Abstract
Background: Accurate methods for cross-sectional incidence estimation are needed for HIV surveillance and prevention research. We developed an avidity assay based on the fourth-generation Genetic Systems HIV Combo Ag/Ab EIA (Bio-Rad Combo assay) and evaluated its performance. Materials and Methods: The Bio-Rad Combo assay was modified incubating samples with and without 0.025 M diethylamine (DEA). The avidity index (AI) was calculated as the ratio of the DEA-treated to untreated result for a specific sample. We analyzed 2,140 samples from 808 individuals from the United States with known duration of HIV infection. The mean duration of recent infection (MDRI) and the false-recent rate (FRR, fraction of samples from individuals known to be infected >2 years misclassified as recent) were calculated for AI cutoffs of 20%–90% for the avidity assay alone and in combination with a viral load assay (VL, limit of detection 400 copies/ml). Factors associated with misclassification of samples collected ≥2 years after infections were also evaluated. Results: The MDRI for the Bio-Rad Combo Avidity assay ranged from 50 days using an AI cutoff of 20% to 276 days using an AI cutoff of 90%; the FRR ranged from 0% to 9%. When samples with a VL <400 copies/ml were classified as nonrecent, the FRRs were reduced approximately twofold and the MDRI estimates were reduced by ∼20%. An AI cutoff of 50% provided an MDRI of 135 days with an FRR of 2.1%. All samples from elite suppressors had an AI >80%. In adjusted analysis, viral suppression and low CD4 cell count were significantly associated with misclassification among individuals infected >2 years. Conclusions: This modified Bio-Rad Combo Avidity assay may be a useful tool for cross-sectional HIV incidence estimation. Further research is needed to evaluate use of this assay in combination with other assays to accurately estimate population-level HIV incidence.
Background
Estimation of HIV incidence from cross-sectional samples is an alternative method for determining the rate of new HIV infections in a population when a longitudinal cohort is not available.1 Accurate methods for HIV incidence estimation are required for general surveillance and HIV prevention research.2 Assays that have been specifically manufactured or developed for this purpose include the Limiting Antigen Avidity assay (LAg-Avidity),3 the BED Capture EIA (BED-CEIA),4 and modified serologic HIV screening tests (see Murphy and Parry for review5). Antibody avidity assays quantify the avidity (binding strength) of anti-HIV antibodies in serum or plasma to HIV antigen.6–9 Antibody avidity usually increases with duration of infection. People with recent HIV infection (e.g., infected for <6 months) tend to have low antibody avidity.
To be useful for cross-sectional HIV incidence estimation, an assay or multi-assay algorithm (MAA) must identify a majority of samples from recently infected individuals without misclassifying samples from long-term infections (e.g., >2 years after HIV seroconversion) as recent infections. Low viral load (VL) (natural10 or drug-induced11), low CD4 cell count,12,13 and HIV subtype D14 are associated with misclassification by cross-sectional incidence assays. The performance of incidence assays can be characterized by two variables: the mean duration of recent infection (MDRI, the average amount of time individuals are considered recently infected for a given assay or algorithm), and the false recent rate (FRR, the frequency that individuals with long-term infection are misclassified as recently infected).15–17 The WHO/UNAIDS Working Group on Incidence Assays recommends that incidence assays have an MDRI of >120 days with an FRR <2%.18–20
A modified version of the third-generation Bio-Rad GS HIV-1/HIV-2 Plus O EIA is used in MAAs to estimate HIV incidence.21–23 However, the GS HIV-1/HIV-2 Plus O EIA is not available in many countries. Fourth-generation HIV screening assays detect HIV p24 antigen as well as anti-HIV antibodies, shortening the window period for detecting HIV infection. In this report, we modified a fourth-generation HIV screening assay [the GS HIV Combo Ag/Ab EIA (Bio-Rad Combo assay)] for measurement of antibody avidity and evaluated the performance of the modified assay by determining the MDRI and FRR at different avidity index (AI) cutoffs. This new incidence assay, referred to as the Bio-Rad Combo Avidity assay, was evaluated alone and in a MAA that included HIV VL.
Materials and Methods
Ethics statement
This study was approved by the Institutional Review Board of the Johns Hopkins University School of Medicine. All trial and cohort studies were conducted according to the ethical standards set forth by the institutional review boards of the participating institutions and the Helsinki Declaration of the World Medical Association. All participants provided written informed consent. This report includes analysis of stored samples and data from those studies. No participants were recruited or followed during the course of this work.
