Abstract
Toll-like receptors (TLRs) are innate immune receptors expressed in all parts of the alimentary tract. However, analyses comparing expression in different segments and data on germ-free animals are lacking. Alimentary tract cancers show increased TLR expression. According to the field effect concept, carcinogenetic factors induce subtle cancer predisposing alterations in the whole organ. We studied TLR1 to TLR9 expression in all segments of the alimentary tract from cancer patients’ tumor-adjacent normal mucosa, healthy organ donors, and conventional and germ-free mice by using immunohistochemistry and quantitative PCR. All TLRs were expressed in all segments of the alimentary tract. Expression was most intensive in the small intestine in humans and conventional mice, but germ-free mice showed less expression in the small intestine. TLR expression levels were similar in cancer patients and organ donors. We provide systematic baseline data on the TLR expression in the alimentary tract. Normal epithelium adjacent to tumor seems to have similar TLR expression compared with healthy tissues suggesting absence of any field effect in TLR expression. Accordingly, specimens from cancer patients’ normal tumor-adjacent tissue can be used as control tissues in immunohistochemical TLR studies in gastrointestinal cancer.
Keywords: alimentary tract, cancer, expression, germ-free mouse, mouse, organ donor, Toll-like receptor, tumor-adjacent normal epithelium
Introduction
Toll-like receptors (TLRs) are innate immune receptors. TLRs recognize microbial structures such as bacterial cell membrane components and DNA and RNA molecules. Recognition of these antigens leads to production of inflammatory cytokines1 and in several cases also in change of cell behavior, such as invasion.2,3 Chronic inflammation is associated with several cancers,4 especially gastrointestinal tract cancers in relation to viral or bacterial infection, such as Helicobacter pylori to gastric cancer5 and hepatitis B and C viruses to hepatocellular carcinoma.6 There are also reports from microbial shift in colorectal cancer and esophageal adenocarcinoma.7–10
TLRs play a key role in the microbe-rich gastrointestinal environment. Normal TLR function in the alimentary tract has been poorly described. TLRs seem to have a role in normal homeostasis of the gut and immune responses modulated by interactions with normal commensal flora.11 TLRs have been shown to affect the pathogenesis and prognosis of gastrointestinal cancers.12–16 TLR2 and TLR5 seem to have a prognostic role in tongue cancer.17,18 TLR1 to TLR9 are overexpressed in Barrett’s high-grade dysplasia, indicating a central role in carcinogenesis.19–22 TLR1, TLR4, TLR8, and TLR9 have effects to esophageal carcinoma progression.21–23 TLR2, TLR3, and TLR4 are associated to high TNM stage in gastric cancer.16,24,25 TLR7 and TLR8 associate to poor survival in colorectal cancer.26 TLR5, TLR7, and TLR8 activation increases proliferation,26–28 and TLR2, TLR4, and TLR9 activation induces invasion in gastrointestinal cancer cells.3,29,30 Despite the abundance of studies showing different TLRs in gastrointestinal cancers, the relative expression between the different organs and gastrointestinal segments is unknown. Neither have the effects of gastrointestinal cancers on immune receptor expression in the tumor-adjacent normal epithelium (TANE) comprehensively been studied.
Mice are extensively used in experimental disease modeling. There are minor differences in TLR gene sequences between humans and mice causing differences in regulation and activation of TLRs between the species.31 However, no comparative analyses of TLR expression in human and mouse alimentary tracts exist. Also, there is a possibility to study the role of bacteria in mice by using germ-free animals.
The first aim of the present study was to systematically characterize the expression of TLR1 to TLR9 in different gastrointestinal organs, including liver and pancreas, in humans and mice. Our second aim was to clarify whether alimentary tract cancer affects TLR expression of tumor-adjacent normal tissues. The third aim was to characterize the effects of commensal microbes of the alimentary tract on TLR expression in gastrointestinal organs by comparing germ-free and conventional animals.
Materials and Methods
Study Material
Patient samples representing tumor-adjacent normal tissues were collected from patients operated in the Department of Surgery, Oulu University Hospital, between the years 2008 and 2014. The primary diagnosis of all patients was adenocarcinoma of the esophagus, stomach, colon, liver, or pancreas. Samples for TANE were collected approximately at the distance of 5 to 10 cm from the primary tumor. Samples from two otherwise healthy organ donors were also collected from the Department of Surgery, Oulu University Hospital, during the removal of other organs. Median age of the study population was 71.5 years (range: 41–88), and the gender distribution was 27 male and 19 female. Mouse samples were collected from the Animal Research Center, Oulu University (CD-1 strains mouse) and germ-free (C57BL/6 strain) mice from the Core Facility for Germ-free Research, Karolinska Institutet, Stockholm, Sweden. Biliary system samples were not available from mice.
The collection of patient material and the organ donor samples was approved by the Oulu University Hospital Ethics Committee. The need to obtain a written or oral consent from the patients for using the samples in research was waived by the Finnish National Authority for Medicolegal Affairs (VALVIRA, Dnro 10832/06.01.03.01/2014).
Five independent samples from five different patients were evaluated from each anatomic segment from the cancer patients. From two organ donors, the whole alimentary tract was evaluated, which acted as the healthy control. We also evaluated samples from eight (four male and four female) conventional CD-1, 8-week-old mice and four (two male and two female) alimentary tracts from microbe-free (germ-free mice) 8-week-old mice.
Assessment of Immunostaining
For immunohistochemical detection of the antibody reaction, we used the Dako EnVision kit (Dako, Copenhagen, Denmark) with high-temperature antigen retrieval in Tris–EDTA buffer for 15 min. Diaminobenzidine (Dako basic DAB-kit) was used as a chromogen. All stainings were done with a Dako Autostainer (Dako). Validation of our immunohistochemical analysis was performed through two series of negative controls (by omitting the primary antibody and by replacing the primary antibody with the mouse primary antibody isotype control). Lymphocytes of the lymph nodes in the sample material were used as an internal positive control for TLR staining. For optimization of the dilution of each primary antibody, a series of dilutions involving the concentration recommended by the manufacturer was used, and dilutions providing expression patterns corresponding to those reported were used for the whole series. The test dilution series was made from the stomach, duodenum, and colon, and staining was optimized for distribution of TLRs expression. Strongest intensity was scored as intensity 3 and weakest positive to intensity 1 within species (humans and normal or germ-free mice). The commercial antibodies and dilutions used in the study are summarized in Table 1.
Table 1.
Used TLR Antibodies With Dilutions, Catalog Numbers, and Manufacturer.
| Antibody | Dilution | Catalog Number | Company |
|---|---|---|---|
| TLR1 | 1:50 (human) 1:75 (mouse) |
ab189337 IMG-5012 |
Abcam Corporation (Cambridge, MA) Imgenex (San Diego, CA) |
| TLR2 | 1:75 (human) 1:75 (mouse) |
MAB0066 IMG-662 |
Abnova Corporation (Walnut, CA) Imgenex |
| TLR3 | 1:25 | IMG-315A | Imgenex |
| TLR4 | 1:1000 | H00007099-M02 | Abnova Corporation |
| TLR5 | 1:75 | IMG-664A | Imgenex |
| TLR6 | 1:750 | PAB3555 | Abnova Corporation |
| TLR7 | 1:750 | IMG-540 | Imgenex |
| TLR8 | 1:850 | NBP2-24917 | Novus Biologicals, LLC (Littleton, CO) |
| TLR9 | 1:150 | NBP2-24729 | Novus Biologicals, LLC |
Abbreviation: TLR, Toll-like receptor.
