Abstract
Dated phylogenies of fossil taxa allow palaeobiologists to estimate the timing of major divergences and placement of extinct lineages, and to test macroevolutionary hypotheses. Recently developed Bayesian ‘tip-dating’ methods simultaneously infer and date the branching relationships among fossil taxa, and infer putative ancestral relationships. Using a previously published dataset for extinct theropod dinosaurs, we contrast the dated relationships inferred by several tip-dating approaches and evaluate potential downstream effects on phylogenetic comparative methods. We also compare tip-dating analyses to maximum-parsimony trees time-scaled via alternative a posteriori approaches including via the probabilistic cal3 method. Among tip-dating analyses, we find opposing but strongly supported relationships, despite similarity in inferred ancestors. Overall, tip-dating methods infer divergence dates often millions (or tens of millions) of years older than the earliest stratigraphic appearance of that clade. Model-comparison analyses of the pattern of body-size evolution found that the support for evolutionary mode can vary across and between tree samples from cal3 and tip-dating approaches. These differences suggest that model and software choice in dating analyses can have a substantial impact on the dated phylogenies obtained and broader evolutionary inferences.
Keywords: tip-dating, divergence dates, phylogenetic comparative methods, theropods
1. Introduction
How fossil organisms are related to each other and to living lineages is a matter of interest both to the general public and the scientific community. This matter surpasses systematic placement, because our estimates of branching relationships and their timing have direct implications on macroevolutionary inferences. Few examples are better than Archaeopteryx, which has long caught public attention as a potential early bird, a position questioned by a recent maximum-parsimony phylogenetic analysis [1] but seemingly reaffirmed by a later maximum-likelihood analysis [2].
Parsimony versus model-based phylogenetics is only one great debate in palaeontological systematics: for decades, there has been disagreement about whether to consider stratigraphic occurrences when inferring relationships [3]. Recently, the oft-criticized parsimony-based ‘stratocladistics’ [4] has been reborn as Bayesian ‘tip-dating’ phylogenetics [5], where non-ultrametric time-scaled phylogenies of extinct fossil tip taxa are inferred as a function of both clock-like models of character change and a tree prior, describing the distributions of divergence dates [6,7]. Most recently, these tree priors belong to the birth–death-serial-sampling (BDSS) family of models, which involve both diversification and sampling processes in the fossil record [8]. Tip-dating with BDSS is implemented in Bayesian phylogenetics applications, such as BEAST2 and MrBayes, including allowing for fossil taxa to be considered as potential sampled ancestors [9,10]. Sampled-ancestor BDSS (‘SA-BDSS’, also known as sampled-ancestor-birth–death or fossilized-birth–death) models [11] differ from non-sampled-ancestor BDSS (‘noSA-BDSS’ or transmission birth–death process), where sampling is synchronous with extinction [12]. Fossilization is unlikely to coincide with extinction, and thus noSA-BDSS may be more fitting to pathogen phylogenetics in epidemiology. Additionally, palaeobiologists often use a posteriori time-scaling (APT) to secondarily date existing cladograms of extinct taxa. While some APT methods are arbitrary rescaling algorithms, the cal3 approach probabilistically dates divergences relative to an SA-BDSS variant [13].
The diversity of approaches, models and software that can be used to obtain a fossil-only time-scaled phylogeny calls for an empirical comparison of tip-dating and probabilistic APT methods. We choose to perform such an examination using the matrix from Xu et al. [1], paired with stratigraphic occurrences. Although this matrix was outdated by later revisions [14], its usage in studies employing different phylogenetic methods makes it an attractive basis for a case study comparing the results of dating approaches, which differ in the model assumed and their implementation. Analysing the original Xu et al. matrix also allows us to test whether Bayesian tip-dating avoids atypical relationships [15,16] inferred by Lee & Worthy [2]. Additionally, the emergence of avian dinosaurs has been a focus for macroevolutionary studies [17], and thus, we can use this dataset to examine how different dating methods impact downstream phylogenetic comparative methods.
