Abstract
Nonalcoholic fatty liver disease (NAFLD/steatosis) is a metabolic disease characterized by the incorporation of fat into hepatocytes. In this study, we developed an in vitro model for NAFLD based on hepatocyte-like cells (HLCs) differentiated from human pluripotent stem cells. We induced fat storage in these HLCs and detected major expression changes of metabolism-associated genes, as well as an overall reduction of liver-related microRNAs. We observed an upregulation of the lipid droplet coating protein Perilipin 2 (PLIN2), as well as of numerous genes of the peroxisome proliferator-activated receptor (PPAR) pathway, which constitutes a regulatory hub for metabolic processes. Interference with PLIN2 and PPARα resulted in major alterations in gene expression, especially affecting lipid, glucose, and purine metabolism. Our model recapitulates many metabolic changes that are characteristic for NAFLD. It permits the dissection of disease-promoting molecular pathways and allows us to investigate the influences of distinct genetic backgrounds on disease progression.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a widespread disease in the western hemisphere. Due to a high-fat diet and a lack of exercise, hepatocytes of NAFLD patients accumulate fat in the form of lipid droplets (LDs) [1]. This is often associated with type 2 diabetes and considered part of the metabolic syndrome [1]. Insulin resistance and obesity-associated chronic inflammation of adipose tissue are critical factors for the development and progression of NAFLD [2,3]. This is often seen as a “first hit” manifesting in the rather benign accumulation of LDs, called steatosis. A “second hit”, frequently due to an increase of reactive oxygen species-mediated stress, induces the progression toward nonalcoholic steatohepatitis (NASH), which is accompanied by liver inflammation and fibrosis [3]. Approximately, 29% of patients with NASH develop cirrhosis. Up to 27% of these further develop hepatocellular carcinoma [1].
Hepatocytes store triacylglycerides (TAGs) in LDs as a reaction to an overload with free fatty acids. These are either derived directly from the diet or result from inflammation induced lipolysis in adipose tissues [2]. The occurrence of LDs in >5% of hepatocytes is the main diagnostic criterion for NAFLD [1].
In LDs, TAGs are enclosed by a lipid monolayer, which is encapsulated by distinct proteins, predominantly from the PAT (Perilipin/ADRP/TIP47) family [4–6]. Perilipins regulate hydrolysis of TAGs by controlling the activity of lipases and their access to LDs [7–9]. Perilipin 2 (PLIN2 or Adipophilin, ADRP) is ubiquitously expressed and plays a major role in the formation of LDs [10–12]. PLIN2 expression correlates with LD content in hepatocytes [13]. A reduction of PLIN2 expression with antisense oligonucleotides reduced liver TAG content and decreased the expression of genes involved in fatty acid and steroid metabolism in mice [14,15]. In addition, PLIN2 knockout mice develop neither obesity nor NAFLD when fed a high-fat diet because they have a higher energy turnover compared to their wild-type counterparts [16].
Nutrition and energy uptake are important factors for the development of NAFLD. However, there exist major differences between humans and mice. Various established diets reproduce effects of NAFLD/NASH in mice. Unfortunately, they fail to mirror the whole spectrum of symptoms observed in humans. While high-fat diets induce obesity and NAFLD, mice generally do not proceed toward NASH even if the diet is supplemented with fructose. To induce NASH, mice are usually fed with a methionine–choline-deficient diet. A major drawback of this diet, however, is the fact that mice do not become obese, which is a major risk-factor for NAFLD in humans [17,18]. In addition, there exist several knockout mouse models, none of which is capable of reflecting all aspects of the disease [17].
Several groups have used human hepatocarcinoma cell lines or immortalized primary hepatocytes to model NAFLD [19,20]. However, cancer-derived cell lines are of limited use for dissecting the molecular basis of NAFLD as they harbor genomic and hence functional aberrations compared to healthy primary liver cells [21,22]. The use of liver biopsy-derived primary human hepatocytes for modeling NAFLD is also limited because they can only be cultivated for a few days before the onset of dedifferentiation [23] or have to be immortalized by virus-mediated transduction with SV40. In addition, liver biopsies, especially those from the early stages, are very rare.
To overcome these limitations, we in this study aimed at dissecting the molecular basis of NAFLD using hepatocyte-like cells (HLCs), which were in vitro derived from human pluripotent stem cells (hPSCs). We used the human embryonic stem cell (ESC) line H1, as well as induced pluripotent stem cells (iPSCs), derived from fetal foreskin fibroblasts of a healthy individual [24,25]. We were able to monitor the accumulation of fat in the HLCs, as well as major biochemical alterations concerning lipid, glucose, and purine metabolism. Our new model system is suitable for the analysis of disease triggering factors, as well as new therapeutics.
Material and Methods
Cell culture
HepG2 cells (ATCC®HB-8065™) were cultured in DMEM low glucose with 10% FCS, 1% Penicillin/Streptomycin, and 1% GlutaMAX (Gibco). For fat induction, cells were induced with 50 μM oleic acid (OA) (Stock solution 100 mM in ethanol). As control, cells were treated with the corresponding amount of ethanol. Fat induction was performed 24 h after passaging.
Differentiation of hPSCs into HLCs
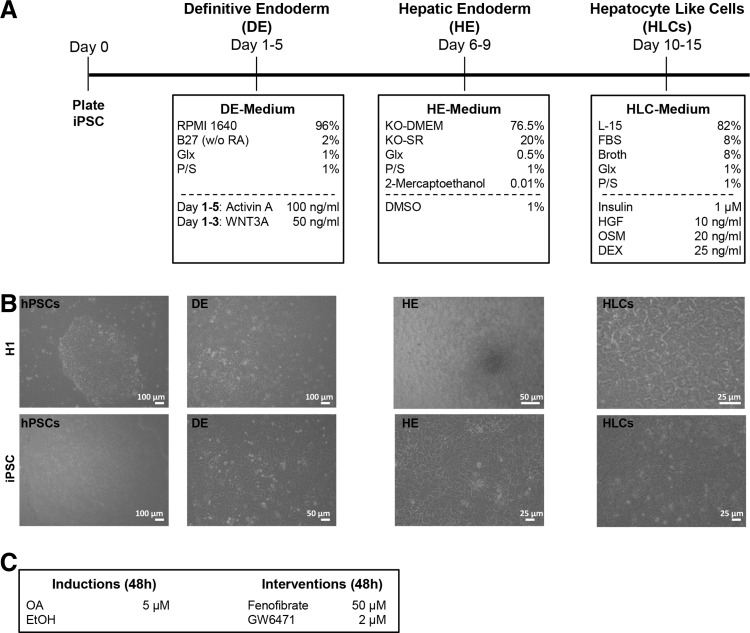
hPSCs were cultured on Matrigel (Corning) coated plates in TeSR E8 medium (STEMCELL Technologies). Medium was changed daily and spontaneously differentiated cells were removed manually before splitting the cells. One or two days after passaging, differentiation into definitive endoderm was induced with definitive endoderm medium: 96% RPMI 1640, 2% B27 (without retinoic acid), 1% GlutaMAX (Glx), 1% Penicillin/Streptomycin (P/S) (all Gibco), 100 ng/mL Activin A (Peprotech), and for the first 3 days, 50 ng/mL WNT3A (R&D). After 5 days, medium was changed toward hepatic endoderm medium as follows: 78% Knockout DMEM, 20% Knockout serum replacement, 0.5% Glx, 1% P/S, 0.01% 2-Mercaptoethanol (all Gibco), and 1% DMSO (Sigma).
After 5 days, differentiation was continued with HLC medium as follows: 82% Leibovitz 15 medium, 8% fetal calf serum, 8% Tryptose Phosphate Broth, 1% Glx, 1% P/S (all Gibco) with 1 μM Insulin (Sigma), 10 ng/mL hepatocyte growth factor (HGF) (Peprotech), 20 ng/mL Oncostatin M (OSM) 209 a.a. (Immunotools), and 25 ng/mL Dexamethasone (DEX) (Sigma) (Fig. 1A). During the whole differentiation period, medium was changed daily.
FIG. 1.
Differentiation of hPSCs into HLCs. hPSCs were differentiated into HLCs using a three-step protocol (see also Materials and Methods section) (A). Morphological changes of the differentiating cells are documented for each stage. At the definitive endoderm (DE) stage, the dense cell–cell contact is lost, and cells acquire the typical endodermal morphology. The characteristic polygonal shape of hepatocytes is already evident at the hepatic endoderm (HE) stage. This morphology is even more pronounced at the HLC stage (B). Two days before the end of differentiation, steatosis was induced with 50 μM OA, and control cells were treated with an equal volume of ethanol. To interfere with LD formation, PPARα modulators were also added on the same day (C). HLCs, hepatocyte-like cells; hPSCs, human pluripotent stem cells; LD, lipid droplet; OA, oleic acid; PPARα, peroxisome proliferator-activated receptor alpha.
