Abstract
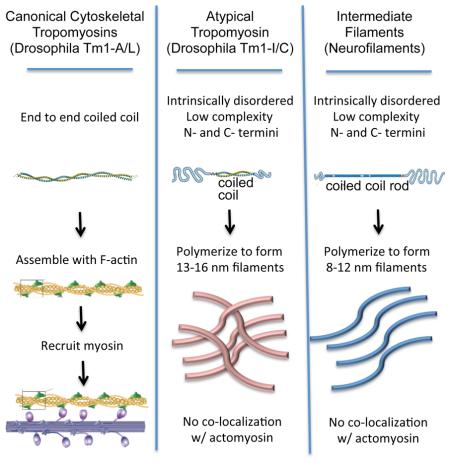
A longstanding mystery has been the absence of cytoplasmic intermediate filaments (IFs) from Drosophila, despite their importance in other organisms. In the course of characterizing the in vivo expression and functions of Drosophila Tropomyosin (Tm) isoforms, we discovered an essential but unusual product of the Tm1 locus, Tm1-I/C, which resembles an IF protein in some respects. Like IFs, Tm1-I/C spontaneously forms filaments in vitro, which are intermediate in diameter between F-actin and microtubules. Like IFs, but unlike canonical Tms, Tm1-I/C contains N- and C-terminal low complexity domains flanking a central coiled coil. In vivo, Tm1-I/C forms cytoplasmic filaments that do not associate with F-actin or canonical Tms. Tm1-I/C is essential for collective border cell migration, in epithelial cells for proper cytoarchitecture, and in the germline for formation of germ plasm. These results suggest that flies have evolved a distinctive type of cytoskeletal filament from Tm.
Graphical abstract
Introduction
The cytoskeleton is composed of three filamentous networks (Huber et al., 2015). Actin and tubulin are globular subunits that require nucleotide triphosphates to polymerize, whereas intermediate filaments (IFs) spontaneously form from proteins composed of a central coiled-coil rod flanked by low complexity domains (Colakoğlu and Brown, 2009; Herrmann et al., 1996; Köster et al., 2015). IFs are so-named because their diameters are intermediate between F-actin (6-8 nm) and microtubules (25 nm) (Fuchs and Cleveland, 1998).
Actins and tubulins are amongst the most highly conserved proteins across animal phyla and are encoded by a handful of genes. In contrast, IF genes are numerous – the human genome codes for >65 – and diverse (Fuchs and Cleveland, 1998). Though divergent in size and sequence, IFs possess a common architecture: central rod domains flanked by N- and C-terminal tails. The rods mediate multimerization whereas the termini are important for end-to- end fibril assembly and side-to-side interactions to form 10 nm filaments (Deek et al., 2013; Guharoy et al., 2013).
The absence of recognizable cytoplasmic IFs in most arthropods, including flies, despite their importance in most other metazoan phyla, is mysterious (Ausmees et al., 2003; Chung et al., 2013; Herrmann and Strelkov, 2011). It has been suggested that other types of proteins might perform crucial functions of IFs (Herrmann and Strelkov, 2011; Mencarelli et al., 2011), however the identities of such proteins if they exist are unknown. Reasonable candidates would be proteins that possess similar domains.
Tropomyosins (Tms) are a large family of coiled coil proteins that regulate actomyosin network structure, stability, and function (Barua et al., 2014; Gunning et al., 2015). Vertebrate genomes encode >40 Tm isoforms. All Tms characterized to date form parallel, coiled coil dimers throughout their length, which polymerize end to end along actin filaments. The Drosophila genome also codes for numerous Tm isoforms, mostly uncharacterized.
While studying the expression and functions of Drosophila Tm1 isoforms, we discovered a Tm1 product with more similarities to IFs than to canonical Tms. Most fly Tm1 gene products, as expected, are end-to-end coiled coil proteins that co-localize with F-actin in vivo. However one product, Tm1-I/C, contains N- and C-terminal intrinsically disordered, low complexity domains. In vitro, the full-length Tm1-I/C protein spontaneously forms 13-16 nm diameter filaments. In vivo, Tm1-I/C forms cytoplasmic filaments that do not co-localize with canonical Tms or F-actin. Loss-of-function studies show that Tm1-I/C is essential in border cells for their collective migration, in epithelial cells for proper cytoarchitecture, and in the germline for formation of germ plasm. These results suggest that flies have evolved an essential and distinctive type of cytoplasmic cytoskeletal filament, intermediate in diameter between F-actin and microtubules.
Results
Expression of canonical and non-canonical Tms
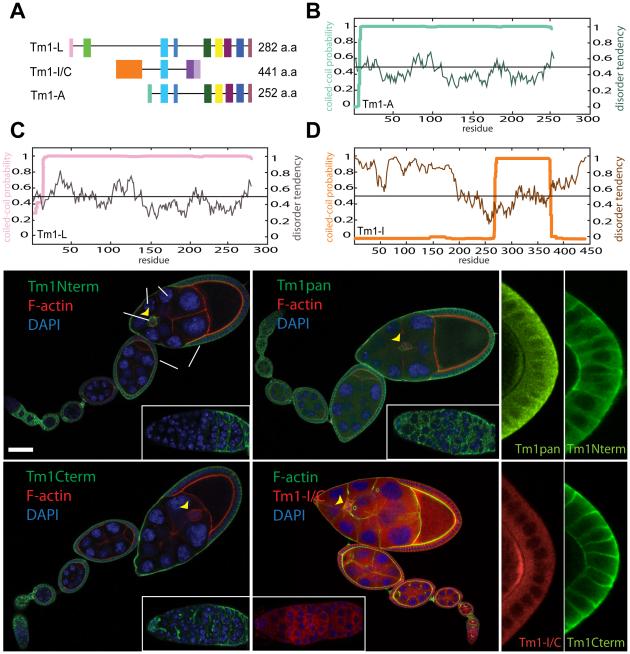
We found expression of three Drosophila Tm1 isoforms (Figure 1A) in the ovary. The fly Tm1 gene annotation predicts 17 mRNA isoforms and 13 distinct polypeptides (Figure S1A), most of which are predicted, as expected for Tms, to form coiled coils from end to end (Figure 1B-1C). However, one set of isoforms possesses unusual N- and C-terminal intrinsically disordered domains (Figure 1D). Expression of only one Tm isoform has previously been documented in vivo (Hanke et al., 1987). Recently three isoforms were studied in S2 cells (Goins and Mullins, 2015; Hsiao et al., 2015), however most remained uncharacterized, and it was unclear whether the atypical isoforms were expressed or required.
Figure 1. Expression of Tm1 isoforms in the Drosophila ovary.
