Abstract
Background
KRAS mutation testing is mandatory in the management of metastatic colorectal cancer prior to treatment with anti-EGFR antibodies as patients whose tumors express mutant KRAS do not benefit from these agents. Although the U.S. Food and Drug Administration has recently approved two in-vitro diagnostics kits for determination of KRAS status, there is generally no consensus on the preferred method and new tests are continuously being developed. Most of these techniques focus on the hotspot mutations at codons 12 and 13 of the KRAS gene.
Methods
We describe a two-step approach to KRAS codon 12/13 mutation testing involving high resolution melting analysis (HRM) followed by pyrosequencing using the Therascreen KRAS Pyro kit (Qiagen) of only those samples that are not clearly identified as KRAS wildtype or mutant by HRM. First, we determined KRAS status in a panel of 61 colorectal cancer samples using both methods to compare technical performance and concordance of results. Subsequently, we evaluated practicability and costs of our concept in an independent set of 120 colorectal cancer samples in a routine diagnostic setting.
Results
HRM and pyrosequencing appeared to be equally sensitive, allowing for clear detection of mutant alleles at a mutant allele frequency ≥12.5 %. Pyrosequencing yielded more exploitable results due to lower input requirements and a lower rate of analysis failures. KRAS codon 12/13 status was called concordantly for 98.2 % (56/57) of all samples that could be successfully analysed by both methods and 100 % (19/19) of samples that were identified mutant by HRM. Reviewing the actual effort and expenses for KRAS mutation testing in our laboratory revealed, that the selective use of pyrosequencing for only those samples that could not be analysed by HRM increased the fraction of valid results from 87.5 % for HRM alone to 99.2 % (119/120) while allowing for a net reduction of operational costs of >75 % compared to pyrosequencing alone.
Conclusions
Combination of HRM and pyrosequencing in a two-step diagnostic procedure constitutes a reliable and economic analysis platform for KRAS mutation testing in colorectal cancer in a clinical setting.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2589-2) contains supplementary material, which is available to authorized users.
Keywords: KRAS mutation, Colorectal cancer, High resolution melting analysis, Pyrosequencing
Background
The anti EGFR-antibodies cetuximab and panitumumab represent well-established treatments for metastatic colorectal cancer (CRC), the third most prevalent cancer entity and fourth most common cause of cancer-related death around the world [1, 2]. Several studies have shown KRAS status to predict outcome under these anti-EGFR targeting agents, with beneficial effects being seen only in patients whose tumors express wildtype (WT) KRAS [3–8]. Thus, testing for KRAS mutations, which are found in approximately 40 % of colorectal cancers, has become routine in the management of metastatic CRC (mCRC) prior to cetuximab or panitumumab treatment [9, 10] and is even required by the responsible regulatory agencies. Notably, current standards regarding oncogenic Ras mutation analysis in mCRC issued by the U.S Food and Drug Administration (FDA) require determination of KRAS status by an FDA-approved test, while the European Medical Agency (EMA) just states application of validated methods by an experienced laboratory [11–15]. Currently available FDA-approved companion diagnostic devices for cetuximab (Erbitux) and panitumumab (Vectibix) comprise the Cobas KRAS Mutation Test (Roche) and Therascreen KRAS RGQ PCR Kit (Qiagen) [16]. Besides these and other commercially available kits, the spectrum of methods for KRAS mutation testing encompasses multiple PCR-derived and sequencing-based techniques. Of note, most of the previously established assays for KRAS mutation detection focus on the hotspot mutations involving codons 12 and 13, which account for >95 % of Ras mutations in CRC [10]. The advantages and limitations of selected methods have been repeatedly evaluated comparatively [17–22], however, beyond the FDA-guideline, there is no consensus on the preferred approach to investigate KRAS status in routine molecular pathological diagnostics [23]. Given the high incidence of CRC resulting in high demand for KRAS mutation testing, an ideal diagnostic assay for this purpose not only needs to be sufficiently sensitive and specific, but, for socio-economic reasons, also should be time- and cost-effective. Therefore, we developed a two-step procedure for KRAS mutation testing including high resolution melting analysis (HRM) followed by pyrosequencing of only those samples that are not clearly identified as KRAS WT or mutant by HRM. HRM is a one-tube qPCR-based technique for DNA-variant detection. The method utilizes alterations in the melting behavior of double-stranded DNA fragments that are conferred by nucleotide exchanges. Melting of qPCR amplicons is monitored in real time using a suitable qPCR instrument capable of time-dense data aquisition and a saturating DNA-intercalating fluorescent dye that does not redistribute during the melting step [24]. Pyrosequencing is a sequencing-by-synthesis approach that involves sequential addition of dNTPs and recording incorporation of a nucleotide based on a light signal that is generated by sulfurylase-catalyzed conversion of the released pyrophosphate to ATP and a subsequent luciferase reaction [25]. Here, we applied a previously described HRM-assay [20] and the Therascreen KRAS Pyro kit (Qiagen) for detection of KRAS codon 12/13 mutations. First we comparatively analysed KRAS status in a panel of 61 colon cancer samples to determine sensitivity, specificity, technical performance and concordance of results of the two methods. Subsequently, we evaluated our two-step approach in the routine setting of our molecular diagnostics laboratory. In summary, we present a reliable, time- and cost-effective operational concept for KRAS mutation testing prior to anti-EGFR antibody treatment in mCRC.
Methods
Tumor samples, control cell lines and DNA isolation
The colorectal cancer samples reported on in this study were obtained from patients with metastatic colorectal cancer (UICC IV) at the University Hospital Marburg, Germany and analysed in a routine diagnostic setting. Tissue samples were fixed, paraffin-embedded, sectioned, hematoxylin-eosin stained and deparaffinated using standard procedures. Tissue sections were reviewed by an experienced pathologist (RM) to establish the diagnosis and to mark regions for microdissections. Microdissection of tumor cells was performed from deparaffinated sections using a scalpel. DNA was isolated from microdissected samples using the QiaAmp DNA Mini kit (Qiagen) as recommended by the manufacturer. KRAS mutant cell lines PL45 (pancreatic adenocarcinoma) and RPMI 8226 (multiple myeloma) were obtained from ATCC and cultured according to standard cell culture methods. Positive control DNA for HRM analyses was isolated from these cell lines using the QiaAmp DNA Mini kit. WT control DNA was extracted from peripheral blood of healthy donors from whom informed consent had been obtained (WT control) with the QiaAmp DNA Mini kit. DNA concentrations were measured using a Nanodrop 1000 spectrophotometer (Peqlab).
