Abstract
Understanding the mechanisms of species coexistence is key to predicting patterns of species diversity. Historically, the ecological paradigm has been that species coexist by partitioning resources: as a species increases in abundance, self-limitation kicks in, because species-specific resources decline. However, determining coexistence mechanisms has been a particular puzzle for sedentary organisms with high overlap in their resource requirements, such as plants. Recent evidence suggests that plant-associated microbes could generate the stabilizing self-limitation (negative frequency dependence) that is required for species coexistence. Here, we test the key assumption that plant–microbe feedbacks cause such self-limitation. We used competition experiments and modelling to evaluate how two common groups of soil microbes (rhizospheric microbes and biological soil crusts) influenced the self-limitation of two competing desert grass species. Negative feedbacks between the dominant plant competitor and its rhizospheric microbes magnified self-limitation, whereas beneficial interactions between both plant species and biological soil crusts partly counteracted this stabilizing effect. Plant–microbe interactions have received relatively little attention as drivers of vegetation dynamics in dry land ecosystems. Our results suggest that microbial mechanisms can contribute to patterns of plant coexistence in arid grasslands.
Keywords: plant–soil feedback, competition and coexistence, stabilizing mechanisms, negative frequency dependence, semiarid grassland, biological soil crust
1. Introduction
What mechanisms allow species to coexist? Historically, species coexistence has been ascribed to niche differentiation [1], whereby species occupy different habitat spaces [2], require divergent nutrition [3] or employ different life-history strategies [4]. These mechanisms promote coexistence by causing species to be more self-limited than they are by their competitors. The degree of self-limitation and potential for stable coexistence depends on the strength of negative frequency dependence experienced by each competitor at high density [5,6]. More recently, the discovery of the ‘unseen majority’—diverse, abundant microbial consortia associated with all macro-organisms—has prompted evaluation of their potential to promote the coexistence of macro-organisms. Pathogens are well known to regulate population dynamics of their hosts [7], and interactions with microbes could be particularly important to sessile organisms, such as plants, that superficially appear to use identical resources [8]. Plant species-specific interactions with beneficial microbes could mediate access to soil resources, increasing niche differentiation. Alternatively, the build-up of soil pathogens as the relative frequency of the host increases can limit the performance of the plant host at high frequencies, promoting self-limitation. Such plant–soil microbial feedbacks have been hypothesized to be important mechanisms of plant species coexistence [9,10].
Past work has shown patterns that are consistent with the hypothesis that plant–soil microbial feedbacks (PSFs) can promote plant coexistence. First, a majority of PSF studies report negative feedbacks [11]. The pathogenic nature of most PSFs indicates that the potential for self-limitation exists, although most studies have been conducted in mesic ecosystems, limiting the geographical scope of inference. Second, in both tropical forests and temperate grasslands, the strength and direction of PSFs is correlated with plant species' relative abundances: rare species are associated with stronger negative feedbacks than common species [12,13] (but see [14]). Third, Janzen–Connell studies, a subset of PSF research originally focused on tropical trees, have demonstrated negative effects of distance from parent or conspecific density on seedling establishment and survival [15]. This could lead to negative frequency dependence between generations: as the relative frequency of a tree species increases, so does the proportion of habitat occupied by its species-specific soil pathogens, thus decreasing the per capita fitness of a population across space. Fourth, experiments have shown that PSFs can alter the outcome of competition in 1 : 1 pairwise combinations [16–18], or under natural gradients of competition [19], albeit with idiosyncratic results.
However, these prior examples do not fully demonstrate PSF as a mechanism of plant coexistence, because a key criterion for coexistence has not been directly assessed: PSFs must have higher demographic costs with increasing relative frequency of the host species in the community, causing negative frequency dependence [20]. For example, this could result from the increased likelihood of an individual encountering species-specific soil pathogens as its conspecific frequency increases in the community with no net change in per capita pathogen load, or from an amplified pathogen load at high host frequency. Under the modern framework for species coexistence [5], this stabilizing effect of niche differentiation (negative frequency dependence) is required for long-term coexistence in the absence of fluctuating temporal or spatial environments. Thus, a comprehensive test of PSF as a stabilizing mechanism of coexistence should meet the following criteria (table 1). (i) Negative PSF occurs and is caused by soil microbiota. (ii) Negative frequency dependence occurs. Stronger negative frequency dependence with higher total plant density indicates plant competition, but is not a necessary criterion. (iii) Negative frequency dependence is stronger (or only occurs) in the presence of microbially driven PSFs than in their absence.
Table 1.
Description of necessary criteria to demonstrate that plant–soil feedbacks are a stabilizing mechanism of coexistence.
| criterion | description | evidence |
|---|---|---|
| 1 | negative PSF occurs and is caused by soil microbiota | species performance is reduced with its own soil microbes relative to those of its competitor(s) |
| 2 | negative frequency dependence occurs | per capita performance of a species declines as its relative frequency in the community increases; stronger effects with higher total plant density indicate strong competition |
| 3 | negative frequency dependence is stronger (or only occurs) in the presence of microbially driven PSF | declines in per capita performance as relative frequency increases are steeper in the presence of PSF than in its absence |
Here, we tested the validity of each criterion for plant–microbe interactions to generate stabilizing mechanisms of coexistence using two common soil microbial groups (species-specific rhizospheric microbes and biological soil crusts) and two dominant desert grasses (Bouteloua gracilis and Bouteloua eriopoda). Plants were competed in a response surface design that allowed independent investigation of frequency dependence and density dependence. This is critical to capturing the frequency dependence of intra- versus interspecific competition. In the greenhouse, we replicated the response surface under a fully reciprocal PSF experiment, where we could control the composition of rhizospheric microbes and presence of biological soil crusts. For PSF to contribute to stabilizing mechanisms of coexistence, we expected that inoculation of live, conspecific, rhizospheric microbes would result in the strongest negative frequency dependence and highest per capita self-limitation. By comparing frequency dependency and modelling competitive interactions and invasion growth rates across treatments, we showed that PSFs caused by rhizospheric microbes can be stabilizing, whereas biological soil crusts partly offset this stabilization through their benefits to both plant species.