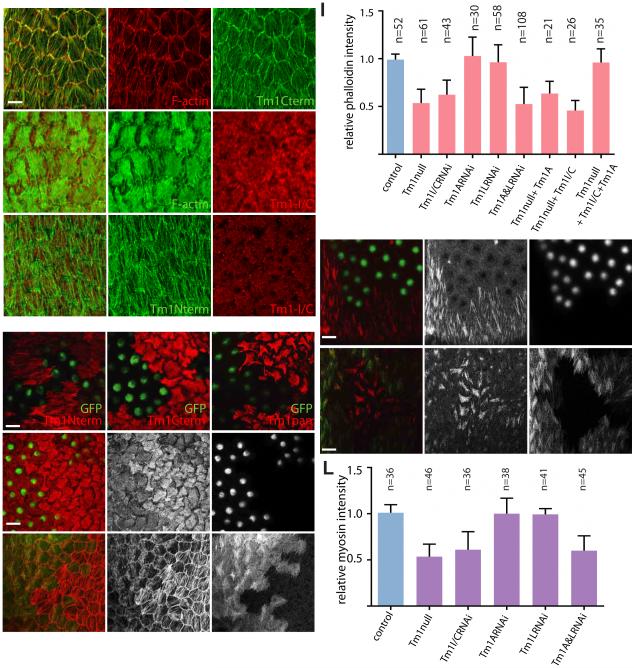
Exon map of the three Tm1 isoforms detected in follicle cells by mRNA tagging (see Figure S1) (A). (B-D) Coiled coil probability (bold lines) and disorder tendency (thin lines) predicted for Tm1-A (B), Tm1-L (C) and Tm1-I/C (D). Tm1 expression (green) in egg chambers of the indicated stages (st.) assessed with antibodies against Tm1-L (Tm1 Nterm) (E), against Tm1-A and –L (Tm1 Cterm) (F), or all isoforms (Tm1pan) (G). Distribution of mCherry-tagged-Tm1-I/C expressed from the endogenous locus (H). Insets show high magnification of germaria. F-actin stained with Alexa568-phalloidin (red) or Alexa488-phalloidin (green), and DNA with DAPI (blue). Tm1-I/C expression in germ plasm at the posterior of the oocyte (arrowheads) stained with Tm1pan antibody (green) (I) and mCherry-Tm1-I/C (red) (J). (K, L) Absence of germ plasm staining with Nterm (K) and Cterm (L) antibodies. Scale bar, 50µm.
Using rtPCR to profile Tm mRNAs in the ovary, many isoforms were detected in total ovary mRNA, while only four were found in follicle cell mRNA. The atypical isoforms Tm1-I and Tm1-C, which encode the same 441 amino acid protein (Figure S1B), were present in both whole ovary and follicle cell mRNA, as were canonical isoforms Tm1-A and Tm1-L (Figure S1C).
To characterize Tm1 developmental expression and subcellular localization, we generated antibodies against full-length Tm1-I/C and Tm1-A proteins, and characterized an antibody that turns out to recognize an N-terminal epitope in Tm1-L (Figure S1D). Tm1-L was present in all somatic follicle cells but not in the germline (Figure 1E). An antibody that recognizes a C-terminal epitope common to canonical isoforms Tm1-A and Tm1-L (Figure S1C), stained follicle cells (Figure 1F) and the germline stem cell niche (cap cells) (Figure 1F inset). An antibody that recognizes all Tm1 isoforms (Tm1pan, Figure S1D) showed both germline and somatic staining (Figure 1G). Since the pan-Tm1 antibody labeled the germline but antibodies that recognize the canonical isoforms did not, the atypical Tm1-I/C protein must be the only one expressed there.
To localize Tm1-I/C specifically, we used a transgenic line in which mCherry was fused in-frame to the N-terminus of endogenous Tm1-I/C (a gift from A. Ephrussi). The tagged protein localized throughout the cytoplasm of germline and somatic cells, including border cells and polar cells (Figure 1H). Tm1-I/C accumulates at the posterior pole of the oocyte (Figure 1I and 1J), where Tm1 is known to promote oskar mRNA localization and pole plasm assembly (Erdélyi et al., 1995), although the specific isoform required was unclear. In contrast, the canonical isoforms were not present in germ plasm (Figure 1K and 1L).
Expression and function of Tm1 isoforms in migratory border cells
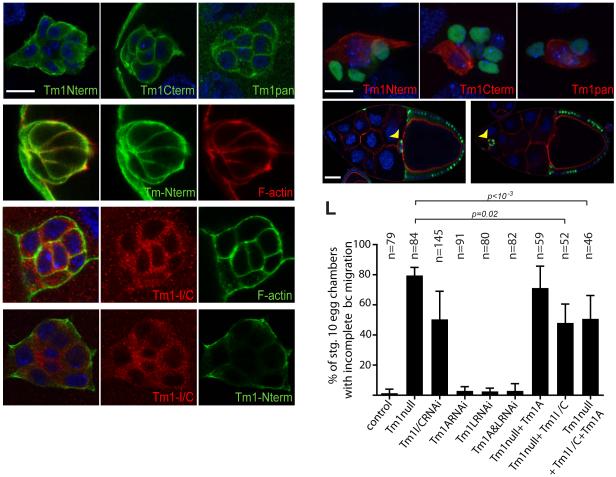
Border cells are follicle cells that migrate collectively as a group of 4-6 motile cells that surround two non-motile cells called polar cells (Montell et al., 2012). Tm1 is required for their motility (Kim et al., 2011), however it was unclear which isoform(s) were expressed or required. All three antibodies labeled migrating border cell clusters (Figures 2A-2C). Tm1-L co-localized with F-actin (Figures 2D-D”), whereas mCherry-Tm1-I/C did not (Figures 2E-E”). Anti-Tm1-L did not stain polar cells (Figure 2A), whereas the other antibodies did (Figures 2B-2C). The mCherry-Tm1-I/C fusion protein was also present in polar cell cytoplasm (Figures 2E). The non-canonical Tm1-I/C protein did not co-localize with F-actin (Figures 2E-E”) or the canonical isoforms (Figures 2F-F”).
Figure 2. Isoform expression and requirements in border cells.