High resolution melting analysis
For HRM analysis, a 92 bp amplicon spanning exons 2 and 3 of the KRAS gene was amplified from 60 ng (or less) of sample DNA using the primers KRAS-92_F 5′-ttataaggcctgctgaaaatgactgaa-3′ and KRAS-92_R 5′-tgaattagctgtatcgtcaaggcact-3′ [20], the DNA-intercalating dye SYTO 9 (Thermo) in a final concentration of 5 μM and Platinum Taq polymerase (Thermo). Amplification and melting analysis was performed on a Rotor Gene 6000 instrument (Corbett Life Sciences) under the following temperature conditions: one cycle 95 °C/2 min, 40 cycles 95 °C/15 sec – 67.5 °C/15 sec - 72 °C/15 sec, one cycle 95 °C/1 sec, pre-melt conditioning at 72 °C/90 sec, HRM-ramp from 72 °C to 95 °C rising at 0.2 °C per step/wait 2 sec each step. Controls in each HRM run included a no-template-control, a WT control (gDNA from healthy donor) and two mutation controls (gDNA from the cell lines RPMI 8226, KRAS codon 12 GGT → GCT/heterozygous, corresponding to G12A and PL45, KRAS codon 12 GGT → GAT/heterozygous, corresponding to G12D). All HRM assays were performed in quadruplicate.
Pyrosequencing
Pyrosequencing of the KRAS codon 12/13 region was performed using the Therascreen KRAS Pyro Kit (Qiagen) as recommended by the manufacturer. 2 ng of DNA were used per analysis. PCR amplification of the target region was performed on a T-100 thermocycler (Biorad). For the pyrosequencing reaction on the PyroMark Q24 platform (Qiagen), amplicons were immobilized to the wells of a PyroMark Q24 plate using streptavidin high performance beads (GE Healthcare). Pyrosequencing results were analysed using the PyroMark Q24 software version 2.0 with the Therascreen KRAS Pyro-plugin report, which already incorporated the thresholds for mutation calls (detection limit for the mutation (LOD) + 3 %).
Statistical analysis
HRM and pyrosequencing results were compared by contingency table analysis test using GraphPad Prism 5 software (GraphPad). Technical performance (1st run success vs. 1st run failure) was evaluated by two-sided Fisher’s exact test at a significance level of 5 %. The agreement between HRM and pyrosequencing results was quantified by kappa using the appropriate Graphpad Prism online calculator (http://graphpad.com/quickcalcs/kappa2).
Results
Sensitivity of HRM and pyrosequencing
In order to test whether pyrosequencing allows for KRAS mutation detection with at least equal sensitivity compared to HRM, we analysed serial dilutions of DNA from a KRAS mutant cell line (PL45, codon 12 GGT → GAT heterozygous mutation) in WT DNA by both HRM and pyrosequencing. For HRM, we found, that the presence of KRAS mutant DNA in the sample was clearly reflected by a shifted or skewed melting curve for a fraction of PL45-DNA exceeding 25 %, which corresponded to a mutant allele frequency of 12.5 % (Fig. 1). Similarly, pyrosequencing definitely yielded a mutation if the sample contained ≥25 % PL45-DNA. On the other hand, samples with 5–10 % cell line DNA were indicated to exhibit a potential low level mutation as the mutant allele frequency was quantified below the threshold for accurate WT/mutant discrimination for the G12D mutation (LOD + 3 %; LOD = 2,2 %) for both the 5 and 10 % samples (Fig. 2). Thus, HRM and pyrosequencing appeared to be equally sensitive methods for the detection of KRAS codon 12/13 mutations.
Fig. 1.
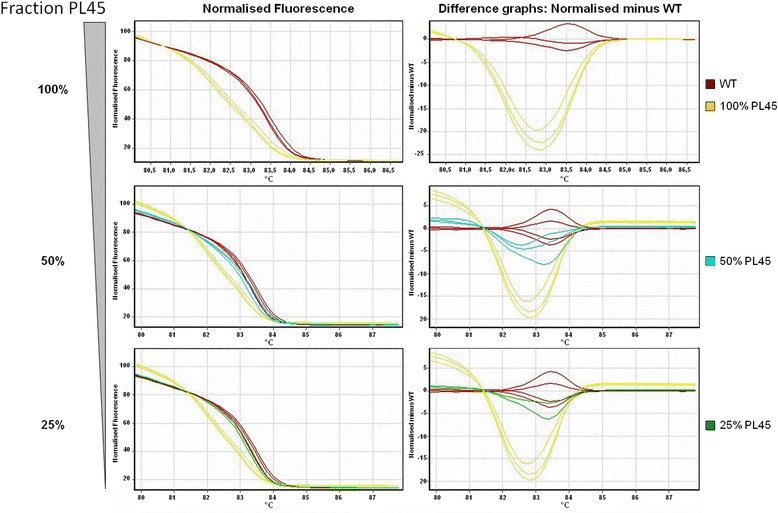
Sensitivity of HRM. Dilutions of genomic DNA from the KRAS mutant cell line PL45 (codon 12 GGT → GAT, heterozygous mutation) in WT genomic DNA was analysed by HRM. Normalised fluorescence and difference graphs are indicated. Clear discrimination of WT and mutant amplicons is possible using either graph if the fraction of PL45-DNA in the sample exceeds 25 %, corresponding to a mutant allele frequency of 12.5 %
Fig. 2.