The evaluation of the immunostainings was separately performed by H.H. and O.H. The intensity of staining (0–3) and the percentage of cells showing expression of TLRs (0–100) of all epithelial cells were assessed. The percentage estimate was separately performed for cytoplasmic, membranous, and nuclear staining. The mean values of the two independent estimates were used if the estimated staining intensity scores did not differ more than by one step or if the difference of proportion of positive cells was less than 30%. In cases with more extensive differences between the assessors, a consensus was reached after reevaluation by T.J.K. (13 of the 2948 samples needed reevaluation). Histoscore was calculated by multiplying the mean of intensity level and mean percentage of positive cells resulting in a value between 0 and 300.
Western Blotting
Western blot was performed using human and conventional mouse liver samples to examine the expression of TLRs. The frozen liver tissue samples were homogenized, and the solution was added to cell lysis buffer (Cell Signaling Technology, Danvers, MA) with protease inhibitors (REF 05892970001; Roche, Mannheim, Germany). Solutions were centrifuged at 12,000 RPM for 15 min at 4C to clarify the supernatants. The samples were boiled for 4 min in reducing sodium dodecyl sulfate (SDS) buffer and loaded equally as 25 µg on the SDS polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA). The electrophoresed proteins were stained with Coomassie blue to confirm equal loading and transferred onto an Immobilon P (polyvinylidene difluoride, size: 0.45 µm) membrane (Merck Millipore, Temecula, CA). After removing the stain with methanol, the membrane was incubated with TLR antibodies and diluted in RT overnight using the same dilution as in immunohistochemistry (Table 1) in Tris-buffered saline with 0.1% Tween 20. After washing the membrane, the secondary antibody was allowed to bind for 1 hr, and it was washed and incubated with ABC complex (Vector Laboratories, Burlingame, CA). Finally, Pierce ECL Western blotting detection reagents (Thermo Fisher Scientific, Waltham, MA) were used to detect the proteins in the membrane.
Real-Time Quantitative RT-PCR
Quantitative PCR (qPCR) analyses were performed from stomach, small intestine, and large intestine wall samples containing mucosa and from other layers of the wall. From humans, a total of six stomach and small intestine samples and a total of 12 large intestine samples were obtained equally from organ donor and cancer patients. From mice, two stomach, six small intestine, and seven large intestine samples were from conventional and two, five, and six, respectively, were from germ-free mice. Total RNA was extracted from examined tissue with a miRNeasy Mini kit (Qiagen, Hilden, Germany) using an automated QIAcube sample preparation instrument (Qiagen) according to the manufacturer’s protocols. A High Capacity cDNA RT kit (Applied Biosystems, Foster City, CA) was used for reverse transcription of the RNAs with random primers according to the manufacturer’s protocol. The complementary DNAs were amplified in duplicates with a Rotor-Gene Q (Qiagen) using gene-specific primers (Sigma, Haverhill, UK) and SYBR Green qPCR mix (Thermo Fisher Scientific). The mean fold change in relative expression of the target gene at each group was calculated using the 2−ΔΔCt method and rbs13 (mice) and beta-actin (humans) as the reference genes. Primers were the same as used by Sanchez-Quintero et al.32 and Zheng et al.,33 except in the case of human TLR3: 5′-AGTGCCGTCTATTTGCCACA-3′ and 5′-GCATCCCAAAGGGCAAAAGG-3′.
Statistical Analysis
We used IBM SPSS Statistics 22.0 (IBM Corp., Armonk, NY) for statistical analyses. To compare TLRs expression between different anatomic segments of the alimentary tract within a species and to compare tissues between the species, we used an independent-sample t-test. We used Spearman’s rho to test the correlations between stomach, small intestine, and large intestine mRNA levels and protein levels.
Results
General
Characteristics of immunohistochemical expression of each TLR type were evaluated from the liver and pancreas, esophagus, stomach, small intestine, and large intestine. Segments of the small intestine were evaluated separately including the duodenum, jejunum, and ileum and large intestine including the cecum and colon (ascending, transverse, descending, sigmoid colon, and rectum), but analyzed as small and large intestines because identical staining in the epithelial cells without significant differences was observed between the segments. Expressions of all TLRs were detectable in all the segments of the gastrointestinal tract in both humans and mice. All TLRs showed cytoplasmic expression. In addition, TLR2, TLR3, and TLR5 showed occasional nuclear expression throughout the alimentary tract in humans, and in mouse samples, nuclear expression was seen with TLR3 to TLR6 and TLR9 in the alimentary tract.
