Abstract
Altered cellular energy metabolism is a hallmark of many diseases, one notable example being cancer. Here, we focus on the identification of the transition from healthy to abnormal metabolic states. To do this, we study the dynamics of energy production in a cell. Due to the thermodynamic openness of a living cell, the inability to instantaneously match fluctuating supply and demand in energy metabolism results in nonautonomous time-varying oscillatory dynamics. However, such oscillatory dynamics is often neglected and treated as stochastic. Based on experimental evidence of metabolic oscillations, we show that changes in metabolic state can be described robustly by alterations in the chronotaxicity of the corresponding metabolic oscillations, i.e. the ability of an oscillator to resist external perturbations. We also present a method for the identification of chronotaxicity, applicable to general oscillatory signals and, importantly, apply this to real experimental data. Evidence of chronotaxicity was found in glycolytic oscillations in real yeast cells, verifying that chronotaxicity could be used to study transitions between metabolic states.
Cellular energy metabolism encompasses many processes, ultimately resulting in the production of adenosine triphosphate (ATP), the fuel continuously used by cells for many essential functions, such as maintenance of ionic balance across the plasma membrane, signalling, and protein synthesis. Every day, we turnover the equivalent of our body weight in ATP1, thus it is important to understand every stage of energy metabolism. Novel imaging techniques have provided insights into the function of metabolic pathways2,3,4,5, and have led to the growing understanding that many diseases can be associated with dysfunctional mechanisms of ATP production6,7,8,9,10. Increasing evidence suggests that metabolic dysfunction plays a key role in carcinogenesis6,7,8,11,12. Moreover, most other properties observed in cancer cells can be explained as consequences of this dysfunction9. Therefore, the observation of the cell in a state of metabolic transition may aid in the understanding of the effects of metabolism on the carcinogenic potential of the cell. Here, we study the dynamics of energy production, and investigate the possibility of identifying robust characteristics which can be (a) used to identify alterations in the metabolic state of a cell, (b) reliably identified from the observed metabolic dynamics.
The hypothesis that such a robust characteristic exists is based on a number of experimental observations of common patterns in metabolic dynamics which are distinctive in different metabolic states, suggesting that we may be able to identify a transition from a healthy or altered states by observing the properties of the dynamics of cellular metabolism.
First of all we focus on the oscillatory and time-dependent nature of the dynamics of energy metabolism. Indeed, the energy produced by a cell continuously fluctuates due to rate constants involved in the production and use of energy. Recently developed experimental techniques for the observation of energy metabolism3,13,14 clearly illustrate these fluctuations, mainly as oscillations, through the measurement of glycolytic intermediates, such as nicotinamide adenine dinucleotide (NADH), and the mitochondrial membrane potential (Δψm). Although the oscillatory nature of energy metabolism is now more often appreciated3,15,16,17,18,19,20,21,22,23, metabolic oscillations can still be overlooked due to intercellular heterogeneity24 or considered as purely stochastic fluctuations25. Oscillations in glycolysis have long been observed in many types of cells, including yeast26, pancreatic β cells27 and muscle cells16. Observing NADH via fluorescence imaging3,20 provides an opportunity to observe the oscillatory dynamics of glycolysis. Mitochondrial oscillations have also been demonstrated. In yeast, in aerobic conditions, oscillations in Δψm were observed, and it was concluded that these were probably entraining the whole metabolic network of the cell28. As well as being oscillatory, energy production within a cell is inherently time-dependent. The contribution of each metabolic pathway to the cellular ATP supply depends on cell type and the current energy requirements of the cell, and is thus necessarily time-varying. This openness of the system of cellular energy metabolism inevitably leads to nonautonomous or time-dependent dynamics29,30.
Energy production via different metabolic processes is tightly regulated. Each metabolic state of the cell will be characterised by different pathways of ATP production. This will result in clear differences between healthy and altered states, arising from the cell switching between glycolytic and mitochondrial ATP production as a primary source of energy. A widely observed example of this is the metabolic switch to glycolysis for an increased proportion of energy production in cancer cells, even in the presence of oxygen. This is known as the Warburg effect31,32. These experimental observations, of metabolic oscillations and switches between metabolic states, suggest that we may be able to identify whether cells are in healthy or altered states by observing the properties of their oscillations.
Metabolic oscillations observed in glycolysis and in the mitochondria are coupled and can influence each other depending on the state of the cell20,21,27,33. This was demonstrated in the form of a driving influence from glycolysis on mitochondrial oscillations in semi-anaerobic conditions20 and at near anoxia21. This driving influence of glycolysis in these ‘altered’ states suggests that changes in this driving would accompany changes in the state of the cell. We propose that these characteristics of metabolic state can be considered under a newly introduced theoretical framework named chronotaxicity34. Chronotaxicity35,36 was recently introduced to describe physical properties of oscillatory systems which are inherently driven, and are capable of resisting perturbations. In doing so, they often generate complex time-dependent behaviour. Their ability to resist perturbation can be robustly identified regardless of the level of complexity, which makes chronotaxic systems an ideal model for cellular metabolic oscillations. We propose that observations of the driven metabolic oscillations imply that they are chronotaxic.
It was previously shown that chronotaxicity can be identified in a single time series37. This allowed us to test this hypothesis in real experimental data recorded by Gustavsson et al.38. Using these recordings of oscillatory glycolysis in single yeast (S. cerevisiae) cells, we investigated the chronotaxic properties of glycolytic oscillations. Yeast cells have been shown to have metabolic similarities with cancer cells39, and their metabolism has received much attention. Therefore we pay particular attention to the properties of yeast in the following work.
Here, we demonstrate that glycolytic oscillations in real yeast cells show clear signs of chronotaxicity. Based on this, we propose chronotaxicity as a robust characteristic whose alterations signify changes in energy metabolism, facilitating identification of altered states in the cell. A theoretical model of different metabolic states is developed and the interplay between glycolytic and mitochondrial involvement in ATP production within a cell is investigated. We also consider how metabolic oscillations may be driven by factors such as substrate availability.
The model shows how the temporal evolution of different states of cellular energy metabolism could be described based on changes in the robustness to external perturbations, or chronotaxicity. This provides a potential method of observing metabolic switches in real cells and investigating possible links to carcinogenesis.
Results
Identifying chronotaxicity in real metabolic oscillations
Glycolysis in yeast cells is one of the most widely studied and well characterised biological oscillators. It was previously thought that glycolytic oscillations only arose as a result of synchronization between yeast cells, but recently Gustavsson et al.38,40 demonstrated glycolytic sustained oscillations in individual isolated yeast cells. Here, to investigate the applicability of chronotaxicity to metabolic oscillations we use data recorded by Gustavsson et al.38 and test it for chronotaxicity using phase fluctuation analysis (PFA), as described in Methods.