Samples used for analysis
Data analyzed in this report were obtained from testing stored plasma or serum from HIV-infected participants from the United States in the AIDS Linked to the Intravenous Experience Study (ALIVE),24 the Multicenter AIDS Cohort Study (MACS),25 the HIV Network for Prevention Trials (HIVNETs) 001/001.1 study,23 the Johns Hopkins Hospital Clinical Cohort (JHHCC),26 and the Johns Hopkins Elite Suppressor (ES) Cohort 27 (Table 1). All samples were from adults who likely had clade B HIV infection; a subset of samples from these cohorts were previously determined to have subtype B HIV, and 99% of HIV infections previously characterized from Baltimore were subtype B.29 HIVNET 001 samples were obtained from HIV seroconverters who were followed up to 4.5 years. Samples from the ALIVE, ES, JHHCC, and MACS cohorts were from individuals who were known to be HIV-infected for at least 2 years before sample collection.21 Individuals from the ES cohort had a VL <400 copies/ml at three study visits and a VL <50 copies/ml at one study visit without being on antiretroviral treatment (ART). JHHCC samples were from: (1) individuals with one visit before ART initiation and three to eight samples (one per year) after ART initiation, or (2) individuals who were virally suppressed (<400 copies/ml) on ART, but experienced viral breakthrough during follow-up. Additionally, 10 samples known to be p24 positive were obtained from a commercial source (ZeptoMetrix, Buffalo, NY).
Table 1.
Characteristics of Samples Used for Analysis by Cohort
| HIVNET 001 | ALIVE | MACS | JHH clinical cohort | Elite suppressors | |
|---|---|---|---|---|---|
| Number of samples | 816 | 489 | 552 | 245 | 38 |
| Number of individuals | 103 | 284 | 360 | 38 | 23 |
| Duration of infection | |||||
| 0–4 months | 73 | 0 | 0 | 0 | 0 |
| 4–8 months | 148 | 0 | 0 | 0 | 0 |
| 8–12 months | 99 | 0 | 0 | 0 | 0 |
| 1–2 years | 260 | 0 | 0 | 0 | 0 |
| >2 years | 236 | 489 | 552 | 245 | 38 |
| Median age in years (IQR)a | 35 (31–41) | 40 (35–45) | 40 (35–47) | 36 (32–42) | — |
| Sex | |||||
| Male | 717 | 362 | 552 | — | — |
| Female | 99 | 127 | 0 | — | — |
| Race | |||||
| Black | 119 | 461 | 43 | — | — |
| Nonblack | 697 | 28 | 509 | — | — |
| CD4 (cells/mm3) | |||||
| >400 | 610 | 208 | 317 | — | — |
| 201–400 | 171 | 158 | 143 | — | — |
| 51–200 | 22 | 94 | 63 | — | — |
| ≤50 | 1 | 27 | 29 | — | — |
| Unknown | 12 | 2 | 0 | 245 | 38 |
| VL (copies/ml) | |||||
| >50,000 | 159 | 125 | 120 | 15 | 0 |
| 10,001–50,000 | 203 | 150 | 92 | 15 | 0 |
| 401–10,000 | 213 | 138 | 92 | 27 | 0 |
| ≤400 | 229 | 51 | 105 | 181 | 38 |
| Unknown | 12 | 25 | 143 | 7 | 0 |
Cohort descriptions are provided in the Methods section.
Age at sample collection for HIVNET 001, ALIVE, and MACS; age at HIV infection for JHHCC.
ALIVE = AIDS Linked to the Intravenous Experience Study; HIVNET = HIV Network for Prevention Trial; IQR = interquartile range; JHHCC = Johns Hopkins Hospital Clinical Cohort; MACS = Multicenter AIDS Cohort Study; VL = viral load.
Laboratory methods
Samples were tested using a modification of the GS HIV Combo Ag/Ab EIA (Bio-Rad, Redmond, WA). Assay procedures were modified to measure HIV antibody avidity, by the addition of a denaturing agent, diethylamine (DEA)6 to disrupt the binding of low-avidity antibodies. In duplicate, 10 μl of sample was mixed with 25 μl of conjugate 1 and 65 μl phosphate-buffered saline (PBS). The samples were incubated for 30 min at 4°C. At all steps, plates were covered with a plastic plate cover rather than a plate sealer. Following sample incubation and the assay protocol wash, 100 μl of 0.025 M DEA (diluted in deionized water) was added to the first well of each sample. Assay wash buffer (100 μl of 1×) was added to the second well of each duplicate pair and the plate was incubated for 30 min at 37°C. The remainder of the assay, beginning with addition of the second conjugate, was run according to the manufacturer's protocol. Each plate included two controls: a sample from an individual with known recent infection (<2 months postinfection, ZeptoMetrix samples 9014–05 or 9014–06) and a sample from an individual with known long-term infection (>2 years postinfection). The results were considered valid if the recent infection control had an AI <40% and the long-term control was >90% AI. Additionally, an HIV-1 antibody-negative and HIV-1 antibody-positive control was included on each plate. The AI was calculated by dividing the optical density (OD) of the DEA-treated well by the OD of the nontreated well for the same sample and multiplying by 100.30 Samples were classified as assay positive or assay negative based on the AI cutoff. Additionally, any sample where the wash buffer well OD was <1.0 was classified as assay positive.
