Abstract
Traditionally, the ability to edit the mammalian genome was inhibited by the inherent low efficiency of homologous recombination (HR; approximately <1 in a million events) and the inability to deliver DNA efficiently to dividing and non-dividing cells/tissue. Despite these limitations, creative selections designed over 20 years ago, clearly demonstrated the powerful implications of gene knock-in and knockout technology for the genetic engineering of mice (Doetschman et al. Nat 330(6148): 576–578, 1987; Thomas and Capecchi. Cell 51(3): 503–512, 1987). The development and application of recombinant vectors based on adeno-associated virus (rAAV) have helped to overcome both of the initial limitations regarding DNA delivery and the frequency of HR. Considering DNA delivery, rAAV infects non-dividing and dividing cultured cells as well as most tissues in mouse and larger animal models (including humans). At the DNA editing level, rAAV genomes have been reported to increase the frequency of HR several orders of magnitude by serving as the repair substrate (Russell and Hirata. Nat Genet 18(4): 325–330, 1998). However, reports on the ability of rAAV genomes to stimulate HR, compared to plasmid DNA and oligonucleotides, are variable, and many labs have found it necessary to augment the frequency of rAAV-induced HR using site-specific endonucleases (Ellis et al. Gene Ther, 2012; Hirsch et al. Gene Ther 17(9): 1175–1180, 2010; Porteus et al. Mol Cell Biol 23(10): 3558–3565, 2003; Radecke et al. Mol Ther 14(6): 798–808, 2006). In this protocol, we describe a method to perform rAAV-mediated double-strand break (DSB) repair for precise genetic engineering in human cells.
Keywords: Adeno-associated virus, Vector production, Gene editing, Double-strand break repair, Self-complementary AAV
1 Introduction
1.1 Endonuclease Selection for AAV-Mediated DSB Gene Editing
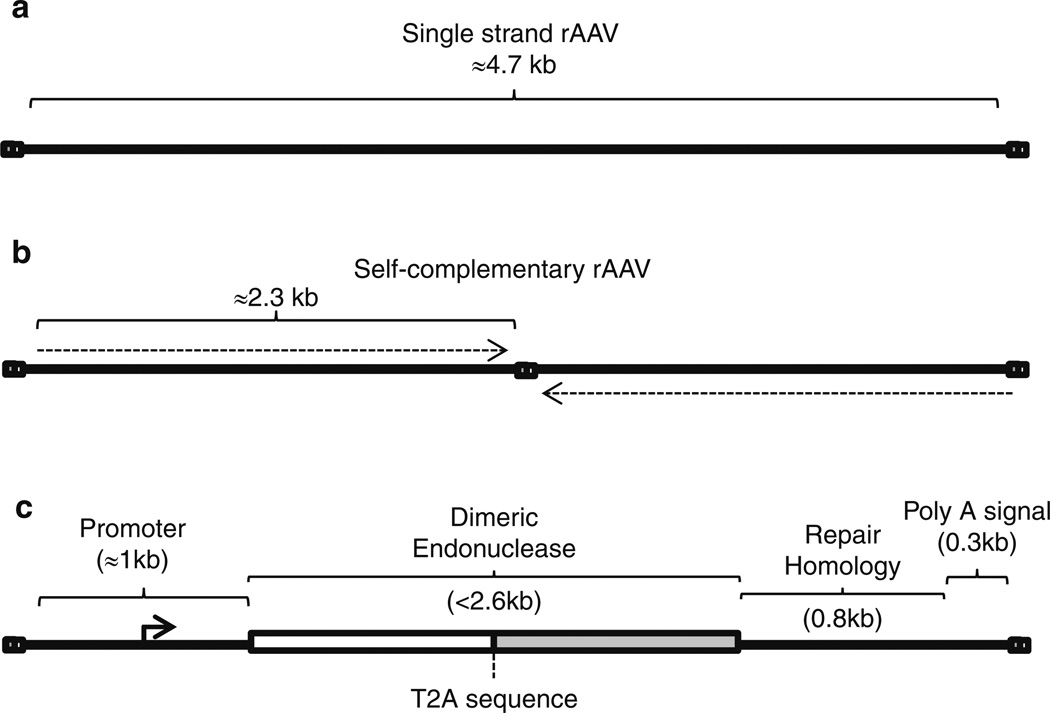
Several endonuclease platforms allow DNA site-specific recognition and strand cleavage events to stimulate DSB repair [5–11]. The design of such enzymes, and their cognate targets, is beyond the scope of this chapter, and alternatively, they are also available for purchase from a variety of manufacturers [12, 13]. However, “designer” endonuclease selection for AAV-mediated gene editing is restricted by the packaging capacity of the viral capsid (≤5 kb) and therefore dependent upon the specific application. For most cases, delivery of the desired endonuclease and the repair substrate to individual cells is necessary, and a single vector format has been described in Fig. 1 [1]. This AAV genome configuration can be employed for single chain or dimeric endonuclease coding sequences, in general, reaching 2.5 kb in size. This context is ideally suited for AAV vectors as it also allows a packaging remainder of approximately 2.2 kb that needs to accommodate a promoter, poly-adenylation signal, a T2A signal (when using a dimeric endonuclease) as well as the repair substrate (Fig. 1c) [1]. Plasticity still exists in the size of these additional sequences as they may be slightly altered without significantly affecting the overall rate of AAV-mediated DSB gene editing (i.e., the length of the repair substrate). Alternatively, the use of multiple AAV vector transduction has demonstrated success in vitro and in vivo eliminating the concern of endonuclease size and or the length of repair sequence homology [2, 3]. In any case, the packaging limitation imposed by the AAV capsid presents a balancing act between all the necessary components, as well as the AAV genome type, single strand or self- complementary, packaged within the capsid shell (discussed below) [14].