2. Methods and material
(a). Study system
We investigated interactions between B. gracilis (Poaceae, blue grama) and B. eriopoda (Poaceae, black grama) with their host-specific rhizospheric microbial communities, as well as the biological soil crusts (biocrusts) that occupy plant interspaces in desert grasslands. B. gracilis and B. eriopoda are perennial, C4 grasses that naturally co-occur in the ecotone between Chihuahuan desert grasslands and the short-grass steppe [21]. Their co-occurrence has been documented for more than 25 years [22] at the Sevilleta National Wildlife Refuge (Sevilleta hereafter), where our field collections occurred. Although prior work shows that the two grass species compete and their coexistence may be facilitated through recruitment niche partitioning [23,24], the mechanisms promoting their long-term coexistence remain elusive.
We focused on biocrusts and rhizosphere microbiota as microbial drivers. Biocrust organisms fix N and C, and engage in exchange with Bouteloua spp. [25]; they also increase soil moisture and surface stability [26]. In arid grasslands, fungal communities in grass rhizospheres are dominated by dark septate endophytes, a polyphyletic group characterized by melanized, septate hyphae [27,28]. They may facilitate host water uptake, increase plant acquisition of organic nitrogen [29,30] or act as plant pathogens [31]. Arbuscular mycorrhizal fungi (AMF) also colonize grass roots at the Sevilleta [32]. While AMF are best known for improving nutrient acquisition, their effects can span the parasitism–mutualism spectrum [33,34]. Other soil microbial taxa associated with Bouteloua spp. include diverse nematodes and bacteria [35,36].
(b). Response surface competition treatments
To quantify frequency-independent and -dependent effects, we employed a response surface plant competition design [37,38] in the greenhouse, where we could control microbial inoculations. We created 15 combinations of B. gracilis and B. eriopoda relative frequencies and total densities by varying the number of individuals of each species per pot (four levels: 0–6 individuals; electronic supplementary material, appendix figure S1). This was preferred over the more commonly used replacement series because it explicitly tests for interactions between total density (total number of plants per pot) and the relative frequency of a given species. We expected the frequency dependence to be stronger at higher total plant density, and our design permits that test.
(c). Soil microbe treatments
We implemented a fully crossed 2 × 2 × 2 factorial design of soil microbial treatments across the response surface. Treatments included all combinations of rhizospheric inoculum sterilization (live or sterilized), rhizospheric inoculum provenance (rhizospheric soil from B. gracilis or B. eriopoda) and biocrust presence (presence or absence). The entire design was fully replicated three times for a total of 360 experimental communities (electronic supplementary material, appendix figure S1).
(i). Rhizospheric microbes
In late October 2012, we collected rhizospheric (immediately around root zone) soil from planted field monocultures of each species at the Sevilleta. Monocultures plots seeded at 9.07 g per m2 (2 × 2.5 m) were established in 2007 (GPS: −106.6089, 34.406136) and weeded to maintain species composition (details at http://sev.lternet.edu/data/sev-174). From each of three plots of B. gracilis and B. eriopoda monocultures, we collected one 5.6 l sample of rhizospheric soil from the root zone (rhizosphere) of two to three mature plants. Inocula were stored at 4°C for one week until application. To isolate the effects of the rhizospheric microbiota, half of the inocula were autoclaved (30 min at 121°C) to reduce the abundance of live biota. Inocula from a different field monoculture plot were used for each replicate to obtain biologically independent replicates.
(ii). Biocrusts
While collections of rhizospheric microbes were designed to maximize plant species-specific differences in microbial composition by taking advantage of existing field monocultures, we had no a priori expectation of host-specific differences in biocrust composition. Therefore, biocrusts were collected from plant interspaces at a Sevilleta location where both Bouteloua spp. co-occur (GPS: −106.7358, 34.3592). We used 9 cm diameter Petri dishes to excise intact biocrust (to 1 cm depth), which were stored at room temperature until use (less than one week).
(d). Greenhouse experiment and harvest
In November 2012, we filled 900 ml pots with spore-free river sand and inoculated with 90 ml live or sterilized soil inoculum added to the top of the pot for optimal seedling colonization. For biocrust additions, each pot received a single Petri dish sample of field-collected biocrust on the soil surface (electronic supplementary material, appendix figure S1). Bouteloua gracilis and B. eriopoda seeds (Curtis & Curtis Inc., Clovis, NM, USA) were sown on the soil surface. After four weeks, we supplemented with additional seedlings of the same age to reach desired treatment structure. All pots were fertilized once (March 2013) with a weak liquid fertilizer (0.2% each FloraMicro and FloraGro, N : P : K = 7 : 1 : 7; General Hydroponics, Sebastopol, CA, USA). After six months, we recorded the number of individuals surviving and separately harvested aboveground biomass by plant species. Because it was not possible to separate roots by plant species, washed root tissue was homogenized by pot. We collected approximately 0.2 g root tissue for microscopic examination of fungal colonization. We also collected surface soil (biocrust, top 5 mm, approx. 5 g) from every pot. All biomass was dried at 60°C then weighed.
(e). Microbial treatment effectiveness
To assess the treatment effect of biocrust additions, samples from 109 pots were analysed for soil carbon and nitrogen content using an ECS 4010 CHNSO analyser (Costech Analytical Technologies, Valencia, CA, USA). To assess the effectiveness of rhizosphere soil sterilization, collected root tissue was cleared and stained following Vierheilig et al. [39], and scored for fungal colonization following McGonigle et al. [40] for 340 pots. Our microbial manipulation worked: total colonization rate was 78% lower under sterile (3.3 ± 0.4% s.e.) than under live soil inoculation (14 ± 1% s.e.).
(f). Statistical analysis
(i). Microbial effects on density and frequency dependence
For each plant species, our experimental design allowed microbial treatments to alter plant performance, regardless of plant total density/frequency, alter slopes of plant density or frequency dependence, and/or alter the potential interaction between density and frequency dependence. To investigate our three criteria, we evaluated six a priori models, representing alternative hypotheses of microbial effects on plant performance (table 2). We used model selection to evaluate the likelihood of each proposed model/hypothesis, given the data collected [41]. We also fitted a null model that included only an intercept (no treatment effects) to examine if candidate models provided more information than the null and to calculate likelihood pseudo-r2 values [41].