Statistical methods
MDRI and FRR were estimated for the avidity assay alone and for the avidity assay in a MAA format that included HIV VL, where samples with VL <400 copies/ml were classified as assay negative. The MDRI was estimated using data obtained from samples from the HIVNET 001 cohort that were from individuals known to have been infected for <2 years. The estimated date of seroconversion was either 13 days postacute (HIV RNA+/antibody negative; 4 individuals) or the midpoint between last antibody-negative and first antibody-positive time points; 97 individuals). MDRIs were estimated using AI cutoffs ranging from 20% to 90%. Methods to calculate the MDRI using T = 2 have been described previously.17,31 The FRR was determined using data obtained from samples from the ALIVE, ES, JHHCC, and MACS cohorts. The FRR (proportion of samples from individuals infected >2 years that were misclassified as recently infected) was determined using AI cutoffs of 20%–90%.
Factors associated with misclassification in the ALIVE and MACS cohorts (AI <80% for individuals infected >2 years) were examined using logistic regression, with general estimating equations and a robust variance estimator as previously described.32 All factors associated with misclassification (p < .1) were included in the multivariate analysis. To determine if viral breakthrough influenced assay results, the change in AI within an individual was measured for all paired time points from the JHHCC group B samples. Changes in AI between longitudinally paired samples were compared by t-tests for individuals for three pair types: (1) sustained viral suppression, (2) viral suppression followed by viral breakthrough, or (3) viral breakthrough followed by viral suppression. Interoperator repeatability was also assessed by the Pearson's correlation coefficient between independent runs performed by two technicians.
Results
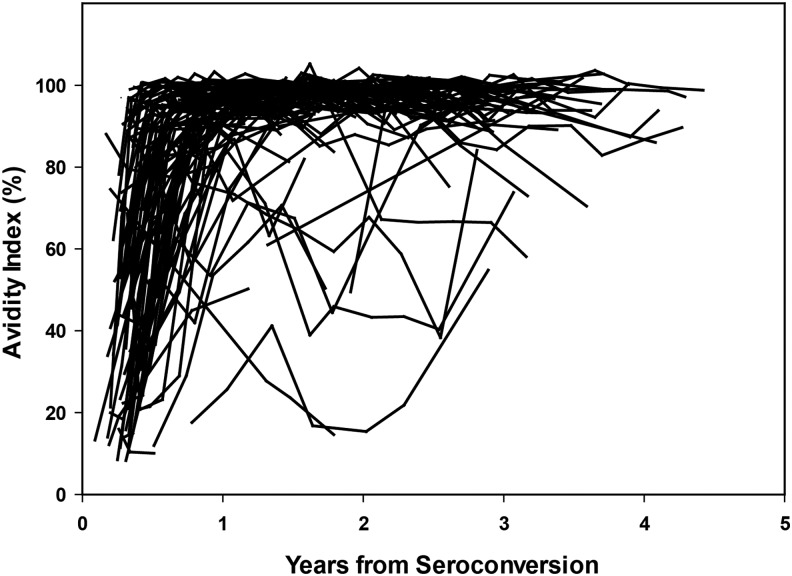
In this report, we analyzed 2,140 antibody-positive samples from 808 individuals with known duration of infection. The first analysis included data from 816 samples from 103 seroconverters in the HIVNET 001 cohort (Fig. 1). By 1 year after HIV seroconversion, AI results for 95% (98/103) of the seroconverters were >40%. Table 2 shows the performance characteristics of the modified Bio-Rad Combo assay alone and in a MAA that includes HIV VL. MDRI estimates ranged from 36.8 days (using an AI of 20% with VL) to 275.6 days (using an AI of 90% without VL). The percentage of samples below each AI cutoff by duration of infection is provided in Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid. FRRs ranged from 0% to 8.5%. The inclusion of VL reduced the MDRI by ∼20% for all AI cutoff values, and reduced the FRR by ∼50% for all AI cutoffs >40%.
FIG. 1.
Change in AI over time. AI values obtained using the Bio-Rad Combo Avidity assay are shown as a function of years after HIV seroconversion (data from 816 samples from 103 seroconverters in the HIVNET 001 cohort). AI, avidity index.
Table 2.