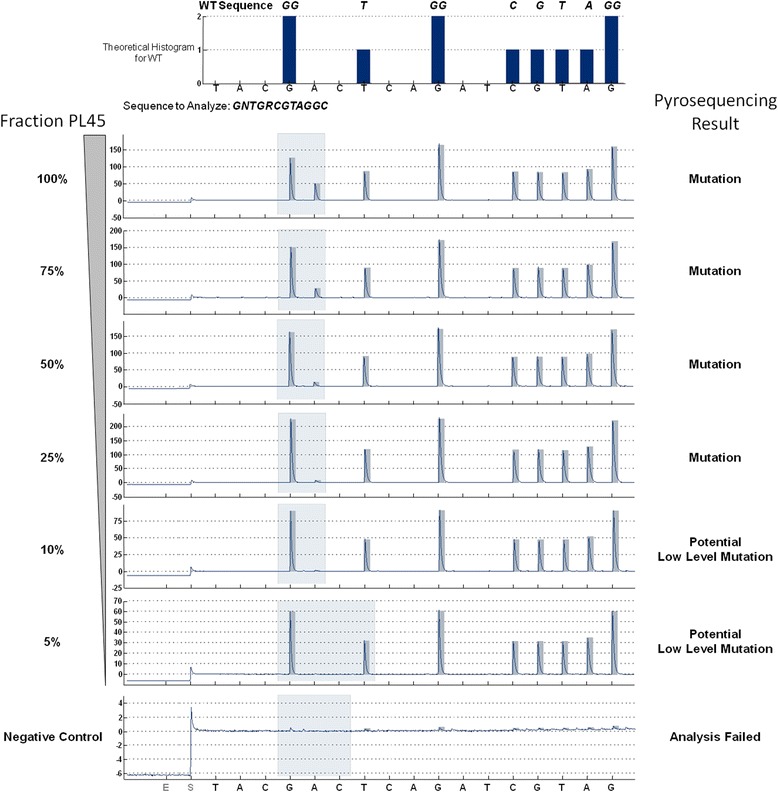
Sensitivity of pyrosequencing. Dilutions of genomic DNA from the KRAS mutant cell line PL45 (codon 12 GGT → GAT, heterozygous mutation) in WT genomic DNA was analysed by pyrosequencing on the Pyromark Q24 platform. The WT sequence and the sequence to analyse including wobble bases at the potentially mutant positions in the region of interest (codons 12 and 13) are indicated in the top panel. Lower panels: KRAS codon 12/13 pyrograms for different dilutions of PL45-DNA in WT DNA. Clear mutation calls are obtained for samples containing ≥25 % of PL45-DNA, corresponding to 12.5 % mutant alleles. For lower concentrations, yielding signals below the LOD of the mutation (2.2 %) + 3 %, presence of a potential low level mutation is suggested, which requires technical and/or biological replication of the analysis
Technical reliability of HRM and pyrosequencing
To further assess the suitability of pyrosequencing to serve as a backup-assay allowing for accurate diagnosis of KRAS mutation status in case of failed HRM analysis, we investigated KRAS status of 61 colorectal cancer samples by both HRM and pyrosequencing and compared the two methods with regard to their technical performance and concordance of results. In a first run of HRM analysis, 11/61 samples (18.0 %) could not be analysed due to PCR-failures or ambiguous melting curves (Table 1). Repetition of the assay for seven samples, which most likely had been compromised technically, allowed for assigning KRAS mutation status in all cases. The remaining four samples were directly subjected to pyrosequencing without a second round of HRM analysis. Indeed, KRAS status could each be determined, although one sample yielded a potential low level mutation. Of the 57 samples that could be definitely classified as KRAS WT or mutant by HRM, 26 WT samples (45.6 % of all samples/68.4 % of WT samples) yielded skewed HRM curves, which, however, did not prevent establishment of a diagnosis (Tables 1 and 2). Moreover, we noted that low DNA content of the samples below the detection limit of the Nanodrop spectrophotometer not necessarily prevented successful HRM analysis. In contrast to HRM, the pyrosequencing assay had to be repeated for only one sample (Table 1). Thus, the failure rate of a first analysis run as a consequence of technical and/or sample-issues was significantly higher for HRM analysis than for pyrosequencing (p = 0.0042). Together, pyrosequencing is technically more reliable than HRM due to lower input requirements and a lower incidence of invalid results.
Table 1.
KRAS codon 12/13 status by HRM and pyrosequencing in 61 CRC samples
| HRM | Pyrosequencing | |||||||
|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | |||||||
| Sample | cDNA [ng/μl] | Run 1 | Run 2 | Result | % mut. Alleles | Result | % mut. Alleles | Final result |
| 1 | 11 | failed | WT a | WT | WT | |||
| 2 | 31 | WT a | WT | WT | ||||
| 3 | 10 | WT a | G12C | 13.4 | mut | |||
| 4 | 93 | WT a | WT | WT | ||||
| 5 | 26 | WT a | WT | WT | ||||
| 6 | 27 | WT a | WT | WT | ||||
| 7 | 10 | WT a | WT | WT | ||||
| 8 | 43 | mut | G13D | 73.9 | mut | |||
| 9 | 85 | failed | WT | WT | WT | |||
| 10 | N/A | mut | G13D | 44.3 | mut | |||
| 11 | N/A | failed | G12V | 2.6 | WT b | |||
| 12 | N/A | WT | WT | WT | ||||
| 13 | N/A | mut | G12V | 41.9 | mut | |||
| 14 | 126 | mut | failed | G12D | 65.7 | mut | ||
| 15 | 133 | mut | G12D | 74.1 | mut | |||
| 16 | 97 | mut | G13D | 52.8 | mut | |||
| 17 | 47 | failed | WT | WT | WT | |||
| 18 | 14 | failed | WT | WT | WT | |||
| 19 | 44 | WT a | WT | WT | ||||
| 20 | 20 | failed | WT a | WT | WT | |||
| 21 | N/A | failed | WT | WT | ||||
| 22 | 138 | mut | G12V | 12.9 | mut | |||
| 23 | 325 | mut | G12D | 56.1 | mut | |||
| 24 | 140 | WT a | WT | WT | ||||
| 25 | 7 | mut | G12C | 61.2 | mut | |||
| 26 | 4 | WT a | WT | WT | ||||
| 27 | 27 | WT a | WT | WT | ||||
| 28 | 13 | WT a | WT | WT | ||||
| 29 | 207 | mut | G12D | 54.5 | mut | |||
| 30 | 10 | failed | WT | G12V | 1.5 | WT b | ||
| 31 | 54 | WT a | WT | WT | ||||
| 32 | 120 | mut | G12V | 29.2 | mut | |||
| 33 | 33 | failed | mut | G12C | 33.5 | mut | ||
| 34 | N/A | mut | G12C | 71.4 | mut | |||
| 35 | N/A | WT | WT | WT | ||||
| 36 | N/A | failed | WT | WT | ||||
| 37 | 67 | WT a | WT | WT | ||||
| 38 | 137 | mut | G12C | 76.7 | mut | |||
| 39 | 63 | mut | G12A | 56 | mut | |||
| 40 | 13 | WT a | WT | WT | ||||
| 41 | 113 | WT a | WT | WT | ||||