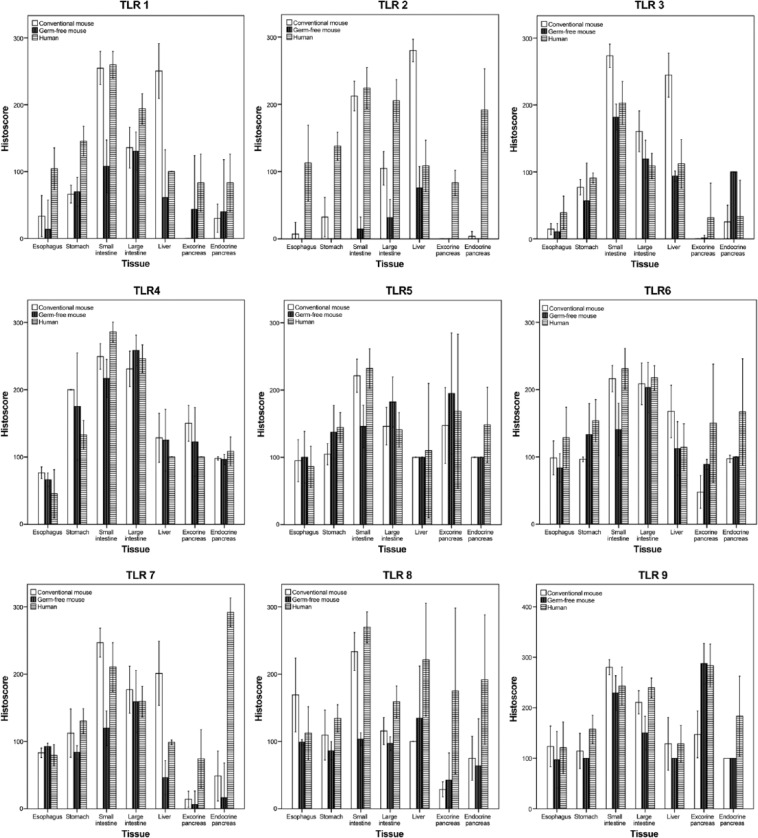
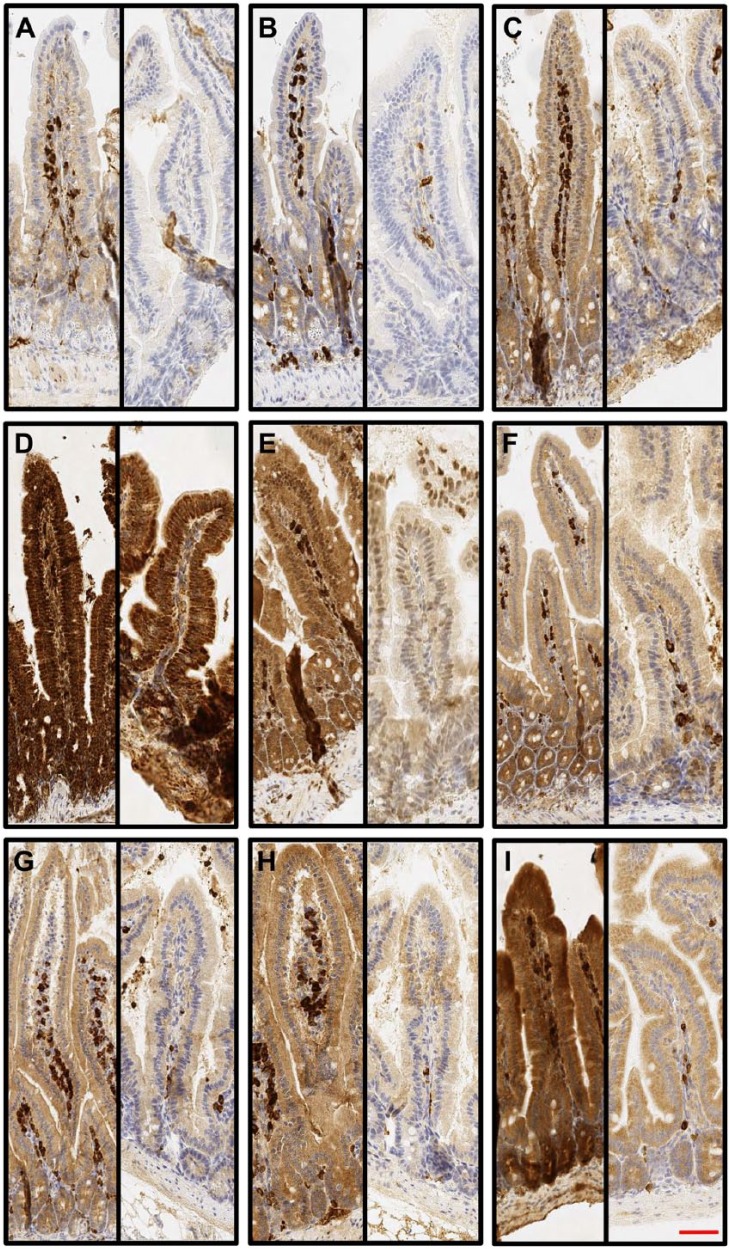
Epithelial expression levels of TLRs in human samples close to tumors and those from the two healthy organ donors were always similar and therefore were analyzed as a single group. Figure 2 shows tumor and organ donors’ similar TLR expression in the colon. Basic data of the immunohistochemical expression of TLR1 to TLR9 in different anatomic segments of the human and mouse gastrointestinal tracts, liver, and pancreas and statistical comparison between the segments are summarized in Table 2. Expression levels are graphically summarized in Fig. 1.
Figure 2.
Typical Toll-like receptor (TLR) expression patterns from the human ascending colon. TLRs are expressed throughout the epithelium with a diffuse manner. Paired figures from ascending colon organ donor (left) and tumor-adjacent normal epithelium (right) are presented: (A) TLR1, (B) TLR2, (C) TLR3, (D) TLR4, (E) TLR5, (F) TLR6, (G) TLR7, (H) TLR8, and (I) TLR9. 20× magnification was used and 50-µm scale bar is in panel I.
Table 2.
TLR1 to TLR9 Protein Expression Detected With Immunohistochemistry From Different Anatomic Segments of the Alimentary Tract.
| Conventional Mouse |
Germ-Free Mouse |
Human |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | 95% CI | Significance | Mean | 95% CI | Significance | Mean | 95% CI | Significance | |
| Esophagus | |||||||||
| TLR1 | 47 | 25–67 | bcde | 14 | 0–55 | abc | 104 | 89–135 | abc |
| TLR2 | 16 | 0–36 | bcd | 0 | 0–0 | acd | 113 | 86–173 | bcf |
| TLR3 | 21 | 13–30 | abcd | 11 | 0–18 | abcdf | 39 | 19–61 | abcd |
| TLR4 | 77 | 72–83 | abcdef | 66 | 60–75 | bcdef | 45 | 13–75 | abcdef |
| TLR5 | 91 | 78–111 | bce | 100 | 83–135 | ce | 86 | 59–110 | abcf |
| TLR6 | 93 | 81–108 | bcde | 84 | 65–94 | abc | 129 | 100–167 | bc |
| TLR7 | 83 | 78–89 | abcdef | 93 | 90–95 | abcdef | 79 | 65–90 | abcdf |
| TLR8 | 144 | 111–178 | bef | 99 | 95–100 | ae | 112 | 88–150 | bcd |
| TLR9 | 125 | 101–148 | bcf | 97 | 50–128 | be | 121 | 84–167 | bce |
| Stomach | |||||||||
| TLR1 | 68 | 55–79 | bcde | 70 | 61–90 | c | 146 | 124–166 | bcdef |
| TLR2 | 33 | 17–48 | bcd | 0 | 0–0 | cde | 138 | 120–157 | bcef |
| TLR3 | 77 | 70–83 | bcde | 57 | 23–85 | bc | 91 | 84–98 | bef |
| TLR4 | 194 | 167–218 | bcdef | 175 | 100–200 | cf | 133 | 115–153 | bcde |
| TLR5 | 103 | 95–114 | bce | 138 | 100–150 | 144 | 123–166 | b | |
| TLR6 | 97 | 95–99 | bcde | 133 | 90–150 | ce | 154 | 127–185 | bc |
| TLR7 | 108 | 94–139 | bcdef | 84 | 75–90 | bcdef | 130 | 114–147 | bdef |
| TLR8 | 107 | 92–137 | bef | 86 | 75–95 | bde | 134 | 114–154 | bd |
| TLR9 | 122 | 100–150 | bcef | 100 | 100–100 | bce | 157 | 132–184 | bce |
| Small intestine | |||||||||
| TLR1 | 253 | 235–269 | ce | 108 | 74–146 | 260 | 241–279 | cdef | |
| TLR2 | 218 | 200–235 | cd | 15 | 1–33 | d | 224 | 197–252 | de |
| TLR3 | 268 | 253–281 | cef | 182 | 162–196 | cdef | 203 | 173–234 | cdef |
| TLR4 | 251 | 236–266 | def | 217 | 193–242 | cdef | 286 | 270–298 | cdef |
| TLR5 | 217 | 198–235 | cde | 146 | 119–179 | df | 232 | 205–258 | cdf |
| TLR6 | 235 | 217–250 | cde | 141 | 110–177 | cef | 231 | 203–257 | de |
| TLR7 | 252 | 237–268 | cef | 120 | 98–144 | def | 210 | 175–242 | cdef |
| TLR8 | 228 | 208–248 | cde | 103 | 98–114 | e | 270 | 246–289 | cf |
| TLR9 | 281 | 271–291 | cde | 229 | 200–258 | cdf | 243 | 205–274 | d |
| Large intestine | |||||||||
| TLR1 | 135 | 109–160 | def | 131 | 105–158 | def | 194 | 172–216 | def |
| TLR2 | 106 | 87–125 | def | 32 | 8–58 | def | 205 | 174–235 | de |
| TLR3 | 172 | 150–193 | def | 119 | 94–148 | e | 109 | 91–127 | ef |
| TLR4 | 238 | 219–257 | def | 258 | 238–279 | def | 246 | 226–265 | def |
| TLR5 | 146 | 127–166 | df | 182 | 148–219 | df | 141 | 117–168 | |
| TLR6 | 206 | 184–227 | ef | 203 | 166–236 | def | 217 | 199–235 | de |