Fat induction in HLCs was performed on day 12 of the differentiation process. Interference with peroxisome proliferator-activated receptor alpha (PPARα) activity was performed by treatment with 50 μM Fenofibrate (agonist) or 2 μM GW6471 (antagonist, both from Cayman Chemical) in parallel with OA induction (Fig. 1C).
Liver-specific biochemical assays of HLCs
The amount of urea produced by the cells over a period of 24 h was determined from the cell culture supernatant using the QuantiChrom™ Urea Assay Kit (BioAssay Systems) according to the manufacturer's recommendations. Cytochrome p450 3A4, 3A5, and 3A7 activity was measured with the P450-Glo™ CYP3A4 Assay Luciferin-PFBE (Promega) using a luminometer (Lumat LB 9507; Berthold Technologies). The presence of active transporter proteins was assessed by the uptake and release of Indocyanine Green dye. Cells were incubated for 30 min with 1 mg/mL Cardiogreen (Santa Cruz Biotechnology, Inc.). Afterward, they were washed with PBS and images were captured with a light microscope (Primo Vert; Zeiss). Subsequently, cells were cultured in their usual medium for 6 h and images were again captured.
Staining of LDs
Paraformaldehyde-fixed cells were incubated for 20 min with either a 60% working solution of Oil Red O (Sigma) or with BODIPY 493/503 (1 μg/mL; Life technologies) in PBS/0.05% Tween. After washing, images were captured with a light microscope (Primo Vert; Zeiss) or a fluorescence microscope, respectively (LSM700; Zeiss).
Immunocytochemistry
For intracellular antibody staining, paraformaldehyde-fixed cells were permeabilized and unspecific binding sites were blocked by incubating for 2 h at room temperature with blocking buffer (1 × PBS with 10% normal goat or donkey serum, 1% BSA, 0.5% Triton, and 0.05% Tween). Afterward, blocking buffer was diluted 1:2 with 1 × PBS and cells were incubated with the primary antibody overnight at 4°C. Cells were washed thrice with 1 × PBS/0.05% Tween and incubated with the secondary antibody, for 2 h at room temperature. Cells were washed as above and images captured using a fluorescence microscope (LSM700; Zeiss).
Extracellular stainings were performed in the same manner without detergents. The following primary antibodies were used: Alpha Fetoprotein, Albumin (Sigma) E Cadherin (CST), HNF4α (Abcam), SOX17 (R&D), and PLIN2 (Proteintech). For details on antibodies, see Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd. DNA was stained with Hoechst 33342. Individual channel images were processed and merged with Photoshop CS6.
Western blot
Cells were lysed in 1 × RIPA buffer (50 mM Tris HCl, pH 8, 150 mM NaCl, 1% IGEPAL (NP-40), 0.1% SDS, 1 mM EDTA, and 0.5% Na-Deoxycholate) with protease inhibitors. Twenty microgram of protein was analyzed by western blot with antibodies against PLIN2 (Proteintech) and ACTIN (CST). HRP coupled secondary antibodies were obtained from Abcam. Chemiluminescence was detected on a Fusion FX instrument (PeqLab) and analyzed with Fusion Capt Advance software (PeqLab) using rolling ball background correction.
RNA isolation and quantitative real-time polymerase chain reaction
For RNA isolation, up to 500,000 cells were lysed in 500 μL TRIzol. RNA was isolated with the Direct-zol™ RNA Isolation Kit (Zymo Research) according to the user's manual, including the DNase digestion step. Reverse transcription of up to 1 μg RNA was performed with the TaqMan Reverse Transcription (RT) Kit (Applied Biosystems). Primers were purchased from MWG; sequences are provided in Supplementary Table S2. Real-time PCR was performed in technical triplicates with Power SYBR Green Master Mix (Life technologies) on a VIIA7 or StepOnePlus (both Life technologies) machine. Mean values were normalized to actin and, subsequently, to the ethanol control. In case of siRNA experiments, data were normalized to the ethanol-treated nt siRNA sample. Experiments were carried out in biological duplicates and are depicted as mean values (log2) with standard error of the mean. Unpaired student's t-tests were performed for calculating significances.
Transcriptome and bioinformatics analysis
Microarray experiments were performed using the Affymetrix PrimeView chip (BMFZ, Düsseldorf). Details of data analysis are given in Supplementary Methods.
Liver-specific microRNA array
cDNA synthesis was carried out using the miScript II RT Kit (QIAGEN) according to the user's manual. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) and data analysis were performed according to the user's manual (miScript miRNA PCR Array; QIAGEN). All values were normalized to six different housekeeping genes (snoRNA/snRNA).
microRNA target gene validation
microRNA target gene validation was performed in HEK293T cells as previously described [26,27]. In brief, 3′ untranslated region (UTR) fragments of putative target genes were cloned at the 3′ end of the Firefly luciferase open reading frame (ORF) in dual-luciferase reporter vector pmirGLO (Promega). Ds-oligonucleotides spanning the predicted microRNA binding sites for ATL3 (hsa-miR-106b) and CPAMD8 (hsa-miR-122) were used (Supplementary Table S3), whereas the 3,406-bp EPHA7-3′ UTR was represented by a 1,384-bp PCR fragment covering two predicted hsa-miR-106b binding sites (Supplementary Table S4). Normalization for effects of endogenous HEK293T microRNAs on the given 3′ UTR was achieved by transfection of both empty pmirGLO, as well as pmirGLO/3′ UTR, into HEK293T cells. Pairwise cotransfections of empty pmirGLO or pmirGLO/3′ UTR with the microRNA mimic of interest (Supplementary Table S5) (Dharmacon) were performed. Firefly and Renilla activities were determined 24 h after transfection. All transfections were performed in at least two independent biological experiments with quadruple transfections each. Mean values with standard deviations are shown. Significances were calculated with unpaired Student's t-test, ***P ≤ 0.001.
PLIN2 knockdown
PLIN2 knockdown was performed by transfecting 5 pmol PLIN2 siRNA or a nontarget (nt) control siRNA (Thermo Scientific) into 100,000 HepG2 cells using lipofectamine RNAiMAX (Life Technologies). Forty-eight hours post transfection, the cells were harvested for further analyses.
Results
Pluripotent stem cells differentiate into HLCs and show characteristic activities of liver cells
We differentiated hPSCs into HLCs using a three-step protocol, which is based on published protocols [28–30], but has been adapted in our laboratory to optimize the individual differentiation steps (Fig. 1A). We used the established ESC line H1 and an iPSC line derived from fetal foreskin fibroblasts [25] to compare two distinct hPSC lines. Morphological changes were monitored at every step of the differentiation process (Fig. 1B). ESCs and iPSCs behaved similarly during differentiation. At the definitive endoderm stage, they adopted a typical flat and elongated shape; then at the hepatic endoderm stage, cells from both sources started to adopt the polygonal shape that is characteristic for hepatocytes. At the maturation step, that is, the HLC stage, the vast majority of the cells had acquired the polygonal morphology.
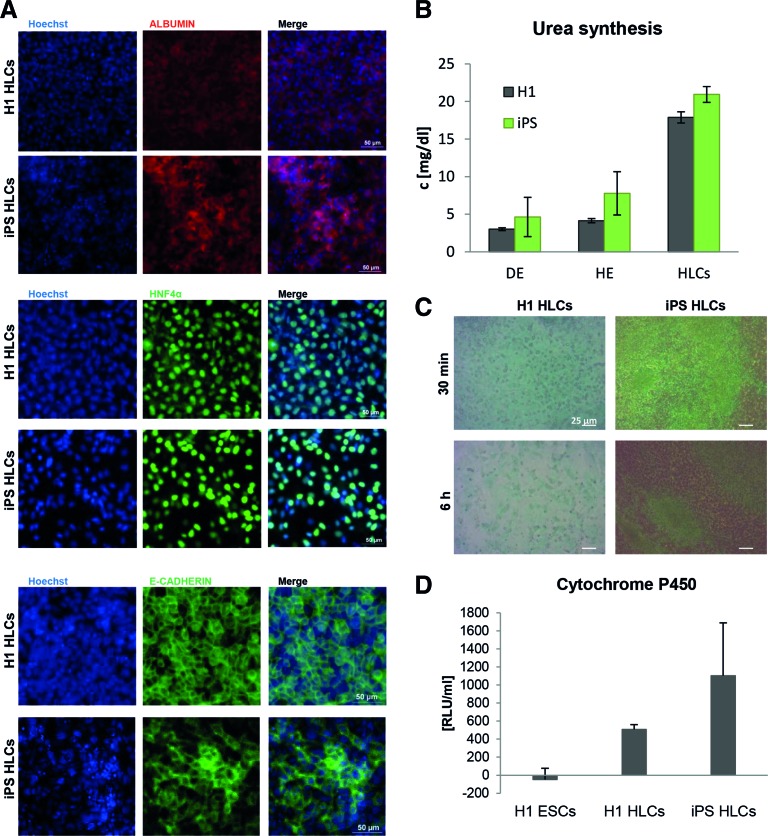
Stainings with antibodies for the hepatocyte markers albumin, HNF4α, and E-Cadherin were all positive for HLCs derived from ESCs and iPSCs (Fig. 2A). In addition, several activity tests indicated that HLCs behave like hepatocytes (Fig. 2B–D). They were able to synthesize urea (Fig. 2B) and expressed active transporters as demonstrated by their ability to take up and release indocyanine green dye (Fig. 2C). Another important characteristic of hepatocytes is the activity of phase I enzymes, for example, members of the Cytochrome p450 family, which was strongly increased in HLCs compared to undifferentiated hPSCs (Fig. 2D). Thus, we can reliably generate functional HLCs from ESCs, as well as from iPSCs, which are suitable for disease modeling.