Fig. 1.
Recombinant AAV genome formats for gene editing. (a) A cartoon depicting a single-strand DNA AAV genome. For these vectors it is important to keep the genome size >3.2 kb yet <4.8 kb. In all panels, the bicolored squares represent inverted repeats necessary for replication and packaging. (b) A single-strand self- complementary AAV genome is depicted with the genomic restriction of >1 kb yet < than 2.4 kb. The dotted arrows indicate the transgenic polarity in attempts to indicate that the scAAV genome has the capacity to form a duplexed molecule. (c) A single-strand AAV context is depicted in which a dimeric endonuclease expression cassette and a DNA repair substrate are included while conforming to the AAV packaging restraint. A T2A sequence allow the production of a bicistronic message encoding distinct ZFNs
1.2 Viral Capsid Selection for AAV-Mediated DSB Gene Editing
Variations in the composition of the AAV protein capsid result in altered transduction efficiencies based on the target cell type [15–17]. Currently, the most studied serotypes are AAV 1–9 which demonstrate dramatic differences in cell tropism. As AAV vectors transduce both dividing and non-dividing cells, this also holds true for tissue transduction following in vivo administration [17]. Therefore, AAV capsid selection for DSB-mediated gene editing should be tailored to the specific target cell type. Numerous reports are available characterizing AAV capsid transduction efficiencies in vitro, ex vivo, and in vivo to aid in capsid selection [15–17]. In general, the AAV2 capsid works well in transformed cell lines, while AAV1 and 6 transduce stem cells most efficiently [1, 18]. In mouse models, AAV1 and 6 are most efficient following muscle injections, while AAV8 and AAV9 result in widespread transduction following intravenous injections or intraperitoneal administration to young animals [16, 19] (data not shown).
1.3 Viral Genome Considerations for rAAV- Mediated DSB Gene Editing
The production of distinct AAV particles that harbor either the endonuclease or the repair substrate, in contrast to the single vector format depicted in Fig. 1c, is often useful at the mechanistic level and has been employed in vivo (i.e., to determine factors limiting transduction) [20]. In these cases, it is also important to keep in mind that AAV genomes can be packaged in two different forms: (1) single-strand DNA (ssAAV; as discussed above) and (2) self-complementary DNA (scAAV) which is also technically single-strand DNA but can form a large stem-loop structure based on intra-strand base pairing [14, 21] (Fig. 1a, b). The advantage of scAAV is that a duplexed molecule can be generated, thereby eliminating the host-mediated second strand DNA synthesis requirement of ssAAV [14]. This enhancement is an important consideration at the levels of endonuclease production and the ability of the repair substrate to function in HR. In regard to endonuclease production levels, scAAV results in earlier and more robust endonuclease production (>tenfold) compared to ssAAV [2]. Considering the AAV genome as a substrate in the actual repair event, the scAAV context was reported to be modestly enhanced, despite an initial conclusion from an indirect experiment which reported that scAAV (dimer) genomes do not contribute to gene targeting [2, 22]. Although a ssAAV genome dependency for gene targeting represents an attractive hypothesis, as a single strand would not require strand separation and could directly strand invade, it remains at odds with a report that directly compared ssAAV and scAAV as substrates in this process [2]. A unique limitation of scAAV vectors is that due to the region of self-complementary sequence, this vector context imposes an additional size constraint on the packaged genome (≤2.4 kb) [14] (Fig. 1b). As such, the utilization of either ssAAV or scAAV genomes in rAAV-mediated gene editing is often based on the overall vector size of the included elements. For instance, the ssAAV genome format depicted in Fig. 1c relies a T2A sequence to allow the synthesis of two ZFNs from a single promoter, and also encodes a repair substrate, to conform to the packaging capacity of ssAAV (<5 kb). This arrangement ensures that all cells containing a nuclease also contain the desired repair substrate and is particularly suited for situations in which the transduction may not be complete (i.e., in vivo applications). However, when transduction is very efficient, such as cultured cells, one may prefer a multiple vector transduction regimen to vary the amount of nuclease and/or repair substrate. In these instances, distinct scAAV vectors can be used for an approximate tenfold increase in nuclease production and, as a repair substrate, modestly increase gene editing twofold.
In the following protocol, the production and application of an ssAAV genome encoding all the necessary requirements for zinc-finger nuclease (ZFN) induced DSB-mediated gene editing is described.
2 Materials
2.1 AAV-ITR Plasmid Construct Generation and Verification
All of the necessary plasmid constructs for rAAV production are offered by the University of North Carolina Vector Core facility.
The adenovirus helper plasmid pXX680. This construct contains adenovirus genes that assist rAAV production including activation of the AAV p5 promoter [23].
An AAV helper plasmid that encodes AAV replication (Rep) genes and one of several AAV capsid serotype coding sequences (often termed the pXR series) [24].
An AAV-ITR plasmid (pAAV-ITR) having the desired sequence to be packaged between the AAV ITR sequences. The AAV-ITR plasmid may contain two WT ITRs for ssAAV production (pSub201) or one WT and one mutant ITR for scAAV production (pHpa-trs-SK; Fig. 1a, b; see Notes 1 and 2) [14, 25].
A universal ssAAV context to express both ZFNs and a repair substrate has also been described for gene editing purposes (Fig. 1c) [1].