| 42 | 82 | mut | G12C | 75.4 | mut | |||
| 43 | 39 | WT | WT | WT | ||||
| 44 | 7 | WT a | G12S | 2 | WT b | |||
| 45 | 23 | WT a | WT | WT | ||||
| 46 | 24 | faileda | G13D | 3.5 | WT b | |||
| 47 | 9 | WT a | WT | WT | ||||
| 48 | 34 | WT a | G12S | 2 | WT b | |||
| 49 | 25 | WT a | WT | WT | ||||
| 50 | 17 | WT | WT | WT | ||||
| 51 | 5 | WT a | WT | WT | ||||
| 52 | 29 | WT a | WT | WT | ||||
| 53 | 31 | mut | G12D | 71.3 | mut | |||
| 54 | 68 | WT | WT | WT | ||||
| 55 | 93 | mut | G12D | 74.2 | mut | |||
| 56 | 221 | WT | WT | WT | ||||
| 57 | 83 | WT | WT | WT | ||||
| 58 | N/A | WT a | G12V | 1.2 | WT b | |||
| 59 | 35 | mut | G12D | 83.3 | mut | |||
| 60 | N/A | WT | WT | WT | ||||
| 61 | 37 | WT a | WT | WT | ||||
aSkewed HRM curve
bLOD/threshold for potential low level mutation (cf. Therascreen KRAS Pyro Kit handbook version 1, July 2011): G12D 2.2 %/5.2 %, G12V 1.0 %/4.0 %, G12C 2.1 %/5.1 %, G12S 1.9 %/4.9 %, G13D 1.9 %/4.9 %
Table 2.
Comparison of HRM and pyrosequencing results in 61 CRC samples
| Run 1 | Run 2 | Summary | ||||
|---|---|---|---|---|---|---|
| Summary of Results | ||||||
| HRM | n | % | n | % | n | % |
| Number of samples | 61 | 100.0 | 7 | 100.0 (11.5) | 61 | 100.0 |
| Analysis passed | 50 | 82.0 | 7 | 100.0 | 57 | 93.4 |
| WT (total) | 32 | 64.0 | 6 | 85.7 | 38 | 66.7 |
| WT (skewed HRM curve) | 24 | 75.0 | 2 | 33.3 | 26 | 68.4 |
| Mutant (total) | 18 | 36.0 | 1 | 14.3 | 19 | 33.3 |
| Mutant (skewed HRM curve) | 0 | 0 | 0 | |||
| Analysis failed | 11 | 18.0 | 0 | |||
| Pyrosequencing | n | % | n | % | n | % |
| Number of samples | 61 | 100.0 | 1 | 100.0 (1.6) | 61 | 100.0 |
| Analysis passed | 60 | 98.4 | 1 | 100.0 | 61 | 100.0 |
| WT (total) | 41 | 68.3 | 0 | 41 | 67.2 | |
| WT (call: WT) | 35 | 58.3 | 35 | |||
| WT (call: potential low level mutation) | 6 | 10.0 | 0 | 6 | ||
| Mutant | 19 | 31.7 | 1 | 100.0 | 20 | 32.8 |
| Analysis failed | 1 | 1.6 | 0 | |||
| Concordance of Results | HRM | Pyrosequencing | ||||
| n | % | n | % | |||
| Number of samples | 57 | 100 | 57 | 100 | ||
| WT (total) | 38 | 66.7 | 37 | 64.9 | ||
| WT (call: WT) | 33 | 57.9 | ||||
| WT (call: potential low level mutation) | 4 | 7.0 | ||||
| Mutant | 19 | 33.3 | 20 | 35.1 | ||
| Concordant | 56 | 98.2 | ||||
| Discordant | 1 | 1.8 | ||||
| Correctly classified WT | 37 | 97.4 | ||||
| Incorrectly classified WT | 1 | 2.6 | ||||
| Correctly classified mutant | 19 | 100 | ||||
| Incorrectly classified mutant | 0 | 0 | ||||
Concordance of HRM and pyrosequencing results
In order to evaluate the diagnostic validity of HRM analysis as a basic test for KRAS mutation detection, we compared the results from this assay to pyrosequencing in the 57 samples that could be successfully analysed by both methods. KRAS status was assigned concordantly for 56 samples (98.2 %; kappa = 0.961), while the result for one sample with a low mutant allele frequency of 13.4 % (#3, Table 1) was inconsistent between HRM and pyrosequencing (Tables 1 and 2). Importantly, pyrosequencing indicated the presence of potential low level mutations (mutant allele frequency < 4.0–5.2 %, cf. Table 1) in four samples that were called WT by HRM. Given that this output is generated due to low signal strength for the potential mutation near the technical detection limit of the pyrosequencing method we finally classified these samples as WT. Conversely, all 19 samples that were clearly identified as mutant by HRM were classified identically by pyrosequencing. Therefore, defining pyrosequencing as the reference method, the specificity of HRM for detection of mutant KRAS alleles was 100 %. On the other hand, the specificity for the detection of WT alleles was slightly reduced (97.4 %) due to erroneous interpretation of the HRM curve for the one sample mentioned (#3, Table 1) with a mutant allele frequency only slightly above the sensitivity threshold of the method. When we applied a different HRM assay for the detection of NRAS codon 61 mutations on an independent set of 19 CRC samples, we found a 100 % concordance of results with reports from a reference laboratory (Additional file 1: Table S1). Of note, sensitivity of the NRAS HRM assay was comparable to the KRAS assay and allowed for reliable identification of mutations at a mutant sample fraction of 20 % (Additional file 2: Figure S1). Taken together, these findings indicate that HRM represents a very reliable basic method for KRAS mutation testing.