FIG. 2.
Characterization of HLCs. The ES cell line H1 and iPS cells were differentiated into HLCs as described in Fig. 1. (A) HLCs derived from H1 (upper row) and iPS cells (lower row) express the liver-specific proteins Albumin, HNF4α (scale bar 50 μm), and E-Cadherin (scale bar 20 μm). (B–D) HLCs have liver-specific activity. (B) Urea production increases during differentiation and reaches its maximum at the HLC. (C) HLCs take up indocyanine green dye (upper row) within 30 min and release it within 6 h (lower row). (D) HLCs have phase I enzyme activity as measured by a luminometric assay detecting Cytochrome P450 (CYP) 3A4, 3A5, and 3A7 activity. In all cases, representative experiments are shown. In (B) and (D), data represent mean value ± standard deviation. HLCs, hepatocyte-like cells. Color images available online at www.liebertpub.com/scd
HLCs can be induced to accumulate LDs
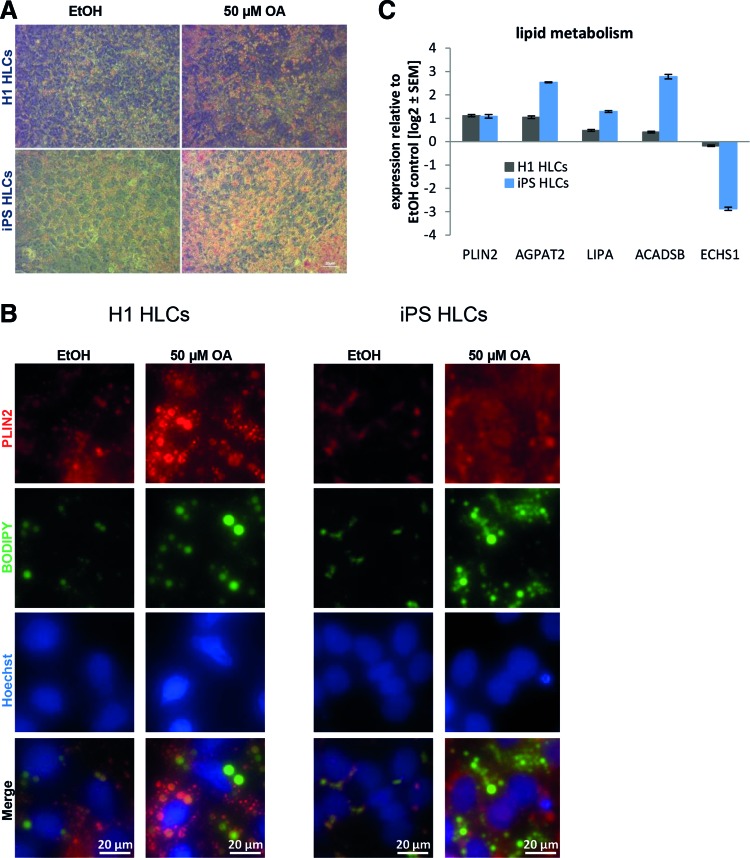
To analyze early steatosis in cell culture, we first established a protocol for the induction of LDs in HLCs, which is based on the addition of 50 μM OA for 48 h into the medium. For both cell types, we observed similar increases in LD accumulation with Oil Red O and BODIPY 493/503 staining (Fig. 3A, B). PLIN2 expression increased consistently after induction with OA (Fig. 3C). In addition, we analyzed the expression of several other genes involved in lipid metabolism to get a first impression of the immediate impact of fatty acid overload (Fig. 3C and Table 1). All factors were selected, because they were significantly regulated in liver cells of patients with high levels of steatosis compared to low-level steatosis [31]. In all cases, except for ACADSB, the expression changes in HLCs mirrored those observed in patient liver biopsies [31].
FIG. 3.
Induction of steatosis in HLCs. hPSCs were differentiated into HLCs. Steatosis was induced by treatment with 50 μM OA for 48 h. Ethanol (EtOH)-treated cells served as controls. Steatosis was monitored by Oil Red O (red) (A) or BODIPY 493/503 (green) (B) staining of LDs. The control cells treated with ethanol show limited LD accumulation, while those treated with 50 μM OA for 48 h have abundant LDs. Scale bars: 20/50 μm. (C) Expression of genes involved in lipid metabolism was analyzed using qRT-PCR (n = 2). Gene expression was normalized to β-actin and, subsequently, to the control samples. For each sample, the mean ± standard error is shown as log2 scale. HLCs, hepatocyte-like cells; hPSCs, human pluripotent stem cells; LDs, lipid droplets; OA, oleic acid; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd
Table 1.
Names and Functions of the Investigated Genes
| Gene symbol | Name | Function | Classification |
|---|---|---|---|
| CPT1A | Carnitine palmitoyltransferase 1A | Transport acyl group of fatty acid-CoA conjugates across the mitochondrial membranes for beta-oxidation | Fatty acid catabolism |
| CPT2 | Carnitine palmitoyltransferase 2 | ||
| ECHS1 | Enoyl coenzyme A hydratase, short chain, 1, mitochondrial | Mitochondrial fatty acid beta-oxidation | |
| HADH | Hydroxyacyl-coenzyme A dehydrogenase | Mitochondrial fatty acid beta-oxidation | |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase | Hydrolase of cholesteryl esters and triglycerides | |
| ACADSB | Acyl-coenzyme A dehydrogenase, short/branched chain | Dehydrogenase of acyl-CoA derivatives | |
| ACAT1 | Acetyl-coenzyme A acetyltransferase 1 | Ketone body metabolism | |
| PRKAA2 | Protein kinase, AMP-activated, alpha 2 catalytic subunit | Regulator of FA and cholesterol biosynthesis | Regulators of metabolism |
| PPARα | Peroxisome proliferator-activated receptor alpha | Regulator of lipid and glucose metabolism | |
| GSK3A | glycogen synthase kinase 3 alpha | Regulator of glucose homeostasis | |
| AGPAT2 | 1-Acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) | Phospholipid biosynthesis | Biosynthesis |
| HMGCR | 3-Hydroxy-3-methylglutaryl-coenzyme A reductase | Cholesterol synthesis | |
| PLIN2 | Perilipin 2 | Coats LDs | LD coating |
| APOC2 | Apolipoprotein C-II | Lipid-binding protein (very low-density lipoprotein) |
LD, lipid droplet.
We decided to further focus on increased PLIN2 expression as an indicator and marker for the induction of steatosis, because its role during progression of the disease has been analyzed extensively [13–16].