Restriction endonuclease digestion for construct generation relies on standard protocols provided by the enzyme manufacturer.
Agarose gels for verification/separation of DNA species should follow standard running and detection procedures (generally 1 % agarose in tris-acetate–EDTA buffer is sufficient).
DNA agarose gel extraction kits (Qiagen) are routinely used following the company’s provided protocol.
For DNA ligations, T4 ligase is often used according to the manufacturer’s protocol.
For bacterial transformations of the ligation reaction, a number of approaches can be used with varying efficiencies. We typically use electroporation relying on the Bio-Rad micropulser and 0.2 mm electroporation cuvettes. Generally, 1 µl of the ligation reaction is mixed with 30 µl of electrocompetent DH10b cells (Invitrogen) thawed on ice. If the plasmid contains the AAV ITRs, then electrocompetent SURE cells (Agilent) are necessary as they are deficient for homologous recombination which helps to preserve the structured ITRs. Then, the mixed is pulsed at 2.5 kV as recommended by the manufacturer.
Following electroporation, cells recover for 1 h at 37° with shaking in 0.5 ml LB, then generally 20–100 µl of the recovery is plated on nutrient broth plates containing the appropriate antibiotic which is dictated by the resistance gene on the transformed plasmid.
DNA mini- and maxi-preparation kits are commercially available and used to recover plasmids from clonal isolates using standard procedures (described by the manufacturer).
Restriction endonuclease digestion, which varies based on the cloning strategy and sequence, is used to screen plasmid isolates for sequencing reactions (carried out by the UNC genome analysis facility). In addition, XmaI (or SmaI) restriction endonuclease digestion is necessary to verify the integrity the ITRs of pAAV-ITR following the desired cloning strategy (following mini-preps and maxi-preps). All ITRs contain two XmaI (or SmaI) sites that need to be maintained for efficient vector production.
2.2 Transfection of HEK293 Cells for rAAV Production
HEK293 cells (ATCC) are maintained at 37 °C in a 5 % CO2 incubator.
DMEM (Cellgro) supplemented with 10 % FBS (Gibco) and 1× penicillin–streptomycin (Gibco) is used at the HEK293 growth medium.
DMEM without any additives is also required for the transfection reaction.
Transfection reagent: Polyethylenimine (PEI) linear MW 25,000 (Polysciences). 1 mg of PEI is dissolved in 1 ml of 1× PBS adjusted to a pH between 4 and 5 with 12 N HCl.
2.3 Isolation of Transfected Cell Nuclei
Hypotonic buffer (500 ml) 10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 175 mg of spermine; filter-sterilize.
10× restore buffer (200 ml) 62.5 % sucrose (wt/vol) in hypotonic buffer filter-sterilized.
15 ml Kontes homogenizer.
2.4 rAAV Vector Purification by Cesium Chloride Gradient Centrifugation
DNase digestion buffer: 10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 U/ml DNase I (made fresh).
Cesium chloride (CsCl), optical grade (Budenheim Gallard-Schlesinger).
Sonicator (Branson Sonifier 250, VWR Scientific).
Beckman Quick-Seal polyallomer (16 mm × 76 mm) centrifuge tubes and tube sealer.
Ultracentrifuge and fixed angle rotor to accommodate tube size.
2.5 rAAV Genome Detection in CsCl Gradient Fractions by Q-PCR
Q-PCR reagents including buffers, enzymes, primers, and detection methods (i.e., SYBR green or probe-based detection) are described by the manufacturer. AAV genome amplification relies on standard cycling protocols/considerations.
Q-PCR machine.
2.6 rAAV Dialysis and Final Titer Determination
1× phosphate buffered saline (PBS).
Slide-A-Lyzer Dialysis Cassette with 7,000 molecular weight cutoff and 0.5–3 ml capacity (Pierce). We routinely use a 21 gauge needle and a 5 ml syringe to inject and remove the viral sample from the cassette.
DNase I digestion buffer (above).
0.5 M EDTA.
Proteinase K solution: 1 M NaCl, 1 % (wt/vol) N-laurylsarcosine (Sarkosyl).
100 mg/ml proteinase K.
Q-PCR reagents and machine (above).
2.7 rAAV Genome Characterization
DNase I digestion buffer.
0.5 M EDTA.
10 % SDS.
6× loading dye for alkaline gel electrophoresis: 0.4 M NaOH, 5 mM EDTA, 18 % Ficoll (wt/vol), xylene cyanol (enough to add color for easy loading).
Alkaline gel (50 ml): Dissolve 1 g agarose in 50 ml water (1 %) and add 300 µl of 5 N NaOH and 50 µl of 0.5 M EDTA.
Alkaline gel running buffer: 50 mM NaOH, 1 mM EDTA in water.
Alkaline transfer buffer: 0.4 M NaOH.
3MM Whatman chromatography paper.
0.45 mm nylon membrane (i.e., GE Hybond-XL).
UV cross-link device.
6× SSC buffer 0.9 M NaCl, 90 mM sodium citrate.
Low-stringency wash buffer 2× SSC, 0.1 % SDS (wt/vol).
High-stringency wash buffer 0.1× SSC, 0.1 % SDS (wt/vol).
Random primed radio-nucleotide labeling kit (Roche).
X-ray film.
2.8 rAAV Cellular Infection
HEK293 cells maintained as described above.
Tittered rAAV prep (viral genomes/vol).
2 % paraformaldehyde in 1× PBS.
Flow cytometer.