Table 2.
Description of candidate models testing microbial effects on frequency- and density dependence and their relationships to coexistence criteria.
| model | description | parameterizationa | fulfils criteria |
|---|---|---|---|
| i | all microbial effects on frequency dependence, density dependence and their interaction | ∼Freq + Dens + Dens : Freq + Crust + Inoc + Inoc : InocSPP + Crust : Freq + Inoc : InocSPP : Freq + Crust : Dens + Inoc : InocSPP : Dens + Crust : Freq : Dens + Inoc : InocSPP : Freq : Dens |
1, 2, 3 |
| ii | only frequency- and density-independent microbial effects | ∼Crust + Inoc + Inoc : InocSPP | 1 |
| iiic | only frequency-dependent microbial effects | ∼Crust : Freq + Inoc : InocSPP : Freq | 1, 2, 3 |
| iv | only density-dependent microbial effects | ∼Crust : Dens + Inoc : InocSPP : Dens | 1 |
| vb | only microbial effects on the interaction between frequency dependence and density dependence | ∼Crust : Freq : Dens + Inoc : InocSPP : Freq : Dens | 1, 2, 3 |
| vi | frequency dependence and density dependence with no microbial effects | ∼Freq + Dens + Freq : Dens | 2 |
aParameter descriptions: Freq, focal species relative frequency; Dens, total plant density; Crust, biocrust presence/absence; Inoc, sterilized or live rhizosphere inoculation; InocSPP, provenance of rhizosphere inoculum. Colons indicate interactions between terms.
bBest model for B. gracilis per capita biomass.
cBest model for B. eriopoda per capita biomass.
Candidate models were evaluated separately for B. eriopoda and B. gracilis using per capita aboveground biomass, ln-transformed to meet normality and homoscedasticity assumptions. We used the number of B. eriopoda and B. gracilis four weeks into the experiment (see ‘Greenhouse experiment and harvest’) for total seedling density and relative frequency predictors. In cases where additional germination occurred after four weeks, we included these additional germinants in the initial number. Briefly, we fitted each linear model to the data, calculated log-likelihood and AICc, ranked candidate models by AICc, and calculated model probabilities (wi) using MuMIn [42] in R v. 3.1.2 [43] following Burnham & Anderson [41]. Because we took an experimental approach, we used parameter estimates from the best model; in all cases, our best models outranked alternatives by AICc > 2.
(ii). Competition model fitting
To further investigate if soil microbial treatment effects on plant competitors altered strengths of per capita intra- and interspecific interactions, we fitted simple two-species competition models, using biomass accumulation to approximate population growth over generations. For each plant species × soil microbial treatment combination, we fitted the modified discrete time difference logistic model [44,45]
| 2.1 |
Mi/Ni indicates the mean per capita biomass of individuals of species i in the population, where positive values indicate growth. λ is equivalent to intrinsic per capita biomass increase, and αii and αij are competition coefficients for the per capita effects of intra- and interspecific competition, respectively. N is the number of individuals of each species in the ‘population’, or experimental pot, at the beginning of the experiment. While logistic difference equations are often used to predict population size at time t + 1 from population sizes at time t, we used it to predict the mean per capita biomass increase of individuals in the population here. This approach assumes that biomass scales linearly with population growth at the same rate for these congener competitors, an assumption that requires future testing, but which allowed us to harness the power of the model fitting approach.
Model parameters were fitted using maximum-likelihood estimation, assuming normally distributed errors, with function mle2 in R v. 3.1.2 [43]. We constrained λ to be positive and the absolute values of all parameters to a maximum of 5 to avoid model fits that could not discriminate between extremely high intrinsic growth and density dependence that was biologically implausible, or weaker intrinsic growth and density dependence. In one model, fitted for B. gracilis inoculated with live B. gracilis rhizospheric microbes and no biocrust, we were unable to find a local fit. Instead, we found very strong intraspecific interactions where the maximum-likelihood parameter was estimated at the upper boundary of 5. Compared with its converged fit at implausibly high intrinsic growth and density dependence, this alternative boundary fit resulted in only a 1% loss in log-likelihood; thus, we used the boundary fit, which was a conservative decision.
We evaluated coexistence by directly calculating the invasion growth rate for each species using the fitted parameters. This expression, the per capita growth rate of species i when it is rare and species j is at single-species equilibrium, can be solved for analytically from equation (2.1) to be (see appendix) [45]
| 2.2 |
Species that coexist are mutually invasible. In this case, both species have positive invasion growth rates. As we assumed that biomass accumulation scaled linearly and at the same rate to population growth rate for both Bouteloua species, relative differences in calculated invasion growth could be compared among microbial treatments and between competitors. While we could not directly translate calculated invasion growth rates to population growth because we lacked information on other vital rates (e.g. survival, recruitment), some positive threshold ‘low density biomass accumulation rate’ must be reached in order to reproduce and contribute to positive invasion population growth. To evaluate the effects of PSF on species coexistence, we compared invasion growth rates for each species when invading their competitor, with either live or sterile competitor soil microbial communities.
3. Results
Competitors showed strong fitness (frequency-independent) differences: B. gracilis was the competitive dominant, achieving on average 890% higher per capita shoot biomass than B. eriopoda. Thus, to increase potential for stable coexistence between these two plant species, inoculation with its own, live, rhizospheric microbes should increase negative frequency dependence and self-limitation for B. gracilis, and plant–microbe interactions should increase invasion (biomass) growth rates for both species.
