Abstract
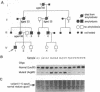
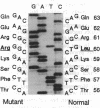
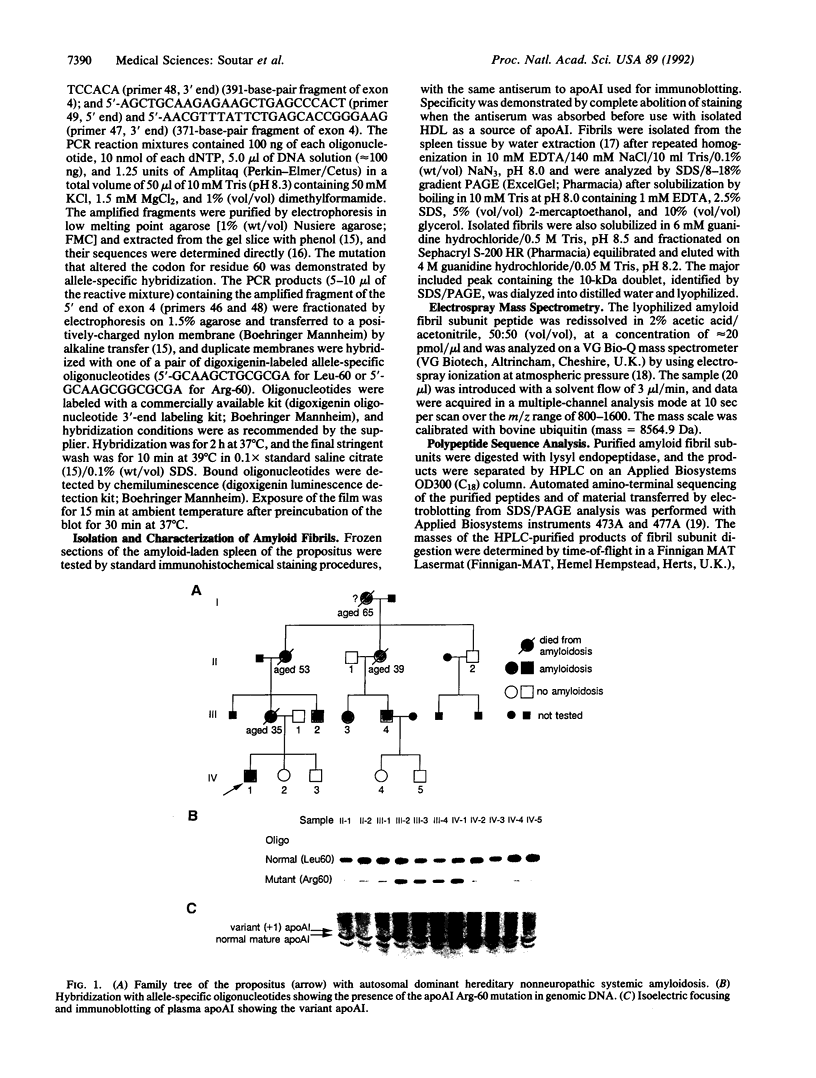
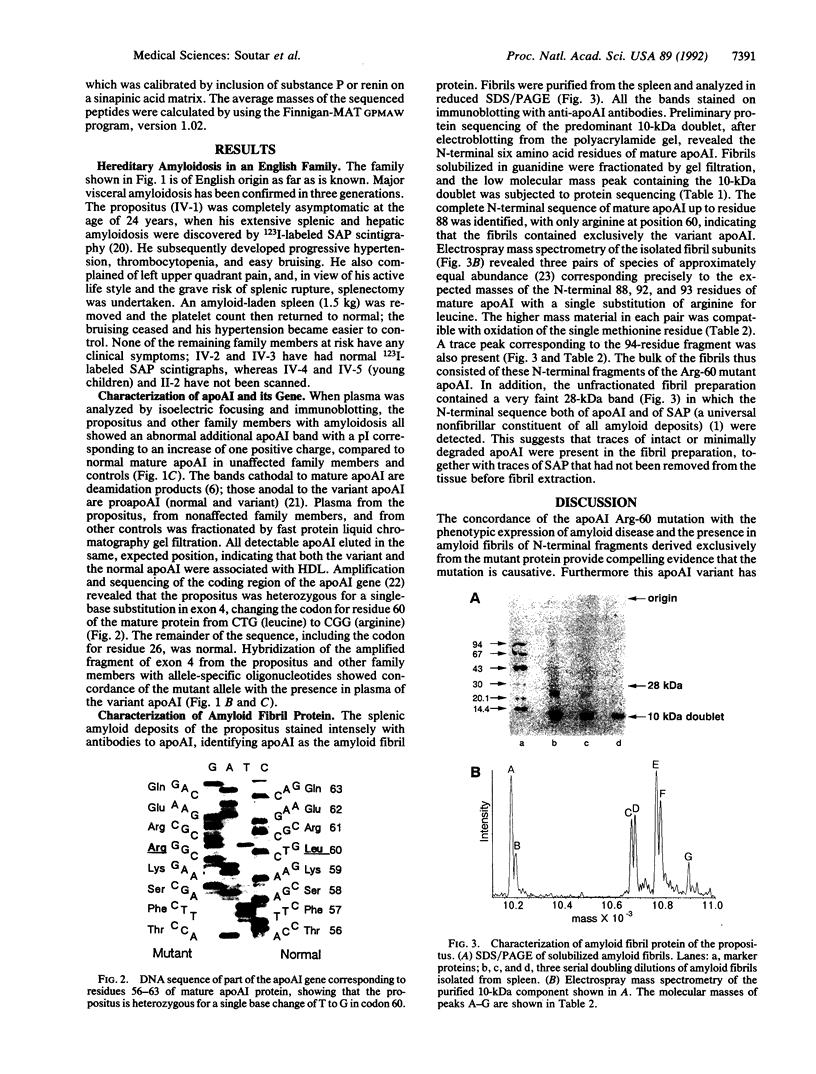
A mutation in the gene for apolipoprotein AI (apoAI) was identified in an English family with autosomal dominant non-neuropathic systemic amyloidosis. The plasma of all affected individuals contained a variant apoAI with one additional charge, as well as normal apoAI. The propositus was heterozygous; the coding region of his apoAI gene contained both the normal sequence and a single-base substitution changing the codon for residue 60 of the mature protein from CTG (leucine) to CGG (arginine). Allele-specific oligonucleotide hybridization showed that the other affected individuals were also heterozygotes and that there was concordance of the mutant allele with the presence of variant plasma apoAI. Amyloid fibrils isolated from the spleen of the propositus consisted of proteins that ran as a doublet with an apparent mass of approximately 10 kDa in SDS/PAGE and a trace band at 28 kDa. Electrospray mass spectrometry of the purified 10-kDa material revealed components with mass corresponding to the N-terminal 88, 92, 93, and 94 residues of apoAI each with substitution of arginine for leucine. These observations were confirmed by direct protein sequencing and laser desorption time-of-flight mass analysis. No material with the normal apoAI sequence was detected. The trace band at 28 kDa yielded the N-terminal sequence of mature apoAI, indicating that intact or minimally degraded apoAI was also present in the fibril preparation. Discovery of this mutation and the detailed characterization of the apoAI fragments that form the amyloid fibrils open additional avenues for investigation of amyloidogenesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Geisow M. J., Oatley S. J., Rérat B., Rérat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J Mol Biol. 1978 May 25;121(3):339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- Casanova J. L., Pannetier C., Jaulin C., Kourilsky P. Optimal conditions for directly sequencing double-stranded PCR products with sequenase. Nucleic Acids Res. 1990 Jul 11;18(13):4028–4028. doi: 10.1093/nar/18.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner G. G. Amyloid deposits and amyloidosis. The beta-fibrilloses (first of two parts). N Engl J Med. 1980 Jun 5;302(23):1283–1292. doi: 10.1056/NEJM198006053022305. [DOI] [PubMed] [Google Scholar]