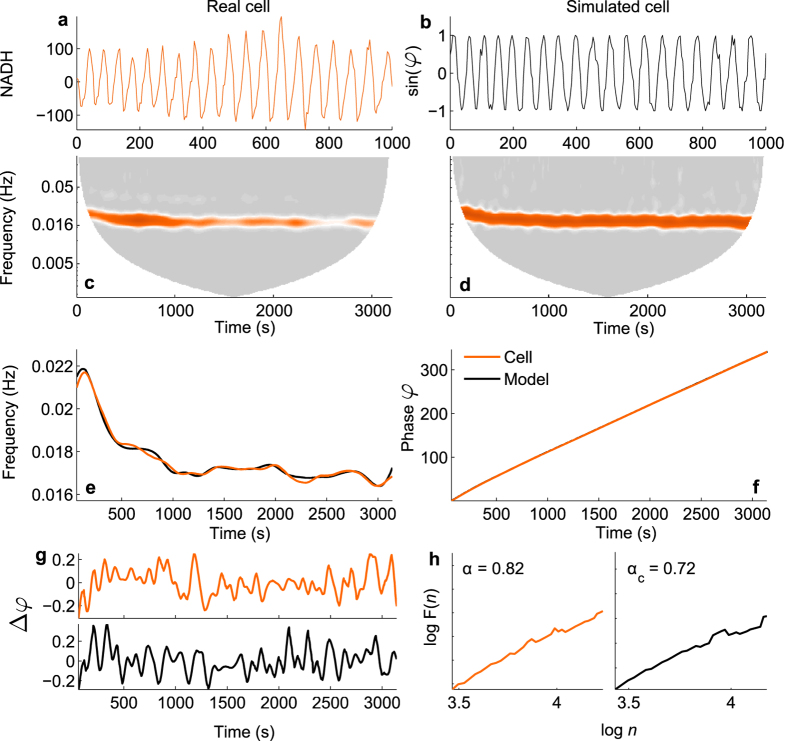
NADH data obtained by Gustavsson and co-authors38 was available from 34 yeast cells, 6 of which did not demonstrate visible oscillations so were excluded from the analysis. An example NADH signal can be seen in Fig. 1a. It was previously demonstrated that at least 30 cycles of oscillation are required to test for chronotaxicity41, therefore all cells which did not meet this requirement were excluded. 16 cells met this criteria. Phase fluctuation analysis (PFA)37 was then applied to the remaining NADH signals to determine whether the oscillations observed in these cells exhibited hallmarks of chronotaxicity. The mean value of α for the 16 included cells was 0.768, which characterises oscillations as chronotaxic. An example analysis on a real yeast cell can be seen in Fig. 1. This demonstrates that real metabolic oscillators are able to be chronotaxic in certain states. Particularly, as shown here, this is the case when metabolic oscillations are sustained and stable despite noise and perturbations. The identification of these biological oscillations as chronotaxic demonstrates that the concept of chronotaxicity, originating from the theory of nonautonomous dynamical systems29,30, can be used to describe real biological oscillations in cellular energy metabolism. Therefore, inverse approach methods may be utilised to detect chronotaxicity with no prior knowledge of the system, as is the case in real experimental data. This result allows us to apply the concept of chronotaxicity to cellular energy metabolism in general.
Figure 1. Testing real experimental data for chronotaxicity.
(a) Example NADH signal from an isolated yeast cell, recorded in38. (b) Time series of sinφGO from the model using the instantaneous frequency extracted from (a) as the driver ωG. (c,d) Continuous wavelet transforms of the time series in (a,b), respectively. (e) The instantaneous frequency of the oscillatory modes were extracted from the wavelet transforms, and smoothed using a moving average. (f) Integrating over the smoothed frequency provides the phase for each case. (g) Subtracting the smoothed phase from the observed phase provides the phase fluctuations, Δφ, in the system. (h) Detrended fluctuation analysis performed on Δφ suggests that the glycolytic oscillations observed in a real yeast cell (orange line) are chronotaxic. α = DFA exponent from yeast cell, αc = DFA exponent from model.
Modelling metabolic oscillations and their chronotaxicity
Based on experimental observations of sustained oscillations, and the evidence of chronotaxicity highlighted in yeast cells, we now consider the chronotaxicity of cellular energy metabolism in healthy and ‘altered’ states, and how this information could be used to observe transitions between these states over time. We consider chronotaxicity during the metabolic transition of a single cell from a state in which aerobic respiration provides the majority of ATP, to one where the cell is increasingly reliant on glycolysis. We consider the former as a healthy state, and the latter as an undesirable state for most cells. An upregulation of glycolysis may be observed temporarily in intermittent hypoxia, or consistently in cancer or yeast cells. Whilst a sign of stress in hypoxic cells, normal yeast cells actually demonstrate a preference for a glycolytic metabolism, displaying many similar metabolic features to cancer cells39,42.
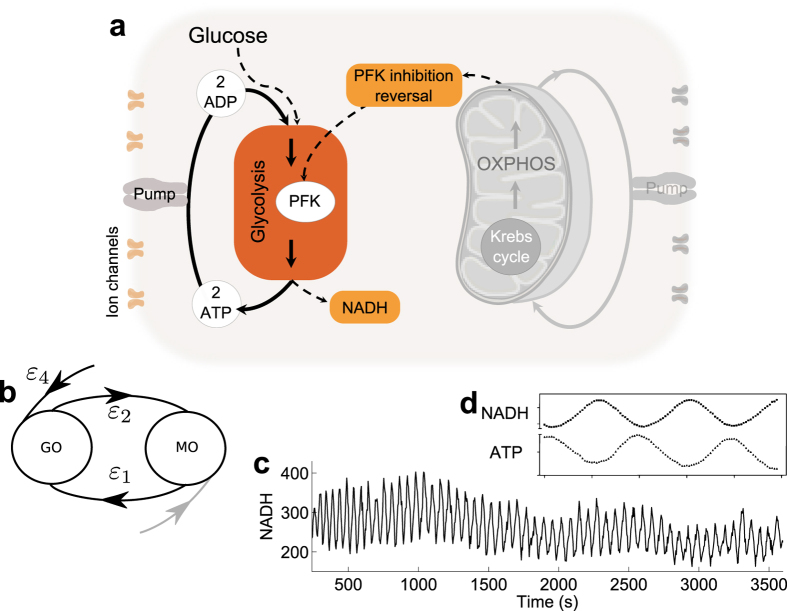
Firstly, we discuss the important features of cellular energy metabolism relevant to the model (see Fig. 2). ATP production begins with glycolysis, which converts glucose to pyruvate and produces ATP and nicotinamide adenine dinucleotide (NADH). Pyruvate is then used during pyruvate decarboxylation, the products of which are used in the Krebs cycle. This releases more ATP and leads to production of substrates (including NADH) which are used to create a hydrogen ion gradient across the inner mitochondrial membrane. This gradient then drives the production of ATP from ADP and inorganic phosphate, in the process of oxidative phosphorylation (OXPHOS). The activity of each of these pathways, and the extent to which the cell relies on them for ATP production depends upon many factors, including the current energy requirements of the cell, and the speed with which each process can match demand. For example, aerobic respiration is much more efficient in terms of amount of ATP produced than glycolysis alone, but glycolysis produces ATP at a faster rate.
Figure 2. Cellular energy metabolism.
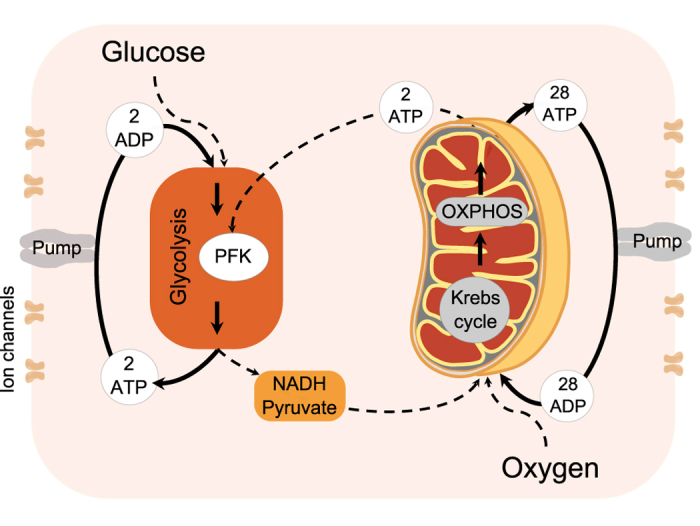
ATP production in a cell (thick lines) occurs via glycolysis using glucose, and when oxygen is present ATP is produced via oxidative phosphorylation (OXPHOS) using the products of glycolysis. Both processes are oscillatory, and can influence each other (for details see main text). Couplings between these processes as well as external influences are shown by dashed lines.