Border cell expression of Tm1-L (A), Tm1-A and –L (B), and all Tm1 (C) isoforms. Tm1-L colocalization with F-actin (red) (D-D”). mCherry-Tm1-I/C protein does not colocalize with F-actin (E-E”) or the canonical isoform (F-F”). (G-I) Mosaic border cell clusters composed of homozygous mutant Tm1 null cells (GFP+, green) and heterozygous cells (GFP-) stained with Tm1-L (Nterm, red) (G), Tm1-A and –L (Cterm, red) (H) and Tm1pan (red) (I) antibodies. Complete border cell migration to the oocyte (arrow) in a wild type stage 10 egg chamber with control clones expressing nuclear GFP (green, arrowhead) (J). Incomplete migration of homozygous Tm1 null mutant border cells (green, arrowhead) (K). Quantification of border cell migration defects for the indicated genotypes (L). Error bars represent s.d. Scale bars in A and G represent 10 μm, in J 50 μm. See also Figure S2.
To probe the functions of the individual isoforms, we generated two null alleles and used isoform specific RNAi and rescue. Homozygous null mutant clones lacked staining for all three antibodies (Figures 2G-2I). The null allele caused border cell migration defects in 80% of fully mutant clusters (Figures 2J-2L). To test the requirement for the individual isoforms we carried out isoform-specific RNAi and rescue. When expressed in border cells with slbo-Gal4, UAS-Tm1-I/C RNAi caused a migration defect nearly as strong as the null mutation (Figure 2L). Knockdown of either canonical isoform or both together, did not cause a defect even though the RNAi was effective (Figures S2A to S2B”). A transgene that re-expressed Tm1-I/C in null mutant border cells provided significant rescue of the null allele whereas Tm1-A did not (Figure 2L). Furthermore, co-expressing Tm1-I/C together with a canonical isoform provided similar rescue as Tm1-I/C alone (Figure 2L). Therefore the unusual isoform I/C is required and the canonical isoforms are dispensable for border cell migration.
Atypical Tm1-I/C forms IF-like filaments in vitro
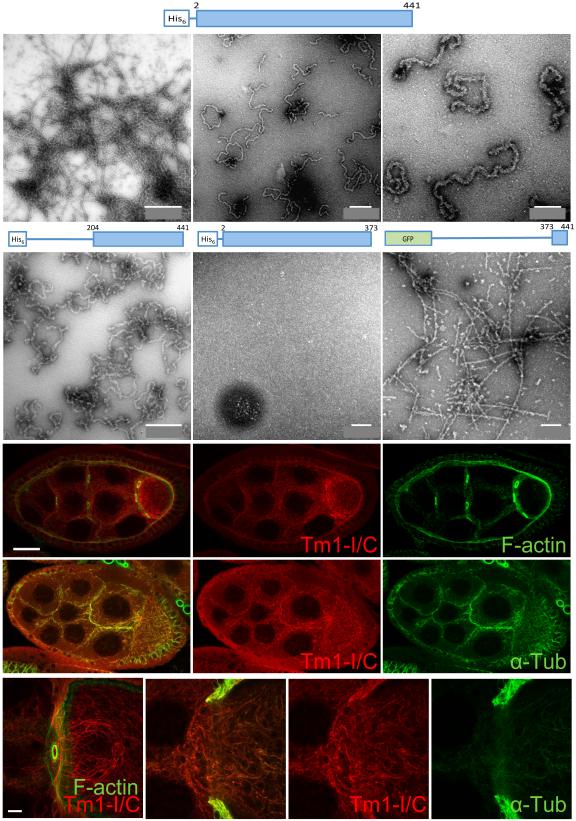
A defining feature of Tms is that they are coiled-coil proteins from end-to-end. By contrast, the Tm1-I/C isoform has a structure composed of a ~100 amino acid coiled coil, flanked by N- and C-terminal intrinsically disordered, low complexity domains (Figure 1D). This architecture is more reminiscent of IFs such as neurofilaments (NFs), which contain central coiled coil domains flanked by intrinsically disordered, low complexity sequences (Deek et al., 2013; Guharoy et al., 2013). To test whether Tm1-I/C forms filaments, we expressed full length Tm1-I/C, as well as N- or C-terminally truncated forms (Figure S3A). Strikingly full length Tm1-I/C formed filaments when dialyzed against the buffer reported for NF formation (Heins et al., 1993)(Figure 3A-C). The filaments were 13-16 nm in diameter, intermediate between F-actin (6-8 nm) and microtutbules (25 nm) and slightly larger than IFs (8-12 nm). The filaments were shorter and curlier than many IFs, however NFs produce short curly filaments like the Tm1-I/C filaments under some conditions (Heins et al., 1993). Deletion of the N-terminal domain had little effect (Figure 3D), whereas deletion of the C-terminus prevented filament formation (Figure 3E), indicating that the C-terminal low complexity domain is crucial for Tm1-I/C filament formation.
Figure 3. In vitro filament formation of atypical Tm1-I/C.
(A-F) Electron microraphs of fibers formed by Tm1-I/C. Three different magnifications of filaments formed by the full-length protein (A-C), the headless protein (D), the tailless protein (E) or the Tm1-I/C C-terminus fused to GFP (F). (G-J”) Confocal images of mCherry-Tm1-I/C filaments. (G-G”) mCherry-Tm1-I/C (red) filament network in stage 7 egg chambers does not overlap with F-actin (green). (H-H”) mCherry-Tm1-I/C (red) filament network co-localizes with tubulin in the germline. (I) High magnification of mCherry-Tm1-I/C (red) filament network and F-actin (green) in germ cells. (J-J”) High magnification of mCherry-Tm1-I/C (red) filament network and tubulin (green) in germ cells. Scale bar in G represents 50 μm, 5 μm in I. See also Figure S3.
The C-terminal low complexity domain polymerizes to form fibers and hydrogels
We then purified the N- and C-terminal Tm1-I/C domains fused to either GFP or mCherry (Figure 3F and Figure S3B), and tested their ability to polymerize on their own. The C-terminal domain, but none of the three N-terminal fragments tested, polymerized into fibers (Figure 3F). When concentrated, these fibers formed hydrogels (Figure S3C) as do some RNA binding proteins (Han et al., 2012; Kato et al., 2012) and IFs (Deek et al., 2013). Prion-like proteins, such as yeast Sup35, form insoluble amyloid-like fibers that cannot dissociate even in sodium dodecyl sulfate (SDS) (Figure S3D). In contrast, Tm1-I/C C-terminal domain fibers were soluble (Figure S3E). The fibers formed by the Tm1-I/C tail, like those formed by RNA binding proteins such as Fused in Sarcoma (FUS), showed the typical X-ray diffraction pattern of a cross-β structure (Figure S3F). These data suggest the Tm1-I/C tail makes dynamic fibers that could facilitate polymerization of the full-length protein.