Two-step KRAS mutation testing in routine diagnostics
To evaluate the actual effectiveness of our two-step analysis platform (Fig. 3) in a routine diagnostic setting, we reviewed effort and outcome of KRAS codon 12/13 mutation testing in 120 independent colorectal cancer samples that were examined consecutively in our laboratory according to this concept (Table 3). We found, that KRAS status could be determined for 87.5 % of samples by HRM and for 99.2 % of samples in total, when pyrosequencing was applied to samples that could not be successfully analysed by HRM (Table 4). However, for both HRM and pyrosequencing, the failure rate was slightly higher than anticipated based on the observations from our initial 61 sample set (Tables 2 and 4). Also of note, the number of samples that were subjected to pyrosequencing in routine diagnostics exceeded the previously estimated need of this analysis (19/120 = 15.5 % vs. 4/61 = 6.6 %) because 15 samples were directly analysed by pyrosequencing after the first failed HRM run in order to utilize otherwise wasted capacities. Yet, in summary, these data strongly support the rationale of our two-step approach to KRAS codon 12/13 mutation analysis, confirming the accuracy of our diagnostic platform.
Fig. 3.
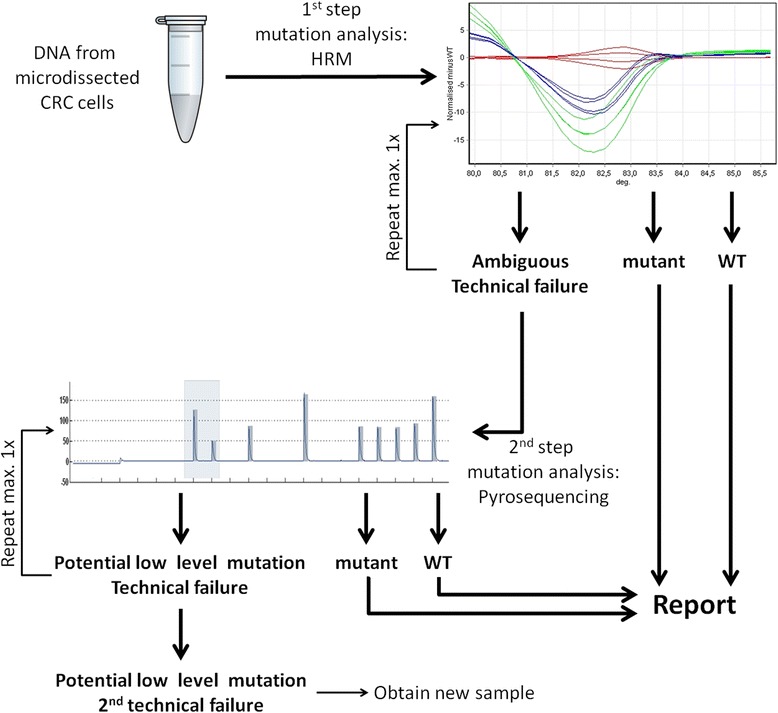
Outline of the two-step procedure for KRAS codon 12/13 mutation analysis. Genomic DNA from microdissected colorectal cancer cells from FFPE samples is subjected to HRM of a PCR amplicon spanning the mutation-bearing region of interest. For samples that are clearly identified as KRAS mutant or, respectively, WT, the HRM result is incorporated in the final diagnostic report. Samples for which HRM analysis fails technically or which yield ambiguous HRM curves are further evaluated by a second round of HRM and, if results are still invalid, to pyrosequencing. Note that samples for which a WT result is obtained by the diagnostic procedure outlined here require further examination for additional KRAS and NRAS mutations
Table 3.
Detailed results of KRAS codon 12/13 mutation testing in 120 CRC samples
| HRM | Pyrosequencing | |||||||
|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | |||||||
| Sample | cDNA [ng/μl] | Run 1 | Run 2 | Result | % mut. Alleles | Result | % mut. Alleles | Final result |
| 62 | 541 | WT a | WT | WT | ||||
| 63 | 29 | WT a | WT | WT | ||||
| 64 | 378 | WT | WT | |||||
| 65 | 342 | WT a | WT | WT | ||||
| 66 | 176 | failed | WT a | G12V | 3.2 | G12V | 5.2 | WT |
| 67 | 139 | mut | mut | |||||
| 68 | 520 | WT | WT | |||||
| 69 | 42 | WT | WT | |||||
| 70 | 202 | WT | WT | |||||
| 71 | 157 | mut | mut | |||||
| 72 | 259 | mut | mut | |||||
| 73 | 145 | WT | WT | |||||
| 74 | 21 | failed | failed | WT | WT | |||
| 75 | 22 | failed | WT | WT | ||||
| 76 | 66 | mut | mut | |||||
| 77 | 55 | mut | mut | |||||
| 78 | 199 | mut | mut | |||||
| 79 | 197 | WT | WT | |||||
| 80 | 171 | WT | WT | |||||
| 81 | 55 | failed | G12V | 41.1 | mut | |||
| 82 | 231 | failed | WT | WT | ||||
| 83 | 250 | failed | WT | WT | ||||
| 84 | 57 | failed | WT a | WT | ||||
| 85 | 21 | mut | mut | |||||
| 86 | 248 | WT | WT | |||||
| 87 | 258 | WT | WT | |||||
| 88 | 83 | failed | failed | failed | N/A | |||
| 89 | 38 | failed | WT | WT | ||||
| 90 | 279 | WT | WT | |||||
| 91 | 122 | failed | WT | WT | ||||
| 92 | 129 | failed | Low Mut. | 5.2 | Low Mut. | 8.2 | mut | |
| 93 | 58 | WT | WT | |||||
| 94 | 129 | WT | WT | |||||
| 95 | 254 | WT | WT | |||||
| 96 | 373 | WT | WT | |||||
| 97 | 158 | WT | WT | |||||
| 98 | 96 | mut | mut | |||||
| 99 | 22 | WT | WT | |||||
| 100 | 30 | mut | mut | |||||
| 101 | 26 | muta | muta | G13D | 7.3 | mut | ||
| 102 | 49 | WT | WT | |||||
| 103 | 47 | WT | WT | |||||
| 104 | 43 | WT | WT | |||||
| 105 | 54 | WT | WT | |||||
| 106 | 363 | failed | mut | mut | ||||
| 107 | 521 | failed | WT | WT | ||||
| 108 | 199 | mut | mut | |||||
| 109 | 260 | WT | WT | |||||
| 110 | 67 | WT | WT | |||||
| 111 | 103 | WT | WT | |||||
| 112 | 24 | mut | mut | |||||
| 113 | 150 | WT | WT | |||||
| 114 | 5 | WT | WT | |||||
| 115 | 25 | WT | WT | |||||
| 116 | 33 | WT | WT | |||||
| 117 | 26 | mut | mut | |||||
| 118 | 72 | WT | WT | |||||
| 119 | 16 | failed | G12V | 12.8 | mut | |||
| 120 | 33 | failed | WT | WT | ||||
| 121 | 48 | WT | WT | |||||
| 122 | 74 | WT | WT | |||||
| 123 | 474 | mut | mut | |||||
| 124 | 431 | mut | mut | |||||
| 125 | 66 | mut | mut | |||||
| 126 | 143 | mut | mut | |||||