In the model which follows, we identify glycolytic and mitochondrial oscillators, and simplify the metabolic pathways and interactions between oscillators to only include features which are necessary to capture chronotaxicity, which can describe subtle changes in dynamics due to the change in the interactions between metabolic oscillators.
Glycolytic oscillator
Oscillations in glycolysis have long been observed in many types of cells, including yeast26, pancreatic β cells27 and muscle cells16. Glycolytic oscillations have been shown to provide an advantage in maintaining a high ATP/ADP ratio43,44. The source of these oscillations is still debated, but evidence suggests that a reaction involving phosphofructokinase (PFK) is responsible for their origin3,33. In addition to PFK, another time-dependent factor which influences the dynamics of glycolysis is the availability of glucose. Reijenga et al. demonstrated that the frequency of glycolytic oscillations is directly affected by sugar transport45, with a maltose induced decrease in sugar transport resulting in a decrease in the frequency of glycolytic oscillations.
The effects of the inevitable fluctuations in the availability of glucose in vivo was further explored46 through simulation of fluctuating external glucose, and it was found that the glycolytic dynamics of yeast cells that do not usually exhibit intrinsic oscillations oscillated at the frequency of extracellular glucose pulsing, demonstrating a clear driving influence. Boiteux et al. investigated the effects on oscillatory glycolysis of stochastic, periodic, and steady sources of substrate47, and found that stochastic variation of the rate of glucose input leads to sustained periodic behaviour, with small variations in period around a preferred frequency, i.e. it exhibits stability but with a time-varying frequency. Periodic variations of glucose in this study revealed instantaneous frequency responses and harmonic entrainment at specific frequencies. More recently, the ability to measure metabolic dynamics in individual yeast cells has allowed the investigation of the effects of periodic external perturbations on existing glycolytic oscillations40. This study found that oscillatory cells synchronize through externally induced phase shifts alone, whilst non-oscillating cells also require amplitude changes to exhibit externally driven oscillations.
Based on the observed oscillations in glycolysis, and the inevitable time-variability of glucose availability in vivo, we consider glycolysis as a single oscillator, the glycolytic oscillator (GO), which may be influenced by external glucose levels.
Mitochondrial oscillator
Mitochondrial oscillations have been associated with cycles of oxidation and reduction of the intracellular NADH pool and extramitochondrial factors. In cases of oxidative stress or substrate deprivation in cardiac myocytes, the inner mitochondrial membrane potential may destabilise, causing depolarization and oscillation14,22, forcing synchronous oscillations within the mitochondrial network13,14,17. Mitochondrial oscillations are usually observed in the form of Δψm oscillations19 using fluorescence20.
As is effectively assumed when observing Δψm as a measure of mitochondrial metabolic activity, we combine the Krebs cycle and OXPHOS, both of which occur in the mitochondria, into a single metabolic oscillator, the mitochondrial oscillator (MO), which may be driven internally or by the extramitochondrial factors such as the plasma membrane potential or Ca2+19.
Interactions between metabolic oscillators
Interactions between mitochondrial and glycolytic oscillations have been experimentally demonstrated under metabolic stress in cardiac myocytes21, and in yeast cells20. Both studies concluded that under conditions in which the cell is relying on glycolysis for the majority of ATP production, which can be considered as an altered state, oscillations in glycolysis drive those in Δψm. They also demonstrated the indirect influence of mitochondrial ATP production on glycolysis even in this state, via sensing of the ATP/ADP ratio, i.e. the lower this ratio, the harder glycolysis has to work. This link is provided by the reversal of the inhibition of PFK in the glycolytic oscillator33, schematically shown Fig. 2. In the healthy state, the majority of ATP will be produced via OXPHOS, and glycolysis will be suppressed to the level necessary to provide substrates for further metabolic reactions. Glycolysis will still be able to influence mitochondrial processes through the availability of pyruvate and NADH, but metabolic oscillations will likely be driven by the mitochondrial oscillations28.
It is the interactions between these oscillators (MO and GO) which would change in a transition to an altered metabolic state. Therefore, our model provides sufficient detail to capture this transition, by considering only these interactions (see Fig. 2). All other possible interactions, and specific enzymatic pathways, are not considered.
Chronotaxicity of metabolic oscillators
In measurements of oscillations in healthy and altered metabolism an apparent stability of glycolytic and mitochondrial oscillations has been observed3,13,33. These oscillations appear to be time-variable yet very stable14,22 despite noise and perturbations, suggesting the presence of chronotaxicity in the system.
The chronotaxicity of GO and MO may be realized in two general configurations. First, the required drive system (see the definition of chronotaxic systems in Methods) could be provided by any external oscillatory influence to GO and MO strong enough to create chronotaxicity. Particularly, oscillators may be chronotaxic because of interaction with other metabolic oscillators20, or due to interaction with other external oscillations such as fluctuating external glucose27,46 or oxygen. In the GO, we consider the time-varying availability of glucose as the potential cause of chronotaxicity, i.e. as the drive system in Fig. 3b. Similarly, we may consider dynamical oxygen availability as the drive system of the MO. However, if glucose and oxygen are abundant within the cell, they are unlikely to be the driver for each process.
Figure 3. Chronotaxic system.
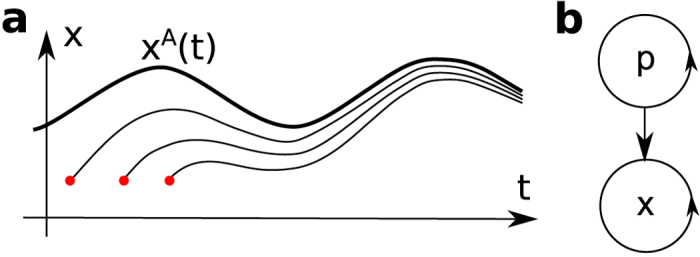
(a) Time-dependent point attractor xA(t), whose trajectory is shown as a thick line, is the main characteristic of a chronotaxic system. Trajectories with different initial conditions (red dots) are approaching xA(t). (b) The simplest configuration of dynamical system where a response system x may be chronotaxic due to a drive system p. Detailed definition of chronotaxic systems is given in Methods.
In the second configuration, the chronotaxicity could be created due to the inner structure of the oscillator, i.e. we assume that due to finite reaction times and interactions within structural elements of the oscillator the stable time-dependent point attractor could appear. Importantly, the oscillations observed by Gustavsson et al.38 in isolated yeast cells, whilst occurring with similar frequencies, were not synchronized. As the external glucose level in these experiments was constant, the oscillations must occur as a result of an internal mechanism, which is presented as a second configuration where chronotaxicity of metabolic oscillators may exist.
Hereafter we do not distinguish between these two configurations, and consider the external resources glucose and oxygen as the drivers which directly or indirectly define chronotaxicity of the GO and MO, independent of the exact mechanism by which this occurs. Our focus will be on the changes in chronotaxicity, i.e. alterations in the stability of metabolic oscillations in different states.