| TLR7 | 169 | 143–193 | def | 159 | 117–202 | def | 159 | 139–182 | def |
| TLR8 | 111 | 98–123 | ef | 97 | 90–106 | e | 159 | 134–180 | |
| TLR9 | 208 | 190–226 | def | 150 | 122–183 | def | 240 | 220–258 | df |
| Liver | |||||||||
| TLR1 | 263 | 236–283 | ef | 61 | 0–95 | 100 | 100–100 | ||
| TLR2 | 287 | 277–297 | ef | 76 | 60–105 | ef | 109 | 88–148 | f |
| TLR3 | 265 | 420–287 | ef | 94 | 90–100 | e | 112 | 93–148 | ef |
| TLR4 | 132 | 109–157 | f | 125 | 100–150 | 100 | 100–100 | ||
| TLR5 | 100 | 100–100 | ef | 100 | 100–100 | e | 110 | 41–193 | |
| TLR6 | 187 | 151–224 | ef | 113 | 100–150 | 114 | 100–150 | ||
| TLR7 | 223 | 191–257 | ef | 46 | 35–70 | e | 99 | 95–100 | f |
| TLR8 | 109 | 100–129 | ef | 134 | 95–200 | e | 221 | 150–283 | |
| TLR9 | 118 | 100–150 | e | 100 | 100–100 | e | 129 | 100–160 | e |
| Exocrine pancreas | |||||||||
| TLR1 | 0 | 0–0 | f | 44 | 0–90 | 83 | 50–100 | ||
| TLR2 | 12 | 0–30 | 0 | 0–0 | 83 | 68–97 | f | ||
| TLR3 | 19 | 4–37 | 1 | 0–5 | f | 32 | 0–73 | ||
| TLR4 | 137 | 114–158 | f | 123 | 93–150 | 100 | 100–100 | ||
| TLR5 | 149 | 115–189 | f | 195 | 143–247 | f | 168 | 70–260 | |
| TLR6 | 49 | 35–64 | f | 89 | 85–95 | f | 150 | 100–220 | |
| TLR7 | 24 | 13–37 | f | 6 | 0–20 | 74 | 37–100 | f | |
| TLR8 | 45 | 28–69 | f | 43 | 16–65 | 175 | 80–275 | ||
| TLR9 | 157 | 130–188 | f | 288 | 250–300 | f | 283 | 243–300 | f |
| Endocrine pancreas | |||||||||
| TLR1 | 45 | 18–83 | 33 | 0–100 | 83 | 43–100 | |||
| TLR2 | 24 | 3–59 | 0 | 0–0 | 192 | 150–250 | |||
| TLR3 | 42 | 22–62 | 100 | 100–100 | 33 | 0–80 | |||
| TLR4 | 99 | 97–100 | 97 | 90–100 | 108 | 100–120 | |||
| TLR5 | 100 | 100–100 | 100 | 100–100 | 148 | 100–196 | |||
| TLR6 | 96 | 83–106 | 100 | 100–100 | 167 | 113–230 | |||
| TLR7 | 53 | 34–72 | 0 | 0–0 | 292 | 270–300 | |||
| TLR8 | 79 | 61–93 | 52 | 0–80 | 192 | 117–263 | |||
| TLR9 | 98 | 94–100 | 100 | 100–100 | 183 | 120–250 | |||
Values are presented as mean histoscore and 95% CI. Statistical comparison was performed with independent samples t-test within species. Abbreviations: TLR, Toll-like receptor; CI, confidence interval.
Compared with stomach, p<0.05.
Compared with small intestine, p<0.05.
Compared with large intestine, p<0.05.
Compared with liver, p<0.05.
Compared with exocrine pancreas, p<0.05.
Compared with endocrine pancreas, p<0.05.
Figure 1.
Histograms of Toll-like receptor (TLR) 1 to 9 histoscores from different anatomic segments of the alimentary tract in humans and in conventional and germ-free mouse.
TLR Expression in the Esophagus and Stomach
The esophageal squamous epithelium showed weak positive expression of all TLRs. The strongest expression was observed in the basal third of the epithelium. However, weak expression was seen throughout the epithelium, and the staining was dominantly diffuse and cytoplasmic. Esophageal staining patterns were similar between the groups (humans, normal mice, and germ-free mice), except for TLR2, which was seen only occasionally in the normal mice esophagus and was absent in the germ-free mice esophagus.
In the gastric mucosa, the expression of all TLRs was seen in the cell cytoplasm on the upper part of the epithelium similarly in all groups. Neck region of the gastric antral glands, including G cells, showed significantly higher expression compared with overall staining in the gastric mucosa, difference being significant in humans for TLR3 (p=0.049); in normal mice for TLR5 (p<0.001), TLR8 (p=0.022), and TLR9 (p=0.034); and in germ-free mice for TLR7 (p=0.022), TLR8 (p=0.007), and TLR9 (p=0.003). Contrasting with cytoplasmic expression in mice, in the human samples TLR9 was present in both cytoplasm and cell membranes.
TLR Expression in the Intestine
In the intestinal epithelium, the TLR expression was cytoplasmic in epithelial cells of the intestine throughout the epithelium in all groups.
