Abstract
Genomic imprinting is an epigenetic phenomenon resulting in parent-of-origin-specific gene expression that is regulated by a differentially methylated region. Gene mutations or failures in the imprinting process lead to the development of imprinting disorders, such as Angelman syndrome. The symptoms of Angelman syndrome are caused by the absence of functional UBE3A protein in neurons of the brain. To create a human neuronal model for Angelman syndrome, we reprogrammed dermal fibroblasts of a patient carrying a defined three-base pair deletion in UBE3A into induced pluripotent stem cells (iPSCs). In these iPSCs, both parental alleles are present, distinguishable by the mutation, and express UBE3A. Detailed characterization of these iPSCs demonstrated their pluripotency and exceptional stability of the differentially methylated region regulating imprinted UBE3A expression. We observed strong induction of SNHG14 and silencing of paternal UBE3A expression only late during neuronal differentiation, in vitro. This new Angelman syndrome iPSC line allows to study imprinted gene regulation on both parental alleles and to dissect molecular pathways affected by the absence of UBE3A protein.
The epigenetic process of genomic imprinting is controlled by differentially methylated regions (DMRs) which results in parent-of-origin-dependent gene expression. Imprinted germ line DMRs are exceptional in three ways: First, they can be properly and entirely established only during female or male gametogenesis. Second, they are protected from demethylation during the global wave of methylation erasure during early embryonic development. Third, methylation of the DMR is invariably preserved during cell division, present in every cell type and independent of gene expression activity1,2. Deletion of a DMR or disturbances in its methylation leads to imprinting disorders, such as Angelman syndrome (AS), exhibiting typical symptoms like absence of speech, movement disorders, happy demeanor and developmental delay3. The cause for these symptoms is the absence of a functional UBE3A protein in the brain4,5. The UBE3A gene is part of the imprinted Prader-Willi/Angelman syndrome locus (PWS/AS locus) on chromosome 15q11q13. At this locus, only the maternal allele is methylated at a CpG island, which encompasses the promoter and exon 1 of the SNRPN gene and represents the DMR of the locus. This DMR is termed PWS-SRO, which stands for Prader-Willi syndrome shortest region of deletion overlap6. DNA methylation silences the protein-coding gene SNURF/SNRPN and several non-coding RNA genes on the maternal allele6 (Fig. 1A). SNURF/SNRPN, the non-coding SNORD gene clusters and the long non-coding RNA, SNHG14 (alternatively named UBE3A-ATS) are expressed from the non-methylated paternal allele. In the brain, SNHG14 overlaps the entire UBE3A gene and promoter in antisense direction, thereby silencing UBE3A expression7. As a consequence, this results in brain-specific monoallelic UBE3A expression from the maternal allele. Hence, UBE3A is vulnerable to mutations occurring on the maternal chromosome 15. So far, large deletions up to several megabases, imprinting defects, paternal uniparental disomy or mutations in the UBE3A gene itself have been described as molecular cause for AS3. The different types of mutations correlate with gradual differences in the severity of the disorder. Large deletions result in loss of several other genes in the same region, and these patients typically present with a more severe phenotype than patients carrying point mutations affecting the UBE3A gene alone8.
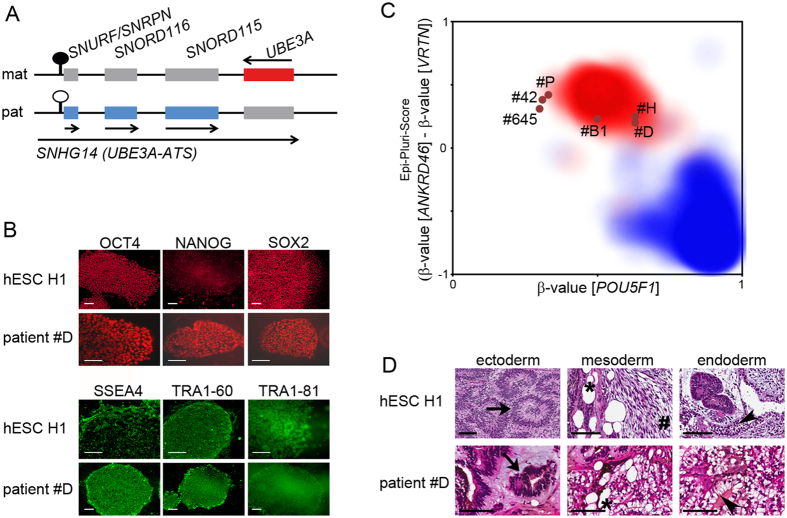
Figure 1. Generation of AS_∆3 iPSCs.