The model
Based on the above discussion, we consider cellular energy metabolism as a system of coupled oscillators, comprising the GO, the MO, and their respective external influences, which we name glucose (G) and oxygen (O), respectively. The inputs to the GO are considered to be glucose and mitochondrial ATP (ATPMO). The end products of glycolysis which are relevant to the model are glycolytic ATP (ATPGO) and NADH. The MO is regulated by the availability of substrates. Here, we consider the availability of NADH as the primary influence on the mitochondrial oscillator (see Fig. 2). The final output of both oscillators represents oscillatory ATP. ATP concentration was previously shown to be oscillatory in yeast cells by Özalp et al.48.
Chronotaxicity is defined by the presence of a time-dependent steady state, and thus does not depend on the particular shape of the oscillations in a signal, only on how their frequency varies in time. Therefore, it can be robustly identified from phase dynamics alone, even when the amplitude dynamics is complex37. We therefore model this system using coupled phase oscillators,
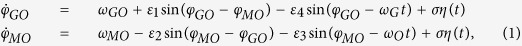 |
where φGO and φMO are the instantaneous phases of the GO and MO, respectively, ωGO and ωMO are the natural frequencies of the GO and MO, ωG and ωO are the frequencies of the external drivers and η(t) is white Gaussian noise. Considering that the GO and MO are continuously interacting, as discussed above, they are represented as bidirectionally coupled oscillators, with coupling strengths ε1 and ε2 which may vary to represent different metabolic states. The inhibitory nature of the influence of ATPMO production on ATPGO production is represented by a repulsive coupling ε1, whilst the excitatory nature of glycolytic oscillations on mitochondrial oscillations is represented by an attractive coupling ε2. The external influences on the GO and MO are represented as unidirectionally coupled drivers, with coupling strengths ε3 and ε4 for O and G respectively, which may also vary.
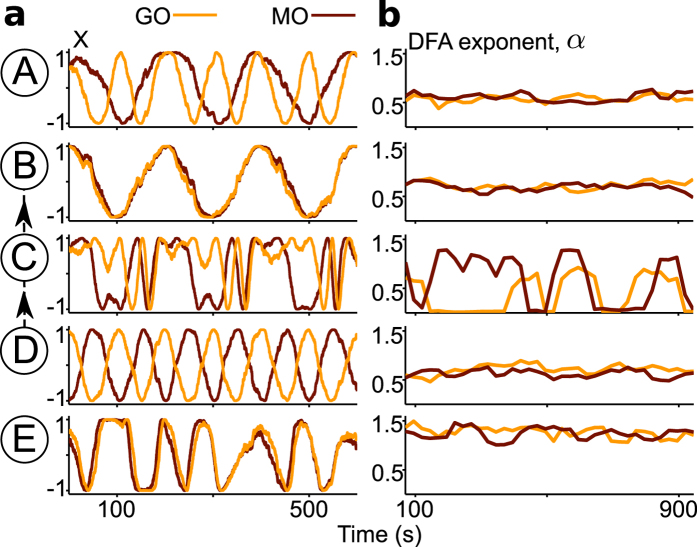
The chronotaxic dynamics of this model was tested numerically. The system (1) was integrated with varying parameters. Numerically, for an oscillator to be chronotaxic, it was required that the observed oscillator was synchronized to one of the unidirectionally coupled drivers G or O. Using this test, 7 different types of dynamics were revealed, the most relevant 5 regions shown in Fig. 4b. Example phase trajectories for all types are shown in Supplementary Fig. S1. Approximate frequencies of these oscillations in different regions are summarised in Table 1.
Figure 4. Model (1) of metabolic oscillations in a cell, and different types of dynamics and the transition from healthy to an altered metabolic state.
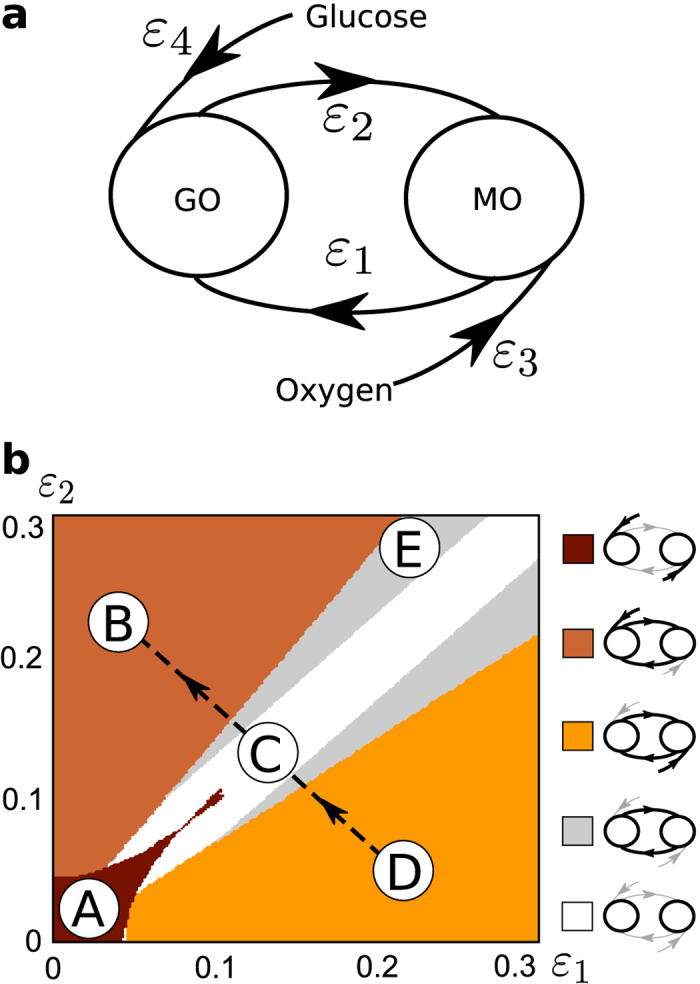
(a) The glycolytic and mitochondrial oscillators (GO and MO, respectively) are represented by two phase oscillators coupled to each other via repulsive ε1 (due to the inhibitory nature of influence of MO on GO) and attractive coupling ε2 (due to excitatory influence of GO on MO). Chronotaxicity arises through attractive couplings ε3 and ε4 to glucose and oxygen drivers. (b) In numerical simulations, couplings ε1 and ε2 were varied to represent different metabolic states. Chronotaxicity of the system was tested for each pair of couplings by checking synchronization between each metabolic oscillator and its potential drivers, see Supplementary Methods. Various types of dynamics of equations (1) with different chronotaxicities were revealed. Region A (dark brown): GO and MO are synchronized with their drivers (Glucose and Oxygen), but not with each other (both GO and MO are chronotaxic); Region B (orange): GO and MO are both chronotaxic and synchronized with the Glucose driver; Region C (white): GO and MO are both not chronotaxic and not synchronized with anything; Region D (yellow): GO and MO are both chronotaxic and synchronized with the Oxygen driver; Region E (light gray) GO and MO are both nonchronotaxic and synchronized only with each other. Regions F and G (not shown, see Supplementary Figs S1 and S3): only GO or only MO is chronotaxic respectively. A potential transition of the system from healthy to a potentially carcinogenic state could correspond to dynamical changes from region D to region B, which are both chronotaxic, but with different drivers, via regions C and E, which are non-chronotaxic. For simplicity, ε3 and ε4 were considered to be equal (ε3 = ε4 = 0.025), but can easily be changed depending on the exact metabolic state one needs to model, e.g. specific substrate deprivations (see Supplementary Fig. S2). Supplementary Figs S3 and S4 show the effects on the system of different constant frequencies and time varying frequencies, respectively.