3 Methods
3.1 AAV Plasmid Construct Generation
Recombinant single-strand DNA genomes packaged in the AAV protein capsid are normally derived from a plasmid sequence flanked by AAV inverted terminal repeats (pAAV-ITR vector; Fig. 2). Such plasmids are commercially available from a variety of sources as well as from the lab that initially created it [24]. Standard cloning procedures (not detailed herein) are used to place a sequence of interest ≤≈5 kb (the AAV capsid packaging capacity) into pAAV-ITR. As the AAV-ITRs undergo “processing” by bacterial DNA repair proteins during replication, a recombination deficient E. coli strain is preferred for plasmid production/purification (i.e., SURE cells from Agilent). Following pAAV-ITR purification, it is then necessary to confirm the integrity of the AAV-ITRs in vitro using restriction endonuclease digestion at sites found within the AAV ITRs. Generally, SmaI or XmaI enzyme digestion is performed according to standard procedures following all plasmid preparations.
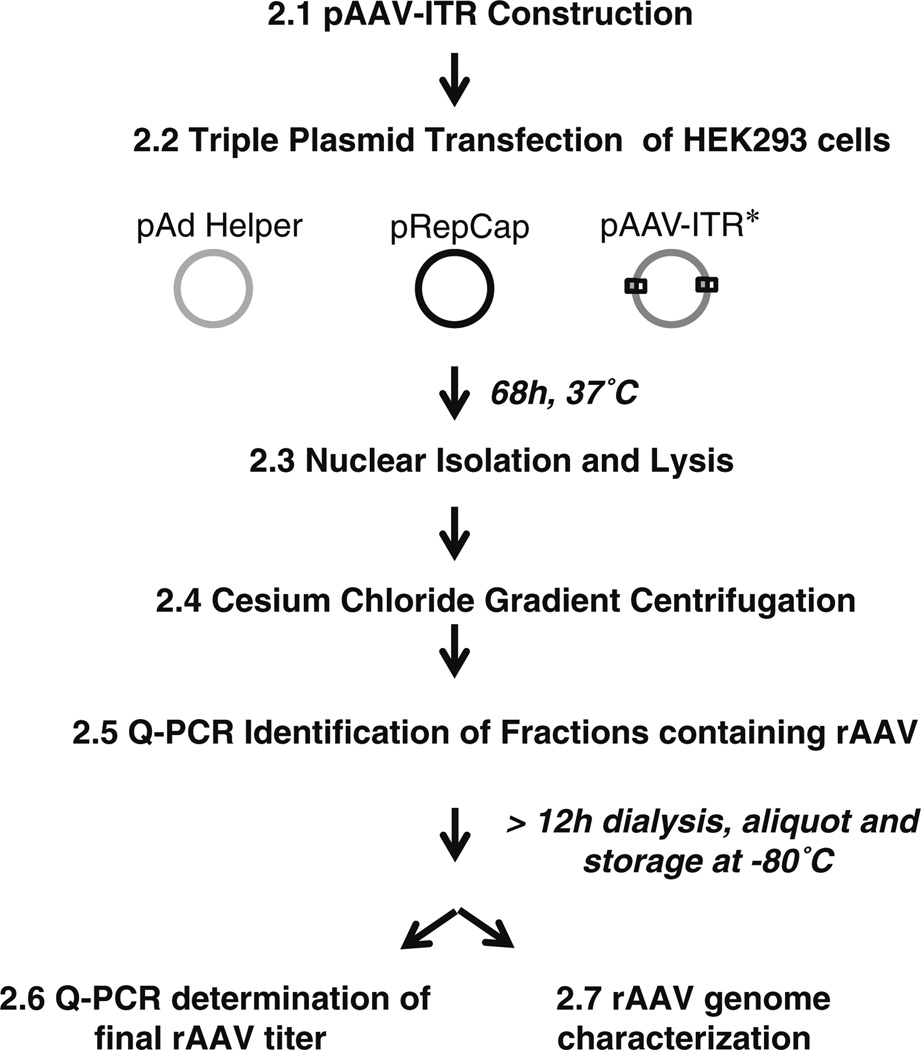
Fig. 2.
AAV vector production flowchart. The number preceding each bolded statement indicates the location within the chapter describing that part of the procedure
For the specific purpose of AAV-mediated gene correction, a vector format has been generated that allows endonuclease and DNA repair substrate delivery in a single AAV particle (Fig. 1c) [1]. For instance, a universal expression cassette that allows AAV-mediated delivery of two ZFN coding sequences and up to a 1 kb repair substrate has been generated that conforms to the AAV capsid packaging capacity of 5 kb [1]. To accommodate this limitation, a ribosomal skipping sequence (T2A) was placed between the two ZFN coding sequences on a bicistronic message [1]. This genetic configuration allowed a 1 kb noncoding repair substrate, positioned upstream of the poly-adenylation sequence, to be included in the packaged genome (Fig. 1c). In a specific published example, which employs the UbqC constitutive promoter, the DNA sequence between the AAV-ITRs on the plasmid vector is 4.7 kb, thus presenting no packaging concerns [1]. This contextual arrangement can be used for ZFNs and other homing endonuclease as long as the ≤5 kb capsid packaging capacity is not exceeded. In the case of larger nucleases, such as TAL effector nucleases, it is currently necessary to use multiple pAAV-ITR vectors, for multiple vector production, purification, and transduction strategies.
3.2 Transfection of Adherent HEK293 Cells for rAAV Production
Adherent HEK293 cells are maintained in a 15 cm culture dish in DMEM supplemented with 10 % FBS and 1× pen–strep.