(A) Schematic and simplified representation of the Prader-Willi/Angelman syndrome locus and its expression status in the brain. Red: maternally expressed genes, blue: paternally expressed genes, grey: non-expressed genes. Lollipop indicates the DMR (black: methylated, white: not methylated). Arrows indicate direction of expression. Scheme is not drawn to scale. (B) Immunofluorescence for nuclear (red) and surface (green) antigens showing expression of pluripotency-associated proteins in patient #D and hESC H1 as a reference. Scale bars indicate 100 μm. (C) Epi-Pluri-Score analysis of generated iPSCs. Clouds represent areas of reference pluripotent (red) or somatic (blue) cells. The six iPSC clones analyzed are shown as dots localizing to the pluripotent area. (D) H&E stained histology of immature teratomas formed by hESC H1 and patient #D. Derivatives of all three germ layers can be identified: neuroectodermal rosettes (arrow); mesodermal loose immature mesenchyme (hash) and fat cells (asterisk), and endodermal glandular tissue with clear and goblet cells (arrowhead). Scale bars indicate 100 μm.
As AS is caused by a lack of UBE3A activity in the brain, access to neurons is needed to study its function at the molecular level. Therefore, the generation of induced pluripotent stem cells (iPSCs) from patient cells and their subsequent directed differentiation into neurons provide a valuable tool for AS research9. The generation of iPSCs from patients with Angelman syndrome has been described by Chamberlain et al.10. These AS iPSC lines carry large deletions on their maternal chromosome, resulting in loss of about 28 genes from NIPA1 to OCA2. This makes it difficult to specifically address the role of UBE3A in neuronal function and disease development.
Here we report the establishment and thorough characterization of a new iPSC line (AS_∆3) of a previously described patient with AS, harboring a defined three-base pair deletion within the maternally inherited UBE3A allele11. The encoded protein is predicted to lack amino acid G538 (based on NM_130838). Using computer modeling of the mutant protein based on the X-ray structure of the wild-type enzyme, a local destabilization around the catalytic cleft of UBE3A was proposed, likely impairing the binding of substrates11. The new iPSC line complements the existing AS iPSCs carrying large chromosomal deletions. It will facilitate the specific attribution of effects observed during neuronal differentiation to the defective UBE3A gene. This will contribute to a deeper understanding of imprinting mechanisms and AS itself.
Results
We reprogrammed primary dermal fibroblasts isolated from a female patient with AS harboring a three-base pair deletion in exon 4 of the UBE3A gene (accession NM_130838)11, and from a normal healthy control person. The reprogramming efficiencies (i.e. the number of isolated colonies per transduced cell number) were similar for patient and control person-derived fibroblasts, ranging from 0.005 to 0.05 percent (Supplementary Table S1). For quality and potency characterization, eight AS_∆3 and seven control iPSC clones were expanded and established as independent lines. As determined by Southern blot analysis, the number of integration sites ranged from one to five in independent clones (Supplementary Fig. S1). For further analysis, only clones containing single vector integrations were chosen: patient-derived AS_Δ3 iPSC clones #B1, #D, #H and #P, and healthy control-derived iPSC clones #42 and #645. The identity of parental fibroblast cells and derived iPSCs was confirmed by high resolution HLA typing (Supplementary Table S2) and the presence of the three-base pair deletion in exon 4 of the UBE3A gene in AS_∆3 iPSCs was confirmed by sequencing (Supplementary Fig. S1). Karyotype analysis revealed a normal female karyotype for three of the four patient lines and both control lines (Supplementary Fig. S1). Patient #H carries an additional marker chromosome present in all metaphases analyzed (Supplementary Fig. S1). This marker chromosome was identified as an isochromosome 12p. Gain of chromosome 12 or i12p has been reported as frequent chromosomal abnormality in hESCs and iPSCs, being probably associated with a proliferation advantage of cells12.
For potency testing, expression of pluripotency markers was determined by different methods. For all assays, human embryonic stem cells (hESCs) H1 were used as a reference for pluripotency. Staining for alkaline phosphatase activity showed expression of the enzyme in all six iPSC clones (Supplementary Fig. S2). Expression of the nuclear proteins OCT4, NANOG, SOX2 and the cell surface antigens SSEA4, TRA1-60 and TRA1-81 was demonstrated by immunofluorescence and, for the surface antigens, by flow cytometry (Fig. 1B, Supplementary Figs S2 and S3). Expression of genes indicative of pluripotency, like endogenous POU5F1 (encoding OCT4), KLF4, SOX2, MYC, NANOG, DNMT3B, LIN28 and REX1 was evaluated by real-time qPCR (Supplementary Fig. S4). All iPSCs expressed these genes at levels similar to hESC H1. In contrast, their expression was not detected in the parental fibroblasts, except for KLF4 and MYC. In addition, we used TaqMan human stem cell pluripotency arrays to assess expression of 96 genes indicative of pluripotency, stemness and differentiated cell lineages. Applying cluster analysis, hESC H1, AS_∆3 and control iPSCs clustered separately from the two parental fibroblast lines, indicating successful reprogramming (Supplementary Fig. S4, Supplementary Table S3). As molecular test for pluripotency, we employed Epi-Pluri-Score analysis, using the DNA methylation levels of three single CpG sites indicative of differentiated and pluripotent cells: pluripotent cells exhibit high levels of DNA methylation at ANKRD46 and low levels at VRTN and POU5F113. All six iPSCs generated in this study, four AS_∆3 and two control clones, mapped to the upper middle area of the plot representative of reference pluripotent cells (Fig. 1C). As the most stringent test for pluripotency of human iPSCs, teratoma assays were performed. Mice were injected with reprogrammed cells of patient #B1, #D and #P, control #645 and also with hESC H1 as a positive control. Tumors formed within three to 10 weeks with 100 percent efficiency (Supplementary Table S4). Histomorphologic analysis revealed that all tumors were immature teratomas containing derivatives of all three germ layers (Fig. 1D, Supplementary Fig. S5). In summary, all tests applied indicated that the newly generated patient and control iPSCs are pluripotent and fully reprogrammed.