Table 1. Approximate characteristic frequencies of GO and MO in the regions shown in Fig. 4b.
| Glycolytic | Mitochondrial | |
|---|---|---|
| A | ωG | ωO |
| B | ωG | ωG |
| C | ωGO | ωMO |
| D | ωO | ωO |
| E | ω* | ω* |
In regions A, B, and D each oscillator is synchronized either to its own driver or to the other oscillator and its driver. In these cases, frequencies are influenced by driving oscillators. In C the oscillators are not synchronized to anything, hence the characteristic frequencies are determined by natural frequencies. In E oscillators are synchronized to each other only, hence the frequency ω* corresponds to the frequency of a synchronized state assuming ε3 = ε4 = 0, and ω* can be found from the condition of synchronization 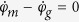 ,
, 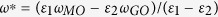 .
.
In real data, utilising chronotaxicity as the defining parameter of the system is superior to the consideration of synchronization alone, as it can be identified experimentally from any single time series, whereas synchronization requires measurements of all interacting oscillators. Therefore, in this case, it would be sufficient to have measurements of only ATPGO or ATPMO to determine their chronotaxicity. To demonstrate this, phase fluctuation analysis was applied to the generated dynamics of GO and MO using parameters from each region shown in Fig. 4b. The chronotaxicity of each metabolic oscillator was tested separately. PFA was used to characterise the phase fluctuations for each oscillator in each region (see Fig. 5). Excellent agreement is shown between the chronotaxicity as calculated by the model using synchronization conditions, and chronotaxicity as calculated via the inverse approach with no prior knowledge. This illustrates that the method is applicable to the observation and identification of chronotaxicity in real systems, where the dynamics are unknown beforehand.
Figure 5. Identification of chronotaxicity via inverse approach methods.
(a) Examples of dynamics from regions A–E, as defined in Fig. 4b. The additive white Gaussian noise with σ = 0.05 was used. (b) Detrended fluctuation analysis (DFA) exponents α characterising chronotaxicity in each region: for α ∈ (0.5, 1) the system can be considered as chronotaxic, for α ∈ (1, 1.5) the system can be considered as non-chronotaxic. In region C the exponent α changes too fast, and the DFA method is not applicable, however such dynamics of α suggest that system is not chronotaxic. This inverse approach shows very good agreement with the chronotaxicity tests directly from the model, revealing that metabolic dynamics in regions A, B and D are chronotaxic, while regions C and E are not. Thus, it may be used to identify chronotaxicity in real data, using a single time series. To demonstrate the method, the time series used here contain at least 100 cycles of oscillation. In reality, this number of cycles is not always feasible. However, this method may still be applied on shorter time series, with reliable results, see Supplementary Fig. S5.
Applicability of the model: modelling chronotaxicity observed in experimental yeast data
The applicability of the model is tested on the data of NADH recorded by Gustavsson et al.38. For this we utilise the closely linked dynamics of intracellular ATP and NADH arising from glycolysis. It has been shown previously that both ATP and NADH are oscillatory in yeast cells during glycolytic activity, and that these oscillations have the same frequency but are out of phase by around 180° 48,49, as demonstrated in Fig. 6d. This means that measurements of NADH in yeast cells may be used to provide an approximation of ATP dynamics. Although the amplitude of these parameters will differ, their phase relationship will remain the same, and can thus be represented by our phase oscillator model. Using this information, the model can be tested for the case of the glycolytic oscillator based on NADH measurements, which are more readily available.
Figure 6. Verifying the glycolytic oscillator (GO) with real experimental data.
(a) The yeast data recorded from individual yeast cells by Gustavsson et al.38 provides our ‘altered’ metabolic state. The addition of glucose followed by cyanide to glucose starved yeast cells induces glycolytic oscillations by preventing respiration. Therefore, glycolysis is upregulated by the reversal of PFK inhibition in response to the reduction in ATPMO. (b) The situation described in (a) can be represented by our model as the dynamics of the system being driven by the glucose driver, causing the system to be chronotaxic. (c) Example NADH time series from an isolated yeast cell exhibiting glycolytic oscillations. (d) Comparison of NADH and ATP oscillations in yeast glycolysis shows that they oscillate 180 out of phase48,49. Modified from49.
In the experiments by Gustavsson et al.38. NADH in individual yeast cells was shown to oscillate following starvation and the addition of cyanide. In this state, glycolysis can be the only means of energy production, as cyanide halts respiration, effectively removing the effects of the mitochondrial oscillator (MO). It is therefore expected that the metabolic state induced in this experimental setup should correspond to our ‘altered’ state, i.e. region B in Fig. 4b, where glycolytic oscillations drive the dynamics of the system, and the GO is chronotaxic.
The model was used to numerically simulate the GO in a chronotaxic (ε1 = 0, ε2 = 0, ε3 = 0, ε4 = 0.25, σ = 0.2) state using the instantaneous frequency extracted from the real experimental data. To make the system chronotaxic, the extracted frequency was used as the driver ωG of the glycolytic oscillator. This provided a chronotaxic oscillator with the same oscillation frequency as the experimental data, allowing the DFA exponent α to be compared between cases (see Fig. 1). Due to the relatively short recording time causing variation between simulations, they were repeated 3 times and the average value of α taken. The mean value of α for the chronotaxic simulations was 0.772 , compared to 0.768 which is the mean value of α for the 16 included cells recorded by Gustavsson et al.38.
The distributions of exponents did not significantly differ between those calculated in the cells and those calculated in the simulated chronotaxic system (p = 0.86) as calculated using the Wilcoxon ranksum test. This shows that our model, although simple, incorporates enough features to allow the calculation of the presented characteristic, chronotaxicity, and that evidence of this characteristic appears to be present in yeast glycolytic dynamics. This verifies the applicability of our model to the glycolytic oscillator. Further investigation is required into the mitochondrial oscillator, and other metabolic states.
Transition between healthy and altered metabolic states in the model
To identify metabolic states, we consider an altered state, such as the glycolysis dependent and potentially carcinogenic case discussed above, to correspond to the dynamics of the model in which the phase of a chronotaxic GO entrains the phase of MO, i.e. region B in Fig. 4b. In contrast, the normal state may correspond to the case where the phase of MO entrains the phase of GO, as discussed in ref. 28. Therefore, the normal state corresponds to region D (where oscillators are chronotaxic with oxygen as a driving system). However, it may also be possible that in the normal state both oscillators are chronotaxic due to only their external influences (from oxygen and glucose), with the interactions between MO and GO not strong enough for entrainment, as shown in region A. In all chronotaxic cases, phase fluctuation analysis will return a DFA exponent around 0.5 < α < 1. The evolution of the system from the normal to the altered state is from D to B via regions C and E, regions which are both non-chronotaxic, and will be characterised by a DFA exponent around 1 < α < 1.5, allowing the observation of the transition between states.
Discussion
This work is based on experimental evidence of metabolic oscillators3,13,15,19,22,23 and their interactions20,21,27,33. In experimental studies oscillations are often overlooked24, as several oscillations may contribute to the same signal thus almost cancelling each other despite existing separately. Alternatively, due to their highly complex nature they are often treated as stochastic25. Even in cases when oscillations and the interactions between them have been studied, the exact characteristics of their amplitude and phase relations have not been considered. Several studies of mitochondrial and glycolytic oscillations have shown that fluctuations of ATP production in a cell are not fully stochastic, but have nonautonomous and deterministic oscillatory components3,13,14,15,16,17,18,19,20,21,22,23,26,27,33,45. Therefore, these oscillations reflect underlying deterministic processes.