2. Material and methods
We used the 374 character matrix for 89 taxa from [1] and age data from the Paleobiology Database for a series of Bayesian tip-dating analyses using BEAST2 and MrBayes. We performed analyses with noSA-BDSS as the tree prior using BEAST v. 2 [12] and SA-BDSS with both programs [9,10]. All tip-dating analyses used the Mkv model of character change [18] and accommodated stratigraphic uncertainties in first appearances of tip taxa as uniform priors. We applied minimum-age and minimum-branch-length APT approaches to 100 randomly selected most-parsimonious trees (MPTs) with first appearance times used as tip dates, including cal3 [13] with input rates taken from the BEAST2 SA posterior estimates to maximize the comparability of our analyses. We compared divergence dates and ancestral placements between samples of 100 APT-dated MPTs to a random selection of 100 post-burn-in trees from the Bayesian analyses. We also used these samples to compare the outcomes of a comparative analysis, mimicking the analyses of [17], fitting models for Ornstein–Uhlenbeck (OU), early burst (EB), and Brownian motion (BM; via geiger [19]). Further details of our methods and convergence assessments for the tip-dating analyses are in the Methods section of the electronic supplementary material.
3. Results
The relationships inferred under the Bayesian methods are similar to previous analyses [1,2]. In the BEAST2 analyses, Archaeopteryx has a posterior probability of 1 of being a member of the branch-defined Avialae (electronic supplementary material, figures S6–S7), in agreement with [2] (and contrary to Xu et al. [1]). However, MrBayes SA gives a posterior probability of 0.68 for the same placement (electronic supplementary material, figure S8). The unexpected relationships found by the maximum-likelihood study [15,16] are avoided, although the placements of the Alvarezsauridae and Scansoriopterygidae can vary considerably with strong support (see electronic supplementary material, Results). For example, all tip-dating analyses find a monophyletic Tyrannosauroidea with high support (no posterior probability less than 0.97).
Although sampling theropod ancestral taxa may seem unlikely, both SA tip-dating analyses generally inferred a median of 1–2 ancestors per tree (this frequency was skewed in MrBayes, with some trees containing up to 33 sampled ancestors). Both BEAST2 and MrBayes SA analyses place similar sets of taxa as ancestors (electronic supplementary material, figure S3), with a strong rank-order correlation of the per-taxon frequencies of ancestor placement (Spearman ρ = 0.69, p-value = 5.31 × 10−14). The cal3 analyses using first appearances never infer any ancestors, but similar correlations were found with ancestor frequencies from cal3 using last appearance times (see electronic supplementary material, Results). While Archaeopteryx is popularly referred to as an ‘ancestral bird’, it is a sampled ancestor in only 5% of the MrBayes posterior (0% for BEAST2 SA), and then only to its close relative Wellnhoferia, not the more nested Avialae.