(a). Plant–soil feedbacks increased self-limitation in Bouteloua gracilis
Bouteloua gracilis experienced the strongest negative frequency dependence in the presence of its own rhizospheric microbes, fulfilling all three criteria to demonstrate that PSF is a viable stabilizing mechanism of coexistence. First, B. gracilis per capita shoot biomass was 28% lower when grown with its own live rhizosphere inocula than with B. eriopoda rhizosphere inocula, indicating negative PSF. This effect weakened when B. gracilis received sterilized inocula from its own or its competitor's rhizosphere, demonstrating that the feedback is microbially driven (criterion 1). Second, we found negative frequency dependence in B. gracilis, which intensified at the highest plant densities, as expected if competition is important (criterion 2). Third, negative frequency dependence was strongest in the presence of host-specific, rhizospheric microbes (criterion 3). The model including microbial effects on the interaction between frequency dependence and density dependence had the highest support (w = 0.768, likelihood pseudo-r2 = 0.23; table 2; electronic supplementary material, appendix table S1). Specifically, B. gracilis inoculated with its own rhizospheric microbes experienced 40–50% stronger negative frequency dependence (difference in slope) than when inoculated with sterilized B. gracilis microbes, and 75–115% stronger negative frequency dependence than when inoculated with live microbes from the competitor, B. eriopoda (figure 1; electronic supplementary material, appendix table S2).
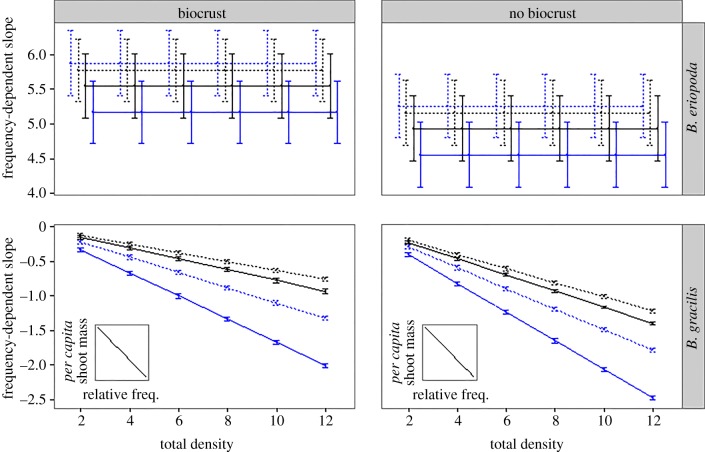
Figure 1.
Each point represents the slope (±s.d.) of per capita biomass regressed on frequency of the plant species in the pot (as shown in insets), calculated for each microbial and density treatment. Frequency-dependent slopes are shown for B. eriopoda (top) and B. gracilis (bottom), with biocrust present/absent. Plants inoculated with B. gracilis microbes are in blue and inoculations with B. eriopoda microbes are in black. Sterile inoculations are indicated by dashed lines and live microbial inoculations by solid lines. B. gracilis showed increasing negative frequency dependence as total density increased, whereas B. eriopoda showed consistent positive frequency dependence regardless of total density. The strongest negative frequency dependence was found in B. gracilis plants inoculated with conspecific microbes, under the highest total densities. Note the difference in y-axis scales between the two species.
Our competition models described similar results. Self-limitation (intraspecific competition coefficient) was stronger for B. gracilis when inoculated with its own live rhizosphere microbes compared with live B. eriopoda microbes (10-fold difference in average α; table 3; electronic supplementary material, appendix figure S2). There was no such difference between competitors when given sterile inoculations. This effect was partially driven by the model for B. gracilis with the high boundary fit (α = 5, no biocrusts added). However, the same rhizosphere treatment but with biocrust added resulted in the second strongest self-limitation for B. gracilis of all treatments (α = 0.84). Thus, we are inclined to conclude that this is a biological result.
Table 3.
Maximum-likelihood estimates of two species competition parameters under different microbial treatments. Standard errors included when estimation was possible. Parameters constrained to [0, 5] for λ and [−5, 5] for α.
| microbial treatment |
model log-likelihood | model log-likelihood | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rhizosphere soil |
biocrust presence | species i (B. eriopoda) fitted parameters ± s.e. |
species j (B. gracilis) fitted parameters ± s.e. |
|||||||
| provenance | sterilize | λi | αii | αij | λj | αjj | αji | |||
| BOER | live | no crust | 0.016 ± 0.004 | −0.203 ± 0.014 | 0.539 ± 0.236 | 78.982 | 0.519 ± 0.128 | 0.374 ± 0.159 | 0.035 ± 0.091 | 40.137 |
| BOGR | live | no crust | 0.028 ± 0.016 | 0.150 ± 0.241 | 0.344 ± 0.382 | 65.778 | 2.659 ± 2.014 | 5.000 ± 4.105 | 1.305 ± 1.178 | 52.491 |
| BOER | sterile | no crust | 0.048 ± 0.027 | 0.190 ± 0.383 | 2.322 ± 2.134 | 76.886 | 0.581 ± 0.287 | 0.306 ± 0.305 | 0.130 ± 0.222 | 9.865 |
| BOGR | sterile | no crust | 0.020 ± 0.006 | −0.106 ± 0.026 | 0.360 ± 0.232 | 82.207 | 0.260 ± 0.074 | 0.123 ± 0.113 | 0.042 ± 0.077 | 39.682 |
| BOER | live | crust | 0.078 ± 0.028 | 0.195 ± 0.295 | 1.691 ± 1.307 | 62.180 | 0.369 ± 0.101 | 0.151 ± 0.106 | 0.044 ± 0.109 | 33.191 |
| BOGR | live | crust | 0.029 ± 0.015 | −0.122 ± 0.104 | 0.350 ± 0.380 | 48.617 | 0.743 ± 0.378 | 0.841 ± 0.631 | 0.366 ± 0.283 | 41.521 |
| BOER | sterile | crust | 0.051 ± 0.016 | −0.112 ± 0.037 | 1.023 ± 0.715 | 62.740 | 0.892 ± 0.286 | 0.601 ± 0.289 | 0.126 ± 0.117 | 36.738 |
| BOGR | sterile | crust | 0.011 | −0.150 | −0.001 | 47.810 | 0.714 ± 0.224 | 0.495 ± 0.290 | 0.219 ± 0.162 | 23.131 |
The subordinate competitor, B. eriopoda, in contrast, experienced very weak, positive PSF (figure 1). The presence of its own rhizospheric microbes resulted in approximately 10% higher positive frequency dependence compared with inoculation with microbes from its competitor (best model included microbial effects on frequency dependence: w = 0.773, likelihood pseudo-r2 = 0.20; table 2; electronic supplementary material, appendix tables S1 and S2). While analysis revealed microbial effects on frequency dependence, the positive direction of frequency dependence did not support criteria 2 or 3 for the subordinate competitor.