- Hawkins P. N., Lavender J. P., Pepys M. B. Evaluation of systemic amyloidosis by scintigraphy with 123I-labeled serum amyloid P component. N Engl J Med. 1990 Aug 23;323(8):508–513. doi: 10.1056/NEJM199008233230803. [DOI] [PubMed] [Google Scholar]
- Higuchi K., Kitagawa K., Naiki H., Hanada K., Hosokawa M., Takeda T. Polymorphism of apolipoprotein A-II (apoA-II) among inbred strains of mice. Relationship between the molecular type of apoA-II and mouse senile amyloidosis. Biochem J. 1991 Oct 15;279(Pt 2):427–433. doi: 10.1042/bj2790427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. L., Morrisett J. D., Gotto A. M., Jr Lipoprotein structure and metabolism. Physiol Rev. 1976 Apr;56(2):259–316. doi: 10.1152/physrev.1976.56.2.259. [DOI] [PubMed] [Google Scholar]
- Keir G., Walker R. W., Johnson M. H., Thompson E. J. Nitrocellulose immunofixation following agarose electrophoresis in the study of immunoglobulin G subgroups in unconcentrated cerebrospinal fluid. Clin Chim Acta. 1982 May 20;121(2):231–236. doi: 10.1016/0009-8981(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Maury C. P. Gelsolin-related amyloidosis. Identification of the amyloid protein in Finnish hereditary amyloidosis as a fragment of variant gelsolin. J Clin Invest. 1991 Apr;87(4):1195–1199. doi: 10.1172/JCI115118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell I. F., Wisdom G. B., Trimble E. R. Apolipoprotein E phenotype determined by agarose gel electrofocusing and immunoblotting. Clin Chem. 1989 Oct;35(10):2070–2073. [PubMed] [Google Scholar]
- Nichols W. C., Dwulet F. E., Liepnieks J., Benson M. D. Variant apolipoprotein AI as a major constituent of a human hereditary amyloid. Biochem Biophys Res Commun. 1988 Oct 31;156(2):762–768. doi: 10.1016/s0006-291x(88)80909-4. [DOI] [PubMed] [Google Scholar]
- Nichols W. C., Gregg R. E., Brewer H. B., Jr, Benson M. D. A mutation in apolipoprotein A-I in the Iowa type of familial amyloidotic polyneuropathy. Genomics. 1990 Oct;8(2):318–323. doi: 10.1016/0888-7543(90)90288-6. [DOI] [PubMed] [Google Scholar]
- Pitkänen P., Westermark P., Cornwell G. G., 3rd Senile systemic amyloidosis. Am J Pathol. 1984 Dec;117(3):391–399. [PMC free article] [PubMed] [Google Scholar]