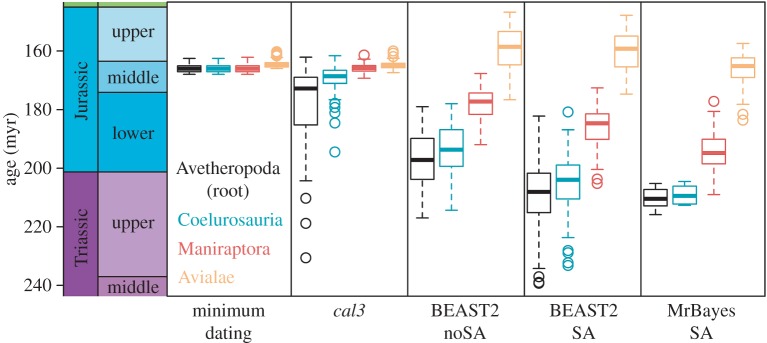
Comparisons of divergence dates for four nested avian clades (using a branch-based definition) show differences in clade age estimates across approaches (figure 1). All APT methods propose similar median ages for all four clades, much younger than tip-dating estimates. This is due to maximum-parsimony analyses placing the early-appearing Epidexipteryx and Epidendrosaurus (i.e. the Scansoriopterygidae) as members of a branch-based Avialae (also observed in [1,2]), which constrains the age of the Avialae to the Middle Jurassic or older. Tip-dating analyses vary in their placement of the Scansoriopterygidae but do not place them with the Avialae (see electronic supplementary material, Results). Divergence date estimates from cal3 for alternative non-Avian clades (Tyrannosauroidea, Therizinosauria) resemble distributions obtained from tip-dating (electronic supplementary material, figure S1), illustrating how APT approaches are ultimately constrained by input topologies. Even among tip-dating methods, there are differences, with BEAST2 noSA estimating earlier root ages than SA analyses, and BEAST2 SA having wider age distributions than MrBayes SA. Comparing age estimates for clades containing identical taxa reveals that tip-dating approaches estimate median divergence dates approximately 4–6 million years (myr) older than the earliest stratigraphic occurrence, although root-ward nodes have median ages as much as 30–40 myr older (see electronic supplementary materials, Results).
Figure 1.
Age estimates for four nested theropod clades, across five different approaches for obtaining a dated phylogeny using the Xu et al. [1] dataset. Plotted boxes represent the first quantile, the median and the third quantile, respectively. From left to right, the approaches used are minimum node age dating and cal3, both applied to a sample of 100 randomly selected maximum-parsimony topologies, and three tip-dating approaches, the noSA and SA with BEAST2, and SA with MrBayes. The four clades examined (ordered left-to-right for each approach) are the root node (essentially, the Avetheropoda) and three clades with ‘branch-based’ definitions: the Coelurosauria (all taxa more closely related to modern birds than Sinraptor and Allosaurus), the Maniraptora (…than Ornithomimus) and the Avialae (…than Troodon or Deinonychus). (Online version in colour.)
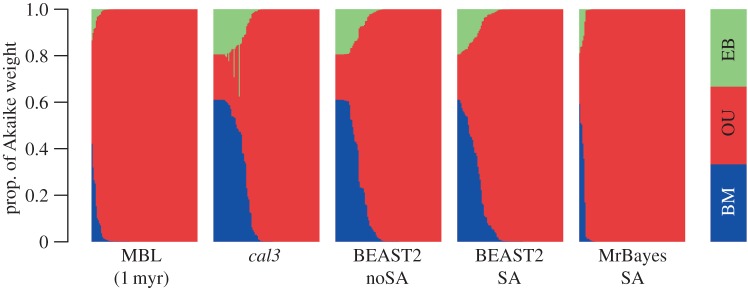
The original body-size analysis [17] used several APT approaches, including the 1 myr minimum-branch-length (MBL) approach. Under all time-scaling variants, they found strong support for single-optima OU for Theropoda and Maniraptora. Our reanalysis with alternative dated phylogenies agrees, with high support for OU across all approaches, particularly MBL (figure 2). However, our analysis reveals that model support varies considerably across trees from the same dating approach, with some phylogenies providing greater support for BM, a pattern that is most evident in cal3 and BEAST2 tree samples.
Figure 2.
The fit of models of body-size evolution across different sets of trees, each from a different approach for obtaining dated phylogenies. The right-most set is trees with edge lengths constrained to a minimum length of 1 myr; for all others, see caption for figure 1. Models are fit to a single dated tree, with Akaike weights for each model, for that tree (which sum to 1), figured as a bar along with other trees from that sample, as a block of 100 stacked barplots with borders removed. The barplots for each sample are reordered relative to their support for Brownian motion (BM), versus Ornstein–Uhlenbeck (OU) and early burst (EB). (Online version in colour.)