Transcriptome and associated pathway analysis of HLCs after steatosis induction
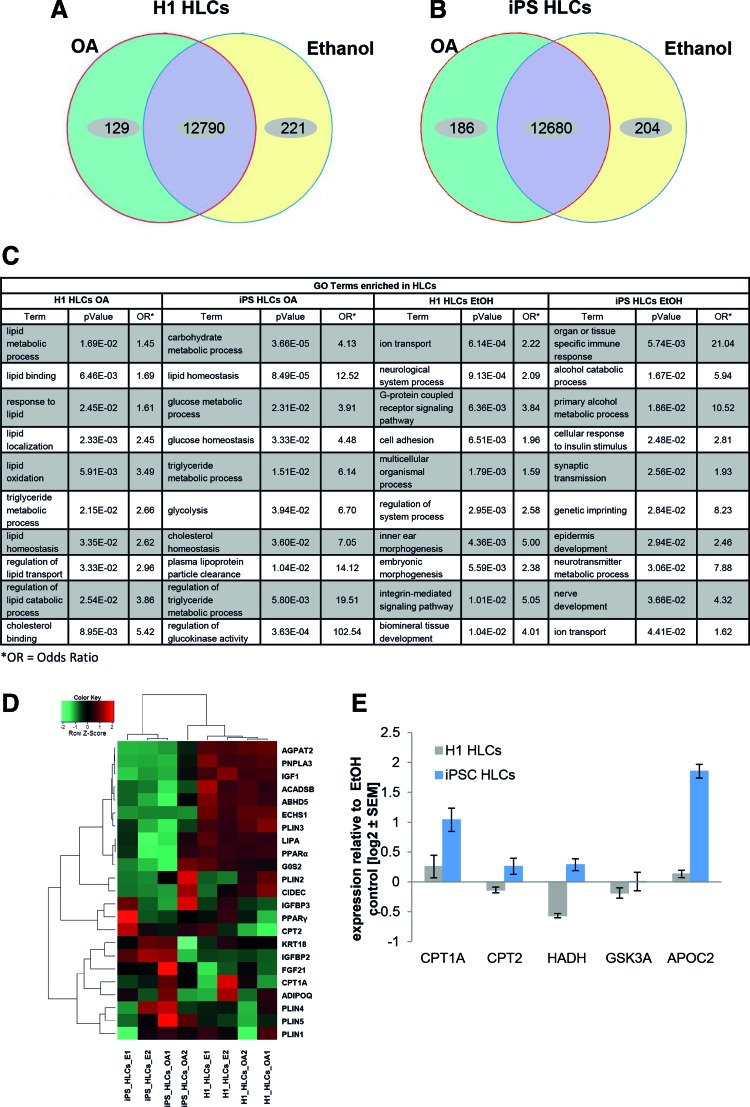
We next wanted to know how the induction of steatosis with OA affects the transcriptomes 48 h post treatment. To achieve this, we induced steatosis in HLCs derived from H1 ESCs and iPSCs as described above. Ethanol-treated cells were used as a solvent control. We found that ∼13,000 genes were significantly expressed in H1- or iPSC-derived HLCs, respectively (Fig. 4A, B). In both cases, about 200 genes were exclusively expressed in the control cells, while 129 (H1) and 186 (iPSC) genes were exclusively expressed in OA-treated cells (Fig. 4A, B).
FIG. 4.
Gene expression profiles of HLCs challenged with OA enable characterization of the early steps in NAFLD. Venn diagrams of genes expressed (detection P value <0.05) in H1- (A) or iPSC- (B) derived HLCs (n = 2). Green and yellow segments represent genes exclusively expressed in OA treated or control cells, respectively. Purple segments comprise genes, which are expressed under both conditions, but not necessarily with the same intensity. (C) GO-analysis of genes upregulated or exclusively expressed under OA treatment reveals an enrichment of NAFLD-related categories, while ethanol control cells predominantly express genes mapping to divergent categories. Shown are preselected significant GO-Terms; for full data set, see Supplementary Tables S6 and S7. (D) Heatmap of genes involved in insulin signaling and lipid or glucose metabolism reveals differences between H1- and iPSC-derived HLCs, which might, in part, be due to their distinct genetic background. However, qualitative changes are similar between both groups, which are reflected by the formation of subclusters connected to OA treatment. (E) Expression of genes involved in lipid and glucose metabolism was analyzed using qRT-PCR (n = 2). Gene expression was normalized to β-actin and, subsequently, to the control samples. For each sample, the mean ± standard error of duplicate experiments is shown as log2 scale. GO, gene ontology; HLCs, hepatocyte-like cells; iPSC, induced pluripotent stem cell; NAFLD, nonalcoholic fatty liver disease; OA, oleic acid; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd
Analysis of the genes that were higher or exclusively expressed in HLCs treated with OA revealed significant enrichment of gene ontology (GO)-terms related to lipid metabolism and transport (Fig. 4C and Supplementary Table S6). As OA is dissolved in ethanol, it is not possible to completely rule out an ethanol-mediated influence on the expression of metabolically relevant genes. However, GO analysis for genes, which were exclusively expressed in ethanol-treated control cells, did not reveal any nonethanol-related metabolic pathways. Instead, pathways connected to signaling and nonhepatic development were enriched (Fig. 4C and Supplementary Table S7).
Overall, the individual regulated genes, as well as the associated GO categories and pathways, differed between both cell lines. This probably reflects innate discrepancies of the differentiation propensity of ES and iPS cells, as well as of course their different genetic background. Nonetheless, numerous genes associated with the PPAR pathway, which plays a major role in the regulation of lipid metabolism, were upregulated after OA treatment of HLCs derived from H1 or iPSCs (Supplementary Fig. S1).
Heatmap analysis of several factors involved in lipid and glucose metabolism, as well as in insulin signaling, revealed that although the absolute transcription levels of many factors differed between H1- and iPSC-derived HLCs, the OA-induced changes were frequently qualitatively similar (Fig. 4D). To get a more detailed insight into the transcriptional changes that occur early in steatosis, we expanded our panel of directly investigated genes (Table 1) and included more factors relevant for lipid metabolism (CPT1A, CPT2, and HADH). We also added APOC2 as another lipid binding protein and GSK3A, which regulates glucose metabolism (Fig. 4E). Interestingly, only CPT1A and APOC2 were consistently upregulated in both samples, while we observed only minor and opposed expression changes for the other factors.
Many liver-specific microRNAs are downregulated in HLCs after induction of steatosis
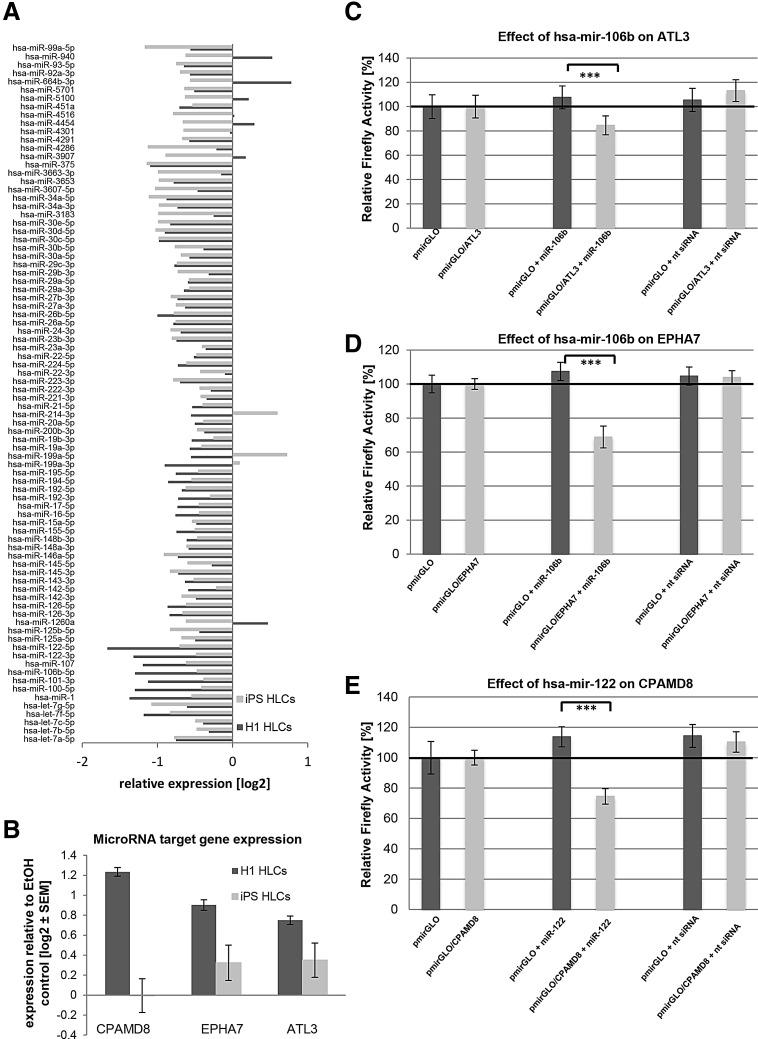
Next, we analyzed whether induction of steatosis in H1- and iPSC-derived HLCs alters expression of liver-specific microRNAs. To this end, we used the liver finder microRNA array composed of 84 liver-related microRNAs. Interestingly, we found most microRNAs downregulated upon steatosis induction and only a few microRNAs upregulated (Fig. 5A). This finding implies that an altered microRNA expression profile is part and parcel of the early events in steatosis-induced cells, especially since the liver-specific microRNA hsa-miR-122 was among the most strongly downregulated miRNAs, together with hsa-miR-106b (Fig. 5A).
FIG. 5.