Performance Characteristics of the Bio-Rad Combo Avidity Assay
| Bio-Rad combo avidity | Bio-Rad combo avidity+HIV VL >400 copies/ml | |||
|---|---|---|---|---|
| Avidity indexCohort (%) | MDRI (95% CI)HIVNET 001 N = 816 | FRR% (95% CI)ALIVE+MACS+JHHCC+ES N = 1324 | MDRI (95% CI)HIVNET 001 N = 804 | FRR% (95% CI)ALIVE+JHHCC+MACS+ES N = 1149 |
| 20 | 50.6 (37.2–71.6) | 0.2 (0.0–0.6) | 36.8 (24.9–54.9) | 0.0 (0.0–0.3) |
| 30 | 84.2 (67.3–104.1) | 0.7 (0.3–1.3) | 63.8 (46.9–82.3) | 0.4 (0.1–0.9) |
| 40 | 109.6 (93.5–125.1) | 1.4 (0.9–2.2) | 94.4 (78.5–110.7) | 0.8 (0.4–1.5) |
| 50 | 135.4 (120.0–150.7) | 2.1 (1.4–3.0) | 108.9 (91.5–124.9) | 1.1 (0.6–1.9) |
| 60 | 150 (133.2–167.2) | 2.9 (2.1–4.0) | 118.8 (102.2–133.9) | 1.7 (1.1–2.7) |
| 70 | 181.5 (164.7–200.2) | 4.5 (3.5–5.8) | 137.5 (123.5–152.9) | 2.6 (1.8–3.7) |
| 80 | 205.5 (187.9–223.3) | 5.3 (4.2–6.7) | 152.2 (137.7–168.3) | 2.9 (2.0–4.0) |
| 90 | 275.6 (254.2–297.4) | 8.5 (7.1–10.2) | 196.7 (180.9–213.5) | 4.4 (3.2–5.7) |
Cohort descriptions are provided in the Methods section.
CI, confidence intervals; ES, elite suppressor; FRR, false-recent rate; MDRI, mean duration of recent infection.
The second analysis included data from 1,041 samples from 644 participants in the ALIVE and MACS cohorts. Factors associated with misclassification were examined using an AI cutoff of 80% (Table 3). Participant age was significantly associated with misclassification in the univariate analysis, but not in the multivariate analysis. Low VL was significantly associated with misclassification in both univariate and multivariate models [adjusted odds ratio (aOR): 3.26; confidence intervals (95% CIs) = 1.57–6.77). Low CD4 cell count (<50 cells/mm3) was also significantly associated with misclassification in both models (aOR: 8.45, 95% CI = 2.99–23.85).
Table 3.
Factors Associated with Misclassification of the Bio-Rad Combo Avidity Assay
| Avidity index <80% | |||
|---|---|---|---|
| % Misclassified (n/N) | OR (95% CI) | aOR(95% CI) | |
| Total | 5.76 (60/1041) | — | — |
| Sex | |||
| Male | 6.13 (56/914) | 1 | — |
| Female | 3.15 (4/127) | 0.52 (0.15–1.79) | — |
| Age (years) | |||
| <35 | 3.10 (8/258) | 1 | 1 |
| 35–40 | 3.09 (8/259) | 0.87 (0.33–2.28) | 0.75 (0.26–2.16) |
| 41–45 | 7.89 (18/228) | 2.10 (0.91–4.86) | 1.90 (0.80–4.53) |
| >45 | 8.78 (26/296) | 2.35 (1.03–5.36) | 1.95 (0.79–4.84) |
| Race | |||
| Black | 7.34 (37/504) | 1 | 1 |
| Nonblack | 4.28 (23/537) | 0.58 (0.32–1.05) | 0.65 (0.31–1.35) |
| HIV VL (copies/ml) | |||
| >50,000 | 6.05 (25/413) | 1 | 1 |
| 10,001–50,000 | 2.89 (7/242) | 0.53 (0.27–1.03) | 0.53 (0.25–1.10) |
| 401–10,000 | 3.48 (8/230) | 0.63 (0.32–1.25) | 0.82 (0.35–1.93) |
| ≤400 | 12.82 (20/156) | 1.93 (1.04–3.58) | 3.26 (1.57–6.77) |
| CD4 cell count (cells/mm3) | |||
| >500 | 3.02 (11/364) | 1 | 1 |
| 201–500 | 5.82 (27/464) | 1.81 (0.97–3.38) | 2.14 (1.02–4.48) |
| 51–200 | 7.64 (12/157) | 2.47 (1.10–5.53) | 2.79 (1.05–7.42) |
| ≤50 | 17.86 (10/56) | 4.90 (1.93–12.42) | 8.45 (2.99–23.85) |
Cohort descriptions are provided in the Methods section. Statistically significant values (p < .05) are shown in bold text.
aOR, adjusted odds ratio; n, number misclassified; N, number tested; OR, odds ratio.
We next compared data obtained from the four cohorts that included individuals infected >2 years. The median AI and interquartile ranges (IQRs) were: ALIVE: 98.4 (94.3–99.8); MACS: 98.7 (94.3–100.1); JHHCC: 99.0 (94.3–100.2); and ES: 97.6 (94.8–99.1). The FRRs using an AI cutoff of 80% were: ALIVE: 7.2%; MACS: 4.4%; JHHCC: 4.1%; and ES: 0.0%.