Here we have proposed a measurable characteristic of metabolic oscillations, chronotaxicity, which is expected to change during the transition of a healthy cell to a state with altered energy metabolism. This was demonstrated using a qualitative phase oscillator model of metabolic oscillations. The model captures only the most general and universal oscillatory dynamics and interactions, and thus provides the advantage that it is applicable to oscillatory metabolism in general, independent of cell type. The model explicitly takes into account the openness of cellular energy metabolism, and therefore its nonautonomous dynamics. The model, based on the theory of chronotaxic systems, introduces a new approach in which the complex metabolic system is considered as a set of functional rather than structural units, these functional units being interacting glycolytic and mitochondrial oscillators with chronotaxic properties.
As a concept within the theory of dynamical systems, describing oscillators which are driven by other oscillators, it is not trivial that chronotaxicity can be applied to biological oscillators, and that the driving in biological oscillators represents itself in the same way as in dynamical systems. In this work, we investigated the applicability of the concept of chronotaxicity to real biological oscillations observed in real experimental studies of cellular metabolism. We proved our assumptions that biological oscillators can be chronotaxic, and that their chronotaxicity can be identified from real experimental observations, and developed a model of cellular energy metabolism.
The interactions of oscillatory processes can be between amplitude and amplitude, phase and phase, or phase and amplitude. These possibilities show how many scenarios of regulations of physiological functions may occur. In this paper, for the sake of simplicity, we have selected phase oscillators and restricted the discussion to phase-phase interactions. Nevertheless, in observations of real life systems the chronotaxicity can be identified from phase dynamics alone, even when amplitude dynamics is complex37, thus making our model verifiable by experimental observations. Moreover, by identifying the characteristics of chronotaxicity we have shown that one in principle can detect a transition to a carcinogenic state of cellular functioning, provided metabolic oscillations are present in both states of the cell. The state in which the glycolytic oscillator determines the dynamics of energy metabolism, or the ‘altered’ state, was experimentally verified using real data from yeast cells exhibiting glycolytic oscillations. The described inverse approach methods found evidence of chronotaxicity in this case, as expected from the numerical simulations of the model.
Cellular metabolism may be affected by many more processes and interactions than those considered here. The simplicity of the model could easily facilitate the inclusion of further couplings, for example the consideration of calcium dynamics or genetic factors in energy production. Calcium has been shown to directly influence mitochondrial dynamics via many pathways50,51, while genetic mutations can have a direct effect on mitochondrial function52. These effects could be included in the model as influences to couplings, extra oscillators, or adaptations of the external drivers.
Results presented in this paper set up bases for experimental verification of the hypothesis that chronotaxicity can be used to identify transitions between metabolic states in a cell, for example the metabolic switch observed in cancer cells. We have demonstrated evidence of chronotaxicity in real metabolic oscillations, which led to a new way of studying metabolic processes inside a cell. The model provides a framework within which the existing understanding of biochemical reactions involved in metabolic processes along with new observations based on recently introduced functional imaging methods2,3,4,5 can be unified in a single picture. Furthermore, focusing on the transitions between metabolic states could facilitate the development of new therapeutic strategies.
Methods
Glycolytic oscillations in isolated yeast cells
As a real life example of driven metabolic oscillations we use glycolytic oscillations in individual isolated yeast cells recorded by Gustavsson et al.38. The brief description of the experiments setup is presented below, for more details see the original work by Gustavsson and co-authors38. Saccharomyces cerevisiae cells were harvested at the time of diauxic shift, starved of glucose for 3 hours and then stored at 0–4 °C until use. The cells were then placed in a microfluidic chamber at a distance of ~10 μm apart using optical tweezers. The cells were covered with 20 mM glucose solution for 4 minutes before flows were increased in order to cover the cells with 20 mM glucose/5 mM KCN solution. NADH fluorescence was monitored for 60 minutes at a sampling frequency of 0.25 Hz.
Characterising driven oscillators with chronotaxicity
The class of chronotaxic systems identifies oscillatory dynamical systems with dynamics ordered in time (chronos – time, taxis – order)34. Such ordering is typical for driven oscillators, where the drive system determines the dynamics of a response system. Chronotaxic systems can sustain their dynamics even with continuous external perturbations. Introduced for low-dimensional and high-dimensional dynamical systems34,35,36, chronotaxic systems have been shown to be useful in studies of living systems, one example being the application to the cardiorespiratory system37.
In addition to living and open systems, which are in continuous contact with the external environment, chronotaxic systems are nonautonomous dynamical systems29, i.e. their dynamics explicitly depends on time, as shown in the equation,
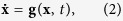 |
where x = (x1, x2, …, xm) is a vector of coordinates which fully determines the state of a system at a given moment of time in phase space x ∈ Rm. Alternatively, chronotaxic systems can be described by drive and response systems, as follows
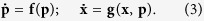 |
The main defining feature of chronotaxic systems is a time-dependent steady state of point attractor xA(t) (see Fig. 3a) which exists in the phase space of a chronotaxic system due to the influence from a drive system p (Fig. 3b). The trajectory xA(t) can be viewed as a uniformly hyperbolic trajectory53 which is linearly attracting in such a way that the distance between a neighboring trajectory and xA(t) can only contract in an unperturbed chronotaxic system. For more details and for relations between chronotaxic and other dynamical systems see ref. [36].
A simple example is given by unidirectionally coupled phase oscillators with phase φX driven by a phase φP as shown in the equation,
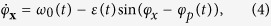 |
where 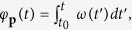 and ω0 is the natural frequency of the observed oscillator,
and ω0 is the natural frequency of the observed oscillator,  is coupling strength, and ω is the frequency of the driving oscillator. The time-dependent point attractor will exist if the condition of chronotaxicity35 is fulfilled, i.e. if
is coupling strength, and ω is the frequency of the driving oscillator. The time-dependent point attractor will exist if the condition of chronotaxicity35 is fulfilled, i.e. if 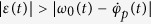 and if the coupling strength ε(t) does not change its sign. In the case of a particular choice
and if the coupling strength ε(t) does not change its sign. In the case of a particular choice 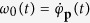 and ε(t) > 0, the time-dependent steady state
and ε(t) > 0, the time-dependent steady state 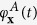 can be found analytically,
can be found analytically, 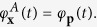 Chronotaxic systems in the simplest case could be considered as a generalization of synchronized unidirectionally coupled phase oscillators (phase-locked loops53).
Chronotaxic systems in the simplest case could be considered as a generalization of synchronized unidirectionally coupled phase oscillators (phase-locked loops53).
Taking into account that the dynamical system x is chronotaxic due to the influence from the driver p, chronotaxicity will change when the external influence from p changes. This makes chronotaxicity a perfect candidate for investigation in the study of metabolic oscillators under changing driving influences.
Detecting chronotaxicity from a single time series
One of main advantages of chronotaxicity is that it can be identified experimentally from a single time series, whereas otherwise the identification of drive-response relationship would require measurements of the driver as well as the response system. To identify chronotaxicity in a time series, a method named phase fluctuation analysis (PFA) was recently developed37. PFA involves the separation of the amplitude and phase of an oscillatory mode by extracting an estimated phase of the point attractor, φA*, from the continuous wavelet transform54,
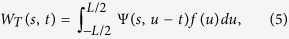 |
where Ψ(s,t) is the mother wavelet which is time-shifted according to t and scaled according to the parameter s. The oscillation can then be traced in WT(s, t). The instantaneous frequency of the oscillation at each time point can be estimated using either the synchrosqueezed wavelet transform55 or a ridge-extraction method56. The phase is then calculated by integrating over the instantaneous frequency in time. The estimation of angular velocity 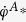 can be found by smoothing over the frequency extracted from the wavelet transform. Then,
can be found by smoothing over the frequency extracted from the wavelet transform. Then,  is integrated over to give φA*.
is integrated over to give φA*.