Regulated expression of microRNAs upon induction of steatosis. (A) Expression of 84 liver-specific microRNAs from H1-HLCs (dark gray) and iPSC-HLCs (light gray) was analyzed after the induction of steatosis. MicroRNA expression was normalized to six different genes of small housekeeping RNAs. For H1-derived HLCs, biological and for iPSC-derived HLCs, technical duplicates were performed. The mean value is shown as log2 expression. With a few exceptions, all miRNAs were downregulated in steatosis-induced cells from both sources. (B) Expression of selected microRNA target genes was analyzed through qRT-PCR (n = 2). Gene expression was normalized to β-actin and, subsequently, to the control samples. For each sample, the mean ± standard error is shown as log2 scale. (C–E) Validation of predicted target genes for hsa-miRNA-106b [ATL3 (C), EPHA7 (D)] and hsa-miRNA-122 [CPAMD8 (E)]. To test the influence of endogenous microRNAs, empty Firefly/Renilla dual-reporter vector pmirGLO and pmirGLO/3′ UTR (containing the 3′UTR of interest at the 3′ end of the Firefly open reading frame) were each transfected into HEK293T cells (n = 4). Normalized Firefly activities were compared with those of pairwise cotransfections of these vectors with the microRNA mimic of interest (miR-106b, -122, and an unspecific siRNA negative control) to test for unspecific effects of the given microRNA-mimic on Firefly/Renilla per se and for validation of the particular target prediction. Dark gray columns show normalized Firefly activities from pmirGLO (mimic-co)-transfections; light gray columns are those from pmirGLO/3′ UTR cotransfections. Percentage reductions of Firefly activities of pmirGLO/3′ UTR compared with pmirGLO are given, as well as their statistical significances (Student's t-test, unpaired, ***P ≤ 0.001). All three predicted interactions were tested positive. HLCs, hepatocyte-like cells; iPSC, induced pluripotent stem cell; qRT-PCR, quantitative reverse transcription polymerase chain reaction; UTR, untranslated region.
Bioinformatic target gene predictions revealed several thousand putative target genes for hsa-miRs-106b and -122 (Supplementary Table S8). Among these, we further analyzed liver-related genes ATL3, EPHA7 (putative miR-106b targets), and CPAMD8 (putative miR-122 target), which were upregulated upon steatosis induction in HLCs derived from H1 and iPSC (Fig. 5B). Remarkably, a significant number of miRNAs from the liver finder array were found among the predictions for these three genes (CPAMD8: 15%, ATL3: 12%, EPHA7: 44% from 84 miRNAs, see Supplementary Table S4). Luciferase reporter-based target gene validation indeed confirmed that hsa-miR-106b downregulates ATL3 and EPHA7, while miR-122 targets CPAMD8 (Fig. 5C–E).
siRNA-based suppression of PLIN2 level and function does not impair LD formation
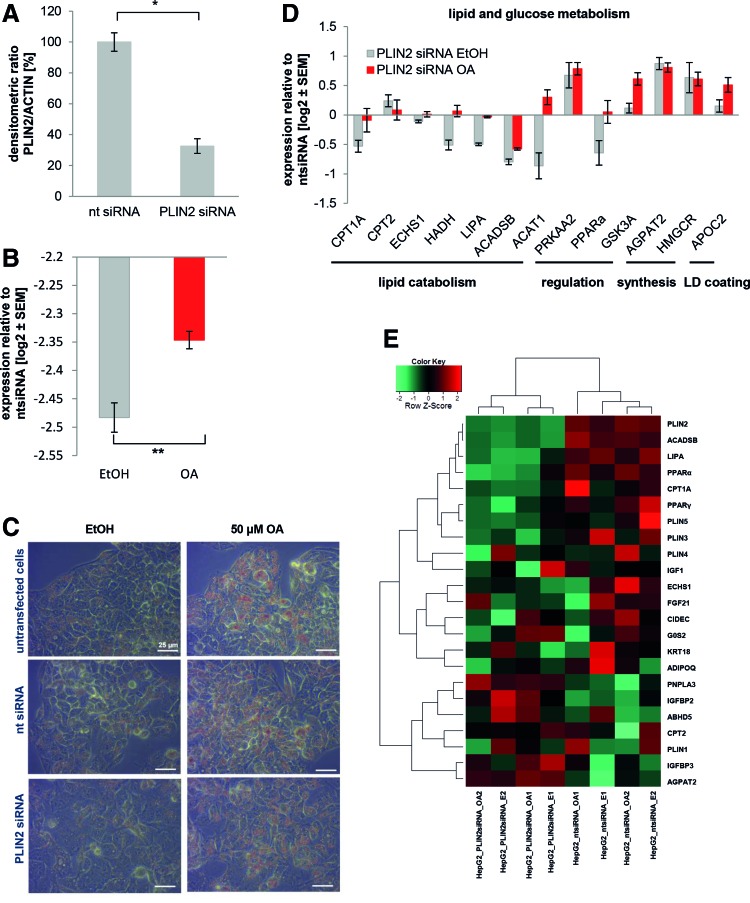
Having characterized the impact of steatosis induction on the global gene expression, we next wanted to interfere with LD accumulation to find indications for putative treatments for NAFLD. First, we reduced PLIN2 expression using siRNA, because it is known that PLIN2 plays a major role during the development of steatosis [13–16]. As the transfection efficiency of in vitro derived HLCs is very low, we decided to focus on HepG2, a hepatocellular carcinoma line, for these experiments. Forty-eight hour treatment with a PLIN2 siRNA reduced its expression on protein level to about 33% (Fig. 6A and Supplementary Fig. S2A). Next, we induced the siRNA-treated HepG2 cells with OA for an additional 48 h. PLIN2 siRNA reduced the expression of PLIN2 on mRNA level to ∼20% in the control and the OA-treated samples (Fig. 6B). However, even under siRNA knockdown conditions, PLIN2 expression was still significantly higher in the induced sample compared to the control (Fig. 6B).
FIG. 6.
Knockdown of PLIN2 in HepG2 cells does not affect LD formation, but alters gene expression. PLIN2 was knocked down in HepG2 cells by lipofectamine-mediated siRNA transfection (n = 2). Cells were either transfected with nt siRNA or with PLIN2 siRNA. (A) After 48 h, PLIN2 expression was assessed by western blot and normalized to ACTIN levels. (B) After 48 h, steatosis was induced with 50 μM OA for an additional 48 h. As control, cells were treated with ethanol (EtOH). Expression of PLIN2 was analyzed on mRNA level using qRT-PCR. Although levels were reduced by ∼80% after transfection, we observed significantly increased levels after OA treatment. Gene expression was normalized to β-ACTIN and, subsequently, to the control samples. For each sample, the mean ± standard error of duplicate experiments is shown as log2 scale. Significances were calculated with an unpaired Student's t-test. (C) LD formation was monitored by Oil Red O staining. In the upper row, untransfected control cells are shown, followed by cells transfected with nt siRNA (middle row) and PLIN2 siRNA, last row. Ethanol-treated control cells show only minor LD accumulation (left column), while those treated with 50 μM OA for 48 h have abundant LDs that are stained by Oil Red O. Scale bar: 25 μm. Expression of genes involved and lipid or glucose metabolism (D) were analyzed using qRT-PCR. (E) Heatmap representation of genes involved in insulin signaling and lipid or glucose metabolism reveals two clusters related to treatment with either PLIN2 or the nt siRNA. LD, lipid droplet; OA, oleic acid; PLIN, Perilipin 2; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd
Contrary to our expectations, PLIN2 knockdown did not alter the accumulation of LDs in the cells after incubation with OA (Fig. 6C). Nonetheless, microarray analysis revealed that transcriptomes of cells treated with PLIN2 siRNA cluster away from those incubated with the nt control siRNA (Supplementary Fig. S2B, left branches). In both clusters, OA treated and control cells form separate subclusters. A detailed view on the expression of lipid and glucose metabolism related genes also confirmed that PLIN2 siRNA treatment induced major transcriptional changes (Fig. 6E).
Numerous factors involved in lipid catabolism such as ACADSB, LIPA, and CPT1A were downregulated after PLIN2 siRNA treatment. These tendencies were confirmed by qRT-PCR (Fig. 6D). However, in this study, the differences between the OA and EtOH-treated cells became more obvious. Interestingly, also PPARα and γ, which regulate lipid metabolism, were downregulated after PLIN2 knockdown (Fig. 6E). This argues in favor of an active PPAR gene regulatory network associated with steatosis. Overall, PLIN2 knockdown modulates the expression of steatosis-related genes, but does not reduce lipid accumulation, at least in the early stage investigated.
Modulation of the lipid metabolism regulating factor PPARα has major impact on numerous metabolism-related pathways
The hepatic nuclear receptor PPARα is activated by a variety of ligands [32,33]. It regulates lipid and glucose metabolism in the liver and PLIN2 is one of its known targets [34,35]. Transcriptome analysis revealed that many members of the PPARα signaling pathway were upregulated either in H1- or iPSC-derived HLCs after treatment with OA (Supplementary Fig. S1). Its expression was also influenced by PLIN2 knockdown (Fig. 6D, E). As we could not transfect HLCs with siRNA against PLIN2 to a significant level, we decided to interfere with PPARα signaling by activating or inhibiting PPARα action with Fenofibrate or GW6471, respectively [36].