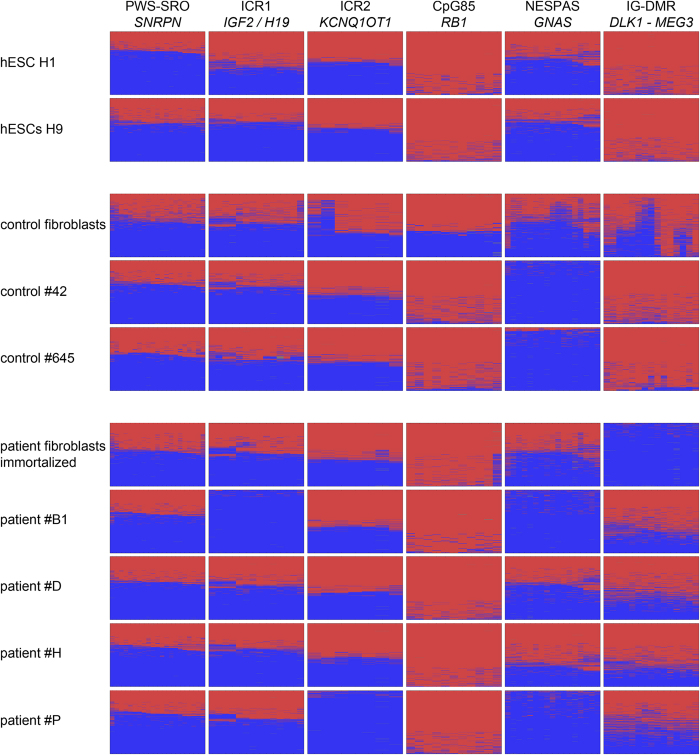
AS_∆3 iPSCs were generated as a tool for modeling the imprinting disorder AS. For proper imprinted gene expression of the AS gene UBE3A, epigenetic stability of the PWS-SRO is mandatory. Using deep bisulfite amplicon sequencing, analysis of DNA methylation was performed in parental fibroblasts, hESCs H1 and H9 and the iPSCs generated in this study at the PWS-SRO and five additional germ line DMRs of imprinted gene clusters or genes: the IG-DMR at the DLK1 - MEG3 locus, the CpG85 DMR at the RB1 gene, the NESPAS DMR at the GNAS locus, and the ICR2 and ICR1 DMRs (Fig. 2, Supplementary Table S5). DNA methylation of the latter two regulates expression at the Beckwith-Wiedemann syndrome locus and the IGF2/H19 locus, respectively. Imprinted DMRs are expected to show a level of about 50% DNA methylation in all types of diploid human cells. This level was observed consistently only for the PWS-SRO in all samples tested. The CpG85 at the RB1 gene showed consistent hypermethylation to a level of almost 100%, except for the fibroblasts derived from the control person. The PWS-SRO and the CpG85 represent the extremes of our analysis, being the most stable and unstable DMRs, respectively. The DMRs ICR1, ICR2 and NESPAS mainly exhibited the expected level of 50% methylation, but showed hypomethylation in one (ICR1, ICR2) and four (NESPAS) clones. The IG-DMR was reported to be prone to a gain of methylation in human pluripotent cells14,15, which we observed in the two hESC lines and both control iPSC clones, but not in the patient-derived clones. In contrast, immortalized fibroblast cells of the patient showed a loss of methylation at the IG-DMR, which was not confirmed in fibroblasts of healthy control persons and blood lymphocytes of the patient (Supplementary Fig. S6). For comparison, we analyzed the methylation status in the previously published AS iPSCs carrying a large type I deletion of the whole PWS/AS locus on the maternal allele (line AGI-0) and control cells (line MCH2-10). As expected, in AGI-0 the PWS-SRO was completely unmethylated as only the paternal allele is present (Supplementary Fig. S6). Methylation levels at the ICR1, ICR2 and NESPAS were normal, whereas the CpG85 and the IG-DMR showed a gain of methylation. Results for MCH2-10 iPSCs were comparable, except for normal 50% methylation at the PWS-SRO and a loss of methylation at the NESPAS (Supplementary Fig. S6).
Figure 2. Epigenetic stability of six imprinted DMRs.
DNA methylation was analyzed by deep bisulfite amplicon sequencing. The heatmaps show each CpG site in columns and individual sequenced reads in rows. Red: methylated, blue: unmethylated. Most heatmaps display a 50% methylation level, which is expected for imprinted DMRs in diploid cells. Exceptions are a loss of methylation at ICR1 (#B1), ICR2 (#P) and NESPAS (#42, #645, #B1, #P). CpG85 shows consistent gain of methylation, except for the control fibroblast sample. IG-DMR exhibits hypermethylation in both hESC lines, #42 and #645 and hypomethylation in the immortalized patient-derived fibroblast sample.