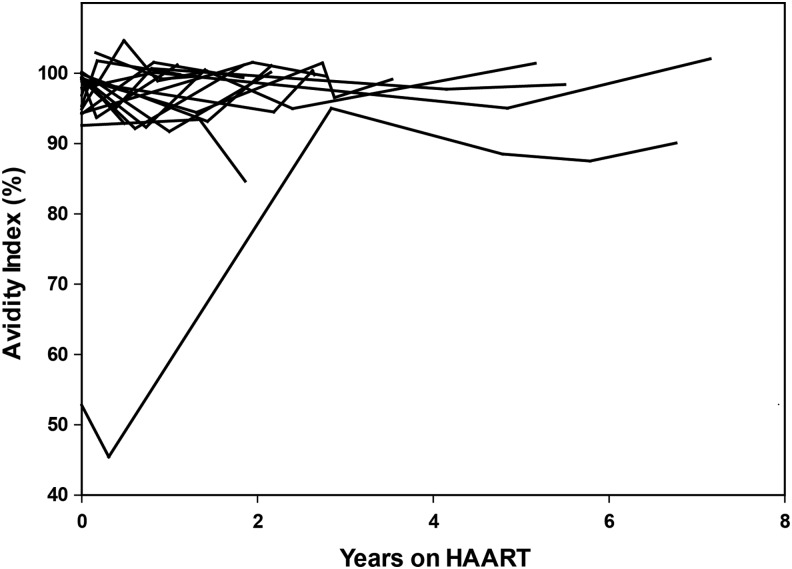
While VL was associated with misclassification, the median AIs for virally suppressed and viremic individuals were comparable. The median AI for samples from individuals with VL <400 was 98.2 (IQR = 93.5–100, N = 375); the median AI for samples from individuals with VL >400 was 98.6 (IQR = 94.6–100, N = 774). There was little change in AI values over time among virally suppressed individuals (0.31% decrease per year on ART, Fig. 2); when the values from one outlier were removed whose AI increased over time, the net decrease was 0.72% per year.
FIG. 2.
Change in AI over time for virally suppressed individuals on ART. AI values obtained using the Bio-Rad Combo Avidity assay are shown as a function of years on ART for a subset of 18 individuals in the JHHCC. These individuals were suppressed on ART with no viral breakthrough events. ART, antiretroviral treatment; JHHCC, Johns Hopkins Hospital Clinical Cohort.
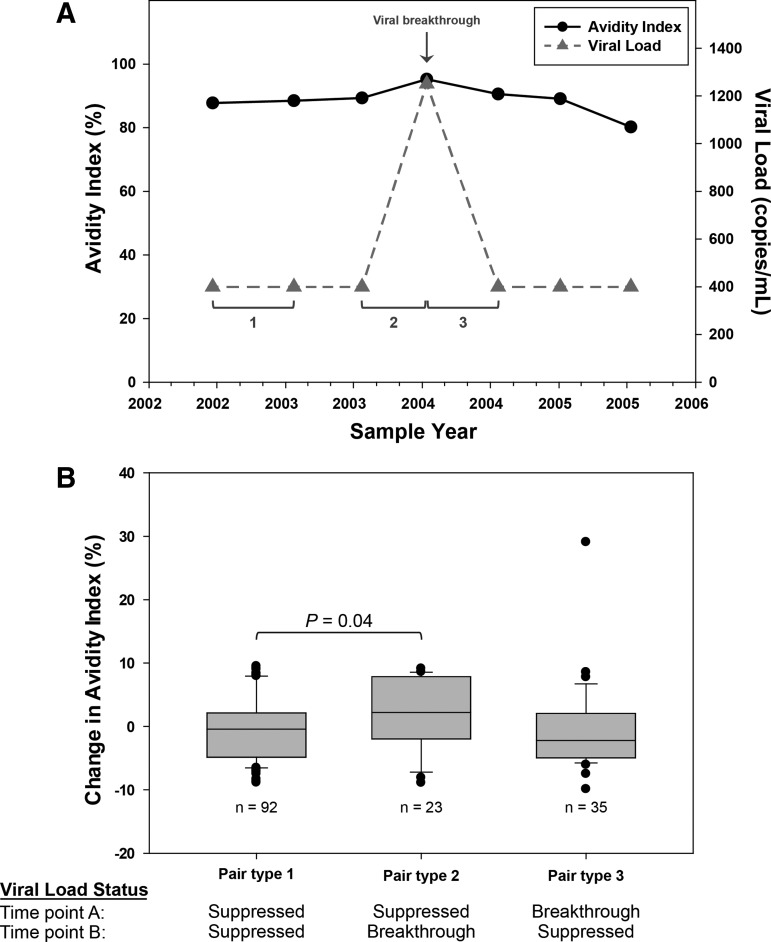
The last analysis assessed the relationship between viral breakthrough and AI (Fig. 3). Representative data from a single individual are shown in Figure 3A. Three different types of paired values were defined and analyzed (Fig. 3A). Results from 20 individuals are shown in Figure 3B. There was mean increase in AI of 2.5% between visits with viral suppression and visits with viral breakthrough (p = .04; Fig. 3B). AI values were significantly correlated between two technologists (r = 0.93; p < .01) when comparing independent runs.