24 h prior to plasmid transfection, two confluent plates are split 1–3 and grown for 24 h to obtain an approximate confluency of 70 % at the time of transfection.
rAAV production relies on a triple transfection of the plasmids listed in Subheading 2.1 (Fig. 2). The amount needed per 15 cm plate is 12 µg of pXX680 (Adenovirus helper), 10 µg of pXR (AAV Replication and Capsid genes), and 8 µg of pAAV- ITR (containing the sequence to be packaged).
For each plate, the plasmids are then diluted in 500 ml of DMEM medium containing no supplements and 110 µl of PEI transfection reagent is added to the solution.
Following the addition of PEI, the mixture is immediately vortexed for 10 s and allowed to complex at room temperature for 5 min.
Then, the solution is added directly to the plate medium in a manner that allows uniform dispersal.
Following the transfection, the cells are cultured under standard conditions for ≈68 h prior to harvest for rAAV particle purification (see Note 3).
3.3 Isolation of Cell Nuclei Post-transfection
After ≈68 h post-transfection, harvest and pool cells from the 15 cm plates. This can be done simply using the force generated from a 10 ml pipette or with a cell scraper if preferred. Then, pellet the cells by centrifugation at 200 × g for 5 min at 4 °C. Discard the growth medium and wash the cell pellet in cold PBS, then pellet the cells again using the same conditions.
Resuspend the cell pellet in a volume of hypotonic buffer approximately five times the packed cell volume. Incubated on ice for 10 min.
Add 0.11 volumes of 10× restore buffer and mix gently.
Homogenize the cell solution in a 15 ml Kontes homogenizer via 12 strokes.
Pellet the cell nuclei by centrifugation at 500 × g for 10 min, routinely performed in a 50 ml conical tube.
3.4 AAV Vector Purification by Cesium Purification by Cesium Gradient Centrifugation
Many different purification methods exist for rAAV production [24]. Herein a simple, serotype-independent, cost-effective strategy is described which is based on a CsCl density gradient established by centrifugation of the nuclear extract derived from the transfected cells. This method relies on the known buoyant density of AAV containing a 4.7 kb genome (1.41 g/cm3). AAV vector recovery using CsCl centrifugation is not the cleanest purification scheme as indicated by silver stains of viral preps also exhibiting contaminating cellular proteins. However, it has the advantage of separating AAV capsids containing genomes from “empty” particles (no packaged genomes), which are assembled during particle production. Empty particles compete for cell surface receptor binding, thus decreasing the overall transduction efficiency of the viral prep, and may generate unwanted immunological concerns in vivo. An extensive review focusing specifically of established rAAV production/characterization methods is available for alternatives not presented herein [24].
Resuspend nuclear pellets in cold ddH2 0 to a final volume of 11.6 ml and disrupt the nuclei for 50 pulses using a Branson Sonifier (duty cycle of 50, output of 5) on ice.
Next, add 100 µl of the DNase solution (10 mg/ml) to the disrupted nuclei solution and incubate at 37 °C for 1 h to remove any “non-packaged” DNA.
Following the DNase digestion reaction, add 6.5 g of CsCl to the sample and vortex. The density of the CsCl gradient should be approximately 1.41 g/cm3 based on the refractive index (fr ¼ 1.3722).
After CsCl addition, sonicate the sample again using the exact conditions mentioned above.
The nuclear lysate is then loaded into Beckman Quick-Seal polyallomer (16 mm × 76 mm) centrifuge tubes, balanced and the tube is then sealed using the Beckman Quick Sealer.
Centrifuge samples at 402,000 × g for a minimum of 10 h (longer is ok) to establish the gradient.
Following centrifugation, 1 ml sample fractions are then collected in individual tubes. This can be done simply by placing a needle in the base of the tube and another in the top to initiate the sample flow. If cleaner vector preparations are desired, the identified peak fractions (see below) can be subjected to an additional CsCl gradient (density 1.41 g/cm3) in the manner described above.
3.5 AAV Genome Detection in CsCl Gradient Fractions by Q-PCR
To determine AAV vector production in the isolated fractions, procedures such as Southern blotting and Q-PCR can be used. The quickest, and perhaps most simple, method to detect which fractions contain viral genomes (indicative of packaged particles) is Q-PCR. It is important to note that at this stage precise particle tittering is not the objective, simply determination of peak gradient fractions containing rAAV. Therefore, we employ a simple protocol that has been shown valid for peak rAAV fraction detection and relies on heat denaturation of the AAV capsid during the PCR reaction. Precise viral tittering is performed after dialysis of the viral preparation and storage at −80 °C.
Dilute 1 µl of each CsCl gradient fraction into 1 ml of water. Vortex.
Use 2 µl of the viral dilution directly as template in a Q-PCR reaction.
Employ Q-PCR using standard protocols for primer construction and one of a variety of different methods to detect DNA amplification. As positive controls a dilution series (50 pg to 5 fg) of the pAAV-ITR plasmid used in production is commonly included in the reaction.
3.6 AAV Dialysis and Final Titer Determination
Following rAAV peak fraction determination, samples containing >1e8 viral genomes can be pooled (dependent upon the desired injection volume and necessary titer) and then dialyzed against 1× PBS. To do this, Slide-A-Lyzer dialysis cassettes are used at 4 °C while stirring the solution. Generally, we use a 1:1,000 ratio of viral sample to PBS for a minimum of 12 h. Following dialysis, the rAAV samples are aliquoted and maintained at −80 °C prior to final titer determination and use. For final titer determination, we describe a method based on Q-PCR, however Southern dot blot remains a viable option and a detailed protocol has been described elsewhere [24].