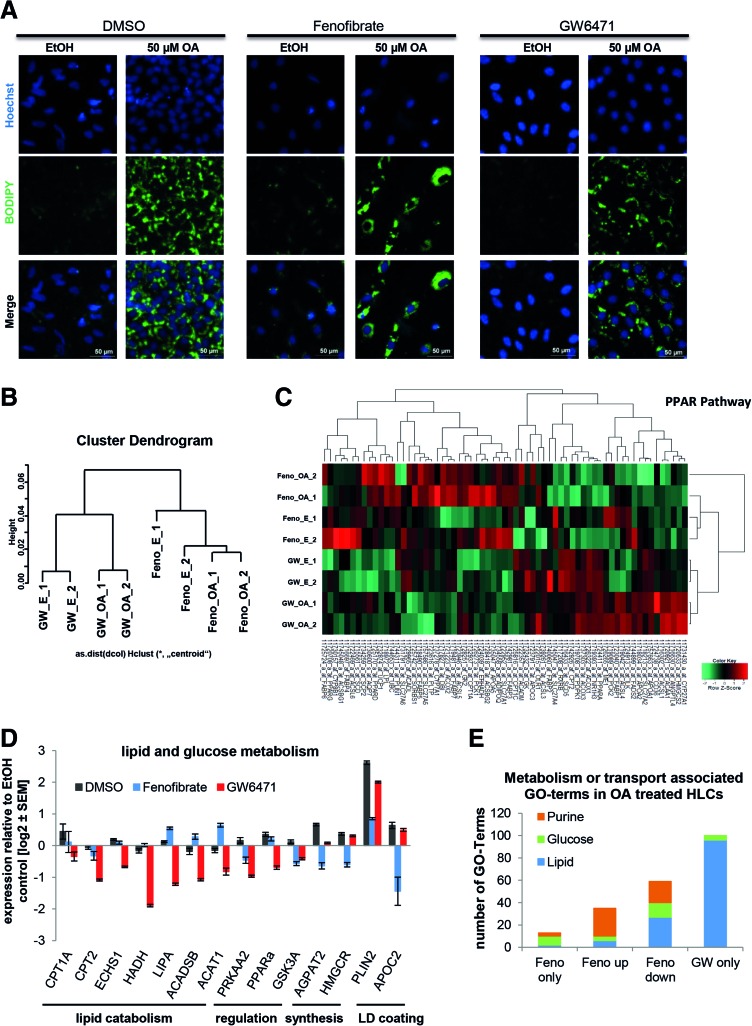
iPSC-derived HLCs were treated for 48 h with OA and either Fenofibrate or GW6471. BODIPY staining revealed that HLCs incorporated LDs regardless of treatment with the PPARα modulators (Fig. 7A). This is in line with results from Rogue et al. who showed that PPARα agonist treatment reduced fat load in HepaRG cells only after a prolonged incubation of 14 days [37]. However, microarray analysis revealed that even short-term treatment with Fenofibrate and GW6471 had an impact on gene expression. The transcriptomes of HLCs incubated with either Fenofibrate or GW6471 clearly clustered away from each other (Fig. 7B).
FIG. 7.
PPARα signaling has major impact on the induction of steatosis. IPSC-derived HLCs were incubated in parallel with either Fenofibrate (Feno, PPARα agonist) or GW6471 (PPARα antagonist) during the 48 h OA induction. (A) Steatosis induction was monitored by BODIPY 493/503 (green) staining of LDs. In every case, LDs increased after OA induction (right columns) compared to the control (left columns), but no differences between the different PPARα treatments are visible. (B) Microarray-based transcriptome analysis revealed two distinct clusters representing Fenofibrate and GW6471 treatment and two subclusters related to OA treatment (n = 2). (C) Heatmap representation of PPAR pathway genes shows distinct gene expression profiles for Fenofibrate (“F”) and GW6471 (“GW”) treated cells with qualitative changes after OA induction. Expression of genes involved in lipid or glucose metabolism (D) was analyzed using qRT-PCR (n = 2). As expected, in most cases, Fenofibrate and GW6471 treatment had opposing effects on gene expression. Gene expression was normalized to β-actin and, subsequently, to the ethanol-treated control samples. For each sample, the mean ± standard error of duplicate experiments is shown as log2 scale. (E) Significantly expressed genes from the global analysis were subdivided into genes only expressed after Fenofibrate treatment, up- or downregulated after Fenofibrate treatment, compared to GW6471 treatment or only expressed after GW6471 treatment. Then they were assigned to GOs (Supplementary Table S9). The numbers of GO-terms that were associated with lipid, glucose, or purine metabolism and transport are displayed. GO, gene ontology; HLCs, hepatocyte-like cells; LDs, lipid droplets; OA, oleic acid; PPARα, peroxisome proliferator-activated receptor alpha; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd
Heatmap-based analysis of the PPAR pathway revealed that cells treated with Fenofibrate or GW6471 behave differently (Fig. 7C). In both cases, there were two subclusters detectable that correspond to the control cells and the OA-treated cells. We monitored the expression levels of genes important for lipid and glucose metabolism in more detail by qRT-PCR. In most cases, treatment with the agonist and the antagonist resulted in opposing changes in gene expression, as expected (Fig. 7D). Inhibition of PPARα with GW6471 resulted in downregulation of genes involved in lipid catabolism, while activation using Fenofibrate reduced expression of AGPAT2 and HMGCR, which are involved in biosynthesis of phospholipids and cholesterol, respectively (Fig. 7D).
When we compared the number of GO-terms associated with distinct metabolic categories that were significantly regulated after PPARα modulation, it became obvious that many were connected with glucose, lipid, and purine metabolism (Fig. 7E and Supplementary Table S9). Interestingly, genes exclusively expressed in GW6471-treated HLCs were predominantly mapped to lipid metabolism or transport, while this category was only marginally upregulated in Fenofibrate-treated cells, indicating that inhibition of PPARα strongly affects lipid metabolism (Fig. 7E). In contrast, expression of genes that are associated with purine metabolism was upregulated after PPARα activation, while none of the genes that were exclusively expressed in GW6471-treated cells mapped onto this GO-category.
A detailed analysis of the purine metabolic pathway demonstrated that genes associated with the early steps of purine metabolism tended to be downregulated under Fenofibrate treatment, while genes for the later steps were either upregulated or exclusively expressed during Fenofibrate treatment (Supplementary Fig. S3). This implies that PPARα activation regulates purine metabolism.
Taken together, we have generated a robust in vitro model for NAFLD based on human PSC-derived HLCs. Upon OA induction, the cells recapitulate many features observed in NAFLD patients (LD accumulation, PLIN2 overexpression, and dysregulation of metabolic pathways) and they react to variation of PPARα activity.
Discussion
In this study, we established a cell culture model for deciphering the molecular basis underlying early steps of LD accumulation in NAFLD, based on HLCs derived from hPSCs. We showed that ESCs and iPSCs both differentiate into HLCs that express hepatocyte-specific proteins and have characteristic hepatocyte-like biochemical functions. HLCs could be induced to accumulate fat in LDs by incubating them with OA. Global gene expression analysis after OA induction revealed upregulation of many GO-categories associated with lipid or glucose metabolism. This effect was qualitatively similar between ESC- and iPSC-derived HLCs, but differed quantitatively due to the different genetic backgrounds of the two cell lines and the usual interdifferentiation variations. Importantly, ethanol-treated control cells had a completely different panel of genes upregulated, which mapped to GO-categories related to signaling or nonhepatic development. This indicates that expression changes observed in the OA group are not induced by the solvent ethanol, but rather by OA itself. However, for the future, it is important to compare effects of different steatosis inducing agents, for example, palmitic acid or steatogenic drugs.
After OA induction, many members of the PPAR signaling pathway, which controls lipid metabolism, were upregulated. Of the different PPAR isoforms, PPARα is most abundant in the liver. It regulates lipid metabolism as a reaction to nutritional status and partly depends on insulin signaling [34]. In general, activation of PPARα with fibrates is a recognized treatment for the metabolic syndrome [38] and its expression levels negatively correlate with the severity of NASH [39].
Upon induction of steatosis, expression of a panel of genes important for lipid metabolism changed in the same manner as previously observed by comparing liver biopsies from patients with high versus low levels of steatosis [31]. Among these factors, we identified PLIN2 upregulation as a suitable marker for successful induction of steatosis. PLIN2 is involved in the synthesis of LDs and plays an important role in the onset of steatosis. It has been shown that PLIN2 knockout mice develop neither obesity nor steatosis [16] and the knockdown of PLIN2 expression with an antisense oligonucleotide protected mice against the development of fatty liver [14].