FIG. 3.
Impact of viral breakthrough on AI. (A) Representative results from a single individual are shown. A viral breakthrough event (VL >400 copies/ml) is indicated by the arrow. Time points were paired based on VL status at two study visits; pair type 1: both time points virally suppressed; pair type 2: viral suppression followed by viral breakthrough; pair type 3: breakthrough followed by viral suppression. (B) Results obtained for 20 individuals. The plot shows the change in AI for three types of paired study visits (A). The p-value for the difference in AI for pair type 1 and pair type 2 is shown. Pairs where both time points had viral breakthrough (six pairs) were excluded. VL, viral load.
Discussion
We developed and evaluated an avidity assay for cross-sectional HIV incidence estimation. This assay, the Bio-Rad Combo Avidity assay, is modified from a commercially available fourth-generation HIV screening test. When used in a single assay format, this assay had performance characteristics that were very close to the minimum performance characteristics recommended by the WHO/UNAIDS Working Group on Incidence Assays (an MDRI of >120 days with an FRR <2%).19 Using an AI cutoff of 50%, the MDRI was 135 days (95% CI: 120–151) with an FRR of 2.1% (95% CI: 1.4–3.0; 26 of 1,324 samples were misclassified as assay positive). Note that this FRR was obtained for a sample set that included virally suppressed individuals (23 elite suppressors and 266 individuals who were virally suppressed on ART); viral suppression is frequently associated with misclassification.
The performance of the Bio-Rad Combo Avidity assay was similar to the performance of an avidity assay based on a third-generation Bio-Rad HIV screening test (the JHU-Bio-Rad Avidity assay). For the previous JHU-modified Bio-Rad Avidity assay, the MDRI was obtained using an AI cutoff of 40% with the same study cohort samples (HIVNET 001); this MDRI is slightly longer than the MDRI of 110 days that we obtained using the same AI cutoff with the new Bio-Rad Combo Avidity assay. The FRRs for these two assays were nearly identical (1.1% vs. 1.3%) for analyses obtained using the same study cohort samples (ALIVE and MACS).
We compared the performance of the Bio-Rad Combo Avidity assay to assays that were recently evaluated by the Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA).17 In the CEPHIA evaluation, the FRR for individuals on ART was 50% for the CDC-modified Bio-Rad Avidity assay and 58.8% for LAg-Avidity assay; the FRR was 12.9% for elite suppressors for both assays. In contrast, none of the 23 elite suppressors evaluated in this report was misclassified as assay positive using the new Bio-Rad Combo Avidity assay. Using the same set of specimens from the ALIVE and MACS cohorts, the Bio-Rad Combo Avidity assay had a similar FRR to the LAg-Avidity assay, using a LAg-Avidity assay cutoff of 1.5 OD-n (FRR: 5.8% and 4.8%, respectively).13 The same factors were associated with misclassification in multivariate analysis for these two assays: viral suppression (VL <400 copies/ml) and low CD4 cell count (<50 cells/mm3).
In another report, the third-generation Bio-Rad GS HIV-1/HIV-2 Plus O EIA was modified for incidence testing using urea as the chaotropic agent, rather than DEA.27 The MDRI of the urea-based assay using an AI of 40% was estimated at 108 days using a CEPHIA qualification panel; the FRR for the assay was 0.89% (95% CI: 0.2%–2.3%). That analysis was performed using 449 samples with different infecting subtypes; samples from virally suppressed individuals were not included. At the same AI cutoff (40%), the Bio-Rad Combo Avidity assay described in this report had an MDRI of 110 days with an FRR of 1.3 (95% CI: 0.7–2.0).
Another avidity assay has also been developed using a different fourth-generation HIV screening assay (the Architect HIV Ag/Ab Combo EIA), which uses an automated system for sample processing.33 In that assay, samples are incubated with a chaotropic agent during the initial sample incubation period; in contrast, in the assay described in this report, the chaotropic agent incubation occurs in a separate step, after sample incubation and a wash. In the Abbott Combo-based avidity assay, the FRR was 6.6% after excluding specimens from individuals on ART.
In this report, the Bio-Rad Combo Avidity assay was evaluated using HIV subtype B samples from the United States. Further evaluation of this assay is needed for samples from other geographic regions with other HIV subtypes. Additionally, the impact of the timing of ART initiation on antibody maturation needs to be evaluated as very early treatment blunts the antibody response to HIV.34 With recent evidence demonstrating the benefit of early ART initiation,35 a greater proportion of individuals infected within the first year of infection will be on ART and this will likely impact most antibody-based measures used from cross-sectional incidence testing. Another problem to address is the capacity to report the presented technology to other laboratories. Assays modified for incidence applications that do not use reagents specifically manufactured for that purpose can be difficult to replicate in different laboratories. Standardized chaotropic agents supplied from commercial vendors would greatly assist in this matter. Further evaluation is also needed to evaluate use of this assay in combination with other assays and biomarkers.21 Because the fourth-generation Bio-Rad GS HIV Combo Ag/Ab EIA assay is widely available, the Bio-Rad Combo Avidity assay may offer an alternative option for HIV incidence testing in settings, where the third-generation Bio-Rad GS HIV-1/HIV-2 Plus O EIA assay is not available.