Tm1-I/C filaments in vivo
To look for Tm1-I/C filaments in vivo, we assessed a variety of buffer and fixation conditions. When fixed at pH6.8, the mCherry-Tm1-I/C fusion protein appeared filamentous in germ cells (Figure 3G-J”). These filaments did not co-localize with F-actin (Figure 3G-G” and 3I) but aligned with microtubules (Figure 3H-H” and 3J-J”). In follicle cells, mCherry-Tm1-I/C was also cytoplasmic but less obviously filamentous.
Canonical Drosophila Tms recruit myosin to F-actin to form stress fibers
In epithelial follicle cells, the canonical Tms co-localized with cortical F-actin and in basal stress fibers (Figures 4A-A” and S4), whereas the mCherry-tagged Tm1-I/C fusion protein did not co-localize with F-actin (Figures 4B-B”) or with the canonical Tms (Figures 4C-C”).
Figure 4. Isoform expression and requirements for stress fiber formation.
Canonical Tm1 localization in stress fibers near the basal surfaces of the follicle cells (A-A”). See also Figure S4. mCherry-Tm1-I/C expression does not colocalize with F-actin (B-B”) or with the canonical isoforms –A and -L (C-C”). Tm1 null epithelial follicle cell clones (green) stained with Tm1-L (Nterm) (D), Tm1-A and –L (Cterm) (E) and Tm1pan antibodies (F). Reduced phalloidin staining of stress fibers in Tm1 null epithelial follicle cells (G-G”). Reduced stress fiber phalloidin staining following double knockdown of Tm1-A and Tm1-L isoforms by RNAi (H-H”). Quantification of stress fiber phalloidin staining (I). Error bars show S.D. Tm1 null epithelial follicle cells show reduced myosin recruitment to stress fibers, detected by sqh-mCherry level (J-J”). Reduced myosin accumulation in Tm1-A and Tm1-L double RNAi knockdown cells (K-K”). Quantification of myosin accumulation (L). Scale bars represent 10 μm.
Many mammalian Tms recruit myosin to F-actin to form contractile stress fibers (Barua et al., 2014; Tojkander et al., 2011). To test the functions of Tm1s in stress fibers, we made null mutant clones in the epithelium, which eliminated labeling with all three antibodies (Figures 4D-4F). Phalloidin labeling of F-actin stress fibers was significantly reduced in clones of homozygous null cells compared to neighboring wild-type cells (Figures 4G-G” and 4I), whereas control clones had no effect (Figure S4). RNAi knockdown of either A or L alone did not affect stress fibers detectably (Figure S4), but simultaneous knockdown of both caused a phenotype similar to the null mutant (Figure 4H-H” and I). Therefore Tm1-A and –L function redundantly to promote basal stress fiber formation.
One function of stress fibers in the follicular epithelium is to serve as a substrate for pulsatile myosin contractions (He et al., 2010; Koride et al., 2014). Myosin accumulation on stress fibers was reduced in Tm1 null clones (Figures 4J-J” and 4L) and following simultaneous knockdown of Tm1-A and –L by RNAi (Figure 4K-K” and 4L). The myosin contractions constrain the growth of the egg chamber to the two poles and thus elongate egg chambers, and thus the eggs that they form (He et al., 2010; Horne-Badovinac and Bilder, 2005). We found that 30-40% of eggs laid by females bearing Tm1 null clones or clones expressing A and L RNAi were round (Figure S5). Therefore canonical isoforms A and L function redundantly and serve typical Tm roles in the assembly and function of contractile actomyosin stress fibers in the follicle cell epithelium.
Non-canonical Tm1 organizes the cytoplasm of germ cells and follicle cells
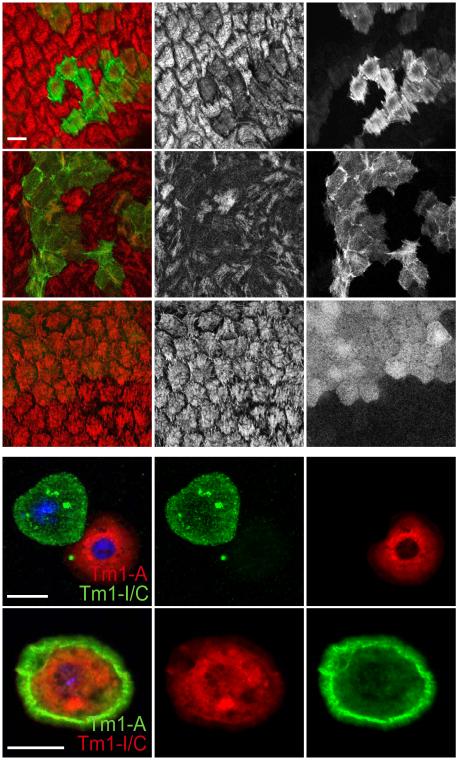
To determine if Tm1-I/C is required in epithelial follicle cells, we knocked it down by RNAi. No defect in overall apical/basal polarity was observed. Surprisingly, although the protein is not present in stress fibers, Tm1-I/C RNAi caused as strong a reduction in stress fiber intensity (Figures 4I and 5A-A”) and myosin accumulation (Figures 4L and 5B-B”) as the null or the double knockdown of A and L. Mosaic females also produced round eggs (Figure S5). No individual Tm1 isoform was sufficient to rescue the Tm1 null phenotype (Figure 4I), however co-expression of isoforms I/C and A together did rescue (Figures 4I and 5C-C”). Thus, epithelial follicle cells require cytoplasmic Tm1-I/C in combination with at least one canonical isoform for basal stress fibers to develop.
Figure 5. Atypical Tm1-I/C required for stress fibers and localization of Tm1-A.
Tm1-I/C RNAi results in reduced stress fiber phalloidin staining (A-A”) and myosin accumulation (B-B”). Rescue of phalloidin staining in Tm1 null follicle cells by re-expression of Tm1-I/C and Tm1-A (C-C”). Tm1-A and Tm1-I/C localize to the cytoplasm of S2 cells when expressed individually (D-D”). Co-expression of Tm1-A and Tm1-I/C results in relocalization of Tm1-A to the periphery of the S2 cell (E-E”). Scale bars represent 10 μm. See also Figure S5.
One hypothesis is that cytoplasmic Tm1-I/C is necessary to localize canonical Tms to the appropriate part of the cell where stress fibers form, as has been described for vimentin (Jiu et al., 2015). To test the effect of Tm1-I/C on the subcellular localization of Tm1-A, we expressed them in S2 cells. When expressed individually Tm1-A or Tm1-I/C each localized throughout the cytoplasm (Figures 5D-D”). However co-expression of both isoforms caused a relocalization of Tm1-A to the cell periphery (Figures 5E-E”). Thus cytosolic Tm1-I can alter the subcellular localization of Tm1-A.