To analyze the influence of PLIN2 expression on the development of steatosis, we knocked down PLIN2 in HepG2 cells using siRNAs. Although the knockdown was highly efficient with only 32% of protein expression remaining, we could not observe any changes in LD accumulation after induction with OA. This might be due to the short time period of only 48 h. There might still be enough PLIN2 left for coverage of some LDs, while the shortage of PLIN2 limits the amount of LDs that can be assembled over the long term. In addition, several other proteins are involved in LD formation. During steatosis, LDs increase in size relative to the abundance of TAGs [36]. The largest LDs are predominantly covered by PLIN1, which competes on the LD surface with PLIN2 [36]. Global gene expression analysis of PLIN2 knockdown cells revealed indeed that PLIN1 and PLIN4 expression was enhanced in at least two of the four samples compared to the nt siRNA samples.
Overall, we observed profound genome-wide transcriptional changes, which are reflected by the fact that the transcriptomes of PLIN2 and nt siRNA-treated cells cluster separately from each other. Interestingly, expression levels of several factors known to have important regulatory functions on metabolism were altered after PLIN2 knockdown. For example, PRKAA2, which is the catalytic subunit of adenosine monophosphate-activated protein kinase (AMPK), a sensor of nutritional level that becomes activated during fasting [40] was strongly upregulated in PLIN2 knockdown cells, but reduced again upon OA induction. This shows that the cells still react to nutritional abundance. The lack of PLIN2, although it does not have a detectable influence on LDs, seems to transfer some kind of fasting signal that activates PRKAA2 transcription. However, our data do not provide any information on PRKAA2 activity, which has to be analyzed in the future.
In addition, PPARα and γ, which regulate lipid metabolism, were downregulated in PLIN2 siRNA-treated cells. This fits well with the predominant downregulation of lipid catabolism genes that are at least, in part, directly regulated by PPARα. Our data point at the existence of a PPAR-gene regulatory network that depends on PLIN2 levels. In addition, expression levels of genes important for insulin signaling were altered after PLIN2 siRNA treatment. Interestingly, the expression of IGFBP2 and 3, two important regulators of IGF1 signaling [41], were induced, while IGF itself was reduced. Overall, these data indicate that although PLIN2 knockdown has no detectable effect on LD formation, it has a major impact on metabolic activities of the cell.
A knockdown of PLIN2 was not possible in HLCs because of the low transfection efficiency in primary cells. As PLIN2 is a target of PPARα signaling [39], we interfered with PPARα activity using two small molecules, Fenofibrate (agonist) and GW6471 (antagonist). Again, we could not observe any changes in LD formation after treating induced HLCs with either Fenofibrate or GW6471. However, we investigated only short-term effects and it has been shown that PPARα activation over a longer time period reduces steatosis in HepaRG cells [37]. Nonetheless, we observed major gene expression changes that occur as an immediate reaction toward PPARα modulation accompanying OA induction. Expressed genes could be mapped to pathways related to lipid, glucose, and purine metabolism. Interestingly, the latter was not present in OA-induced HLCs, where PPARα activity was reduced by GW6471. In this study, the most prominently upregulated GOs belonged to lipid metabolism. This indicates that the PPARα agonist and antagonist do not simply influence gene expression in opposing directions, although we observed this for a panel of key metabolic genes, but they also modify metabolic pathways from different angles.
Genes of the purine metabolism were generally regulated in Fenofibrate-treated cells. A recent study by Asby et al. demonstrated that Aminoimidazole carboxamide ribonucleotide (AICAR), an intermediate product in de-novo purine synthesis, activates AMPK [42], which is a key regulator of metabolism that senses the nutritional state of the cell and induces PPARα activity and catabolic pathways [40]. We observed that genes, which directly precede the synthesis of AICAR, were either upregulated or exclusively expressed during Fenofibrate treatment. Thus, it is possible that in addition to a direct influence on fat and glucose metabolism, PPARα also indirectly enhances these pathways by regulating purine metabolism, and thus, AMPK activity.
For most of the genes that were analyzed in more detail, treatment with the PPARα agonist and antagonist resulted as expected in opposing changes in expression. While GW6471-mediated inhibition of PPARα resulted in downregulation of genes involved in lipid catabolism, activation using Fenofibrate reduced expression of AGPAT2 and HMGCR, which are involved in biosynthesis of phospholipids and cholesterol, respectively. Thus, our HLC model can, in part, reproduce the beneficial role of PPARα enhancement in patients with metabolic syndrome at least with regard to the reduced expression of lipid and cholesterol synthesizing enzymes.
In addition to major gene expression changes, we observed that the expression of most liver-specific microRNAs was downregulated as an early reaction to steatosis induction. Among the most strongly downregulated miRNAs, miR-122 is a key factor in liver development, differentiation, and homeostasis. It is elevated in the serum of NAFLD patients, while its expression levels in hepatocytes are concomitantly reduced, which is corroborated by our observations [43,44]. MiR-122a knockout mice develop steatosis and have altered levels of enzymes important for lipogenesis, LD formation, and lipid transport [45,46]. MiR-106b has not yet been associated with NAFLD, but its overexpression has been reported during development of cirrhosis and hepatocellular carcinoma [47].
Among the putative miR-106b and -122 targets predicted by at least four of the algorithms implemented in miRWalk, we confirmed CPAMD8, ATL3, and EPHA7 as new miR-122 and miR-106b targets, respectively. None of these three proteins have been functionally associated with NAFLD. However, the validated, as well as the predicted, impact of downregulated liver-related miRNAs on their expression together with their concordant upregulation in induced HLCs point to a functional role of these factors, but mechanisms remain to be elucidated.
We are aware of the fact that the two analyzed hPSC lines can only give a first impression of the general mechanisms of NAFLD development. To dissect these in more detail, we have generated several patient-specific iPSC lines, which are currently being characterized and which will be used in the future for more detailed analyses of the disease [48].
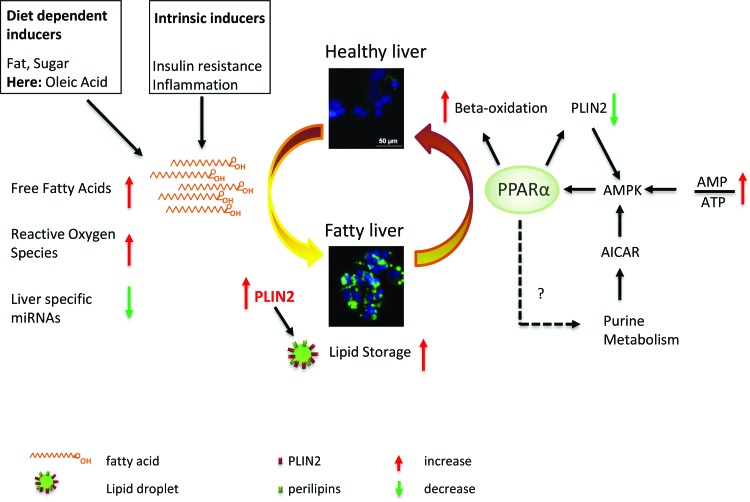
To summarize, our results show that hPSC-derived HLCs are a valuable in vitro model for investigating the molecular basis of the early steps of NAFLD. They accumulate LDs, and expression changes of metabolically relevant genes mirror those observed in liver biopsies of steatosis patients [31]. In addition, lipid metabolism can be regulated by modulating the activity of PPARα. A short overview of the different processes is given in Fig. 8.
FIG. 8.
Schematic overview of inducers of NAFLD and potential intervention points. The transition from a healthy liver (upper panel) toward a fatty liver (lower panel) is shown. Factors that promote NAFLD are shown on the left. Besides diet-related inducers that are in this study mimicked by OA addition to the medium, intrinsic factors like insulin resistance promote NAFLD. From a global perspective, the triggers result in increased free fatty acids and reactive oxygen species. We also observed a global downregulation of hepatic miRNAs. Eventually, every trigger results in an upregulation of PLIN2 expression and an increase in lipid storage within hepatocytes. Beneficial processes that might reduce fat content of hepatocytes are shown on the right side. In general, PPARα activation (in our case by application of Fenofibrate) decreases the amount of stored fat by inducing β-oxidation. Besides fibrates, which activate PPARα, it is also regulated by AMPK, which senses the low energy state of a cell as measured by the ratio between AMP and ATP. AMPK expression increases in response to PLIN2 knockdown conditions and its activity might be regulated by intermediates of purine metabolism, which is in turn regulated by PPARα. This indicates a possible positive feedback loop that has to be verified by additional experiments. Besides PPARα modulation, a direct reduction of PLIN2 expression could also be beneficial. AMP, adenosine monophosphate; AMPK, adenosine monophosphate-activated protein kinase; ATP, adenosine triphosphate; NAFLD, nonalcoholic fatty liver disease; OA, oleic acid; PLIN, Perilipin 2; PPARα, peroxisome proliferator-activated receptor alpha. Color images available online at www.liebertpub.com/scd
Supplementary Material
Acknowledgments
J.A. acknowledges support from the Medical faculty of the Heinrich Heine University Düsseldorf. The authors thank M. Bohndorf and S. Wehrmeyer for technical support.