Our competition modelling confirmed positive per capita intraspecific interactions among B. eriopoda (shown by a negative α) in five out of eight microbial treatments. Of the three treatments that did not result in intraspecific facilitation, standard errors of parameter estimates were large and overlapped zero (table 3). These results suggest that there are facilitative interactions among B. eriopoda individuals, and that positive frequency dependence is unlikely to be caused solely by release from strong interspecific competition (electronic supplementary material, appendix figure S3). Interestingly, B. eriopoda experienced stronger average self-limitation when inoculated with live, rhizospheric microbes from B. gracilis compared with live microbes from its conspecifics. These results should be interpreted with caution, however, owing to the combinations of positive and negative intraspecific interactions fitted in B. eriopoda.
(b). Biological soil crusts destabilize coexistence
The effects of biocrust additions were smaller than those of rhizospheric microbes, were generally positive rather than negative and showed potential to destabilize, rather than stabilize, coexistence. Destabilization emerged through two pathways: increasing positive frequency dependence in B. eriopoda and decreasing negative frequency dependence in B. gracilis. Biocrust addition decreased negative frequency dependence of B. gracilis at the highest total plant densities by approximately 30%, and increased positive frequency dependence in B. eriopoda approximately 20% (figure 1; electronic supplementary material, appendix table S2). This result was also mirrored in the competition modelling, where biocrust addition resulted in 50% more intraspecific facilitation in B. eriopoda and 64% less intraspecific competition in B. gracilis (table 3; electronic supplementary material, appendix figures S2 and S3). Elemental analyses indicated that biocrust addition increased soil nitrogen by nearly 30% (F1,107 = 48.93, p < 0.001) and soil carbon by 20% (F1,107 = 34.45, p < 0.001).
(c). Potential for plant–microbial interactions to promote coexistence between plant competitors
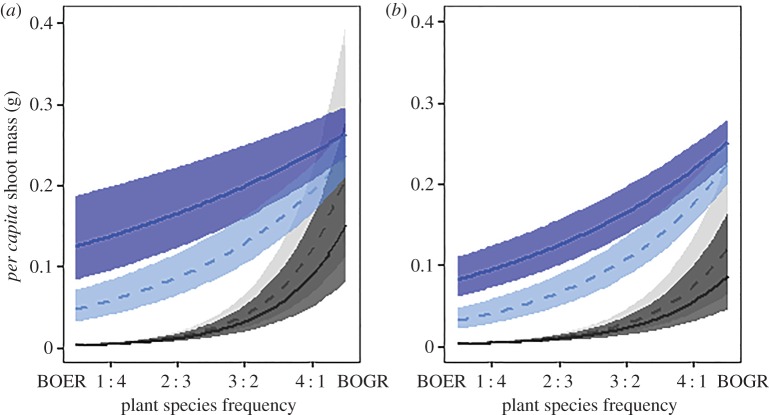
Our results showed that negative PSF from species-specific, rhizospheric microbes strengthened negative frequency dependence and self-limitation in B. gracilis, the competitive dominant (figure 2). Positive (destabilizing) microbial effects were small compared with the microbially driven increases in B. gracilis negative frequency dependence (10% increase in positive frequency dependence versus 75–115% stronger negative frequency dependence; figure 2).
Figure 2.
Frequency dependence of fitted per capita biomass of B. gracilis (light blue/blue) and B. eriopoda (grey/black) under rhizospheric microbe treatments and (a) biocrust present versus (b) biocrust absent. Dashed lines with pale ribbons (s.d.) indicate inoculation with conspecific rhizospheric microbes and solid lines with dark ribbons (s.d.) are with heterospecific microbes. Only live inoculations are shown here for clarity.
Invasion growth rate calculations, best interpreted here as ‘low-frequency biomass accumulation rates’, indicated that the presence of live PSF allowed mutual invasibility, a coexistence criterion. Particularly for B. eriopoda, invasion growth rates were more positive when the resident B. gracilis experienced live PSF compared with sterile PSF (figure 3a). In comparison, B. gracilis invasion growth rates were one to two orders of magnitude larger than those of B. eriopoda and positive, regardless of whether the resident B. eriopoda population experienced live PSF (figure 3b). We caution that our invasion growth rates only considered biomass accumulation and no other vital rates, such as reproduction or germination. Therefore, positive invasion rates for both species under live PSF provide a foundation for coexistence, but do not ensure coexistence.
Figure 3.

Calculated invasion growth rates for (a) B. eriopoda/BOER and (b) B. gracilis/BOGR with different microbial treatments. Grey bars indicate biocrust addition treatments, whereas white bars received no biocrust addition. Rhizosphere soil microbial inocula were live or sterilized, from the resident host plant species.
4. Discussion
Under the modern species coexistence framework, plant–microbe interactions can mediate plant species coexistence via two pathways: decreasing the fitness difference between plant competitors or increasing stabilization by increasing self-limitation. In this study, there were strong fitness differences between competitors: B. gracilis was the competitive dominant and B. eriopoda the competitive subordinate in all scenarios. Thus, to increase the chances of coexistence between these two plant species, plant–microbe interactions should increase self-limitation of B. gracilis (stabilizing) and/or decrease average fitness differences between the two species (equalizing). As our results showed little evidence of strong frequency-independent microbial effects, we focus our discussion on the former.
(a). Plant–soil feedbacks increased self-limitation in Bouteloua gracilis
Our study demonstrated experimentally that negative PSFs are capable of generating negative frequency dependence in a plant species. While there may be other mechanisms (such as resource partitioning between plants) that accounted for the weakly negative frequency dependence of B. gracilis inoculated with sterilized rhizospheric microbes, it was the presence of its own rhizospheric microbes that resulted in the strongest negative frequency dependence. Our competition modelling focusing on per capita effects also confirmed this result, showing the strongest self-limitation in B. gracilis inoculated with its own, live, rhizospheric microbes. Prior work correlating the strength of PSF with species relative abundance [12,13] as well as pairwise studies examining PSF under competitor present/absent scenarios [16–18,46] set the stage for PSF as a potential driver of plant community dynamics, but do not wholly demonstrate it as a stabilizing mechanism that increases self-limitation. In addition, while negative conspecific density- or distance-dependent effects on seedling survival found in Janzen–Connell studies may result in negative frequency dependence when considered intergenerationally, we found that soil communities are able to intensify intraspecific interactions within the same generation. Our results established the importance of PSF by comparing the strength of negative frequency dependence/self-limitation in the presence versus absence of host-specific soil microbes. In addition, our response surface design demonstrated the interactive effect of total plant density and relative species frequency, which has not yet been shown for any plant–microbe interaction, but is probably a prevalent phenomenon.