- Pras M., Schubert M., Zucker-Franklin D., Rimon A., Franklin E. C. The characterization of soluble amyloid prepared in water. J Clin Invest. 1968 Apr;47(4):924–933. doi: 10.1172/JCI105784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saraiva M. J. Recent advances in the molecular pathology of familial amyloid polyneuropathy. Neuromuscul Disord. 1991;1(1):3–6. doi: 10.1016/0960-8966(91)90037-s. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974 Jan 15;38(3):247–258. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- Shoulders C. C., Kornblihtt A. R., Munro B. S., Baralle F. E. Gene structure of human apolipoprotein A1. Nucleic Acids Res. 1983 May 11;11(9):2827–2837. doi: 10.1093/nar/11.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud P., Tybjaerg-Hansen A., Bhatnagar D., Mbewu A., Miller J. P., Durrington P., Humphries S. Rapid screening for specific mutations in patients with a clinical diagnosis of familial hypercholesterolaemia. Atherosclerosis. 1991 Aug;89(2-3):137–141. doi: 10.1016/0021-9150(91)90053-6. [DOI] [PubMed] [Google Scholar]
- Turnell W., Sarra R., Glover I. D., Baum J. O., Caspi D., Baltz M. L., Pepys M. B. Secondary structure prediction of human SAA1. Presumptive identification of calcium and lipid binding sites. Mol Biol Med. 1986 Oct;3(5):387–407. [PubMed] [Google Scholar]
- Van Dorsselaer A., Bitsch F., Green B., Jarvis S., Lepage P., Bischoff R., Kolbe V. J., Roitsch C. Application of electrospray mass spectrometry to the characterization of recombinant proteins up to 44 kDa. Biomed Environ Mass Spectrom. 1990 Nov;19(11):692–704. doi: 10.1002/bms.1200191108. [DOI] [PubMed] [Google Scholar]
- Zannis V. I., Karathanasis S. K., Keutmann H. T., Goldberger G., Breslow J. L. Intracellular and extracellular processing of human apolipoprotein A-I: secreted apolipoprotein A-I isoprotein 2 is a propeptide. Proc Natl Acad Sci U S A. 1983 May;80(9):2574–2578. doi: 10.1073/pnas.80.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eckardstein A., Funke H., Walter M., Altland K., Benninghoven A., Assmann G. Structural analysis of human apolipoprotein A-I variants. Amino acid substitutions are nonrandomly distributed throughout the apolipoprotein A-I primary structure. J Biol Chem. 1990 May 25;265(15):8610–8617. [PubMed] [Google Scholar]