| 127 | 49 | failed | WT | WT | ||||
| 128 | 143 | WT | WT | |||||
| 129 | 122 | mut | mut | |||||
| 130 | 139 | WT | WT | |||||
| 131 | 21 | failed | WT | WT | ||||
| 132 | 39 | failed | WT | WT | ||||
| 133 | 128 | failed | G12D | 26.7 | mut | |||
| 134 | 60 | mut | mut | |||||
| 135 | 330 | failed | mut | mut | ||||
| 136 | 165 | mut | mut | |||||
| 137 | 213 | mut | mut | |||||
| 138 | 31 | failed | mut | mut | ||||
| 139 | 156 | mut | mut | |||||
| 140 | 59 | WT | WT | |||||
| 141 | 68 | WT | WT | |||||
| 142 | 164 | WT | WT | |||||
| 143 | 238 | mut | mut | |||||
| 144 | 12 | mut | mut | |||||
| 145 | 33 | failed | WT | WT | ||||
| 146 | 81 | WT | WT | |||||
| 147 | 11 | WT | WT | |||||
| 148 | 13 | mut | mut | |||||
| 149 | 71 | WT | WT | |||||
| 150 | 11 | mut | mut | |||||
| 151 | 40 | failed | WT | WT | ||||
| 152 | 50 | WT | WT | |||||
| 153 | 128 | mut | mut | |||||
| 154 | 146 | WT | WT | |||||
| 155 | 69 | WT | WT | |||||
| 156 | 182 | WT | WT | |||||
| 157 | 32 | failed | WT | WT | ||||
| 158 | 142 | WT | WT | |||||
| 159 | 53 | WT | WT | |||||
| 160 | 91 | failed | WT | WT | ||||
| 161 | 334 | WT | WT | |||||
| 162 | 86 | failed | WT | WT | ||||
| 163 | 61 | WT | WT | |||||
| 164 | 64 | WT | WT | |||||
| 165 | 141 | WT | WT | |||||
| 166 | 271 | WT | WT | |||||
| 167 | 40 | WT | WT | |||||
| 168 | 34 | failed | WT | WT | ||||
| 169 | 29 | failed | failed | WT | WT | |||
| 170 | 354 | WT | WT | |||||
| 171 | 66 | WT | WT | |||||
| 172 | 43 | failed | WT | WT | ||||
| 173 | 114 | WT | WT | |||||
| 174 | 268 | WT | WT | |||||
| 175 | 107 | WT | WT | |||||
| 176 | 170 | mut | mut | |||||
| 177 | 65 | failed | mut | mut | ||||
| 178 | 31 | failed | mut | mut | ||||
| 179 | 61 | WT | WT | |||||
| 180 | 659 | mut | mut | |||||
| 181 | 34 | WT | WT | |||||
aSkewed HRM curve
Table 4.
Operational analysis of two-step KRAS mutation testing of 120 CRC samples
| Run 1 | Run 2 | Summary | ||||
|---|---|---|---|---|---|---|
| HRM | n | % | n | % | n | % |
| Number of samples | 120 | 100.0 | 20 | 100.0 (16.7) | 120 | 100.0 |
| Analysis passed | 89 | 74.2 | 18 | 90.0 | 105 | 87.5 |
| WT (total) | 60 | 67.4 | 12 | 66.7 | 71 | 67.6 |
| WT (skewed HRM curve) | 3 | 5.0 | 2 | 16.7 | 4 | 5.6 |
| Mutant (total) | 29 | 32.6 | 6 | 33.3 | 34 | 32.4 |
| Mutant (skewed HRM curve) | 1 | 3.4 | 1 | 16.7 | 1 | 2.9 |
| Analysis failed | 31 | 25.8 | 2 | 10.0 | ||
| Pyrosequencing | n | % | n | % | n | % |
| Number of samples | 19 | 100.0 (15.8) | 3 | 100.0 (2.5) | 19 | 100.0 |
| Analysis passed | 18 | 94.7 | 2 | 66.7 | 18 | 94.7 |
| WT | 12 | 66.7 | 0 | 0.0 | 13 | 72.2 |
| Potential low level mutation | 2 | 11.1 | 1 | 50.0 | 0 | 0.0 |
| Mutant | 4 | 22.2 | 1 | 50.0 | 5 | 27.8 |
| Analysis failed | 1 | 5.3 | 1 | 33.3 | 1 | |
| Combined HRM + Pyrosequencing | n | % | ||||
| Number of samples | 120 | 100.0 | ||||
| Number of HRM runs | 140 | 116.7 | ||||
| Number of pyrosequencing runs | 22 | 18.3 | ||||
| Analysis passed | 119 | 99.2 | ||||
| WT | 81 | 68.1 | ||||
| Mutant | 38 | 31.9 | ||||
| Analysis failed | 1 | 0.8 | ||||
Assay costs
In order to estimate the economic benefits of our two-step approach to KRAS mutation testing, we compared analysis costs in our routine setting to a pyrosequencing-only platform. Based on current list prices for reagents and consumables, we estimated the assay costs for HRM analysis and pyrosequencing at approximately € 7.50 and € 100, respectively (Table 5). The costs for the essential technical devices for both methods have not been converted to per-sample costs because operation expenses are highly dependent on sample throughput, including not only the KRAS mutation assay but also other applications. Moreover, investments for technical equipment are in the same range for pyrosequencing and HRM. Considering the failure rates of each assay in our set of 120 routine samples (23.6 % for HRM and 9.1 % for pyrosequencing), leading to repeated testing of some samples, our two-step approach allows for net reduction of operational costs of approximately 75 % compared to pyrosequencing alone. Moreover, according to our experience, hands-on time for processing the maximum number of samples for one HRM-run (14 + 4 controls) is only half of the time required to prepare and perform a pyrosequencing run at full capacity (22 + 2 controls) (Table 5). Therefore, our concept to maintain two sequential assays for KRAS codon 12/13 mutation testing represents cost- and time-effective approach for routine diagnostics.