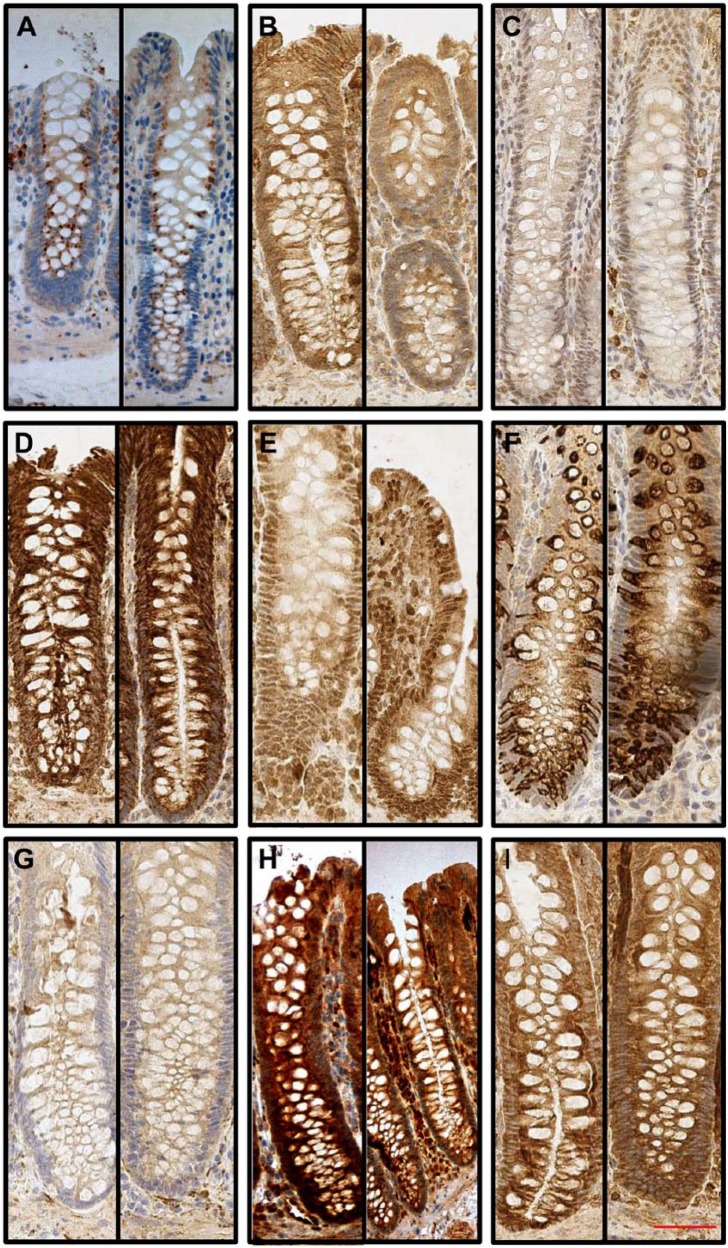
All TLRs were strongly expressed in the small intestine, and the expression was stronger in the villi, with the crypt zone showing only a weak expression. The intensity of TLRs varied from moderate to strong in humans and in normal mice (2.0–2.8; Table 2). However, in germ-free mice, TLRs mean intensity was only 1.0 to 1.8 in the small intestine (Table 2). Figure 3 demonstrates difference between conventional and germ-free mice small intestine TLRs expression levels. In humans, the small intestine showed significantly (p<0.05) higher TLR expression compared with the large intestine for TLR3, TLR4, TLR5, TLR7, and TLR8, and in normal mice, similar difference was seen for TLR1 to TLR9 (Table 2). In contrast, in germ-free mice, higher TLR expression in the small intestine compared with the large intestine was seen only for TLR3 and TLR9 (Table 2).
Figure 3.
Typical Toll-like receptor (TLR) expression patterns from the conventional and germ-free mouse small intestines. TLRs are expressed throughout the epithelium with a diffuse manner. Paired figures from small intestine conventional (left) and germ-free mice (right) are presented: (A) TLR1, (B) TLR2, (C) TLR3, (D) TLR4, (E) TLR5, (F) TLR6, (G) TLR7, (H) TLR8, and (I) TLR9. 10× magnification was used and 50-µm scale bar is in panel I.
In the large intestine, TLR expression was similar in crypts and on the upper part of the epithelium. TLRs expression was dominantly diffuse and cytoplasmic (Fig. 2). Variation in the expression between the different TLRs was more pronounced in the large intestine (Table 2). In germ-free mice, TLRs expression levels were similar between small and large intestines. Interestingly, TLR4 and TLR6 showed significantly higher expression in the germ-free mice large intestine compared with that in the small intestine (Table 2). In humans, TLR8 and TLR9 were expressed also occasionally on the cell membrane, but in mice, membrane expression was not detected.
TLR Expression in the Liver and Pancreas
In humans, TLRs showed weak diffuse cytoplasmic expression in the whole liver. Mean intensity of TLR expression was from 1.0 to 1.3, except for TLR8, which showed stronger expression (Table 2). In mice, liver TLR expression was cytoplasmic and diffuse. In addition to cytoplasmic TLR expression, strong membrane expression was seen, however except for TLR5 and TLR9.
In the human pancreas, the expression of all TLRs was similar to that in the liver and diffuse and cytoplasmic throughout the whole pancreas, and mean intensity was weak. In the mice pancreas, TLR expression was not convincingly seen in the exocrine pancreas. However, TLR4, TLR5, and TLR9 were expressed throughout exocrine pancreas in normal mice and intensity for them was moderate, and the germ-free mice pancreas showed strong diffuse expression for TLR5 and TLR9.
Interestingly, in the human pancreas, TLR2 and TLR7 showed significantly higher (p<0.05) expression in the endocrine pancreas compared with that in the exocrine pancreas. Especially, TLR7 showed a strong staining in the Langerhans’ islets highlighting them. TLR7 was also strongly expressed in the autonomic nerve ganglia. Similar difference between endocrine and exocrine parts was seen with TLR1, TLR6, TLR7, and TLR8 in the normal mice pancreas and with TLR3, TLR5, and TLR6 in germ-free mice pancreas. Conversely, TLR9 expression was significantly higher in the exocrine pancreas in all groups and TLR4 and TLR5 also in normal mice.
Comparison of TLR Expression Levels Between Normal and Germ-Free Mice
The small intestine showed higher TLRs expression in conventional mice than in germ-free mice with all TLRs, p<0.001, except TLR4 and TLR9, p<0.05 (Fig. 3, Table 2). The normal mouse large intestine had higher TLR expression for TLR2, TLR3, and TLR9. Liver expression was significantly higher in normal mice than in germ-free mice for TLR1 to TLR3 and TLR7 (p<0.001) and TLR6 (p=0.039). The stomach showed significantly higher (p<0.05) TLR2 and TLR5 expressions in normal mice and TLR2, TLR3, and TLR9 in the large intestine. In the liver, TLR1 to TLR3, TLR6, and TLR7 expressions were significantly higher in conventional mice. Expressions of TLR6 and TLR9 in the exocrine pancreas and TLR3 in the endocrine pancreas were significantly stronger in normal mice compared with those in germ-free mice.
Western Blot Analyses From Human and Mouse Liver Samples
We performed Western blot analyses from human and conventional mouse livers to confirm specificity of commercial antibodies for each TLR protein. Liver tissue was selected due to homogeneous structure and low amount of connective tissue. Western blot detection of TLR1 to TLR9 revealed similar expressions of TLRs as reported by the manufacturers and is presented in Supplementary Fig. 1.