Discussion
Despite the prevalence of IFs in metazoans, they are lacking from the largest phylum, the arthropods, with one known exception (Mencarelli et al., 2011). It has been suggested that some of the mechanical support provided by IFs, such as the protection from skin blistering by epidermal keratins, might be unnecessary in animals with an exoskeleton, whereas other IF functions might have been subsumed by distinct proteins (Herrmann and Strelkov, 2011). The nature of the proteins that might form cytoplasmic filaments and perform IF-like functions however, has been mysterious. In retrospect, evolution of a filament forming protein with IF-like properties from a coiled coil, cytoplasmic, cytoskeletal protein like Tm seems reasonable.
Keratin IFs, Tms, and myosins were the earliest recognized alpha helical, coiled coil proteins (Herrmann and Strelkov, 2011). IFs and Tms have some intriguing similarities and some clear differences. In contrast to actin and tubulin, which must bind nucleotide triphosphates to polymerize, Tms and IFs polymerize without this energy source. Tms are end-to-end coiled coil proteins that polymerize head to tail in a highly cooperative manner with actin filaments, and influence myosin recruitment and activity. IFs, like Tms, form parallel, coiled coil dimers and polymerize into filaments. Unlike Tms, which require N-terminal acetylation to polymerize, IFs possess low complexity head and tail domains of highly variable size and sequence at their N- and C-termini, which mediate polymerization. Unlike Tms, IF protofibrils assemble into 8-12 nm fibers, which can further form higher-order liquid crystals and hydrogels. Overall IFs have arguably more in common with Tms than with actin and tubulin.
Canonical structures, localizations, and redundant functions of Tm1-A and Tm1-L
Tm1-A and Tm1-L exhibit the expected end-to-end coiled coil structure. In addition, they co-localize with F-actin within follicle cells, both at the cortex and in basal stress fibers, making it likely that they bind F-actin directly. Consistent with that idea, Tm1-A and Tm1-L function canonically in these cells to promote myosin recruitment and stress fiber assembly and function. Given their extensively overlapping sequences, their redundancy is not surprising.
There were however two unexpected findings. First due to the dependence of border cell migration on the F-actin cytoskeleton, we anticipated that canonical Tm would be required, yet both Tm1-A and –L proved dispensable. There is a second Tm gene so it is possible that a canonical Tm expressed from Tm2 is necessary for border cell migration. Alternatively it may be that Tms are more important for stabilizing actomyosin assemblies and that the dynamics required for border cell migration do not require canonical Tms. Functional analysis of Tm2 will be required to distinguish between these possibilities. In contrast to Tm1-A and -L, we detected no co-localization of Tm1-I/C with F-actin or with the canonical isoforms. The highly divergent structure and localization of Tm1-I/C calls into question its identity as a Tm at all.
Similarities between Tm1-I/C and IFs
Tm1-I/C resembles an IF in that it assembles into filaments that are intermediate in diameter between F-actin and microtubules in the absence of cofactors. Tm1-I/C, like cytoplasmic IFs, is present throughout the cytoplasm and does not co-localize with F-actin. In germ cells mCherry-Tm1-I/C filaments are evident, which closely associate with microtubules, similar to vimentin and NFs (Huber et al., 2015; Liao and Gundersen; Yuan et al., 2012).
Tm1-I/C contains a central coiled coil flanked by N- and C-terminal low complexity sequences, the shorter one of which is essential for filament formation. The 60-amino-acid C-terminus of Tm1-I/C may serve a function similar to the typical N-terminal “head” domain of IFs. These domains are shorter than the IF C-terminal tails and mediate end-to-end associations into protofibrils. The long N-terminus of Tm1-I/C is perhaps analogous, though not homologous, to the C-terminal tails of IFs. It is a 260-residue intrinsically disordered, low complexity domain enriched in serine (16%). Although Tm1-I/C shares a single exon with the canonical Tm isoforms, Tm1-I/C has a domain architecture, filament forming ability, and subcellular localization more typical of an IF than a Tm.
Differences between Tm1-I/C and IFs
Tm1-I/C nevertheless lacks some of the conserved features of the IF protein family as currently defined. Although the primary amino acid sequences of IFs are not highly conserved, there are common structural features. All IF proteins have a ~300 amino acid rod domain that is interrupted by flexible linkers, which divide it into three separate coiled coils. In addition, although the sequences are highly variable, there are a few consensus motifs (Herrmann and Strelkov, 2011). In contrast, Tm1-I/C contains a single unbroken coiled coil domain of ~100 amino acids and lacks the consensus motifs.
The fibers formed by the Tm1-I/C C-terminal domain alone possess a cross-β structure that is somewhat similar to beta amyloid (Kato et al., 2012), although pathogenic fibers are locked in an irreversible state. Although this property initially seemed like a difference between Tm1-I/C and IFs, recent work demonstrates that the head domains of IFs also form cross-β filaments (Lin et al., 2016). This feature is thus an additional similarity between the IF head and the Tm1-I/C tail.
Relationship between the in vivo functions of Tm1-I/C and IFs
Tm1-I/C promotes collective border cell migration whereas the canonical Tms are dispensable. Although canonical Tms have been extensively implicated in cell migration, this has been true primarily for individual cells, migrating on two-dimensional (2D) substrates coated with extracellular matrix (ECM) proteins in vitro, and thus containing obvious stress fibers. In contrast, border cells migrate collectively, in between other cells, and in a compliant 3-dinmensional (3D) environment without obvious stress fibers. IFs promote cell migration in a variety of contexts (Chung et al., 2013), and intriguingly the intermediate filament K14 was recently established as a critical factor in collective breast cancer migration in 3D (Cheung et al., 2013).
Tm1-I/C is also required in vivo for proper epithelial follicle cell cytoarchitecture, specifically establishment of basal F-actin stress fibers. Though most studies of the eukaryotic cytoskeleton treat the three filamentous networks as independent entities, crosstalk amongst them clearly occurs and is under active investigation (Chang and Goldman, 2004; Huber et al., 2015). Specifically multiple studies show interactions of vimentin with focal adhesions and stress fibers. For example, association of vimentin with focal adhesions stabilizes them (Burgstaller et al., 2010). Mutual dependence of vimentin and nestin IFs and F-actin stress fibers also occurs in osteosarcoma cells (Jiu et al., 2015).