Our results suggest that rhizospheric microbes associated with B. gracilis plants increased self-limitation. Past work at our study site has found diverse fungal taxa associated with B. gracilis, including AMF species [32] as well as putative pathogens [27]. Work from other sites has also reported bacteria and nematode species that may affect B. gracilis growth and performance [35,36]. Fungi such as AMF could potentially increase niche partitioning via alternative resource acquisition [47], and species-specific phytopathogens are well known to drive plant community dynamics [20]. Future molecular work characterizing the microbial community associated with experimental populations could shed more light on potential mechanisms behind the observed phenomena.
(b). Biocrusts: a destabilizing mechanism
Our results suggest that biocrusts generally benefited Bouteloua grasses. While the effects of biocrust additions were smaller than those of rhizospheric microbes, biocrusts were destabilizing through two pathways: increasing positive intraspecific interactions in B. eriopoda and decreasing negative intraspecific interactions in B. gracilis. Elemental analyses indicated that biocrust addition increased C and N in surface soils. These effects probably arose from the dominant microorganisms found in the light biocrusts of our desert grasslands: photosynthesizing cyanobacteria such as Microcoleus spp., which can contribute to soil organic carbon, and Nostoc spp., which can fix nitrogen [26,48]. Whether benefits were due directly to the presence of biocrust microbes or indirectly to their effects on edaphic characteristics remains unresolved. A greenhouse biocrust-addition experiment using the perennial grass Elymus elymoides found similar gains in biomass; moreover, Elymus grown with biocrust showed higher tissue nutrient concentration, a potential mechanism for increased per capita performance [49]. Few studies have investigated biocrust mediation of plant competition and coexistence. However, a comparison of spatially paired plots that were naturally biocrusted versus naturally bare found that biocrusted soils supported 4–9-fold greater percentage of exotic plant species, which were probably the stronger competitors, lending support to potential destabilizing effects of biocrusts [50].
(c). Can plant–microbial interactions contribute to stabilizing coexistence?
Under our experimental conditions, results indicated that B. gracilis would probably have competitively excluded B. eriopoda under all treatment scenarios due to its much faster growth rate. However, we found that negative PSF resulted in stronger negative frequency dependence/self-limitation in the dominant competitor, B. gracilis, which could contribute to coexistence via stabilizing forces. In addition, positive invasion growth rates were present for both species (mutual invasibility) only when the resident species experienced live PSF (figure 3). Our results suggest that while the two competitors were asymmetrically matched in their competitive abilities, feedbacks between plants and rhizospheric microbes could potentially decrease the time to competitive exclusion for species in mixture and increase the potential for coexistence in our experimental communities. Stable, long-term coexistence in the field could require additional mechanisms. For example, others have found that plant-scale spatial heterogeneity in PSF can further promote coexistence [51]. Temporal variance in climate, such as interannual variability in precipitation, could also promote species coexistence via fluctuation-dependent mechanisms such as the storage effect [5,52].
Our results are probably a conservative test of coexistence. Here, we measured per capita aboveground biomass as an indicator of individual plant fitness. However, competition outcomes are population-level phenomena, which additionally depend on adult survival, reproduction and recruitment. In our modelling, we assumed that per capita biomass accumulation of both Bouteloua species scaled linearly to population growth at the same rate. Previous work at this site has shown that B. eriopoda has much higher fecundity per gram of vegetative biomass compared with B. gracilis [23]. Therefore, it is possible that the fitness difference between the two species at the population level is smaller than measured in this study, owing to the higher reproductive capacity of B. eriopoda. Additional information on survival, germination and recruitment where the two species co-occur will increase our ability to generalize our results based on plant growth.
In the Chihuahuan desert grasslands of New Mexico, USA, where their ranges overlap and field collections for this study took place, B. gracilis and B. eriopoda are the dominant species and are known to coexist based on long-term monitoring [22]. Results of experimental removals at this site suggest that it is probably competitive dominance of B. gracilis that results in B. eriopoda subordination, whereas other factors, such as the abiotic environment, may determine areas where B. eriopoda dominates and B. gracilis is subordinate [24]. Our greenhouse experiment uncovered a competitive hierarchy similar to that demonstrated experimentally at our field site, which lends confidence that it is applicable to this ecosystem. However, it remains to be explored under what circumstances PSF is an important mechanism in driving species coexistence in the field, relative to unexamined alternatives including fluctuation-dependent mechanisms [5].
5. Conclusion
For the first time, to the best of our knowledge, we experimentally quantified the contributions of two classes of plant–microbe interactions to the mechanisms of coexistence between foundational plant species. Our study demonstrates that conspecific plant–soil feedbacks from rhizospheric microbes enhance the possibility of species coexistence through stabilizing effects that increase self-limitation of the dominant competitor. These findings also suggest a role for plant–microbe interactions in structuring plant communities in arid and semiarid ecosystems, where such interactions have not been well studied.
Supplementary Material
Acknowledgements
We thank J. Avritt and undergraduate assistants for help with the study. T. E. X. Miller, S. L. Collins and R. L. Sinsabaugh gave advice throughout the design, analysis and write-up. We also thank P. B. Adler, K. M. Crawford, A. Porras-Alfaro and M. A. Bowker for comments on an earlier draft, as well as comments from R. A. Lankau and one anonymous reviewer that greatly improved this manuscript.
Data accessibility
The data supporting this article have been deposited on Dryad at http://dx.doi.org/10.5061/dryad.780hc.