Relative mRNA Levels in Human and Mouse Alimentary Tracts
We measured the mRNA expression of TLR1 to TLR9 from the stomach, small intestine, and large intestine using qPCR. In agreement with immunohistochemical observations, all TLR types were expressed in all studied tissues. Based on low TLR expression levels in the immunohistochemical evaluation, the stomach was set to a baseline for comparisons. There was no statistical correlation between protein and mRNA levels. Statistical tests comparing mRNA levels within a species and between the species were not done due to small sample size. Relative mRNA expression mean with 95% confidence interval is presented in Supplementary Table 1.
Discussion
TLRs have been previously shown to be expressed through whole alimentary tract.16,19–22,34 For the first time, we describe systematically the differential expression levels of TLR1 to TLR9 in each anatomic segment of the human and mouse alimentary tracts. We have also evaluated the TLR expression in the microbe-free gastrointestinal tract with germ-free mice. In addition, we investigated the effect of alimentary tract cancer on the expression of TLRs in the adjacent normal epithelial cells.
We determined the expression of TLR1 to TLR9 throughout the human alimentary tract by using two sets of normal mucosa samples. The first set came from patients suffering from adenocarcinoma of the alimentary tract and was taken 5 to 10 cm from the tumor. The second set was taken from healthy organ donors. There were no differences in the TLR expression between tissues obtained from cancer patients and organ donors. Cancers are systemic diseases,34 with subtle predisposing alterations in the whole organ according to the field effect concept,35 and thus could affect the TLR expression in the closely located mucosa. Our results indicate that the expression of TLRs in the tumor-adjacent normal mucosa is similar to that in healthy subjects. There are no previous studies systematically comparing the cancer-adjacent tissue with completely healthy tissues. Although our material is small with only two organ donors, it answers to an important question. However, as regulation of TLR expression is complex and depends on several factors including age, gender, and nutrition, a comprehensive analysis would need much more extensive case series of both tumor-adjacent and healthy tissues.
All TLRs were expressed in all parts of the alimentary tract. Interestingly, all studied TLRs showed higher expression in the small intestine compared with that in the large intestine. This was evident in both humans and in normal mice. Surprisingly, all studied TLRs were detected in the germ-free mouse gastrointestinal tract. However, in conventional mice, all TLRs showed significantly higher expression in the small intestine compared with that in germ-free mice (Fig. 3, Table 2), but only for TLR2, TLR3, and TLR9, such difference was evident in the colon (Table 2). Our study showing TLR expression in the microbe-free alimentary tract indicates that some constant TLR expression is maintained by epithelial cells without any need for interaction with the TLR ligands. The large intestine is subject to a major bacterial exposure9 but, according to our observations, shows less TLR expression than the small intestine. The large intestine may even be less dependent on the overall microbial load as shown by our observation with germ-free animals with expression levels mostly comparable with conventional mice. It has been shown that TLRs are important in the regulation of homeostatic interaction with the intestinal microbiome. TLRs participate in immunomodulation and mediate protective effects of probiotics.36 TLRs also have effects on epithelial cell proliferation and immunomodulation.37
Our observations disclosed evidence for some novel specific locations and functional roles of some TLRs. In the human, base of the gastric glands had groups of strongly positive cells expressing TLR2, TLR3, TLR4, and TLR9. However, statistically significant difference was seen only with TLR3. With normal mice samples, similar finding was seen with TLR5 and TLR7 to TLR9. We have previously reported that TLR4 is expressed in antral G and D cells and suggested TLR4 is involved in the regulation of gastrin secretion.38 Cell-specific localization of TLR2, TLR3, and TLR9 in the human gastric glands needs additional studies. However, as gastrin response with induction of acid secretion would be a reasonable antimicrobial response by the innate immunity system, their colocalization within the G and D cells in a way similar to TLR4 would be biologically plausible. In the human pancreas, the expression of TLR2, TLR7, and TLR9 was significantly different between the endocrine and exocrine compartments. Especially, TLR7 spotted Langerhans’ islets in human samples. TLR7-positive dendritic cells have been observed in Langerhans’ islets in newly onset type 1 diabetes patients, in insulitis lesions, and in nonobese diabetic mice, supporting importance of TLR7-mediated T-lymphocyte-mediated insulitis.39 Our current observations of endocrine cells of the pancreas and stomach suggest that TLRs have a role in the neuroendocrinology of the alimentary tract.
Nuclear expression was seen in human and mouse alimentary tract epithelial cells and especially for TLR3 to TLR5 and occasional with other TLR types. We have previously reported nuclear expression of TLR1, TLR3, TLR4, TLR5, and TLR8 in Barrett’s esophagus and esophageal adenocarcinoma.19–22 Others have shown nuclear expression of TLR2 and TLR4 in oral squamous cell carcinoma,18 TLR4 in laryngeal squamous cell carcinoma,40 and TLR5 in adenoid cystic carcinoma of the salivary glands.41 Using the freely available NucPred tool, which predicts the nuclear localization of proteins, we obtained the following scores for the different TLRs (TLR1: 0.61, TLR2: 0.70, TLR3: 0.55, TLR4: 0.43, TLR5: 0.36, TLR6: 0.53, TLR7: 0.67, TLR8: 0.88, and TLR9: 0.55).42 This suggests that it is somewhat likely that different TLRs translocate to the nucleus. TLRs have two optional signaling pathways, either MyD88 dependent and/or MyD88 independent. Based on previous studies,18,40,41 it seems possible that TLRs have also an alternative signaling pathway where TLR goes straight to nuclei after ligand interaction without any secondary adaptor molecule. However, the role and function of nuclear translocation of TLRs remain speculative.
Occasional cell membrane expression of TLR9 in the gastric mucosa and of TLR8 and TLR9 in the colon epithelium was seen. TLR8 and TLR9 have been originally characterized as endosomal receptors. There is, however, increasing number of the studies where these endosomal receptors located in the cell membrane. We have previously shown TLR9 membrane expression on esophageal gastric metaplasia.20 Nojiri et al. have also reported membrane expression of TLR9 in the colon mucosa,43 and recently, they are reported also in cell nuclei.21,22,40,41 Taken together, these results suggest that, in addition to endosomes, TLRs might have alternative localization in cells.
We could confirm synthesis of all TLR types in the gastrointestinal tract by using qPCR. TLR expression on the protein level was highest in the small intestine (Fig. 1, Table 2), but in mice, mRNA levels for nearly all TLRs were higher in the large intestine than in the small intestine (Supplementary Table 1). In human samples, gene expression levels did not differ between the small and large intestines. We found no correlation between mRNA and protein expression levels. Significant correlation between mRNA and protein levels is known to be rare,44 related with posttranscriptional and posttranslational regulation, variable half-life of the proteins, and error and noise in both protein and mRNA experiments.45,46 Accordingly, no quantitative conclusions can be made from the TLR mRNA expression data.