A further function of Tm1 is to localize oskar mRNA to the posterior oocyte pole, which is essential for germ plasm formation. Intrinsically disordered domains are common in RNA and DNA binding proteins. The precise role of Tm1-I/C in oskar mRNA localization and germ plasm assembly is not known, however IFs have long been implicated in asymmetrically localizing Vg1 mRNA in Xenopus oocytes (Pondel and King, 1988). The observation that Tm1-I/C is required in multiple cell types for disparate functions raises the possibility that it is a multifunctional protein. Differences in post-translational modifications and binding partners may result in distinct structures and functions in germline versus somatic cells. The structural flexibility of intrinsically disordered protein domains might enable distinct functions in different cell types. Alternatively Tm1-I/C may function to localize mRNAs, and thus support functional polarization, of migrating border cells, epithelial follicle cells, and the oocyte.
The results presented here suggest that the Tm1 locus produces both canonical Tms such as Tm1-A and Tm1-L, as well as an atypical protein Tm1-I/C that can polymerize and form a cytosplasmic filament network. Tm1-I/C possesses some properties in common with IFs. This could be an example of convergent evolution of proteins that can form filaments, by combining coiled coil and low complexity, filament forming modules. Perhaps the Tm1-I/C protein allows flies to survive in the absence of traditional cytoplasmic IF proteins. An alternative, which is not mutually exclusive, is that Tm1-I/C modulates the interaction between microtubules and microtubule motors, analogous to the relationship between canonical Tms and myosin. Tm1-I/C is widely expressed, and thus may be the sole cytoskeletal, filament-forming protein other than actin and tubulin in Drosophila; however it’s also possible that there are other, as-yet-unrecognized proteins that form filaments. Tm1-I/C provides a search motif for such proteins in arthropods. The combination of one or more coiled coil domains flanked by low complexity domains of intrinsic structural disorder may predict filament-forming potential.
Experimental Procedures
Clonal analyses
Tm1null alleles generated by homologous recombination and CRISPR as described in Supplemental Experimental Procedures were recombined with FRT82B (BL# 2050). Hsp70-FLP,tub-Gal4,UAS-GFP-nls; FRT82B, tub-Gal80 flies were crossed with FRT82B, Tm1null/TM3 to generate GFP-positive, homozygous mutant clones. Heat shock was performed either using 3rd instar larvae, to generate large clones, or 2- to 3-day old female adults. When larvae were used, the vials were kept at 25°C until the adult flies enclosed. Then theywere transferred to a new vial with yeast paste and kept at 29°C overnight before dissection. Larvae were heat shocked by incubating at 37°C for two hours. Adults were treated with either three heat shocks per day for two days or two heat shocks per day for three days at 37°C. The heat shocked flies were kept at 25°C and dissected 5~7 days after the first heat shock. For RNAi knockdown in the border cell cluster, slbo-Gal4 or C306-Gal4 drivers were used. To generate clusters composed of both mutant and wild type cells, hsp70-FLP; Ay17bGal4, UAS-moesin-GFP was used to generate Flp-out clones in the epithelial follicle cells or the border cells. Heat shock was performed with 2-day old adult females, twice a day for two days and the flies were kept at 29°C until dissection 2~3 days later. mCherry tagged sqh protein under control of endogenous sqh promoter (sqh::sqh-mcherry, BL#59024) was recombined with the RNAi lines and the Tm1null line to detect myosin accumulation in the epithelial follicle cells.
mRNA tagging method to isolate follicle cell specific mRNA
The mRNA tagging method was adapted from (Yang et al., 2005). P[UAS-hPABP-FLAG] (BL#9419) was crossed to Slbo-Gal4/CyO or c306-Gal4 to drive FLAG tagged poly-A binding protein expression in subsets of follicle cells. 400 ovaries were dissected and fixed with 1 ml of PBS containing 1% formaldehyde and 0.5% NP-40 for 30 minutes at 4°C. After fixation, 140µl of 2M glycine was added for 5 minutes. The ovaries were washed three times with 1x PBS and then in 0.8ml of homogenization buffer (HB : 150 mM NaCl, 50 mM HEPES buffer pH 7.6, 1 mM EGTA, 15 mM EDTA, 10% glycerol). Immediately before homogenization, 50U/ml SUPERase-In (Ambion) was added and 1 tablet of Protease Inhibitor Cocktail (Roche) was added per 10ml of HB. The ovaries were sonicated for 1min using 30% intensity (Fisher Sonic Dismembrator, Model 500). The supernatant was collected after centrifuging for 10 minutes at 13,000 ×g and added to 100µl of Anti-FLAG-M2 affinity agarose beads (Sigma-Aldrich) equilibrated with HB. The supernatant and bead mix were gentle rotated at 4°C for 2 hours. The beads were washed with HB four times and the mRNA was eluted by incubating the beads with the elute buffer (50mM Tris-HCl pH 7.0, 10mM EDTA, 1.3% SDS, 50U/ml SUPERase-IN) at 65°C for 30 minutes. mRNA from the eluate was isolated using the Trizol protocol (Ambion). The isolated mRNA was reverse transcribed to cDNA using Superscript III following the manufacturer’s protocol (Invitrogen).
Immunohistochemistry and Imaging
Ovaries were dissected in Schneider’s medium (GIBCO) with 10% FBS (Sigma) and fixed for 15 minutes in 4% paraformaldehyde (Prasad and Montell, 2007). After three 15 minute washes with PBT (1x PBS, 0.1% Triton x-100), egg chambers were blocked with PBT block (1x PBT, 5% goat serum) for 2 hours at room temperature or overnight at 4°C. Primary antibodies were incubated overnight at 4°C followed by secondary antibody incubation at room temperature for 1 hour. Primary antibodies were used at the following dilutions: anti-Tm1Nterm antibody (1:5000,mac141, abcam), anti-Tm1Cterm (1:1000), anti-Tm1pan (1:1000). Alexa Fluor conjugated goat anti-rabbit or anti-rat IgG antibodies were used as secondary antibodies (Molecular Probes). Alexa phalloidins, 488 or 568 (1:200) were used to stain the actin filaments. DAPI (1:10000) was used to mark nuclei. After three 15 minute washes, the samples were mounted with Vectashield (Vector Laboratories). A Zeiss 780 confocal was used for imaging.
In order to preserve and image mCherry-Tm1-I/C filaments, egg chambers were first incubated in 80mM PIPES buffer (pH 6.8) containing 0.1% Triton X-100, 1mM EGTA and1mM MgCl2 for 30 minutes to 1 hour. The samples were fixed for 15 minutes with 4% paraformaldehyde, followed by the same washing and blocking procedure stated above. Rabbit mCherry antibody (novus, 1:1000) was used to visualize the mCherry-Tm1-I/C filaments. For microtubule staining, the egg chambers were fixed with methanol at −20°C for 15 minutes after the PIPES buffer incubation. The fixed samples were rehydrated in PBT for 4 hours at RT or 4°C followed by 1hr block with PBT containing 2% goat serum. Rabbit FITC-α-Tubulin antibody was used to visualize microtubules in the egg chamber.