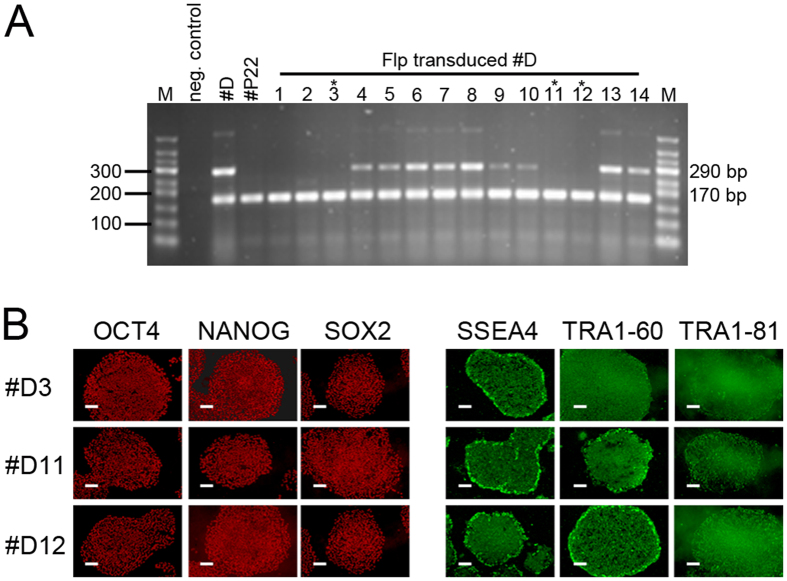
For the generation of iPSCs from dermal fibroblasts, we used a self-inactivating lentiviral reprogramming vector, which contains two FRT sites in the ∆U3 regions within the LTRs that allow for excision of the viral vector by Flp recombinase after successful reprogramming16,17. To eliminate the risk of reactivation of the integrated vector, which could lead to inhibition of differentiation18, we delivered Flp recombinase into patient #D and #P and control #42 and #645 by lentiviral-mediated protein transfer to excise the reprogramming vector. Successful excision was screened for by PCR (Fig. 3A, Supplementary Fig. S7). Two to three subclones were analyzed of each parental iPSC clone and all maintained their pluripotency characteristics, as determined by expression analysis of alkaline phosphatase and selected pluripotency-associated genes (Fig. 3B, Supplementary Fig. S7). Pluripotency of the excised subclone #P22 was also functionally proven by successful teratoma formation in mice (Supplementary Fig. S5).
Figure 3. Virus excision from patient #D iPSCs.
(A) PCR screening for successful virus excision indicated by absence of the 290 bp product, the 170 bp product serves as internal control. Clones marked by an asterisk (#D3, #D11 and #D12) were analyzed further. (B) Positive immunofluorescence of three excised clones of parental patient #D for pluripotency-associated nuclear (OCT4, NANOG, SOX2) and surface (SSEA4, TRA1-60, TRA1-81) antigens. Scale bars indicate 100 μm.
Imprinted expression of UBE3A is observed only in the brain. To study its function and regulation, neurons are needed. Only AS_Δ3 #D iPSCs passed all quality and pluripotency assessments and were chosen for in vitro differentiation into mature neurons. The generated AS_Δ3 iPSCs contain both parental UBE3A alleles, whose expression can be distinguished by single-nucleotide primer extension analysis using the three-base pair deletion in exon 4 (accession NM_130838) as a SNP. This offers the unique possibility to observe silencing of the paternal allele and persistent maternal UBE3A expression in the same cell.