Our study has several limitations. The study material was heterogeneous. Studied groups were standardized only by group size in humans, and the number of subjects was low. Similarly, the mice group size was small. However, the immunohistochemical staining results were very consistent, and interobserver agreement was excellent. Normal and germ-free mice in our study were from different strains. However, there are no known differences between these strains relating with expression or function of TLRs.47,48 Finally, our main conclusions of the role of germ-free environment were based on comparisons within one strain. We have validated the functionality of immunohistochemistry with several methods. Also, previously published studies have reported the expression of these TLRs in the gastrointestinal tract,16,19–22,34 which is in line with the present results.
In conclusion, TLR1 to TLR9 were expressed in human and murine gastrointestinal organs and followed a similar general expression pattern. The expression of TLRs was the most intensive in the small intestine in both normal mice and humans. In germ-free mice, the expression of TLRs was downregulated in the small intestine in particular and emphasis of small intestinal expression was largely lost. The normal epithelium adjacent to tumor seems to have similar TLR expression compared with the normal healthy alimentary tract and thus can be used as a control tissue in immunohistochemical TLR studies in gastrointestinal cancer.
Supplementary Material
Acknowledgments
We thank Erja Tomperi, Riitta Vuento, Helmi Konola, and Emilia Kiljander for their excellent technical assistance and Pasi Ohtonen for advice in statistical analyses.
Footnotes
Author Contributions: HH had full access to all of the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: HH, OH, JHK, PPL, JS, TJK. Acquisition of data: HH, OH, JHK, PPL, JS, TJK. Analysis and interpretation of data: HH, OH, JHK, KP, TS, TJK. Drafting of the manuscript: HH, OH, JHK, KP, TS, PPL, JS, TJK. Critical revision of the manuscript for important intellectual content: HH, OH, JHK, KP, PPL, JS, TJK. Statistical analysis: HH, OH, TJK. Administrative, technical, or material support: HH, OH, JHK, KP, PPL, JS, TJK. Study supervision: PPL, JS, TJK.
Competing Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the following grants. Päivikki and Sakari Sohlberg Foundation, the Emil Aaltonen Foundation, Thelma Mäkikyrö Foundation, Georg C. and Mary Ehrnrooth Foundation, and the Finnish Medical Foundation.
Literature Cited
- 1. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. [DOI] [PubMed] [Google Scholar]
- 2. Nurmenniemi S, Kuvaja P, Lehtonen S, Tiuraniemi S, Alahuhta I, Mattila RK, Risteli J, Salo T, Selander KS, Nyberg P, Lehenkari P. Toll-like receptor 9 ligands enhance mesenchymal stem cell invasion and expression of matrix metalloprotease-13. Exp Cell Res. 2010;316:2676–82. [DOI] [PubMed] [Google Scholar]
- 3. Kauppila JH, Karttunen TJ, Saarnio J, Nyberg P, Salo T, Graves DE, Lehenkari PP, Selander KS. Short DNA sequences and bacterial DNA induce esophageal, gastric, and colorectal cancer cell invasion. APMIS. 2013;121:511–22. [DOI] [PubMed] [Google Scholar]
- 4. Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park). 2002;16:217–26, 229; discussion 230–2. [PubMed] [Google Scholar]
- 5. Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, Kim YR, Kim JS, Kim JM, Kim JM, Lee DH, Jung HC. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than helicobacter pylori in the gastric carcinogenesis. Helicobacter. Epub 2016. February 24. doi: 10.1111/hel.12293. [DOI] [PubMed] [Google Scholar]
- 6. Mazzanti R, Arena U, Tassi R. Hepatocellular carcinoma: where are we? World J Exp Med. 2016;6:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pei Z, Yang L, Peek RM, Levine SM, Jr, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett’s esophagus. World J Gastroenterol. 2005;11:7277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–82. [DOI] [PubMed] [Google Scholar]
- 9. Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Philos Soc. 2012;87:701–30. [DOI] [PubMed] [Google Scholar]
- 10. Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, Boyd NH, Gazarik KE, Heit MI, Nistico L, Kasi PM, Spirk TL, Byers B, Lloyd EJ, Landreneau RJ, Jobe BA. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer. 2016;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, Bercik P, Verdu EF, McCoy KD, Macpherson AJ. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grimm M, Kim M, Rosenwald A, Heemann U, Germer CT, Waaga-Gasser AM, Gasser M. Toll-like receptor (TLR) 7 and TLR8 expression on CD133+ cells in colorectal cancer points to a specific role for inflammation-induced TLRs in tumourigenesis and tumour progression. Eur J Cancer. 2010;46:2849–57. [DOI] [PubMed] [Google Scholar]
- 13. Ronkainen H, Hirvikoski P, Kauppila S, Vuopala KS, Paavonen TK, Selander KS, Vaarala MH. Absent Toll-like receptor-9 expression predicts poor prognosis in renal cell carcinoma. J Exp Clin Cancer Res. 2011;30:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pimentel-Nunes P, Goncalves N, Boal-Carvalho I, Afonso L, Lopes P, Roncon-Albuquerque R, Jr, Henrique R, Moreira-Dias L, Leite-Moreira AF, Dinis-Ribeiro M. Helicobacter pylori induces increased expression of Toll-like receptors and decreased Toll-interacting protein in gastric mucosa that persists throughout gastric carcinogenesis. Helicobacter. 2013;18:22–32. [DOI] [PubMed] [Google Scholar]
- 15. Pimentel-Nunes P, Teixeira AL, Pereira C, Gomes M, Brandao C, Rodrigues C, Goncalves N, Boal-Carvalho I, Roncon-Albuquerque R, Jr, Moreira-Dias L, Leite-Moreira AF, Medeiros R, Dinis-Ribeiro M. Functional polymorphisms of Toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Dig Liver Dis. 2013;45:63–69. [DOI] [PubMed] [Google Scholar]
- 16. Fernandez-Garcia B, Eiro N, Gonzalez-Reyes S, Gonzalez L, Aguirre A, Gonzalez LO, Del Casar JM, Garcia-Muniz JL, Vizoso FJ. Clinical significance of toll-like receptor 3, 4, and 9 in gastric cancer. J Immunother. 2014;37:77–83. [DOI] [PubMed] [Google Scholar]
- 17. Kauppila JH, Mattila AE, Karttunen TJ, Salo T. Toll-like receptor 5 (TLR5) expression is a novel predictive marker for recurrence and survival in squamous cell carcinoma of the tongue. Br J Cancer. 2013;108:638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Makinen LK, Atula T, Hayry V, Jouhi L, Datta N, Lehtonen S, Ahmed A, Makitie AA, Haglund C, Hagstrom J. Predictive role of Toll-like receptors 2, 4, and 9 in oral tongue squamous cell carcinoma. Oral Oncol. 2015;51:96–102. [DOI] [PubMed] [Google Scholar]