Table 5.
Per-sample costs and hands-on time for HRM and pyrosequencing analyses
| HRM | Pyrosequencing | |
|---|---|---|
| Costs (Euro) | ||
| Reagents | 3.40 | 90.00 |
| Consumables | 2.40 | 3.50 |
| Controls | 1.70 | 8.50 |
| Total | 7.50 | 102.00 |
| Time (minutes) | ||
| 14 samples + 4 controls | 60 | |
| 22 samples + 2 controls | 120 | |
Costs for the controls were estimated based on the maximum number of samples that can be processed in one HRM- or pyrosequencing run, respectively. Costs for HRM controls also include DNA isolation from KRAS WT and mutant cell lines. Hands-on time is indicated for full capacity runs
Discussion
Here we present a two-step approach to KRAS codon 12/13 mutation testing for mCRC employing HRM analysis and pyrosequencing using the Therascreeen KRAS Pyro Kit. Comparing the performance of the two methods in a panel of 61 samples, we observed a 98.2 % concordance of results with a 100 % specificity of HRM for the detection of mutant alleles. Thus, HRM analysis needs methodically independent confirmation of results by pyrosequencing only in exceptional cases and therefore can serve as a filter assay to exclude clearly WT or mutant samples from the more expensive and more laborious pyrosequencing analysis. Specifically, based on our observations reported here, this approach can reduce throughput of the pyrosequencing assay by >85 %, resulting in a >75 % cost reduction compared to using pyrosequencing only. We emphasize, that our comparison of the two methods in the first place aimed on diagnostic accuracy for sequential application in order to establish a reliable and economized platform for KRAS mutation testing. Of note, we reached this goal in spite we were able to detect mutant KRAS alleles only at a frequency >12.5 % instead of 5 % as reported in the literature [20, 26].
With respect to technical performance, although we successfully applied HRM to very low input samples, we state a clear advantage for the pyrosequencing assay due to lower input requirements and an apparently relatively high susceptibility of HRM to artifacts. More precisely, previous authors have pointed out, that especially WT HRM curves show a certain degree of variation resulting from poor quality of FFPE-derived template DNA, differing salt- or inhibitor concentrations or unspecific amplification [20, 27], that may complicate correct determination of KRAS status. Consistent with this notion, 6 of the 7 samples in our 61-sample validation set that were subjected to a second round of HRM analysis due to poor interpretability of first round results were eventually called WT by this method. Conversely, we did not obtain false positive results by HRM, i.e., none of our samples that had been identified as mutant by HRM was found to be WT according to pyrosequencing. Yet, we state that the mutation frequency of KRAS codon 12/13 observed in our study was slightly lower than reported in the literature [9, 10], which may be explained by our homogenous patient population from a single center (Marburg, Germany).
Concerning diagnostic value of results from our sequential KRAS mutation analysis procedure, it is important to point out that pyrosequencing results include information on the site, type and frequency of the nucleotide exchange, while HRM only allows for categorical discrimination of WT and mutant tumors. According to current standards, such a dual output is actually sufficient to establish the indication for anti-EGFR treatment, although certain authors have suggested that not all KRAS mutations are equal regarding outcome in mCRC patients treated with cetuximab [3]. Consequently, as clinical routine testing at present in principle does not require sequence-based analysis, the more differentiated output of the pyrosequencing assay does not warrant the higher costs for this analysis. Therefore, the two-step procedure for KRAS mutation testing presented here represents a reasonable diagnostic approach not only from a technical-practical and economical, but also from a clinical perspective. More specifically, using our diagnostic platform focused on KRAS codon 12/13 mutation testing, even small diagnostic laboratories can provide accurate and clinically meaningful results within a short processing time for the most relevant genetic alteration that determines a treatment decision for mCRC patients. Consequently, only a small fraction of patient samples has to be sent to an external reference laboratory for further molecular studies in accordance with the current EMA standards and recommendations by the American Society of Clinical Oncology, which state that Ras mutation testing prior to initiation of treatment with cetuximab and panitumumab has to include analysis of both KRAS and NRAS exons 2, 3 and 4 (codons 12, 13, 59, 61, 117 and 146). Also of note, besides KRAS and NRAS mutations, alterations in several other genes such as BRAF and PIK3CA have been proposed to predict outcome with EGFR antibody treatment [28–30]. Thus, identification of patients eligible for cetuximab or panitumumab treatment in fact requires either a broad panel of single mutation tests or a multiplex approach. Optimized methods for DNA melting analysis of short PCR amplicons have been suggested to allow for comprehensive hot spot mutation testing in a clinical setting as they require only standard qPCR equipment. However, each assay requires careful optimization, implying considerable efforts for a diagnostic laboratory to set up all tests on site [31]. Alternatively, next generation sequencing (NGS) with a targeted resequencing approach appears to be a suitable technology for extensive clinically relevant mutation testing in the future, which has already been evaluated for the molecular diagnostics of colorectal cancer [32, 33]. Given the high frequency of KRAS codon 12/13 mutations compared to other KRAS- or NRAS mutations and the fact that these mutations occur mutually exclusive [10], it still seems reasonable, to filter the samples that actually need advanced testing method as proposed here. Thus, a two-step approach including HRM analysis of KRAS codon 12/13 mutations followed by next generation targeted resequencing might be the most attractive implementation for routine KRAS mutation diagnostics in the future.
Conclusion
We present a diagnostically reliable and cost-effective two-step approach to KRAS codon 12/13 mutation testing of CRC samples prior to initiation of treatment with anti-EGFR antibodies. The platform appears to be especially attractive for small to medium diagnostic laboratories that don’t have the capacities to maintain an extensive spectrum of rare mutation tests according to regulatory standards for diagnostic laboratories [34] or to adopt NGS-technology with its complex associated infrastructure including bioinformatics.