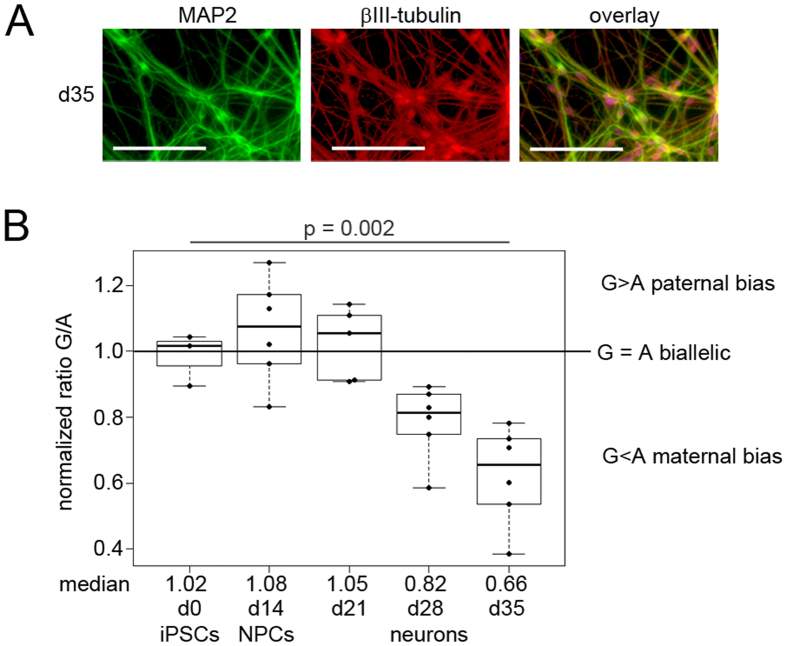
Successful differentiation into neurons within 35 days was confirmed by staining for MAP2 and βIII-TUBULIN, which are both markers of terminally differentiated, postmitotic neurons (Fig. 4A, Supplementary Fig. S8). Using qPCR, we observed a consistent upregulation of the early neural marker NESTIN and the neural progenitor marker PAX6 from day 14 on (Supplementary Fig. S8). Expression of βIII-TUBULIN was detectable at day 14 and increased up to day 35. Total expression of UBE3A increased during differentiation and a slight induction of the brain-specific non-coding RNA SNHG14 was observed at day 14 which subsequently strongly increased during terminal differentiation (Supplementary Fig. S8). To determine at which time point the expression of SNHG14 induces imprinted expression of UBE3A, single-nucleotide primer extension analysis in differentiated AS_∆3 iPSCs was employed. In the analysis, the paternal wildtype allele is represented by a G. The maternal allele carrying the mutation is represented by an A. As expected, undifferentiated iPSCs showed biallelic expression of UBE3A, indicated by a G/A ratio of about 1. Expression of UBE3A remained biallelic up to day 21 of neuronal differentiation. Starting with day 28, a reduction in the G/A ratio was observed, indicating higher expression from the maternal (A) than from the paternal (G) allele. At day 35 of differentiation, the mean G/A ratio, was about 0.6. This can be attributed to the onset of UBE3A silencing by the expression of SNHG14 from the paternal allele. Despite the onset of paternal UBE3A silencing in neurons, the total level of UBE3A protein was comparable between undifferentiated iPSCs and neurons at d35 (Supplementary Fig. S9).
Figure 4. Paternal UBE3A expression is silenced late in neuronal differentiation.
(A) Neurons at d35 of differentiation stained positive for MAP2 and βIII-tubulin. Scale bars indicate 100 μm. (B) SNaPshot analysis of the allelic expression ratio of the paternal (G) and the maternal (A) allele of UBE3A. The G/A ratio decreased at day 28 and day 35 in the time course of neuronal differentiation, indicating that silencing of the paternal allele is a late event.
Discussion
We present here the generation and detailed characterization of iPSCs from a patient with AS carrying a mutation in the UBE3A gene. With these cells it is possible to follow the onset of imprinted UBE3A expression by analyzing expression of both parental alleles in one cell. Deep bisulfite amplicon sequencing was used to determine the epigenetic stability of DMRs at six imprinted gene loci, showing that only the PWS-SRO, the DMR of the PWS/AS locus, was stable in all clones analyzed. Excision of the reprogramming vector did not compromise pluripotency, resulting in the derivation of AS_∆3 iPSCs with only a minimum of ectopic DNA. During differentiation of iPSCs into neurons, we observed strong induction of SNHG14 expression, which accompanied silencing of paternal UBE3A expression. Our results indicate that silencing of paternal UBE3A expression by SNHG14 is a late event during neuronal differentiation.
DMRs are the regulators of imprinted gene expression and in general their epigenetic status is stable in established hESC lines19. However, failures in imprint maintenance during reprogramming to iPSCs or due to prolonged time in culture have been widely discussed and described14,15. It is therefore mandatory for iPSCs to be used as models for imprinting disorders to analyze the status of DNA methylation of at least the DMR addressed in the study. Using deep bisulfite amplicon sequencing, we demonstrated that the PWS-SRO was the only DMR showing stable differential DNA methylation in all pluripotent cells and fibroblasts analyzed (Fig. 2, Supplementary Fig. S6). As expected, the AS iPSC line AGI-0, carrying a large type I deletion on the maternal chromosome 15, showed only unmethylated reads resulting from the retained paternal allele. This exceptional stability of differential DNA methylation at the PWS-SRO during reprogramming is in agreement with earlier reports10,15. In contrast, we observed a loss of differential DNA methylation at the germ line DMRs ICR1, ICR2 and NESPAS in at least one sample (Fig. 2, Supplementary Fig. S6). The IG-DMR has been described to be susceptible to hypermethylation in human pluripotent cells and in agreement with that we report a gain of DNA methylation in 7 of 12 pluripotent samples14,15. However, we observed a loss of DNA methylation at the IG-DMR in the immortalized fibroblasts of the patient, which was not seen in another three fibroblast samples analyzed (Fig. 2, Supplementary Fig. S6). We therefore assume that this loss of methylation is the result of immortalization, as changes in imprinted gene expression and imprint methylation have been observed in immortalized lymphoblastoid cell lines and fibroblasts20,21. The CpG85 was the least stable DMR, showing maintenance of differential methylation only in two fibroblast samples from independent healthy control subjects and a blood sample from the patient (Supplementary Fig. S6).