Supplementary Material
Acknowledgments
This work was supported by the HIV Prevention Trials Network (HPTN) sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute on Drug Abuse (NIDA), the National Institute of Mental Health (NIMH), and the Office of AIDS Research, of the National Institutes of Health (NIH), Department of Health and Human Services (DHHS) (UM1AI068613), and by R01 AI095068 (NIAID). Additional funding was provided by the Division of Intramural Research, NIAID, NIH.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Mastro TD, Kim AA, Hallett T, Rehle T, Welte A, Laeyendecker O, et al. : Estimating HIV incidence in populations using tests for recent infection: Issues, challenges and the way forward. J HIV AIDS Surveill Epidemiol 2010;2:1–14 [PMC free article] [PubMed] [Google Scholar]
- 2.Brookmeyer R: Measuring the HIV/AIDS epidemic: Approaches and challenges. Epidemiol Rev 2010;32:26–37 [DOI] [PubMed] [Google Scholar]
- 3.Duong YT, Qiu M, De AK, Jackson K, Dobbs T, Kim AA, et al. : Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: Potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012;7:e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobbs T, Kennedy S, Pau CP, McDougal JS, Parekh BS: Performance characteristics of the immunoglobulin G-capture BED-enzyme immunoassay, an assay to detect recent human immunodeficiency virus type 1 seroconversion. J Clin Microbiol 2004;42:2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy G, Parry JV: Assays for the detection of recent infections with human immunodeficiency virus type 1. Euro Surveill 2008;13:pii [PubMed] [Google Scholar]
- 6.Masciotra S, Dobbs T, Candal D, Hanson D, Delaney K, Rudolph D, et al. : Antibody avidity-based assay for identifying recent HIV-1 infections based on genetic systems TM 1/2 plus O EIA. In: 17th Conference on Retroviruses and Opportunistic Infections, abstract 937, San Francisco, CA; February16–19; 2010 [Google Scholar]
- 7.Keating SM, Hanson D, Lebedeva M, Laeyendecker O, Ali-Napo NL, Owen SM, et al. : Lower-sensitivity and avidity modifications of the vitros anti-HIV 1 + 2 assay for detection of recent HIV infections and incidence estimation. J Clin Microbiol 2012;50:3968–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suligoi B, Massi M, Galli C, Sciandra M, Di Sora F, Pezzotti P, et al. : Identifying recent HIV infections using the avidity index and an automated enzyme immunoassay. J Acquir Immune Defic Syndr 2003;32:424–428 [DOI] [PubMed] [Google Scholar]
- 9.Curtis KA, Longosz AF, Kennedy MS, Keating S, Heitman J, Laeyendecker O, et al. : Inter-laboratory assessment of a prototype multiplex kit for determination of recent HIV-1 infection. PLoS One 2013;8:e77765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laeyendecker O, Rothman RE, Henson C, Horne BJ, Ketlogetswe KS, Kraus CK, et al. : The effect of viral suppression on cross-sectional incidence testing in the Johns Hopkins Hospital Emergency Department. J Acquir Immune Defic Syndr 2008;48:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marinda ET, Hargrove J, Preiser W, Slabbert H, van Zyl G, Levin J, et al. : Significantly diminished long-term specificity of the BED capture enzyme immunoassay among patients with HIV-1 with very low CD4 counts and those on antiretroviral therapy. J Acquir Immune Defic Syndr 2010;53:496–499 [DOI] [PubMed] [Google Scholar]
- 12.Hayashida T, Gatanaga H, Tanuma J, Oka S: Effects of low HIV type 1 load and antiretroviral treatment on IgG-capture BED-enzyme immunoassay. AIDS Res Hum Retroviruses 2008;24:495–498 [DOI] [PubMed] [Google Scholar]
- 13.Longosz AF, Mehta SH, Kirk GD, Margolick JB, Brown J, Quinn TC, et al. : Incorrect identification of recent HIV infection in adults in the United States using a limiting-antigen avidity assay. AIDS 2014;28:1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longosz AF, Serwadda D, Nalugoda F, Kigozi G, Franco V, et al. : Impact of HIV subtype on performance of the limiting antigen-avidity enzyme immunoassay, the Bio-Rad Avidity Assay, and the BED capture immunoassay in Rakai, Uganda. AIDS Res Hum Retroviruses 2014;30:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welte A, McWalter TA, Barnighausen T: A simplified formula for inferring HIV incidence from cross-sectional surveys using a test for recent infection. AIDS Res Hum Retroviruses 2009;25:125–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassanjee R, McWalter TA, Barnighausen T, Welte A: A new general biomarker-based incidence estimator. Epidemiology 2012;23:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassanjee R, Pilcher CD, Keating SM, Facente SN, McKinney E, Price MA, et al. : Independent assessment of candidate HIV incidence assays on specimens in the CEPHIA repository. AIDS 2014;28:2439–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Incidence Assay Critical Path Working Group: More and better information to tackle HIV epidemics: Towards improved HIV incidence assays. PLoS Med. 2011;8:e1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UNAIDS/WHO working group on global HIV/AIDS and STI surveillance: When and how to use assays for recent infection to estimate HIV incidence at a population level. 2011. Available at www.who.int/diagnostics_laboratory/hiv_incidence_may13_final.pdf, accessed May29, 2015