Abbreviations
CRC, colorectal cancer; EMA. European medical agency; FDA, U.S. food and drug administration; HRM, high resolution melting analysis; LOD, limit of detection; mCRC, metastatic colorectal cancer; NGS, next generation sequencing; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; WT, wildtype; UICC, Union internationale contre le cancer.
Acknowledgements
We like to thank Viktoria Wischmann, Department of Pathology, University Hospital Marburg for microdissection of CRC specimens. Petra Ross, Lisa-Marie Koch and the staff of the Molecular Biology Laboratory, Department of Hematology and Oncology are acknowledged for excellent technical assistance and helpful discussion.
Funding
This work was supported by DFG Klinische Forschergruppe KFO210 (AN, CB) and the José Carreras Leukemia Foundation (AH06-01, to AN).
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article and its supplementary material.
Authors’ contributions
CB, EM and RM conceived and designed the study. RM reviewed tissue sections. KS established the method and performed KRAS mutation analysis. KS, CB and EM analysed data. AN and JRK contributed to the analysis and interpretation of the data. EM wrote the manuscript, which was critically revised by CB, AN, RM and JRK. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was conducted in a routine diagnostic setting for internal quality control purposes and did not require formal ethics approval according to the guidelines of the local ethics comitee. Verbal informed consent to perform routine pathological examinations on their samples as needed was obtained from all patients.
Additional files
Supplementary method and Table S1. HRM analysis of NRAS codon 61 and Results of NRAS codon 61 mutation testing in 19 CRC samples. (DOCX 22 kb)
HRM analysis of NRAS codon 61. (TIF 553 kb)
Contributor Information
Elisabeth Mack, Email: elisabeth.mack@staff.uni-marburg.de.
Kathleen Stabla, Email: stablak@staff.uni-marburg.de.
Jorge Riera-Knorrenschild, Email: rierakno@med.uni-marburg.de.
Roland Moll, Email: mollr@med.uni-marburg.de.
Andreas Neubauer, Email: neubauer@staff.uni-marburg.de.
Cornelia Brendel, Email: brendelc@staff.uni-marburg.de.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Stein A, Bokemeyer C. How to select the optimal treatment for first line metastatic colorectal cancer. World J Gastroenterol. 2014;20(4):899–907. doi: 10.3748/wjg.v20.i4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 4.De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 5.Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, Bastit L, Killian A, Sesboue R, Tuech JJ, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96(8):1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 9.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 10.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50(5):307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 11.05/12/2014 Erbitux -EMEA/H/C/000558 -N/0071 - Erbitux : EPAR - Product Information [http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000558/WC500029119.pdf]
- 12.31/03/2015 Vectibix -EMEA/H/C/000741 -II/0065 - Vectibix: EPAR Product Information [http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000741/WC500047710.pdf]
- 13.Label approved on 03/11/2015 (PDF) for VECTIBIX, BLA no. 125147 [http://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125147s200lbl.pdf]
- 14.Label approved on 04/10/2015 (PDF) for ERBITUX, BLA no. 125084 [http://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125084s262lbl.pdf]
- 15.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 16.List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools) [http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm]
- 17.Bolton L, Reiman A, Lucas K, Timms J, Cree IA. KRAS mutation analysis by PCR: a comparison of two methods. PLoS One. 2015;10(1):e0115672. doi: 10.1371/journal.pone.0115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez De Castro D, Angulo B, Gomez B, Mair D, Martinez R, Suarez Gauthier A, Shieh F, Velez M, Brophy VH, Lawrence HJ, et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer. 2012;107(2):345–351. doi: 10.1038/bjc.2012.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heideman DA, Lurkin I, Doeleman M, Smit EF, Verheul HM, Meijer GA, Snijders PJ, Thunnissen E, Zwarthoff EC. KRAS and BRAF mutation analysis in routine molecular diagnostics: comparison of three testing methods on formalin-fixed, paraffin-embedded tumor-derived DNA. J Mol Diagn. 2012;14(3):247–255. doi: 10.1016/j.jmoldx.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai K, Tsurutani J, Yamanaka T, Yoneshige A, Ito A, Togashi Y, De Velasco MA, Terashima M, Fujita Y, Tomida S, et al. Extended RAS and BRAF Mutation Analysis Using Next-Generation Sequencing. PLoS One. 2015;10(5):e0121891. doi: 10.1371/journal.pone.0121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuononen K, Maki-Nevala S, Sarhadi VK, Wirtanen A, Ronty M, Salmenkivi K, Andrews JM, Telaranta-Keerie AI, Hannula S, Lagstrom S, et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer. 2013;52(5):503–511. doi: 10.1002/gcc.22047. [DOI] [PubMed] [Google Scholar]
- 23.Herreros-Villanueva M, Chen CC, Yuan SS, Liu TC, Er TK. KRAS mutations: analytical considerations. Clin Chim Acta. 2014;431:211–220. doi: 10.1016/j.cca.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 24.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49(6 Pt 1):853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 25.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281(5375):363. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 26.Qiagen: Therascreen® KRAS Pyro® Kit handbook version 1, German edition. Hilden, Germany: Qiagen; 2011.
- 27.Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget. 2012;3(5):546–558. doi: 10.18632/oncotarget.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 29.Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2011;23(6):1518–1525. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 30.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26(1):13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 31.Botezatu IV, Nechaeva IO, Senderovich AI, Kondratova VN, Shelepov VP, Lichtenstein AV, Stroganova capital A CEMC Optimization of melting analysis with TaqMan probes for detection of KRAS, NRAS, and BRAF mutations. Anal Biochem. 2015;491:75–83. doi: 10.1016/j.ab.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Betge J, Kerr G, Miersch T, Leible S, Erdmann G, Galata CL, Zhan T, Gaiser T, Post S, Ebert MP, et al. Amplicon sequencing of colorectal cancer: variant calling in frozen and formalin-fixed samples. PLoS One. 2015;10(5):e0127146. doi: 10.1371/journal.pone.0127146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malapelle U, Vigliar E, Sgariglia R, Bellevicine C, Colarossi L, Vitale D, Pallante P, Troncone G. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol. 2015;68(1):64–8. [DOI] [PubMed]
- 34.Allegra CJ, Rumble RB, Hamilton SR, Mangu PB, Roach N, Hantel A, Schilsky RL. Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol. 2016;34(2):179–185. doi: 10.1200/JCO.2015.63.9674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article and its supplementary material.