Overall, our observations indicate a non-random pattern of deviating methylation at most ICRs: the CpG85 and the IG-DMR exhibited only a gain of methylation, whereas the NESPAS and the ICR2 showed only a loss of methylation. The exception was the ICR1, where we observed one loss and one gain of methylation. We did not observe preferred combinations of DMRs in clones having more than one aberrantly methylated DMR. The molecular mechanisms predisposing DMRs to a gain or a loss of methylation in hESCs or iPSCs are not clear yet. Both types of cells are derived from somatic cells, which are expected to carry stable methylation imprints1,2, like we observed for the fibroblast sample of the control person: except for the CpG85 at RB1, all imprinted DMRs displayed the expected pattern of 50% DNA methylation (Fig. 2). This leads to the assumption that aberrant imprint methylation in hESCs and iPSCs is due to a failure of imprint maintenance either during reprogramming or during cultivation of fibroblasts and the derived iPSCs or hESCs14,15,22. Indeed, we observed a gain of methylation at ICR1 in a sample of hESCs H9 at a later passage (Fig. 2, Supplementary Fig. S6). Differences in stability of paternal and maternal imprints have been discussed22 but our observation of consistent gain of methylation at both maternal and paternal DMRs, the CpG85 and the IG-DMR, respectively, is not in line with this hypothesis.
Angelman syndrome is caused by the absence of functional UBE3A protein in the brain. Although microcephaly is a frequent finding in patients with AS and delayed myelinization was repeatedly observed in infant patients, gross structural abnormalities of their brains have not been reported3,23. The normal brain structure of patients with AS suggests that the defect in UBE3A protein expression or function does not restrict neuronal development and differentiation, but might interfere with neuron function. Since no patient-derived consecutive brain samples can be obtained at different stages of development, iPSCs and their subsequent differentiation into neurons are a valuable tool for the understanding of imprinting regulation at the PWS/AS locus during neuronal development. We differentiated AS_Δ3 iPSCs into mature postmitotic neurons and monitored allelic expression of UBE3A using the three-base pair deletion as a SNP (Fig. 4, Supplementary Fig. S8). We observed strong upregulation of SNHG14 and a decrease in the ratio of paternal to maternal UBE3A expression only late during neuronal differentiation. This indicates that UBE3A imprinted expression possibly occurs only in postmitotic neurons, as it was described in mice24,25. It is noteworthy that UBE3A expression per se shows an increase in neurons, despite the silencing of the paternal allele, indicating enhanced expression from the maternal allele in neurons. This is in line with our observation of stable UBE3A protein levels in neurons, which has also been described by Chamberlain et al.10.
The AS_Δ3 iPSCs generated in this study complement the AS iPSCs carrying large deletions on the maternal chromosome that were generated by Chamberlain et al.10. This new iPSC line has the advantage that it does not carry large genomic deletions and will express a UBE3A protein that differs by only one missing amino acid from its normal counterpart. This opens the possibility to assign aberrant protein-protein interactions and functional processes specifically to the defective UBE3A protein. In addition, the mechanisms leading to UBE3A imprinted expression can be studied on both parental alleles in an almost normal cellular environment. Further characterization of neuronal differentiation using the two available AS iPSC lines will help to define the time and the cell type at and in which silencing of paternal UBE3A expression occurs. Such analyses would greatly benefit from their verification in iPSC lines generated from additional patients with AS. The knowledge gained by neuronal differentiation of AS iPSCs is urgently needed to develop efficient potential therapeutic approaches for AS.
Methods
An additional and extended Materials and Methods section is included in the supplement.
Statements
The use of hESCs H1 and H9 is in compliance with the German Stem Cell Law and covered by permission AZ:3.04.02/0099.
This study on reprogramming human dermal fibroblasts into induced pluripotent stem cells and all applied experimental protocols were approved by the local ethics committee of the University Hospital Essen. All applied methods were performed in accordance with the approved guidelines. We confirm that informed consent for skin biopsies were obtained from all subjects, that is the healthy control persons and the legal representative of the patient with Angelman syndrome.
The teratoma formation assays performed in this study were carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the German Government and it was approved by the Committee on the Ethics of Animal Experiments of the responsible authorities (Landesamt für Natur, Umwelt und Verbraucherschutz, LANUV AZ 8.87-50.10.37.09.187 and AZ 84-04.04.2013.A350).
Cell culture, reprogramming and virus excision
hESC and iPSC lines were kept on proliferation-arrested, CF1 feeder cells in KSR medium (DMEM/F12 Glutamax, 20% knockout serum replacement, 4 ng/μl bFGF, 50 U/ml Penicillin/Streptomycin, non-essential amino acids, 40 ng/ml heparin), or feeder-independent in mTeSR1 on Vitronectin-coated tissue-culture dishes. Cells were kept at 37 °C and 5% CO2 in a humidified incubator. Skin biopsies were cultivated in vitro and obtained fibroblasts were reprogrammed using a lentiviral vector co-expressing the coding sequences of POU5F1, KLF4, MYC and SOX2 together with the gene encoding tomato fluorescent protein17. Seven days post transduction, cells expressing the tomato fluorescent protein were isolated by flow cytometric cell sorting. Colonies were picked approximately four weeks post transduction and expanded on irradiated feeder cells in KSR medium. Virus excision was mediated by direct protein transduction of Flp recombinase into iPSCs16. Isolated single clones were screened by PCR for absence of integrated virus genome.
