Fattore L. Spice drugs are more than harmless herbal blends: a review of the pharmacology and toxicology of synthetic cannabinoids. Prog Neuropsychopharmacol Biol Psychiat. 2012;39:234–243. doi: 10.1016/j.pnpbp.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanks KG, Winston D, Heidingsfelder J, Behonick G. Case reports of synthetic cannabinoid XLR-11 associated fatalities. Forensic Sci Int. 2015;252:e6–e9. doi: 10.1016/j.forsciint.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Tse R, Kodur S, Squires B, Collins N. Sudden cardiac death complicating acute myocardial infarction following synthetic cannabinoid use. Intern Med J. 2014;44:934–936. doi: 10.1111/imj.12521. [DOI] [PubMed] [Google Scholar]
- 10.Uchiyama N, Shimokawa Y, Kikura-Hanajiri R, Demizu Y, Goda Y, Hakamatsuka T. A synthetic cannabinoid FDU-NNEI, two 2H-indazole isomers of synthetic cannabinoids AB-CHMINACA and NNEI indazole analog (MN-18), a phenethylamine derivative N-OH-EDMA, and a cathinone derivative dimethoxy-α-PHP, newly identified in illegal products. Forensic Toxicol. 2015;33:244–259. doi: 10.1007/s11419-015-0268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchiyama N, Shimokawa Y, Matsuda S, Kawamura M, Kikura-Hanajiri R, Goda Y. Two new synthetic cannabinoids, AM-2201 benzimidazole analog (FUBIMINA) and (4-methylpiperazin-1-yl)(1-pentyl-1H-indol-3-yl)methanone (MEPIRAPIM), and three phenethylamine derivatives, 25H-NBOMe 3,4,5-trimethoxybenzyl analog, 25B-NBOMe, and 2C-N-NBOMe, identified in illegal products. Forensic Toxicol. 2014;32:105–115. doi: 10.1007/s11419-013-0217-2. [DOI] [Google Scholar]
- 12.DeRuiter J, Smith FT, Abdel-Hay K, Clark CR. Analytical differentiation of 1-alkyl-3-acylindoles and 1-acyl-3-alkylindoles: isomeric synthetic cannabinoids. Anal Chem. 2014;86:3801–3808. doi: 10.1021/ac500316x. [DOI] [PubMed] [Google Scholar]
- 13.Scheidweiler KB, Jarvis MJ, Huestis MA. Nontargeted SWATH acquisition for identifying 47 synthetic cannabinoid metabolites in human urine by liquid chromatography-high-resolution tandem mass spectrometry. Anal Bioanal Chem. 2015;407:883–897. doi: 10.1007/s00216-014-8118-8. [DOI] [PubMed] [Google Scholar]
- 14.Diao X, Scheidweiler KB, Wohlfarth A, Pang S, Kronstrand R, Huestis MA. In vitro and in vivo human metabolism of synthetic cannabinoids FDU-PB-22 and FUB-PB-22. AAPS J. 2016;18:455–464. doi: 10.1208/s12248-016-9867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao X, Wohlfarth A, Pang S, Scheidweiler KB, Huestis MA. High-resolution mass spectrometry for characterizing the metabolism of synthetic cannabinoid THJ-018 and its 5-fluoro analog THJ-2201 after incubation in human hepatocytes. Clin Chem. 2016;62:157–169. doi: 10.1373/clinchem.2015.243535. [DOI] [PubMed] [Google Scholar]
- 16.Wen B, Zhu M. Applications of mass spectrometry in drug metabolism: 50 years of progress. Drug Metab Rev. 2015;47:71–87. doi: 10.3109/03602532.2014.1001029. [DOI] [PubMed] [Google Scholar]
- 17.Gao R, Li L, Xie C, Diao X, Zhong D, Chen X. Metabolism and pharmacokinetics of morinidazole in humans: identification of diastereoisomeric morpholine N+-glucuronides catalyzed by UDP glucuronosyltransferase 1A9. Drug Metab Dispos. 2012;40:556–567. doi: 10.1124/dmd.111.042689. [DOI] [PubMed] [Google Scholar]
- 18.Diao X, Deng P, Xie C, Li X, Zhong D, Zhang Y, Chen X. Metabolism and pharmacokinetics of 3-n-butylphthalide (NBP) in humans: the role of cytochrome P450s and alcohol dehydrogenase in biotransformation. Drug Metab Dispos. 2013;41:430–444. doi: 10.1124/dmd.112.049684. [DOI] [PubMed] [Google Scholar]
- 19.Zhu M, Ma L, Zhang H, Humphreys WG. Detection and structural characterization of glutathione-trapped reactive metabolites using liquid chromatography-high-resolution mass spectrometry and mass defect filtering. Anal Chem. 2007;79:8333–8341. doi: 10.1021/ac071119u. [DOI] [PubMed] [Google Scholar]
- 20.Soars MG, McGinnity DF, Grime K, Riley RJ. The pivotal role of hepatocytes in drug discovery. Chem Biol Interact. 2007;168:2–15. doi: 10.1016/j.cbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Baranczewski P, Stanczak A, Sundberg K, Svensson R, Wallin A, Jansson J, Garberg P, Postlind H. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol Rep. 2006;58:453–472. [PubMed] [Google Scholar]
- 22.Diao X, Pang X, Xie C, Guo Z, Zhong D, Chen X. Bioactivation of 3-n-butylphthalide via sulfation of its major metabolite 3-hydroxy-NBP: mediated mainly by sulfotransferase 1A1. Drug Metab Dispos. 2014;42:774–781. doi: 10.1124/dmd.113.056218. [DOI] [PubMed] [Google Scholar]