In this approach, perturbations are assumed to be due to an uncorrelated Gaussian process. In chronotaxic systems, perturbations decay due to the influence of the point attractor and the divergence from the attractor is similar to the original Gaussian process37. In contrast, in non-chronotaxic systems, where the phase of oscillation is neutrally stable, the perturbations are integrated over, resulting in a random walk (i.e. Brownian noise). To distinguish these two cases, detrended fluctuation analysis (DFA)57 is performed on the phase fluctuations extracted from the time series. First, the difference Δφ = φ* − φA* is calculated to obtain information about the divergence of the system from the point attractor due to perturbations. The DFA technique explores the fractal self-similarity of fluctuations at different timescales in Δφ. The scaling of fluctuations is determined by the self-similarity parameter α.
To estimate α the time series is integrated in time and divided into sections of length n. The local trend is removed for each section by subtracting a fitted polynomial, usually a first order fit57,58. The root mean square fluctuation F(n) for the scale equal to n is then defined by the following equation,
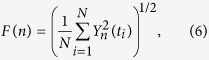 |
where Yn(t) is the integrated and detrended time series of length N. The self-similarity parameter is given by the gradient of the line of the plot of logF(n) against logn. The self-similarity parameter α for Δφ for uncorrelated Gaussian noise (as expected in chronotaxic systems) gives a value of 0.5; in contrast, for integrated white Gaussian noise (as expected in non-chronotaxic systems) the parameter α returns a value of 1.5. In Fig. 5 the lowest length n was chosen to be equal to 2 cycles of oscillations, and the largest length n was chosen to be equal to 15 cycles of oscillations. Supplementary Fig. S5 shows how this method may be applied for an example ATP signal obtained from the model.
In order to reliably test for chronotaxicity, it should be noted that the time series should be sufficiently long, i.e. contain at least 30 cycles of oscillation (may vary depending on the characteristics of the data), be evenly sampled, and have a sampling frequency which is high enough to capture the dynamics at the frequency of interest41.
Additional Information
How to cite this article: Lancaster, G. et al. Modelling chronotaxicity of cellular energy metabolism to facilitate the identification of altered metabolic states. Sci. Rep. 6, 29584; doi: 10.1038/srep29584 (2016).
Supplementary Material
Acknowledgments
The authors are grateful to Mattias Goksör for providing the data from 34 yeast cells recorded in ref. [38] and for useful comments on the manuscript. The authors would also like to thank Phil Clemson for helpful discussions and Peter V. E. McClintock, Stephen Roberts, Tomislav Stankovski, Jolanta Tarasiuk and Jose J. Suárez-Vargas for constructive comments on the manuscript. This study was supported by the Engineering and Physical Sciences Research Council (UK) (Grant Nos EP/100999X1, EP/M006298/1, PhD grant 1060895 (GL)) and the ITN COSMOS (funded by the EU Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant Agreement No. 642563).
Footnotes
Author Contributions A.S. conceived and supervised the study. Y.F.S. and K.J. formulated the model and performed preliminary simulations. Y.F.S. established the link between chronotaxicity and the state of a cell, and devised the theoretical background. G.L. implemented the numerical simulations of the model (Fig. 4 and Supplementary Information). Y.F.S. and G.L. applied inverse approach methods. G.L. analysed the experimental yeast data. G.L. and Y.F.S. drafted the manuscript. All authors edited and commented on the manuscript, and discussed the results of the model and their interpretation.
References
- Törnroth-Horsefield S. & Neutze R. Opening and closing the metabolite gate. Proc. Nat. Acad. Sci. 105, 19565–19566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarias G. & Chatton J. Y. Selective ion changes during spontaneous mitochondrial transients in intact astrocytes. PloS One 6, e28505, doi: 10.1371/journal.pone.0028505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrins M. J., Van Dyke A. R., Mapp A. K., Rizzo M. A. & Satin L. S. Direct measurements of oscillatory glycolysis in pancreatic islet β-cells using novel fluorescence resonance energy transfer (FRET) biosensors for pyruvate kinase M2 activity. J. Biol. Chem. 288, 33312–33322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Sokabe M. & Grygorczyk R. Real-time luminescence imaging of cellular ATP release. Methods 66, 330–344 (2014). [DOI] [PubMed] [Google Scholar]
- Rieger B., Junge W. & Busch K. B. Lateral pH gradient between OXPHOS complex IV and F0F1 ATP-synthase in folded mitochondrial membranes. Nat. Commun. 5, 3103, doi: 10.1038/ncomms4103 (2014). [DOI] [PubMed] [Google Scholar]
- Ralph S. J., Rodrìguez-Enrìquez S., Neuzil J., Saavedra E. & Moreno-Sànchez R. The causes of cancer revisited: ‘Mitochondrial malignancy’ and ROS-induced oncogenic transformation–Why mitochondria are targets for cancer therapy. Mol. Aspects Med. 31, 145–170 (2010). [DOI] [PubMed] [Google Scholar]
- Cloonan S. M. & Choi A. M. K. Mitochondria: commanders of innate immunity and disease? Curr. Opin. Immunol. 24, 32–40 (2012). [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried T. N. & Shelton L. M. Cancer as a metabolic disease. Nutr. Metab. 7, 7, doi: 10.1186/1743-7075-7-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G. & Pouyssegur J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 13, 472–482 (2008). [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. & Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S., Galluzzi L. & Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discovery 9, 447–464 (2010). [DOI] [PubMed] [Google Scholar]
- Aon M. A., Cortassa S., Marbán E. & O’Rourke B. Synchronized whole cell oscillations in mitochondrial metabolism triggered by a local release of reactive oxygen species in cardiac myocytes. J. Biol. Chem. 278, 44735–44744 (2003). [DOI] [PubMed] [Google Scholar]
- Kurz F. T., Aon M. A., O'Rourke B. & Armoundas A. A. Wavelet analysis reveals heterogeneous time-dependent oscillations of individual mitochondria. Am. J. Physiol. Heart Circ. Physiol. 299, H1736–H1740 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke B., Ramza B. M. & Marban E. Oscillations of membrane current and excitability driven by metabolic oscillations in heart cells. Science 265, 962–966 (1994). [DOI] [PubMed] [Google Scholar]
- Goldbeter A. Computational approaches to cellular rhythms. Nature 420, 238–245 (2002). [DOI] [PubMed] [Google Scholar]
- Cortassa S., Aon M. A., Winslow R. L. & O'Rourke B. A mitochondrial oscillator dependent on reactive oxygen species. Biophys. J. 87, 2060–2073 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B. P. & McKnight S. L. Metabolic cycles as an underlying basis of biological oscillations. Nat. Rev. Mol. Cell Biol. 7, 696–701 (2006). [DOI] [PubMed] [Google Scholar]