After thawing at room temperature, combine 10 µL of rAAV sample with 90 µL of the DNaseI solution and vortex (perform in triplicate). Incubate the reaction for 1 h at 37 °C to degrade any unpackaged viral genomes.
Then, add 6 µL of 0.5 M EDTA and vortex to stop the DNase digestion reaction.
To degrade the AAV protein capsid, 120 µL of the Proteinase K solution is added, vortexed, and incubated at 55 °C for ≥2 h (sample can be left at 55 °C overnight).
Following the Proteinase K digest, samples are then heated for 10 min at 95 °C to inactivate Proteinase K.
Dilute the sample at least 100× in water or 10 mM Tris (pH 8) and use this solution as the DNA template in a standard Q-PCR reaction. A pAAV-ITR standard is run in parallel using a duplicate dilution series ranging from 50 pg to 0.5 fg. rAAV final titers are generally expressed as the number of viral genomes (calculated from the plasmid standard) in a given volume. Common rAAV titers range from 1e8 to 5e9 viral genomes per microliter (see Note 4).
3.7 rAAV Genome Characterization
rAAV genomes are primarily homogenous if the size is >3.5 kb but <5 kb and are prepared in the single-strand ITR plasmid context. scAAV preparations are often contaminated by the packaging of single-strand monomer genomes, in addition to the scAAV dimer genomes, which can decrease the overall vector potency and confuse result interpretations, depending on the application. Therefore, it is wise to characterize the packaged genomic species under denaturing conditions via Southern blotting.
10 µl of dialyzed rAAV sample is incubated with 10 µl of the DNase solution (as described above). The reaction is then terminated using 5 µl of 0.5 M EDTA.
2 µl of 10%SDS solution is then added followed by addition of 6× NaOH loading buffer, vortexed for 10 s then incubated at 20 min at RT.
The samples are then spun at max speed in a tabletop centrifuge for 10 min and loaded into a 1 % denaturing gel that has been soaked in alkaline gel running buffer for >10 min at RT (see Note 5).
Alkaline gel electrophoresis is then performed at 15 V for approximately 16 h to separate the AAV genomic species (see Note 6).
Next, the gel is incubated in the alkaline transfer buffer under mild agitation at RT for 10 min.
The gel is then placed atop an absorbent stack consisting of the following (from bottom to top): 1 in. of paper towels, three pieces of Whatman paper, and a nitrocellulose membrane (paper and membrane are sized slightly bigger than the gel). A bridge is then constructed of Whatman paper with a width equivalent to the width of the gel and a length long enough to accommodate the gel length and positioning of the other end in a reservoir containing the alkaline transfer buffer. The reservoir is positioned approximately 3 in. higher than the gel stack to allow downward transfer flow of the DNA from the gel onto the membrane. This transfer event is performed at RT for a minimum of 10 h.
Following DNA transfer, the genomes are cross-linked to the nitrocellulose membrane using a UV photo-cross-linker “auto-crosslink” function.
The membranes are then placed in a glass hybridization tube containing Church buffer at a volume that is sufficient enough to cover the length of the tube when placed on its side (dependent upon the membrane and tube size).
Then, a 32P-radioactive labeled probe is used for genome hybridization. Commonly, a labeled random probe library is generated using a Roche random labeling kit as detailed by the manufacturer (see Note 7). The radiolabeling reaction should be cleaned using, for example, a PCR purification kit to remove non-incorporated radio-nucleotides (as described by the kit manufacturer). The labeled probe preparation is then added to 100 µl of salmon sperm DNA and the mixture is boiled for 5 min to denature the DNA and then maintained on ice.
The denatured, radiolabeled probe is added to the hybridization tube (containing the Church buffer and membrane) and incubated for >12 h at 65 °C.
Next, the membrane is washed twice in the high-salt wash buffer for 10 min at 65 °C, then using the low-salt wash buffer for >20 min at the same temperature.
The washed membrane is then placed in plastic wrap, exposed to film and developed. Genome size and purity is determined based on the loaded DNA standards.