S2 expression of Tropomyosin isoforms
S2 cells were plated into 6 well plates with a coverslip in each well. HA-Tm1-I/C and V5/His-Tm1-A under UAS promoter along with actin-Gal4 were transfected into S2 cells using Effectene (Qiagen). The cells were incubated at room temperature for 48 hours before further processing. Briefly, 4% paraformaldehyde in PBS was used to fix for 10 minutes. After three washes using PBST (1x PBS, 0.1% Triton-X100), the cells were incubated with mouse anti-HA antibody (1:1000) and rabbit anti-V5 antibody. Secondary antibodies conjugated with Alexa-568 (mouse) and Alexa-488 (rat) were diluted 1:400. Vectashield was used to mount coverslips on slides for imaging.
Image Analysis
For quantification of phalloidin staining and myosin accumulation, WT and mutant cells that were immediately adjacent were selected and the ratio (mutant/WT) was calculated using ImageJ. At least 4 independent experiments were analyzed for each genotype. Graphs were generated with Graphpad Prism, using mean ± standard deviation. Statistical tests were done using unpaired t-test.
Recombinant protein expression and purification
DNA fragments encoding the N-terminal LC domains (N1: 2-291, N2: 2-260, N3: 2-214), C-terminal LC domain (C: 373-441), full length (FL: 2-441), headless (HL: 204-441) or tailless (TL: 2-373) of Tm1-I/C were amplified by PCR using a parental plasmid encoding a full-length Tm1-I/C as a template. The DNA fragments of FL, HL or TL were inserted into the multiple cloning sites of the pHis-parallel vector (Sheffield et al., 1999), and the fragments of all the LC domains were sub-cloned into the pHis-parallel-GFP or pHis-parallel-mCherry plasmid (Kato et al., 2012). The resulting vectors were confirmed by DNA sequencing. All proteins were overexpressed in E. Coli BL21(DE3) cells with 0.5 mM IPTG at 20 °C for overnight. Harvested cells for the GFP- or mCherry-fusion proteins were lysed with 0.4 mg/mL lysozme in a lysis buffer containing 50 mM Tris-HCl pH7.5, 500 mM NaCl, 20 mM BME, 1% Triton X-100 and protein inhibitor cocktail (Sigma-Aldrich, USA) for 30 min on ice, and then sonicated. The cell lysates were centrifuged at 35,000 rpm for 1 hour. The supernatants were mixed with Ni-NTA resin (Qiagen, USA) for 30 min at 4°C. The Ni-NTA resin was packed in a glass column and washed with a buffer containing 20 mM Tris-HCl pH7.5, 500 mM NaCl, 20 mM imidazole, 20 mM BME, and 0.1 mM PMSF. The bound proteins were eluted from the resin with a buffer containing 20 mM Tris-HCl pH7.5, 500 mM NaCl, 250 mM imidazole, 20 mM BME, and 0.1 mM PMSF. EDTA was added to the elutions at a final concentration of 0.5mM. Urea was further added to the mCherry fusion protein at a final concentration of 2 M. The purified proteins were concentrated with Amicon Ultra centrifugal filters (Millipore, USA). Glycerol was added to the GFP fusion proteins at the final concentration of 50%. The protein solutions were kept at −20°C. The mCherry fusion protein was kept at −80°C. The FL, HL and TL proteins were purified in the same way mentioned above except that all the buffers contain 2 M guanidine hydrochloride. The purities of the purified proteins were checked by SDS-PAGE (Figure 4A), and the concentrations were determined by absorbance at UV280.
Formation of cross-β fibers and a hydrogel of Tm1-I/C LC domains
The purified GFP fused LC domains (N1, N2, N3 and C) were dialyzed against a gelation buffer containing 20 mM Tris-HCl pH7.5, 200 mM NaCl, 20 mM BME, 0.5 mM EDTA and 0.1 mM PMSF for overnight. The protein solutions were concentrated to ~ 100 mg/ml, and then incubated at 4°C for a couple of days. To inspect fiber formation, the protein solution was directly deposited on a surface of a TEM grid (CF-400-Cu from Electron Microscopy Sciences, USA). The surface of the grid was washed three times with 10 μl of distilled water to remove excess of hydrogel material. The grid was subsequently stained for a few seconds with a 5μl drop of 2% uranyl acetate. After the uranyl solution was blotted, the grid was dried in air. TEM images were obtained with a JEOL 1200EX electron microscope at 120 kV. The fiber solution of GFP-Tm1-I/C C-terminal LC domain prepared as above was diluted 100-fold with the gelation buffer, and sonicated to make short fiber seeds. mCherry-Tm1-I/C C-terminal LC domain was dialyzed against the gelation buffer for overnight. The protein solution was centrifuged to remove precipitates. To the supernatant, the GFP seeds prepared above were added at a ratio of 1/1000. This protein solution was concentrated to the final protein concentration of over 200 mg/ml, and then filled into 2-cm silicon tubes (3.6 mm diameter) followed by sealing the tube ends with parafilm. The tubes were incubated at 4 °C for overnight. The formed hydrogel was carefully squeezed out from the tubes for photography.
SDD-AGE and X-ray diffraction of cross-β fibers of Tm1-I/C C-terminal LC domain
The stability of cross-β fibers of Tm1-I/C C-terminal low complexity domain was examined by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE). A hydrogel of mCherry-fused Tm1-I/C C-terminal LC domain was resuspended in the gelation buffer and sonicated briefly to make fibers short. The short fibers were incubated in the gelation buffer containing indicated concentrations of SDS (0 – 2%) at 37 °C for 20 min. As a control, amyloid fibers of yeast Sup35NM protein were treated in the same way. The reaction mixtures were loaded on 1.5% agarose gel to separate fibers and monomers. The agarose gel was scanned by a fluorescent imager (Typhoon 9200, GE) to visualize mCherry-fusion proteins. Subsequently, proteins were transferred on a cellulose membrane from the agarose gel and analyzed by western blot with a His-tag antibody to visualize Sup35NM bands as described before (Kato et al., 2012). For fiber X-ray diffraction, the hydrogel was resuspended in milli-Q water and dialyzed overnight in 2 L milli-Q water twice. The dialyzed sample was lyophilized and then exposed to an X-ray beam to obtain cross-β diffraction as described before (Kato et al., 2012).