Immunofluorescence
Immunofluorescent antibody staining of adherent iPSC colonies was conducted using the StemLite Pluripotency kit from Cell Signaling Technology, providing antibodies for the nuclear antigens OCT4, NANOG, SOX2 and surface antigens TRA1-60, TRA1-81 and SSEA4. Stainings were done according to manufacturer’s instructions. Antibodies and dilutions are listed in Supplementary Table S7.
Teratoma formation assay
Mice were kept under standard conditions (12 hours light and dark cycle, food and water ad libitum) in the Central Animal Facility of the University Hospital Essen. Cells were resuspended in DMEM/F12 supplemented with 50% growth-factor reduced Matrigel and 1 × 106 cells/ml were injected subcutaneously into both hind limbs of immunodeficient NMRI nu/nu mice (Harlan Laboratories). Mice were monitored daily for teratoma growth. Tumors were explanted when they reached a critical size, when skin lesions appeared, or latest 67 days after injection. For teratoma histopathology, 5 μm paraffin cross-sections were stained with hematoxylin and eosin.
Deep bisulfite sequencing and Epi-Pluri-Score analysis
Deep bisulfite sequencing was performed on the 454 GS junior platform (Roche) as described previously26. Data analysis was done using the Amplikyzer software27 and results are depicted as methylation heatmaps showing single CpG sites in columns and single reads in rows. The percentage of overall methylation is given in Supplementary Table S6. Amplicon-specific tagged primer sequences are given in Supplementary Table S8, as well as an example for a MID-primer pair. For the Epi-Pluri-Score analysis, pyrosequencing of three CpGs of interest (cg 23737055 in ANKRD46, cg22247240 in VRTN, cg13083810 in POU5F1) was performed and analyzed as described by Lenz et al.13. The Epi-Pluri-Score is calculated as the difference of β-value(ANKRD46) minus β-value(VRTN). A positive value is indicative of pluripotent cells, a negative value of differentiated cells. The Epi-Pluri-Score is plotted against the β-value measured for POU5F1. In the plot, reference sets of 265 pluripotent and 1,951 somatic cells are indicated as clouds. Primer sequences are listed in Supplementary Table S8.
Neuronal differentiation
Neuronal differentiation was started from a monolayer culture of iPSCs at 50–60% confluency by changing to induction medium (50% DMEM/F12/Glutamax, 50% Neurobasal, 0.5% N2, 1% B27 without retinoic acid, 20 μM SB431543, 100 nM LDN193189). Induction medium was changed every other day. At day 10, medium was changed to neural expansion medium (DMEM/F12/Glutamax, 1% N2, 2% B27 without retinoic acid, 20 ng/ml FGF2, 20 ng/ml EGF, 200 μM ascorbic acid). At day 13, neural progenitor cells were enriched by positive selection using magnetic anti-PSA-NCAM MicroBeads (Miltenyi Biotec). Cells were passaged every three to four days as single-cell suspension. Terminal differentiation was induced by plating a single cell suspension onto culture vessels coated with poly-D/L-ornithine and laminin at a density of 3 × 104/cm2 in terminal differentiation medium (Neurobasal, 2% B27, 2 mM Glutamax, 1% non-essential amino acids, 200 μM ascorbic acid, 20 ng/ml BDNF, 20 ng/ml GDNF). 50% of medium was changed every other day up to the final time point of differentiation. Cells were harvested at the day of seeding (undifferentiated iPSCs), at day 14 (neural progenitor cells) and days 21, 28 and 35 during the terminal differentiation phase for RNA extraction and immunofluorescence.
SNaPshot analysis
The ABI Prism SNaPshot ddNTP Primer Extension Kit (Life Technologies) was used to determine allelic ratios of mRNA transcripts (after reverse transcription into cDNA) following the manufacturer’s instructions. Genomic DNA of the respective iPSC clones was used as a reference. The reaction products were analyzed on an ABI 3130XL sequencer and electropherograms were analyzed using Gene Mapper 4.0 software (Applied Biosystems, Life Technologies). The “area” value in the Gene Mapper graphical output was used as indicator for the amount of PCR products, amplified from cDNA, extended by a G (product of the paternal allele) or an A (product of the maternal allele). Next, the G/A ratio was calculated and normalized to the G/A ratio of genomic DNA. Analysis was done in three biological and two technical replicates. All data points are included in the bee swarm boxplots, which were calculated using the respective packages provided by The R Project for Statistical Computing. In the boxplots, the median and quartiles are indicated. The p-values were calculated in R using Welch’s unequal variances t-test. Primer sequences are listed in Supplementary Table S8.
Additional Information
How to cite this article: Stanurova, J. et al. Angelman syndrome-derived neurons display late onset of paternal UBE3A silencing. Sci. Rep. 6, 30792; doi: 10.1038/srep30792 (2016).