- 19. Helminen O, Huhta H, Takala H, Lehenkari PP, Saarnio J, Kauppila JH, Karttunen TJ. Increased Toll-like receptor 5 expression indicates esophageal columnar dysplasia. Virchows Arch. 2014;464:11–18. [DOI] [PubMed] [Google Scholar]
- 20. Huhta H, Helminen O, Kauppila JH, Takala H, Metsikko K, Lehenkari P, Saarnio J, Karttunen T. Toll-like receptor 9 expression in the natural history of Barrett mucosa. Virchows Arch. 2015;467:9–18. [DOI] [PubMed] [Google Scholar]
- 21. Helminen O, Huhta H, Lehenkari Petri P, Saarnio J, Karttunen Tuomo J, Kauppila Joonas H. Nucleic acid-sensing Toll-like receptors 3, 7 and 8 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. OncoImmunology. 2015;5:e1127495–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huhta H, Helminen O, Lehenkari PP, Saarnio J, Karttunen TJ, Kauppila JH. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget. 7:23658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kauppila JH, Takala H, Selander KS, Lehenkari PP, Saarnio J, Karttunen TJ. Increased Toll-like receptor 9 expression indicates adverse prognosis in oesophageal adenocarcinoma. Histopathology. 2011;59:643–49. [DOI] [PubMed] [Google Scholar]
- 24. Yuan X, Zhou Y, Wang W, Li J, Xie G, Zhao Y, Xu D, Shen L. Activation of TLR4 signaling promotes gastric cancer progression by inducing mitochondrial ROS production. Cell Death Dis. 2013;4:e794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang H, Wang B, Yan J, Wang T, Zhou XN, Wen HY, Zhu XM. Toll-like receptor 2 promotes invasion by SGC-7901 human gastric carcinoma cells and is associated with gastric carcinoma metastasis. Ann Clin Lab Sci. 2014;44:158–66. [PubMed] [Google Scholar]
- 26. Grimmig T, Matthes N, Hoeland K, Tripathi S, Chandraker A, Grimm M, Moench R, Moll EM, Friess H, Tsaur I, Blaheta RA, Germer CT, Waaga-Gasser AM, Gasser M. TLR7 and TLR8 expression increases tumor cell proliferation and promotes chemoresistance in human pancreatic cancer. Int J Oncol. 2015;47:857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Z, Yu X, Zhang J, Tian Z, Zhang C. TLR7/8 agonists promote NK-DC cross-talk to enhance NK cell anti-tumor effects in hepatocellular carcinoma. Cancer Lett. 2015;369:298–306. [DOI] [PubMed] [Google Scholar]
- 28. Omar AA, Korvala J, Haglund C, Virolainen S, Hayry V, Atula T, Kontio R, Rihtniemi J, Pihakari A, Sorsa T, Hagstrom J, Salo T. Toll-like receptors -4 and -5 in oral and cutaneous squamous cell carcinomas. J Oral Pathol Med. 2015;44:258–65. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y, Wang Q, Ma A, Li Y, Li R, Wang Y. Functional expression of TLR9 in esophageal cancer. Oncol Rep. 2014;31:2298–304. [DOI] [PubMed] [Google Scholar]
- 30. Wang N, An D, Li Q, Li C, Yang G. Effect of Toll-like receptor 4 signaling activation on the biological behavior of gastric cancer cell lines. Zhonghua Yi Xue Za Zhi. 2015;95:2104–8. [PubMed] [Google Scholar]
- 31. Rehli M. Of mice and men: species variations of Toll-like receptor expression. Trends Immunol. 2002;23:375–8. [DOI] [PubMed] [Google Scholar]
- 32. Sanchez-Quintero MJ, Torres MJ, Blazquez AB, Gomez E, Fernandez TD, Dona I, Ariza A, Andreu I, Melendez L, Blanca M, Mayorga C. Synergistic effect between amoxicillin and TLR ligands on dendritic cells from amoxicillin-delayed allergic patients. PLoS ONE. 2013;8:e74198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zheng B, Morgan ME, van de Kant HJ, Garssen J, Folkerts G, Kraneveld AD. Transcriptional modulation of pattern recognition receptors in acute colitis in mice. Biochim Biophys Acta. 2013;1832:2162–72. [DOI] [PubMed] [Google Scholar]
- 34. Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27:234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, Ogino S. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;28:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strobel S, Mowat AM. Oral tolerance and allergic responses to food proteins. Curr Opin Allergy Clin Immunol. 2006;6:207–13. [DOI] [PubMed] [Google Scholar]
- 37. Johnston DG, Corr SC. Toll-like receptor signalling and the control of intestinal barrier function. Methods Mol Biol. 2016;1390:287–300. [DOI] [PubMed] [Google Scholar]
- 38. Pohjanen VM, Koivurova OP, Huhta H, Helminen O, Makinen JM, Karhukorpi JM, Joensuu T, Koistinen PO, Valtonen JM, Niemela SE, Karttunen RA, Karttunen TJ. Toll-like receptor 4 wild type homozygozity of polymorphisms +896 and +1196 is associated with high gastrin serum levels and peptic ulcer risk. PLoS ONE. 2015;10:e0131553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uno S, Imagawa A, Okita K, Sayama K, Moriwaki M, Iwahashi H, Yamagata K, Tamura S, Matsuzawa Y, Hanafusa T, Miyagawa J, Shimomura I. Macrophages and dendritic cells infiltrating islets with or without beta cells produce tumour necrosis factor-alpha in patients with recent-onset type 1 diabetes. Diabetologia. 2007;50:596–601. [DOI] [PubMed] [Google Scholar]
- 40. Ilmarinen T, Hagstrom J, Haglund C, Auvinen E, Leivo I, Pitkaranta A, Aaltonen LM. Low expression of nuclear Toll-like receptor 4 in laryngeal papillomas transforming into squamous cell carcinoma. Otolaryngol Head Neck Surg. 2014;151:785–90. [DOI] [PubMed] [Google Scholar]
- 41. Hirvonen K, Back L, Haglund C, Leivo I, Jouhi L, Makitie AA, Hagstrom J. Toll-like receptor 5 and 7 expression in adenoid cystic carcinoma of major salivary glands. Tumour Biol. 2016. Epub 2016. February 18. doi: 10.1007/s13277-016-4971-8. [DOI] [PubMed] [Google Scholar]
- 42. Brameier M, Krings A, MacCallum RM. NucPred—predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–60. [DOI] [PubMed] [Google Scholar]
- 43. Nojiri K, Sugimoto K, Shiraki K, Tameda M, Inagaki Y, Kusagawa S, Ogura S, Tanaka J, Yoneda M, Yamamoto N, Okano H, Takei Y, Ito M, Kasai C, Inoue H, Takase K. The expression and function of Toll-like receptors 3 and 9 in human colon carcinoma. Oncol Rep. 2013;29:1737–43. [DOI] [PubMed] [Google Scholar]
- 44. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szallasi Z. Genetic network analysis in light of massively parallel biological data acquisition. Pac Symp Biocomput. 1999;4:5–16. [DOI] [PubMed] [Google Scholar]
- 46. Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. [DOI] [PubMed] [Google Scholar]
- 47. Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat Genet. 2005;37:1181–86. [DOI] [PubMed] [Google Scholar]
- 48. Sellers RS, Clifford CB, Treuting PM, Brayton C. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol. 2012;49:32–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.