- Aon M. A., Cortassa S. & O’Rourke B. Mitochondrial oscillations in physiology and pathophysiology. Adv. Exp. Med. Biol. 641, 98–117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. F., Andersen A. Z., Lunding A., Brasen J. C. & Poulsen A. K. Regulation of glycolytic oscillations by mitochondrial and plasma membrane H+-ATPases. Biophys. J. 96, 3850–3861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganitkevich V., Mattea V. & Benndorf K. Glycolytic oscillations in single ischemic cardiomyocytes at near anoxia. J. Gen. Physiol. 135, 307–319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz F. T., Aon M. A., O'Rourke B. & Armoundas A. A. Spatio-temporal oscillations of individual mitochondria in cardiac myocytes reveal modulation of synchronized mitochondrial clusters. PNAS 107, 14315–14320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra F. A., Buzi G. & Doyle J. C. Glycolytic oscillations and limits on robust efficiency. Science 333, 187–192 (2011). [DOI] [PubMed] [Google Scholar]
- Spiller D. G., Wood C. D., Rand D. A. & White M. R. H. Measurement of single-cell dynamics. Nature 465, 736–745 (2010). [DOI] [PubMed] [Google Scholar]
- Kiviet D. J. et al. Stochasticity of metabolism and growth at the single-cell level. Nature 514, 376–379 (2014). [DOI] [PubMed] [Google Scholar]
- Higgins J. A chemical mechanism for oscillation of glycolytic intermediates in yeast cells. PNAS 51, 989–994 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. G., Mosekilde E., Polonsky K. S. & Luciani D. S. Complex patterns of metabolic and Ca2+ entrainment in pancreatic islets by oscillatory glucose. Biophys. J. 105, 29–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aon M. A., Cortassa S., Lemar K. M., Hayes A. J. & Lloyd D. Single and cell population respiratory oscillations in yeast: A 2–photon scanning laser microscopy study. FEBS Lett. 581, 8–14 (2007). [DOI] [PubMed] [Google Scholar]
- Kloeden P. E. & Rasmussen M. Nonautonomous Dynamical Systems (American Mathematical Soc, 2011). [Google Scholar]
- Clemson P. T. & Stefanovska A. Discerning non-autonomous dynamics. Phys. Rep. 542, 297–368 (2014). [Google Scholar]
- Warburg O. Metabolism of tumours. Biochem. Z. 142, 317–333 (1923). [Google Scholar]
- Koppenol W. H., Bounds P. L. & Dang C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- Bertram R., Satin L. S., Pedersen M. G., Luciani D. S. & Sherman A. Interaction of glycolysis and mitochondrial respiration in metabolic oscillations of pancreatic islets. Biophys. J. 92, 1544–1555 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprunenko Y. F., Clemson P. T. & Stefanovska A. Chronotaxic systems: a new class of self-sustained nonautonomous oscillators. Phys. Rev. Lett. 111, 024101, doi: 10.1103/PhysRevLett.111.024101 (2013). [DOI] [PubMed] [Google Scholar]
- Suprunenko Y. F., Clemson P. T. & Stefanovska A. Chronotaxic systems with separable amplitude and phase dynamics. Phys. Rev. E 89, 012922, doi: 10.1103/PhysRevE.89.012922 (2014). [DOI] [PubMed] [Google Scholar]
- Suprunenko Y. F. & Stefanovska A. Generalized chronotaxic systems: time-dependent oscillatory dynamics stable under continuous perturbation. Phys. Rev. E 90, 032921, doi: 10.1103/PhysRevE.90.032921 (2014). [DOI] [PubMed] [Google Scholar]
- Clemson P. T., Suprunenko Y. F., Stankovski T. & Stefanovska A. Inverse approach to chronotaxic systems for single-variable time series. Phys. Rev. E 89, 032904, doi: 10.1103/PhysRevE.89.032904 (2014). [DOI] [PubMed] [Google Scholar]
- Gustavsson A.-K. et al. Sustained glycolytic oscillations in individual isolated yeast cells. FEBS J. 279, 2837–2847 (2012). [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R., Uribe-Carvajal S., Devin A. & Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim. Biophys. Acta 1796, 252–265 (2009). [DOI] [PubMed] [Google Scholar]
- Gustavsson A.-K., Adiels C. B., Mehlig B. & Goksör M. Entrainment of heterogeneous glycolytic oscillations in single cells. Sci. Rep. 5, 9404, doi: 10.1038/srep09404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster G., Clemson P. T., Suprunenko Y. F., Stankovski T. & Stefanovska A. Detecting chronotaxic systems from single-variable time series with separable amplitude and phase. Entropy 17, 4413–4438 (2015). [Google Scholar]
- Natter K. & Kohlwein S. D. Yeast and cancer cells–common principles in lipid metabolism. Biochim. Biophys. Acta 1831, 314–326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornheim K. & Lowenstein J. M. The purine nucleotide cycle: Interactions with oscillations of glycolytic pathway in muscle extracts. J. Biol. Chem. 249, 3241–3247 (1974). [PubMed] [Google Scholar]
- Tornheim K., Andrés V. & Schultz V. Modulation by citrate of glycolytic oscillations in skeletal muscle extracts. J. Biol. Chem. 266, 15675–15678 (1991). [PubMed] [Google Scholar]
- Reijenga K. A. et al. Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae. Biophys. J. 80, 626–634 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijenga K. A., Bakker B. M., van der Weijden C. C. & Westerhoff H. V. Training of yeast cell dynamics. FEBS J. 272, 1616–1624 (2005). [DOI] [PubMed] [Google Scholar]
- Boiteux A., Goldbeter A. & Hess B. Control of oscillating glycolysis of yeast by stochastic, periodic, and steady source of substrate: a model and experimental study. Proc. Nat. Acad. Sci. 72, 3829–3833 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özalp V. C., Pedersen T. R., Nielsen L. J. & Olsen L. F. Time-resolved measurements of intracellular ATP in the yeast Saccharomyces cerevisiae using a new type of nanobiosensor. J. Biol. Chem. 285, 37579–37588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoke H. S. et al. Tight coupling of metabolic oscillations and intracellular water dynamics in Saccharomyces cerevisiae. PLOS One 10, e0117308, doi: 10.1371/journal.pone.0117308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B., Willis W. T., Chess D. J. & Balaban R. S. Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry 52, 2793–2809 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrins M. J. et al. Metabolic oscillations in pancreatic islets depend on the intracellular Ca2+ level but not Ca2+ oscillations. Biophys. J. 99, 76–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M., Baldi P. & Wallace D. C. Mitochondrial mutations in cancer. Oncogene 25, 4647–4662 (2006). [DOI] [PubMed] [Google Scholar]
- Bishnani Z. & MacKay R. S. Safety criteria for aperiodically forced systems. Dynam. Sys. 18, 107–129 (2003). [Google Scholar]
- Kaiser G. A Friendly Guide to Wavelets (Birkhäuser, Boston, 1994). [Google Scholar]
- Daubechies I., Lu J. & Wu H. Synchrosqueezed wavelet transforms: An empirical mode decomposition-like tool. Appl. and Comput. Harmon. Anal. 30, 243–261 (2011). [Google Scholar]
- Delprat N. et al. Asymptotic wavelet and Gabor analysis: Extraction of instantaneous frequencies. IEEE Trans. Inf. Theory 38, 644–664 (1992). [Google Scholar]
- Peng C. K. et al. Mosaic organisation of DNA nucleotides. Phys. Rev. E 49, 1685–1689 (1994). [DOI] [PubMed] [Google Scholar]
- Shiogai Y., Stefanovska A. & McClintock P. V. E. Nonlinear dynamics of cardiovascular ageing. Phys. Rep. 488, 51–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.