- 20.Bill & Melinda Gates Foundation Letters of Inquiry (LOI): New biomarkers for HIV incidence measurement. 2012. Available at https://docs.gatesfoundation.org/Documents/hiv-incidence-rules-and-guidelines.pdf, accessed May29, 2015
- 21.Laeyendecker O, Brookmeyer R, Cousins MM, Mullis CE, Konikoff J, Donnell D, et al. : HIV incidence determination in the United States: A multiassay approach. J Infect Dis 2013;207:232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laeyendecker O, Brookmeyer R, Mullis C, Donnell D, Lingappa J, Celum C, et al. : Specificity of four laboratory approaches for cross-sectional HIV incidence determination: Analysis of samples from adults with known non-recent HIV infection from five African countries. AIDS Res Hum Retroviruses 2012;28:1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laeyendecker O, Kulich M, Donnell D, Komarek A, Omelka M, Mullis CE, et al. : Development of methods for cross-sectional HIV incidence estimation in a large, community randomized trial. PLoS One 2013;8:e78818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlahov D, Anthony JC, Munoz A, Margolick J, Nelson KE, Celentano DD, et al. : The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: Description of methods and characteristics of participants. NIDA Res Monogr 1991;109:75–100 [PubMed] [Google Scholar]
- 25.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR, Jr: The Multicenter AIDS Cohort Study: Rationale, organization, and selected characteristics of the participants. Am J Epidemiol 1987;126:310–318 [DOI] [PubMed] [Google Scholar]
- 26.Celum CL, Buchbinder SP, Donnell D, Douglas JM, Jr., Mayer K, Koblin B, et al. : Early human immunodeficiency virus (HIV) infection in the HIV Network for Prevention Trials Vaccine Preparedness Cohort: Risk behaviors, symptoms, and early plasma and genital tract virus load. J Infect Dis 2001;183:23–35 [DOI] [PubMed] [Google Scholar]
- 27.Moore RD. Understanding the clinical and economic outcomes of HIV therapy: The Johns Hopkins HIV clinical practice cohort. J Acquir Immune Defic Syndr Hum Retrovirol 1998;17 Suppl 1:S38–S41 [DOI] [PubMed] [Google Scholar]
- 28.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, et al. : Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol 2007;81:2508–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr JK, Osinusi A, Flynn CP, Gilliam BL, Maheshwari V, Zhao RY: Two independent epidemics of HIV in Maryland. J Acquir Immune Defic Syndr 2010;54:297–303 [DOI] [PubMed] [Google Scholar]
- 30.Shepherd SJ, McAllister G, Kean J, Wallace LA, Templeton KE, Goldberg DJ, et al. : Development of an avidity assay for detection of recent HIV infections. J Virol Methods 2015;217:42–49 [DOI] [PubMed] [Google Scholar]
- 31.Kassanjee R, McWalter TA, Welte A: Short Communication: Defining optimality of a test for recent infection for HIV incidence surveillance. AIDS Res Hum Retroviruses 2014;30:45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laeyendecker O, Brookmeyer R, Oliver AE, Mullis C, Eaton KP, Mueller AC, et al. : Factors associated with incorrect identification of recent HIV infection using the BED capture immunoassay. AIDS Res Hum Retroviruses 2012;28:816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suligoi B, Rodella A, Raimondo M, Regine V, Terlenghi L, Manca N, et al. : Avidity Index for anti-HIV antibodies: Comparison between third- and fourth-generation automated immunoassays. J Clin Microbiol 2011;49:2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hare CB, Pappalardo BL, Busch MP, Karlsson AC, Phelps BH, Alexander SS, Bentsen C, Ramstead CA, Nixon DF, Levy JA, Hecht FM: Seroreversion in subjects receiving antiretroviral therapy during acute/early HIV infection. Clin Infect Dis 2006;42:700–708 [DOI] [PubMed] [Google Scholar]
- 35.INSIGHT START Study Group: Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med. 2015;373:795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.