Supplementary Material
Acknowledgments
We thank Prof. Bernhard Horsthemke and Prof. Peter A. Horn for hosting our groups and their support in this project. We especially thank the patient’s family for their support and willingness to provide the biopsy sample and Prof. Dagmar Wieczorek for taking the biopsy. We acknowledge Sabine Kaya and Melanie Heitmann for excellent technical assistance in conducting the Roche 454 runs and Elke Jürgens for karyotyping. Stefan Radtke performed immortalization of the patient’s fibroblasts. We also thank Prof. Ludger Klein-Hitpass for help in performing the TaqMan human stem cell pluripotency arrays. Prof. Wolfgang Wagner (RWTH Aachen) gave us access to the new technique of Epi-Pluri-Score analysis. Prof. Stormy Chamberlain provided us with the AS iPSC line AGI-0 and the control iPSC line MCH2-10, which facilitated our start with iPSCs. This work was supported by the “IFORES Sonderforschungsprogramm” of the Medical Faculty at the University of Duisburg-Essen.
Footnotes
Author Contributions J.S. and A.N. performed or were involved in preparation and analysis of most of the experiments; M.H. assisted in cell culture maintenance, RNA and DNA preparation and PCR applications; H.d.O.K. performed FACS experiments and assisted in microscopy and western blot analyses; K.S. conducted reprogramming of fibroblasts; R.G. performed Epi-Pluri-Score experiments; D.K. performed teratoma assays; A.B. analyzed paraffin sections of teratomas; H.K. conceived and supervised experiments; L.S. conceived, supervised, performed and analyzed experiments and wrote the manuscript. All authors read and reviewed the manuscript.
References
- Hanna C. W. & Kelsey G. The specification of imprints in mammals. Heredity 113, 176–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G. & Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli A., Buiting K. & Williams C. A. Molecular and Clinical Aspects of Angelman Syndrome. Mol Syndromol 2, 100–112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T., Lalande M. & Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15, 70–73 (1997). [DOI] [PubMed] [Google Scholar]
- Rougeulle C., Glatt H. & Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 17, 14–15 (1997). [DOI] [PubMed] [Google Scholar]
- Horsthemke B. & Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A 146A, 2041–2052 (2008). [DOI] [PubMed] [Google Scholar]
- Meng L. et al. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS genetics 9, e1004039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz L. G. et al. Neurodevelopmental outcome in Angelman syndrome: genotype-phenotype correlations. Res Dev Disabil 35, 1742–1747 (2014). [DOI] [PubMed] [Google Scholar]
- Chamberlain S. J., Li X. J. & Lalande M. Induced pluripotent stem (iPS) cells as in vitro models of human neurogenetic disorders. Neurogenetics 9, 227–235 (2008). [DOI] [PubMed] [Google Scholar]
- Chamberlain S. J. et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA 107, 17668–17673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B. et al. Parental origin and functional relevance of a de novo UBE3A variant. Eur J Med Genet 54, 19–24 (2011). [DOI] [PubMed] [Google Scholar]
- Mayshar Y. et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7, 521–531 (2010). [DOI] [PubMed] [Google Scholar]
- Lenz M. et al. Epigenetic biomarker to support classification into pluripotent and non-pluripotent cells. Sci Rep 5, 8973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson B. et al. Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell 15, 634–642 (2014). [DOI] [PubMed] [Google Scholar]
- Ma H. et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel C. et al. Protein transduction from retroviral Gag precursors. Proc Natl Acad Sci USA 107, 7805–7810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warlich E. et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther 19, 782–789 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Mejia V. et al. Residual expression of the reprogramming factors prevents differentiation of iPSC generated from human fibroblasts and cord blood CD34+ progenitors. PLoS One 7, e35824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn P. J., Ferguson-Smith A. C. & Pedersen R. A. Epigenetic status of human embryonic stem cells. Nature genetics 37, 585–587 (2005). [DOI] [PubMed] [Google Scholar]
- Saferali A. et al. Cell culture-induced aberrant methylation of the imprinted IG DMR in human lymphoblastoid cell lines. Epigenetics 5, 50–60 (2010). [DOI] [PubMed] [Google Scholar]
- Okamura K., Ohno M. & Tsutsui T. Possible involvement of loss of imprinting in immortalization of human fibroblasts. International journal of oncology 38, 903–910 (2011). [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P. J., Ferguson-Smith A. C. & Pedersen R. A. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Human molecular genetics 16 Spec No. 2, R243–R251 (2007). [DOI] [PubMed] [Google Scholar]
- Harting I. et al. Abnormal myelination in Angelman syndrome. Eur J Paediatr Neurol 13, 271–276 (2009). [DOI] [PubMed] [Google Scholar]
- Judson M. C., Sosa-Pagan J. O., Del Cid W. A., Han J. E. & Philpot B. D. Allelic specificity of Ube3a expression in the mouse brain during postnatal development. The Journal of comparative neurology 522, 1874–1896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. & Stryker M. P. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci USA 107, 5611–5616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenpass L. et al. Human PPP1R26P1 functions as cis-repressive element in mouse Rb1. PLoS One 8, e74159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmann S. et al. Amplikyzer: automated methylation analysis of amplicons from bisulfite flowgram sequencing. Peer J PrePrints (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.