4. Discussion
While the Bayesian tip-dating analyses return broadly similar phylogenies, the contrast in topology, divergence dates and model support patterns between approaches suggests that workers need to carefully evaluate the models and priors applied, and the plausibility of complex models when datasets are limited [20]. Tip-dating methods appear to favour divergence dates that are several Ma older than the minimum age, sometimes tens of millions of years (figure 1 and electronic supplementary material, figures S1–S2). One explanation may be that by treating taxa in tip-dating analyses as single tips (i.e. a single point occurrence), even though more than 20% are known from multiple occurrences across millions of years, the inferred level of sampling may be so low that the average morphological clock rate dominates, swamping increases in the rate of character change and erroneously leading to older dates. The differences between MrBayes and BEAST2 SA-BDSS analyses are difficult to explain given their congruence in a previous comparison (electronic supplementary material, table S3 in [10]). As that study had both extant and extinct taxa, our discrepancy might be due to MrBayes having poor MCMC mixing when all tips are extinct.
Our comparative analyses support previous findings of constrained body-size evolution [18], but there is variation among dating methods in the relative support for OU across trees. Variation in model support among sampled posterior trees reinforces the importance of not taking a single point estimate of phylogeny for downstream analyses [21], and highlights the need to evaluate dated phylogenies from multiple approaches. Future studies should investigate body-size evolution through additional analyses than model choice [22], particularly given the known bias of some dating methods toward supporting OU [23]. The similarity of cal3 and the BEAST2 comparative analyses suggests that cal3 may be a suitable alternative when tip-dating is inapplicable.
Palaeobiologists will likely become major users of tip-dating and probabilistic APT approaches to generate dated phylogenies, replacing the arbitrary APT approaches. However, these techniques are still maturing. Careful consideration and applying multiple dating approaches may be necessary to isolate artefacts and identify what consensus does exist across models and implementations.
Supplementary Material
Acknowledgements
This is Paleobiology Database publication #266.
Data accessibility
All data, input files and programming scripts for recreating all analyses and figures can be found (separate from the electronic supplementary material) at the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.n2g80.
Authors' contributions
G.L. compiled the data and performed parsimony analyses; A.W. and N.M. performed tip-dating analyses; D.B. performed statistical comparisons and created figures; D.B., A.W., N.M. and G.L. wrote the manuscript and all authors agree to be held accountable for the content therein and approve the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
D.B. was supported by NSF EAR-1147537; A.W. was supported by NSF DEB-1256993; N.M. was supported by NSF EFJ-0832858 and ARC grant no. DE150101773; and G.L. was supported by ARC grant no. DE140101879.
References
- 1.Xu X, et al. 2011. An Archaeopteryx-like theropod from China and the origin of Avialae. Nature 475, 465–470. ( 10.1038/nature10288) [DOI] [PubMed] [Google Scholar]
- 2.Lee MSY, Worthy TH. 2012. Likelihood reinstates Archaeopteryx as a primitive bird. Biol. Lett. 8, 299–303. ( 10.1098/rsbl.2011.0884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AB, et al. 1998. Is the fossil record adequate? In Nature online debates (ed. Smith AB.). [Google Scholar]
- 4.Fisher DC. 2008. Stratocladistics: integrating temporal data and character data in phylogenetic inference. Annu. Rev. Ecol. Evol. Syst. 39, 365–385. ( 10.1146/annurev.ecolsys.38.091206.095752) [DOI] [Google Scholar]