3.8 rAAV Cellular Infection
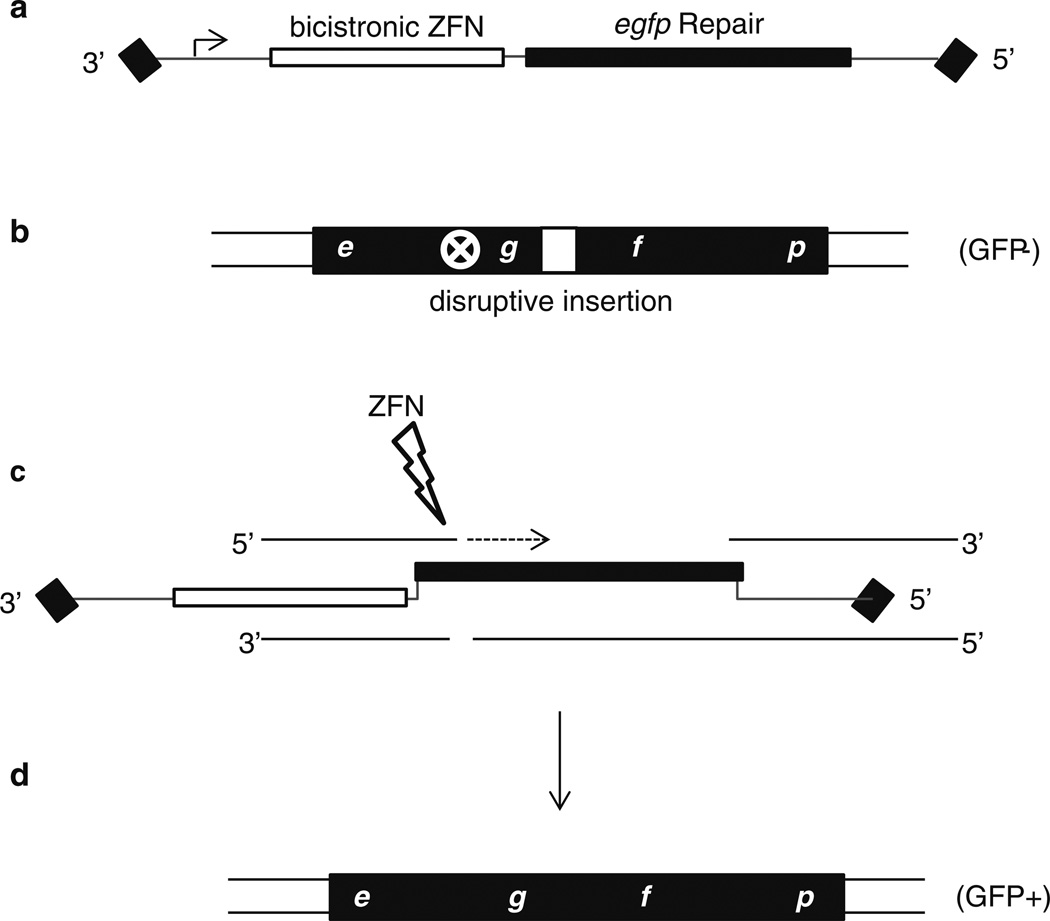
The protocol described herein focuses on rAAV-mediated DSB gene editing in HEK293 cells containing a defective egfp reporter system (Fig. 3) [3, 4]. A described egfp -specific ZFN in the single vector context is also outlined to induce a DSB-mediated HR event to “correct” the defective fluorescent reporter (Figs. 1c and 3) [1]. For HEK293 cell, the AAV serotype 2 capsid is the most efficient for transduction and is used herein. However, considering other cell types, as well as for in vivo tissue tropism, the literature should be consulted regarding the most efficient capsid serotype for transduction [15–17]. In general, we have found that AAV2 works best for most cell lines, while AAV1, 3 and 6 should be considered for different stem cell types [26, 27]. For in vivo applications, AAV8 and 9 are efficient for systemic mouse transduction (especially if administer via the intraperitoneal route prior to day 12 post-birth), while AAV1 and 6 transduce efficiently following intramuscular administration [19].
HEK293 cell cultures are maintained under the conditions described above and the desired viral genome number per cell is added directly to the growth medium (see Note 8). Transduction is often performed in a 24 well context and a viral particle to cell ratio of 10,000 is sufficient for most applications. Depending on the experimental design, multiple rAAV preps can be added to the cells at the same time (i.e., for delivery of the endonuclease and repair substrate using separate rAAV particles).
Three days post-infection, the cells are harvested for determination of gene editing (see Note 9).
In this example, a single rAAV particle (encoding both the nuclease and repair substrate) was used to generate a specific ZFN-induced DSB in the egfp coding sequence to induce HR with the rAAV delivered repair substrate [1] (Figs. 1c, 3). In this extensively reported gene correction system, the desired gene editing removes a disruptive insertion in a defective egfp reporter creating an easily tallied GFP+ phenotype [3, 5]. Therefore, harvested cells are pelleted at 1,000 × g for 3 min and resuspended in 2 % paraformaldehyde in PBS.
Flow cytometry for detection of GFP+ cells is then performed using the fixed cells and standard flow cytometry detection protocols for GFP (see Notes 10 and 11).
Fig. 3.
AAV vector mediated gene editing. In the depicted cartoon, the transduced AAV genome (a) encodes a promoter, an eGFP-specific ZFN (target site is indicated by a white circled X), and an eGFP repair sequence to correct the defective chromosomal fluorescent reporter (b). In this system, the endonuclease generates a specific double-strand break inducing a host DNA damage response (c). Multiple repair mechanisms can result in correction of the defective reporter using the repair sequence provided by the AAV vector. In this cartoon, we demonstrate an example based on gene conversion in which the AAV repair homology serves as a synthesis template for the free 3′ end generated by the ZFN (c). This new synthesis integrates the desired genetic modification (in this case egfp coding sequence devoid of the insertion represented by the white box) creating, in this example, a functional fluorescent reporter (d)
3.9 Detection of the AAV-Mediated DSB Gene Editing Event
As stated above, the gene editing event described herein is based on a previously reported egfp gene correction system [3, 5]. In this system, a single vector AAV context was used to deliver the egfp repair substrate and the genes encoding egfp directed ZFNs [1]. This correction event is easily determined by a cellular phenotypic change to GFP+ which is sensitively detected by flow cytometry in a high-throughput manner [3, 5]. Verification of the edited locus following extended cell growth to dilute rAAV episomal genomes allows for clonal isolation of GFP+ cells, and DNA sequencing across the edited region has been described elsewhere [2].
Acknowledgments
This work was supported by the Northwest Genome Engineering Consortium (NIH) and by R01AI072176 awarded to Hirsch and Samulski.