Formation of intermediate filaments of recombinant proteins
The FL, HL and TL proteins of Tm1-I/C (1 mg/ml) were denatured in 6 M guanidine hydrochloride and then dialyzed in a filament formation buffer containing 50 mM MES pH 6.25, 170 mM NaCl and 1 mM DTT at 37 °C for over night. The dialyzed proteins were directly deposited on EM grids as described above, and then visually inspected by electron microscopy.
Supplementary Material
HIGHLIGHTS and ETOC BLURB.
(a) HIGHLIGHTS
1. Flies express Tm1-I/C, an atypical Tropomyosin that forms 13-16 nm filaments in vitro
85 characters including spaces
2. Canonical isoforms Tm1-A and –L redundantly recruit myosin to stress fibers in vivo
83 characters including spaces
3. Tm1-A and L localize with actin in vivo but Tm1-I/C filaments localize with microtubules
88 characters including spaces
4. Tm1-I/C is essential in migratory border cells, germ cells, and epithelial follicle cells
89 characters including spaces
(b) ETOC BLURB
Most arthropods lack cytoplasmic intermediate filaments (IFs). While characterizing Drosophila Tropomyosins, Cho et al find an essential but unusual isoform that resembles IFs: it contains a central coiled coil flanked by low complexity domains, forms filaments in vitro and in vivo, and does not co-localize with F-actin or canonical Tropomyosins.
Acknowledgements
This work was supported by NIH R01GM73164 to DJM. We thank Steven L McKnight for inspiration, for coordinating the collaboration, and for insightful comments on the manuscript. SLM is supported by NIH grant U01GM10762301 and unrestricted funding from an anonymous donor. We thank Imre Gaspar and Anne Ephrussi for sharing observations and reagents prior to publication, especially the mCherry-Tm1-I/C knock-in. We thank Micha Kornreich for his insights into NF structure and biochemistry and for analyzing the Tm1-I/C sequence.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J. H. Kim contributed Figure 5D-E”. M. Kato contributed the data shown in Fig. 3A-F and Fig. S3, experimental design, interpretation of the data and writing. A. Cho did all of the other work presented in this paper with technical assistance from T. Whitwam. D.J. Montell conceived of and coordinated the project, advised J.H.K, A.C., and T. W., and wrote the manuscript.
References
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochemistry. 2014;53:4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller G, Gregor M, Winter L, Wiche G. Keeping the vimentin network under control: cell-matrix adhesion-associated plectin 1f affects cell shape and polarity of fibroblasts. Mol Biol Cell. 2010;21:3362–3375. doi: 10.1091/mbc.E10-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BM, Rotty JD, Coulombe PA. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol. 2013;25:600–612. doi: 10.1016/j.ceb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colakoğlu G, Brown A. Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. J Cell Biol. 2009;185:769–777. doi: 10.1083/jcb.200809166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deek J, Chung PJ, Kayser J, Bausch AR, Safinya CR. Neurofilament sidearms modulate parallel and crossed-filament orientations inducing nematic to isotropic and re-entrant birefringent hydrogels. Nat Commun. 2013;4:2224. doi: 10.1038/ncomms3224. [DOI] [PubMed] [Google Scholar]
- Erdélyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Cleveland DW. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Goins LM, Mullins RD. A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila. Mol Biol Cell. 2015;26:2491–2504. doi: 10.1091/mbc.E14-12-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guharoy M, Szabo B, Contreras Martos S, Kosol S, Tompa P. Intrinsic structural disorder in cytoskeletal proteins. Cytoskeleton (Hoboken) 2013;70:550–571. doi: 10.1002/cm.21118. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci. 2015;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hanke PD, Lepinske HM, Storti RV. Characterization of a Drosophila cDNA clone that encodes a 252-amino acid non-muscle tropomyosin isoform. J Biol Chem. 1987;262:17370–17373. [PubMed] [Google Scholar]
- He L, Wang X, Tang HL, Montell DJ. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol. 2010;12:1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins S, Wong PC, Müller S, Goldie K, Cleveland DW, Aebi U. The rod domain of NF-L determines neurofilament architecture, whereas the end domains specify filament assembly and network formation. J Cell Biol. 1993;123:1517–1533. doi: 10.1083/jcb.123.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV. History and phylogeny of intermediate filaments: now in insects. BMC Biol. 2011;9:16. doi: 10.1186/1741-7007-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Häner M, Brettel M, Müller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Hsiao JY, Goins LM, Petek NA, Mullins RD. Arp2/3 Complex and Cofilin Modulate Binding of Tropomyosin to Branched Actin Networks. Curr Biol. 2015;25:1573–1582. doi: 10.1016/j.cub.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber F, Boire A, López MP, Koenderink GH. Cytoskeletal crosstalk: when three different personalities team up. Curr Opin Cell Biol. 2015;32:39–47. doi: 10.1016/j.ceb.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Jiu Y, Lehtimäki J, Tojkander S, Cheng F, Jäälinoja H, Liu X, Varjosalo M, Eriksson JE, Lappalainen P. Bidirectional Interplay between Vimentin Intermediate Filaments and Contractile Actin Stress Fibers. Cell Rep. 2015;11:1511–1518. doi: 10.1016/j.celrep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cho A, Yin H, Schafer DA, Mouneimne G, Simpson KJ, Nguyen KV, Brugge JS, Montell DJ. Psidin, a conserved protein that regulates protrusion dynamics and cell migration. Genes Dev. 2011;25:730–741. doi: 10.1101/gad.2028611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koride S, He L, Xiong LP, Lan G, Montell DJ, Sun SX. Mechanochemical regulation of oscillatory follicle cell dynamics in the developing Drosophila egg chamber. Mol Biol Cell. 2014;25:3709–3716. doi: 10.1091/mbc.E14-04-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. Intermediate filament mechanics in vitro and in the cell: from coiled coils to filaments, fibers and networks. Curr Opin Cell Biol. 2015;32:82–91. doi: 10.1016/j.ceb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Gundersen G. Kinesin Is a Candidate for Cross-bridging Microtubules and Intermediate Filaments. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016 doi: 10.1016/j.cell.2016.10.003. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C, Ciolfi S, Caroti D, Lupetti P, Dallai R. Isomin: a novel cytoplasmic intermediate filament protein from an arthropod species. BMC Biol. 2011;9:17. doi: 10.1186/1741-7007-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondel MD, King ML. Localized maternal mRNA related to transforming growth factor beta mRNA is concentrated in a cytokeratin-enriched fraction from Xenopus oocytes. Proc Natl Acad Sci U S A. 1988;85:7612–7616. doi: 10.1073/pnas.85.20.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Yang Z, Edenberg HJ, Davis RL. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments at a glance. J Cell Sci. 2012;125:3257–3263. doi: 10.1242/jcs.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.