- 5.O'Reilly JE, dos Reis M, Donoghue PCJ. 2015. Dating tips for divergence-time estimation. Trends Genet. 31, 637–650. ( 10.1016/j.tig.2015.08.001) [DOI] [PubMed] [Google Scholar]
- 6.Pyron RA. 2011. Divergence time estimation using fossils as terminal taxa and the origins of Lissamphibia. Syst. Biol. 60, 466–481. ( 10.1093/sysbio/syr047) [DOI] [PubMed] [Google Scholar]
- 7.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadler T. 2010. Sampling-through-time in birth-death trees. J. Theor. Biol. 267, 396–404. ( 10.1016/j.jtbi.2010.09.010) [DOI] [PubMed] [Google Scholar]
- 9.Gavryushkina A, Welch D, Stadler T, Drummond AJ. 2014. Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration. PLoS Comput. Biol. 10, e1003919 ( 10.1371/journal.pcbi.1003919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C, Stadler T, Klopfstein S, Heath TA, Ronquist F. 2016. Total-evidence dating under the fossilized birth–death process. Syst. Biol. 65, 228–249. ( 10.1093/sysbio/syv080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath TA, Huelsenbeck JP, Stadler T. 2014. The fossilized birth–death process for coherent calibration of divergence-time estimates. Proc. Natl Acad. Sci. USA 111(29), E2957–E2966. ( 10.1073/pnas.1319091111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadler T, Yang Z. 2013. Dating phylogenies with sequentially sampled tips. Syst. Biol. 62, 674–688. ( 10.1093/sysbio/syt030) [DOI] [PubMed] [Google Scholar]
- 13.Bapst DW. 2013. A stochastic rate-calibrated method for time-scaling phylogenies of fossil taxa. Methods Ecol. Evol. 4, 724–733. ( 10.1111/2041-210X.12081) [DOI] [Google Scholar]
- 14.Turner AH, Makovicky PJ, Norell MA. 2012. A review of dromaeosaurid systematics and paravian phylogeny. Bull. Am. Museum Nat. Hist. 371, 1–206. ( 10.1206/748.1) [DOI] [Google Scholar]
- 15.Spencer MR, Wilberg EW. 2013. Efficacy or convenience? Model-based approaches to phylogeny estimation using morphological data. Cladistics 29, 663–671. ( 10.1111/cla.12018) [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Pol D. 2013. Archaeopteryx, paravian phylogenetic analyses, and the use of probability-based methods for palaeontological datasets. J. Syst. Palaeontol. 12, 323–334. ( 10.1080/14772019.2013.764357) [DOI] [Google Scholar]
- 17.Benson RBJ, Campione NE, Carrano MT, Mannion PD, Sullivan C, Upchurch P, Evans DC. 2014. Rates of dinosaur body mass evolution indicate 170 million years of sustained ecological innovation on the avian stem lineage. PLoS Biol. 12, e1001853 ( 10.1371/journal.pbio.1001853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925. ( 10.1080/106351501753462876) [DOI] [PubMed] [Google Scholar]
- 19.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. ( 10.1093/bioinformatics/btu181) [DOI] [PubMed] [Google Scholar]
- 20.Wright AM, Lloyd GT, Hillis DM. 2016 Modeling character change heterogeneity in phylogenetic analyses of morphology through the use of priors. Syst. Biol. 65, 602–611. ( 10.1093/sysbio/syv122) [DOI] [PubMed] [Google Scholar]
- 21.Wright AM, Lyons KM, Brandley MC, Hillis DM. 2015. Which came first: the lizard or the egg? Robustness in phylogenetic reconstruction of ancestral states. J. Exp. Zool. B 324, 504–516. ( 10.1002/jez.b.22642) [DOI] [PubMed] [Google Scholar]
- 22.Slater GJ, Pennell MW. 2014. Robust regression and posterior predictive simulation increase power to detect early bursts of trait evolution. Syst. Biol. 63, 293–308. ( 10.1093/sysbio/syt066) [DOI] [PubMed] [Google Scholar]
- 23.Bapst DW. 2014. Assessing the effect of time-scaling methods on phylogeny-based analyses in the fossil record. Paleobiology 40, 331–351. ( 10.1666/13033) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, input files and programming scripts for recreating all analyses and figures can be found (separate from the electronic supplementary material) at the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.n2g80.