Footnotes
When designing an AAV genome it is imperative to remember that for ssAAV the packaged genome should be <5 kb yet >2.7 kb in size. Therefore, depending on the desired particles, it may be necessary to adjust the sequence between the ITRs (of pAAV-ITR) to adhere to these restrictions. For scAAV, the DNA sequence to be packaged (the sequence between the ITRs of pHpa-trs-sk) needs to be <2.2 kb yet >1 kb.
The restriction endonucleases necessary for repair substrate and endonuclease gene cloning will vary based on the cloning strategy.
During rAAV production, approximately 50 h post-transfection the treated cells should appear smaller, spherical, with partial loss of adherence due to the pXX680 plasmid. Such morphological changes suggest only an efficient transfection, not necessarily rAAV production. However, if such changes are not observed the researcher should be suspicious that something is wrong with the transfection conditions.
Precise viral tittering of scAAV preparations by Q-PCR is often difficult, compared to ssAAV, due to the nature of the duplexed DNA molecule. Currently several labs are working out protocol conditions to overcome the complications for scAAV tittering by Q-PCR. Therefore, scAAV tittering is validated by additional methods including Southern dot blotting and by the Southern alkaline gel electrophoresis described elsewhere and above [24]. In these cases, a plasmid dilution series is included (ranging from 50 to 0.5 ng) to generate a standard curve from which the number of rAAV genomes can be calculated [24].
It is important to also include DNA size standards while performing Southern alkaline gel electrophoresis which can be obtained by enzymatic digestion and gel isolation of the pAAV-ITR plasmid fragments. Approximately 10 ng of the standards are incubated in the NaOH sample buffer, vortexed and incubated for 30 min at RT prior to alkaline gel loading.
When preparing the alkaline gel, the gel solution is heated in the microwave until the agarose dissolves. Then, the solution is allowed to cool (not solidify) prior to NaOH and EDTA addition, as described above.
As a radioactive probe template, a gel isolated DNA fragment, corresponding to the entire transgenic cassette (the region to be packaged on the pAAV-ITR plasmid), is preferred. This fragment can also be used as a size standard during Southern blotting.
As rAAV infects both dividing and non-dividing cells, cell confluency will not dramatically affect transduction at similar viral genome to cell ratios. However, as HR recombination is restricted to or more prevalent in mammalian dividing cells, then the use of arrested or differentiated cell populations for gene editing will result in very few or no events (Hirsch unpublished).
Longer culture times, following rAAV infection, generally result in the increased frequency of the desired host chromosome modification.
As stated above, the gene editing event described herein is based on a previously reported egfp gene correction system [3, 5]. In this system, a single vector AAV context was used to deliver the egfp repair substrate and the genes encoding egfp directed ZFNs [1] (Fig. 3). This correction event is easily determined by a cellular phenotypic change to GFP+ which is sensitively detected by flow cytometry in high-throughput manner. Verification of the edited locus has also been described following extended cell growth to dilute rAAV episomal genomes, clonal isolation of GFP+ cells, and DNA sequencing across the edited region [2].
References
- 1.Ellis BL, et al. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther. 2012;20(1):35–42. doi: 10.1038/gt.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirsch ML, et al. Self-complementary AAV mediates gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Ther. 2010;17(9):1175–1180. doi: 10.1038/gt.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porteus MH, et al. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23(10):3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radecke F, et al. Targeted chromosomal gene modification in human cells by single-stranded oligodeoxynucleotides in the presence of a DNA double-strand break. Mol Ther. 2006;14(6):798–808. doi: 10.1016/j.ymthe.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 6.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29(18):3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlowski J, Boniecki M, Bujnicki JM. I-Ssp6803I: the first homing endonuclease from the PD-(D/E)XK superfamily exhibits an unusual mode of DNA recognition. Bioinformatics. 2007;23(5):527–530. doi: 10.1093/bioinformatics/btm007. [DOI] [PubMed] [Google Scholar]
- 9.Heath PJ, et al. The structure of I-Crel, a group I intron-encoded homing endonuclease. Nat Struct Biol. 1997;4(6):468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61(1):176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell Mol Life Sci. 2007;64(22):2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8(16):1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell AM, et al. AAV’s anatomy: roadmap for optimizing vectors for translational success. Curr Gene Ther. 2010;10(5):319–340. doi: 10.2174/156652310793180706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asokan A, Schaffer DV, Samulski RJ. The AAV vector toolkit: poised at the clinical crossroads. Mol Ther. 2012;20(4):699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zincarelli C, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16(6):1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 18.Ellis BL, et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23(3):321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 20.Li H, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samulski RJ, et al. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A. 1982;79(6):2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata RK, Russell DW. Design and packaging of adeno-associated virus gene targeting vectors. J Virol. 2000;74(10):4612–4620. doi: 10.1128/jvi.74.10.4612-4620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72(3):2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1(3):1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 25.Samulski RJ, et al. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33(1):135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch ML, et al. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS One. 2011;6(11):e27520. doi: 10.1371/journal.pone.0027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira JR, et al. Three-dimensional multipotent progenitor cell aggregates for expansion, osteogenic differentiation and ‘in vivo’ tracing with AAV vector serotype 6. Gene Ther. 2013;20(2):158–168. doi: 10.1038/gt.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011;117(12):3311–3319. doi: 10.1182/blood-2010-08-302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, et al. AAV vectors containing rDNA homology display increased chromosomal integration and transgene persistence. Mol Ther. 2012;20(10):1902–1911. doi: 10.1038/mt.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donsante A, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317(5837):477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]