Prior Conference Presentation: Part of the work was presented as a poster at the 8th International Meeting of the Stem Cell Network North Rhine-Westphalia, April 21–22, 2015, Bonn, Germany.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Cohen JC, Horton JD. and Hobbs HH. (2011). Human fatty liver disease: old questions and new insights. Science 332:1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry RJ, Camporez JP, Kursawe R, Titchenell PM, Zhang D, Perry CJ, Jurczak MJ, Abudukadier A, Han MS, et al. (2015). Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basaranoglu M, Basaranoglu G. and Senturk H. (2013). From fatty liver to fibrosis: a tale of “second hit”. World J Gastroenterol 19:1158–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okumura T. (2011). Role of lipid droplet proteins in liver steatosis. J Physiol Biochem 67:629–636 [DOI] [PubMed] [Google Scholar]
- 5.Thiele C. and Spandl J. (2008). Cell biology of lipid droplets. Curr Opin Cell Biol 20:378–385 [DOI] [PubMed] [Google Scholar]
- 6.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H. and Mashek DG. (2011). The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121:2102–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ. and Brown DA. (2007). Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res 48:2751–2761 [DOI] [PubMed] [Google Scholar]
- 8.Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR. and Londos C. (2003). Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol 161:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyoshi H, Souza SC, Zhang HH, Strissel KJ, Christoffolete MA, Kovsan J, Rudich A, Kraemer FB, Bianco AC, Obin MS. and Greenberg AS. (2006). Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem 281:15837–15844 [DOI] [PubMed] [Google Scholar]
- 10.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ. and Londos C. (1997). Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res 38:2249–2263 [PubMed] [Google Scholar]
- 11.Targett-Adams P, Chambers D, Gledhill S, Hope RG, Coy JF, Girod A. and McLauchlan J. (2003). Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem 278:15998–16007 [DOI] [PubMed] [Google Scholar]
- 12.Imamura M, Inoguchi T, Ikuyama S, Taniguchi S, Kobayashi K, Nakashima N. and Nawata H. (2002). ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am J Physiol Endocrinol Metab 283:E775–E783 [DOI] [PubMed] [Google Scholar]
- 13.Motomura W, Inoue M, Ohtake T, Takahashi N, Nagamine M, Tanno S, Kohgo Y. and Okumura T. (2006). Up-regulation of ADRP in fatty liver in human and liver steatosis in mice fed with high fat diet. Biochem Biophys Res Commun 340:1111–1118 [DOI] [PubMed] [Google Scholar]
- 14.Imai Y, Boyle S, Varela GM, Caron E, Yin X, Dhir R, Dhir R, Graham MJ. and Ahima RS. (2012). Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression. Physiol Genomics 44:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai Y, Varela GM, Jackson MB, Graham MJ, Crooke RM. and Ahima RS. (2007). Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology 132:1947–1954 [DOI] [PubMed] [Google Scholar]
- 16.McManaman JL, Bales ES, Orlicky DJ, Jackman M, MacLean PS, Cain S, Crunk AE, Mansur A, Graham CE, Bowman TA. and Greenberg AS. (2013). Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res 54:1346–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y, Soejima Y. and Fukusato T. (2012). Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol 18:2300–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machado MV, Michelotti GA, Xie G, de Almeida TP, Boursier J, Bohnic B, Guy CD. and Diehl AM. (2015). Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One 10:e0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, et al. (2009). Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol 24:830–840 [DOI] [PubMed] [Google Scholar]
- 20.De Gottardi A, Vinciguerra M, Sgroi A, Moukil M, Ravier-Dall'Antonia F, Pazienza V, Pugnale P, Foti M. and Hadengue A. (2007). Microarray analyses and molecular profiling of steatosis induction in immortalized human hepatocytes. Lab Invest 87:792–806 [DOI] [PubMed] [Google Scholar]
- 21.Currie E, Schulze A, Zechner R, Walther TC. and Farese RV., Jr. (2013). Cellular fatty acid metabolism and cancer. Cell Metab 18:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor JR. and Sabatini DM. (2012). Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2:881–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, et al. (2013). Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87:1315–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. and Jones JM. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y. and Adjaye J. (2011). A cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev 7:331–341 [DOI] [PubMed] [Google Scholar]
- 26.Iwaniuk KM, Schira J, Weinhold S, Jung M, Adjaye J, Muller HW, Wernet P. and Trompeter HI. (2011). Network-like impact of MicroRNAs on neuronal lineage differentiation of unrestricted somatic stem cells from human cord blood. Stem Cells Dev 20:1383–1394 [DOI] [PubMed] [Google Scholar]
- 27.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Schira J, Muller HW. and Wernet P. (2011). MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One 6:e16138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R. and Cui W. (2008). Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 26:894–902 [DOI] [PubMed] [Google Scholar]
- 29.Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, et al. (2008). Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A 105:12301–12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jozefczuk J, Prigione A, Chavez L. and Adjaye J. (2011). Comparative analysis of human embryonic stem cell and induced pluripotent stem cell-derived hepatocyte-like cells reveals current drawbacks and possible strategies for improved differentiation. Stem Cells Dev 20:1259–1275 [DOI] [PubMed] [Google Scholar]
- 31.Wruck W, Kashofer K, Rehman S, Daskalaki A, Berg D, Gralka E, Jozefczuk J, Drews K, Pandey V, et al. (2015). Multi-omic profiles of human non-alcoholic fatty liver disease tissue highlight heterogenic phenotypes. Sci Data 2:150068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMullen PD, Bhattacharya S, Woods CG, Sun B, Yarborough K, Ross SM, Miller ME, McBride MT, LeCluyse EL, Clewell RA. and Andersen ME. (2014). A map of the PPARalpha transcription regulatory network for primary human hepatocytes. Chem Biol Interact 209:14–24 [DOI] [PubMed] [Google Scholar]
- 33.Grygiel-Gorniak B. (2014). Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications—a review. Nutr J 13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawlak M, Lefebvre P. and Staels B. (2015). Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 62:720–733 [DOI] [PubMed] [Google Scholar]
- 35.Targett-Adams P, McElwee MJ, Ehrenborg E, Gustafsson MC, Palmer CN. and McLauchlan J. (2005). A PPAR response element regulates transcription of the gene for human adipose differentiation-related protein. Biochim Biophys Acta 1728:95–104 [DOI] [PubMed] [Google Scholar]
- 36.Pawella LM, Hashani M, Eiteneuer E, Renner M, Bartenschlager R, Schirmacher P. and Straub BK. (2014). Perilipin discerns chronic from acute hepatocellular steatosis. J Hepatol 60:633–642 [DOI] [PubMed] [Google Scholar]
- 37.Rogue A, Antherieu S, Vluggens A, Umbdenstock T, Claude N, de la Moureyre-Spire C, Weaver RJ. and Guillouzo A. (2014). PPAR agonists reduce steatosis in oleic acid-overloaded HepaRG cells. Toxicol Appl Pharmacol 276:73–81 [DOI] [PubMed] [Google Scholar]
- 38.Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, et al. (2006). International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev 58:726–741 [DOI] [PubMed] [Google Scholar]
- 39.Francque S, Verrijken A, Caron S, Prawitt J, Paumelle R, Derudas B, Lefebvre P, Taskinen MR, Van Hul W, et al. (2015). PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol 63:164–173 [DOI] [PubMed] [Google Scholar]
- 40.Fogarty S. and Hardie DG. (2010). Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta 1804:581–591 [DOI] [PubMed] [Google Scholar]
- 41.Yamada PM, Mehta HH, Hwang D, Roos KP, Hevener AL. and Lee KW. (2010). Evidence of a role for insulin-like growth factor binding protein (IGFBP)-3 in metabolic regulation. Endocrinology 151:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asby DJ, Cuda F, Beyaert M, Houghton FD, Cagampang FR. and Tavassoli A. (2015). AMPK activation via modulation of de novo purine biosynthesis with an inhibitor of ATIC homodimerization. Chem Biol 22:838–848 [DOI] [PubMed] [Google Scholar]
- 43.Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ. and Sookoian S. (2015). Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 64:800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN. and Beretta L. (2011). Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One 6:e23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, et al. (2012). Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 122:2871–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, et al. (2012). MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122:2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan W, Li Y, Lim SG. and Tan TM. (2014). miR-106b-25/miR-17–92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol 20:5962–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jozefczuk J, Kashofer K, Ummanni R, Henjes F, Rehman S, Geenen S, Wruck W, Regenbrecht C, Daskalaki A, et al. (2012). A systems biology approach to deciphering the etiology of steatosis employing patient-derived dermal fibroblasts and iPS cells. Front Physiol 3:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.