Authors' contributions
Y.A.C. and J.A.R. designed the experiment and wrote the manuscript. Y.A.C. conducted the experiment and analyses under the guidance of J.A.R.
Competing interests
We declare that we have no competing interests.
Funding
This project was supported by the University of New Mexico Biology Graduate Research Allocations Committee, the Garden Club of America Desert Studies Award and NSF DEB-1456955.
References
- 1.Hutchinson GE. 1959. Homage to Santa Rosalia; or why are there so many kinds of animals? Am. Nat. 93, 145–159. ( 10.1086/282070) [DOI] [Google Scholar]
- 2.MacArthur RH. 1958. Population ecology of some warblers of northeastern coniferous forests. Ecology 39, 599–619. ( 10.2307/1931600) [DOI] [Google Scholar]
- 3.Tilman D. 1982. Resource competition and community structure. Princeton, NJ: Princeton University Press. [PubMed] [Google Scholar]
- 4.Grime J. 1977. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194. ( 10.1086/283244) [DOI] [Google Scholar]
- 5.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 6.Levine JM, HilleRisLambers J. 2009. The importance of niches for the maintenance of species diversity. Nature 461, 254–257. ( 10.1038/nature08251) [DOI] [PubMed] [Google Scholar]
- 7.Holt RD, Pickering J. 1985. Infectious disease and species coexistence: a model of Lotka–Volterra form. Am. Nat. 126, 196–211. ( 10.1086/284409) [DOI] [Google Scholar]
- 8.Van Der Heijden MG, Bardgett RD, Van Straalen NM. 2008. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. ( 10.1111/j.1461-0248.2007.01139.x) [DOI] [PubMed] [Google Scholar]
- 9.Bever JD, Westover KM, Antonovics J. 1997. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85, 561–573. ( 10.2307/2960528) [DOI] [Google Scholar]
- 10.Bever JD et al. 2010. Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 25, 468–478. ( 10.1016/j.tree.2010.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulmatiski A, Beard KH, Stevens JR, Cobbold SM. 2008. Plant–soil feedbacks: a meta-analytical review. Ecol. Lett. 11, 980–992. ( 10.1111/j.1461-0248.2008.01209.x) [DOI] [PubMed] [Google Scholar]
- 12.Klironomos JN. 2002. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70. ( 10.1038/417067a) [DOI] [PubMed] [Google Scholar]
- 13.Mangan SA, Schnitzer SA, Herre EA, Mack KM, Valencia MC, Sanchez EI, Bever JD. 2010. Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755. ( 10.1038/nature09273) [DOI] [PubMed] [Google Scholar]
- 14.Reinhart KO. 2012. The organization of plant communities: negative plant–soil feedbacks and semiarid grasslands. Ecology 93, 2377–2385. ( 10.1890/12-0486.1) [DOI] [PubMed] [Google Scholar]
- 15.Comita LS, Queenborough SA, Murphy SJ, Eck JL, Xu K, Krishnadas M, Beckman N, Zhu Y. 2014. Testing predictions of the Janzen–Connell hypothesis: a meta-analysis of experimental evidence for distance- and density-dependent seed and seedling survival. J. Ecol. 102, 845–856. ( 10.1111/1365-2745.12232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pendergast TH, Burke DJ, Carson WP. 2013. Belowground biotic complexity drives aboveground dynamics: a test of the soil community feedback model. New Phytol. 197, 1300–1310. ( 10.1111/nph.12105) [DOI] [PubMed] [Google Scholar]
- 17.Kardol P, Cornips NJ, Kempen MMLV, Bakx-Schotman JMT, Putten WHVD. 2007. Microbe-mediated plant–soil feedback causes historical contingency effects in plant community assembly. Ecol. Monogr. 77, 147–162. ( 10.1890/06-0502) [DOI] [Google Scholar]
- 18.Petermann JS, Fergus AJF, Turnbull LA, Schmid B. 2008. Janzen–Connell effects are widespread and strong enough to maintain diversity in grasslands. Ecology 89, 2399–2406. ( 10.1890/07-2056.1) [DOI] [PubMed] [Google Scholar]
- 19.Bagchi R, Gallery RE, Gripenberg S, Gurr SJ, Narayan L, Addis CE, Freckleton RP, Lewis OT. 2014. Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature 506, 85–88. ( 10.1038/nature12911) [DOI] [PubMed] [Google Scholar]
- 20.Mordecai EA. 2011. Pathogen impacts on plant communities: unifying theory, concepts, and empirical work. Ecol. Monogr. 81, 429–441. ( 10.1890/10-2241.1) [DOI] [Google Scholar]
- 21.Kröel-Dulay G, Ódor P, Peters DP, Hochstrasser T. 2004. Distribution of plant species at a biome transition zone in New Mexico. J. Veget. Sci. 15, 531–538. ( 10.1111/j.1654-1103.2004.tb02292.x) [DOI] [Google Scholar]
- 22.Collins SL, Xia Y. 2015. Long-term dynamics and hotspots of change in a desert grassland plant community. Am. Nat. 185, E30–E43. ( 10.1086/679315) [DOI] [PubMed] [Google Scholar]
- 23.Peters DPC. 2002. Plant species dominance at a grassland–shrubland ecotone: an individual-based gap dynamics model of herbaceous and woody species. Ecol. Model 152, 5–32. ( 10.1016/S0304-3800(01)00460-4) [DOI] [Google Scholar]
- 24.Peters DP, Yao J. 2012. Long-term experimental loss of foundation species: consequences for dynamics at ecotones across heterogeneous landscapes. Ecosphere 3, 1–23. ( 10.1890/ES11-00273.1) [DOI] [Google Scholar]
- 25.Green LE, Porras-Alfaro A, Sinsabaugh RL. 2008. Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J. Ecol. 96, 1076–1085. ( 10.1111/j.1365-2745.2008.01388.x) [DOI] [Google Scholar]
- 26.Belnap J, Lange OL. 2002. Biological soil crusts: structure, function, and management. New York, NY: Springer. [Google Scholar]