- 23.Wohlfarth A, Castaneto MS, Zhu M, Pang S, Scheidweiler KB, Kronstrand R, Huestis MA. Pentylindole/pentylindazole synthetic cannabinoids and their 5-fluoro analogs produce different primary metabolites: metabolite profiling for AB-PINACA and 5F-AB-PINACA. AAPS J. 2015;17:660–677. doi: 10.1208/s12248-015-9721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castaneto MS, Wohlfarth A, Pang S, Zhu M, Scheidweiler KB, Kronstrand R, Huestis MA. Identification of AB-FUBINACA metabolites in human hepatocytes and urine using high-resolution mass spectrometry. Forensic Toxicol. 2015;33:295–310. doi: 10.1007/s11419-015-0275-8. [DOI] [Google Scholar]
- 25.Ellefsen KN, Wohlfarth A, Swortwood MJ, Diao X, Concheiro M, Huestis MA. 4-Methoxy-α-PVP: in silico prediction, metabolic stability, and metabolite identification by human hepatocyte incubation and high-resolution mass spectrometry. Forensic Toxicol. 2016;34:61–75. doi: 10.1007/s11419-015-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swortwood MJ, Carlier J, Ellefsen KN, Wohlfarth A, Diao X, Concheiro-Guisan M, Kronstrand R, Huestis MA. In vitro, in vivo and in silico metabolic profiling of α-pyrrolidinopentiothiophenone, a novel thiophene stimulant. Bioanalysis. 2016;8:65–82. doi: 10.4155/bio.15.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNaney CA, Drexler DM, Hnatyshyn SY, Zvyaga TA, Knipe JO, Belcastro JV, Sanders M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev Technol. 2008;6:121–129. doi: 10.1089/adt.2007.103. [DOI] [PubMed] [Google Scholar]
- 28.Diao XX, Zhong K, Li XL, Zhong DF, Chen XY. Isomer-selective distribution of 3-n-butylphthalide (NBP) hydroxylated metabolites, 3-hydroxy-NBP and 10-hydroxy-NBP, across the rat blood-brain barrier. Acta Pharmacol Sin. 2015;36:1520–1527. doi: 10.1038/aps.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obach RS. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos. 1999;27:1350–1359. [PubMed] [Google Scholar]
- 30.Lave T, Dupin S, Schmitt C, Valles B, Ubeaud G, Chou RC, Jaeck D, Coassolo P. The use of human hepatocytes to select compounds based on their expected hepatic extraction ratios in humans. Pharmaceut Res. 1997;14:152–155. doi: 10.1023/A:1012036324237. [DOI] [PubMed] [Google Scholar]
- 31.Zhong S, Jones R, Lu W, Schadt S, Ottaviani G. A new rapid in vitro assay for assessing reactivity of acyl glucuronides. Drug Metab Dispos. 2015;43:1711–1717. doi: 10.1124/dmd.115.066159. [DOI] [PubMed] [Google Scholar]
- 32.Uldam HK, Juhl M, Pedersen H, Dalgaard L. Biosynthesis and identification of an N-oxide/N-glucuronide metabolite and first synthesis of an N-O-glucuronide metabolite of Lu AA21004. Drug Metab Dispos. 2011;39:2264–2274. doi: 10.1124/dmd.111.040428. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim H, Couderc F, Perio P, Collin F, Nepveu F. Behavior of N-oxide derivatives in atmospheric pressure ionization mass spectrometry. Rapid Commun Mass Spectrom. 2013;27:621–628. doi: 10.1002/rcm.6493. [DOI] [PubMed] [Google Scholar]
- 34.Ismail IM, Dear GJ, Roberts AD, Plumb RS, Ayrtont J, Sweatman BC, Bowers GD. N-O glucuronidation: a major human metabolic pathway in the elimination of two novel anti-convulsant drug candidates. Xenobiotica. 2002;32:29–43. doi: 10.1080/00498250110081623. [DOI] [PubMed] [Google Scholar]
- 35.Freijo TD, Jr, Harris SE, Kala SV. A rapid quantitative method for the analysis of synthetic cannabinoids by liquid chromatography-tandem mass spectrometry. J Anal Toxicol. 2014;38:466–478. doi: 10.1093/jat/bku092. [DOI] [PubMed] [Google Scholar]
- 36.Skopp G, Potsch L. Stability of 11-nor-Δ9-carboxy-tetrahydrocannabinol glucuronide in plasma and urine assessed by liquid chromatography-tandem mass spectrometry. Clin Chem. 2002;48:301–306. [PubMed] [Google Scholar]
- 37.Scheidweiler KB, Schwope DM, Karschner EL, Desrosiers NA, Gorelick DA, Huestis MA. In vitro stability of free and glucuronidated cannabinoids in blood and plasma following controlled smoked cannabis. Clin Chem. 2013;59:1108–1117. doi: 10.1373/clinchem.2012.201467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li XD, Xia SQ, Lv Y, He P, Han J, Wu MC. Conjugation metabolism of acetaminophen and bilirubin in extrahepatic tissues of rats. Life Sci. 2004;74:1307–1315. doi: 10.1016/j.lfs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Gao C, Zhang H, Guo Z, You T, Chen X, Zhong D. Mechanistic studies on the absorption and disposition of scutellarin in humans: selective OATP2B1-mediated hepatic uptake is a likely key determinant for its unique pharmacokinetic characteristics. Drug Metab Dispos. 2012;40:2009–2020. doi: 10.1124/dmd.112.047183. [DOI] [PubMed] [Google Scholar]
- 40.Xie C, Zhou J, Guo Z, Diao X, Gao Z, Zhong D, Jiang H, Zhang L, Chen X. Metabolism and bioactivation of famitinib, a novel inhibitor of receptor tyrosine kinase, in cancer patients. Br J Pharmacol. 2013;168:1687–1706. doi: 10.1111/bph.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]