- 27.Khidir HH, Eudy DM, Porras-Alfaro A, Herrera J, Natvig DO, Sinsabaugh RL. 2010. A general suite of fungal endophytes dominate the roots of two dominant grasses in a semiarid grassland. J. Arid Environ. 74, 35–42. ( 10.1016/j.jaridenv.2009.07.014) [DOI] [Google Scholar]
- 28.Porras-Alfaro A, Herrera J, Natvig DO, Lipinski K, Sinsabaugh RL. 2011. Diversity and distribution of soil fungal communities in a semiarid grassland. Mycologia 103, 10–21. ( 10.3852/09-297) [DOI] [PubMed] [Google Scholar]
- 29.Kivlin SN, Emery SM, Rudgers JA. 2013. Fungal symbionts alter plant responses to global change. Am. J. Bot. 100, 1445–1457. ( 10.3732/ajb.1200558) [DOI] [PubMed] [Google Scholar]
- 30.Newsham KK. 2011. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 190, 783–793. ( 10.1111/j.1469-8137.2010.03611.x) [DOI] [PubMed] [Google Scholar]
- 31.Tellenbach C, Grünig CR, Sieber TN. 2011. Negative effects on survival and performance of Norway spruce seedlings colonized by dark septate root endophytes are primarily isolate-dependent. Environ. Microbiol. 13, 2508–2517. ( 10.1111/j.1462-2920.2011.02523.x) [DOI] [PubMed] [Google Scholar]
- 32.Johnson NC, Rowland DL, Corkidi L, Egerton-Warburton LM, Allen EB. 2003. Nitrogen enrichment alters mycorrhizal allocation at five mesic to semiarid grasslands. Ecology 84, 1895–1908. ( 10.1890/0012-9658(2003)084%5B1895:NEAMAA%5D2.0.CO;2) [DOI] [Google Scholar]
- 33.Johnson NC, Graham JH, Smith FA. 1997. Functioning of mycorrhizal associations along the mutualism–parasitism continuum. New Phytol. 135, 575–585. ( 10.1046/j.1469-8137.1997.00729.x) [DOI] [Google Scholar]
- 34.Hoeksema JD et al. 2010. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol. Lett. 13, 394–407. ( 10.1111/j.1461-0248.2009.01430.x) [DOI] [PubMed] [Google Scholar]
- 35.Stanton NL. 1983. The effect of clipping and phytophagous nematodes on net primary production of blue grama, Bouteloua gracilis. Oikos 40, 249–257. ( 10.2307/3544589) [DOI] [Google Scholar]
- 36.Ingham RE, Trofymow JA, Ingham ER, Coleman DC. 1985. Interactions of bacteria, fungi, and their nematode grazers: effects on nutrient cycling and plant growth. Ecol. Monogr. 55, 119–140. ( 10.2307/1942528) [DOI] [Google Scholar]
- 37.Inouye BD. 2001. Response surface experimental designs for investigating interspecific competition. Ecology 82, 2696–2706. ( 10.1890/0012-9658(2001)082%5B2696:RSEDFI%5D2.0.CO;2) [DOI] [Google Scholar]
- 38.Law R, Watkinson A. 1987. Response-surface analysis of two-species competition: an experiment on Phleum arenarium and Vulpia fasciculata. J. Ecol. 75, 871–886. ( 10.2307/2260211) [DOI] [Google Scholar]
- 39.Vierheilig H, Coughlan AP, Wyss U, Piché Y. 1998. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGonigle T, Miller M, Evans D, Fairchild G, Swan J. 1990. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. ( 10.1111/j.1469-8137.1990.tb00476.x) [DOI] [PubMed] [Google Scholar]
- 41.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer. [Google Scholar]
- 42.Barton K. 2014. MuMIn: multi-model inference. R package version 1.10.5. See http://cran.r-project.org/package=MuMin.
- 43.R Development Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 44.Hassell M, Comins H. 1976. Discrete time models for two-species competition. Theor. Popul. Biol. 9, 202–221. ( 10.1016/0040-5809(76)90045-9) [DOI] [PubMed] [Google Scholar]
- 45.Adler PB, HilleRisLambers J, Levine JM. 2007. A niche for neutrality. Ecol. Lett. 10, 95–104. ( 10.1111/j.1461-0248.2006.00996.x) [DOI] [PubMed] [Google Scholar]
- 46.Kulmatiski A, Heavilin J, Beard KH. 2011. Testing predictions of a three-species plant–soil feedback model. J. Ecol. 99, 542–550. ( 10.1111/j.1365-2745.2010.01784.x) [DOI] [Google Scholar]
- 47.Gustafson DJ, Casper BB. 2006. Differential host plant performance as a function of soil arbuscular mycorrhizal fungal communities: experimentally manipulating co-occurring Glomus species. Plant Ecol. 183, 257–263. ( 10.1007/s11258-005-9037-8) [DOI] [Google Scholar]
- 48.Evans R, Lange O. 2003. Biological soil crusts and ecosystem nitrogen and carbon dynamics. In Biological soil crusts: structure, function, and management (J Belknap, OL Lange), pp. 263–279. New York, NY: Springer. [Google Scholar]
- 49.Pendleton RL, Pendleton BK, Howard GL, Warren SD. 2003. Growth and nutrient content of herbaceous seedlings associated with biological soil crusts. Arid Land Res. Manage. 17, 271–281. ( 10.1080/15324980301598) [DOI] [Google Scholar]
- 50.DeFalco L, Detling J, Tracy CR, Warren S. 2001. Physiological variation among native and exotic winter annual plants associated with microbiotic crusts in the Mojave Desert. Plant Soil 234, 1–14. ( 10.1023/A:1010323001006) [DOI] [Google Scholar]
- 51.Burns JH, Brandt AJ. 2014. Heterogeneity in plant–soil feedbacks and resident population dynamics affect mutual invasibility. J. Ecol. 102, 1048–1057. ( 10.1111/1365-2745.12258) [DOI] [Google Scholar]
- 52.Angert AL, Huxman TE, Chesson P, Venable DL. 2009. Functional tradeoffs determine species coexistence via the storage effect. Proc. Natl Acad. Sci. USA 106, 11 641–11 645. ( 10.1073/pnas.0904512106) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article have been deposited on Dryad at http://dx.doi.org/10.5061/dryad.780hc.