Abstract
Glioblastoma multiforme (GBM) is the most common and lethal cancer of the adult brain, remaining incurable with a median survival time of only 15 months. In an effort to identify new targets for GBM diagnostics and therapeutics, recent studies have focused on molecular phenotyping of GBM subtypes. This has resulted in mounting interest in microRNAs (miRNAs) due to their regulatory capacities in both normal development and in pathological conditions such as cancer. miRNAs have a wide range of targets, allowing them to modulate many pathways critical to cancer progression, including proliferation, cell death, metastasis, angiogenesis, and drug resistance. This review explores our current understanding of miRNAs that are differentially modulated and pathologically involved in GBM as well as the current state of miRNA‐based therapeutics. As the role of miRNAs in GBM becomes more well understood and novel delivery methods are developed and optimized, miRNA‐based therapies could provide a critical step forward in cancer treatment.
Keywords: cancer therapy, microRNA, glioblastoma multiforme
Introduction
Glioblastoma multiforme (GBM) is the most common and lethal cancer of the brain, with approximately 10,000 newly diagnosed cases each year in the United States 1. Despite recent advances in understanding of its pathogenesis, GBM remains incurable with standard treatment options contributing little to survival time. Currently, GBM has a 5‐year survival rate of only 10% and a median survival time of 15 months following treatment 1, 2. Contributing to its poor prognosis are numerous therapeutic challenges including aggressive growth rates, tumor heterogeneity, drug resistance, and obstacles to drug delivery such as the blood–brain barrier.
In an effort to find novel approaches to GBM treatment, recent studies have focused on molecular phenotyping of GBM subtypes to identify new targets for biomarkers and therapeutics. Of particular interest have been microRNAs (miRNAs), an abundant class of endogenously expressed 18–25 nucleotide noncoding RNAs. These molecules can inhibit gene expression by binding to target messenger RNA (mRNA), thereby inducing translational silencing or degradation based on complementarity to targets 3.
miRNAs are encoded by nuclear DNA and are transcribed by RNA polymerase II to generate capped and polyadenylated long primary transcripts, called pri‐miRNAs (Fig. 1) 4. Each pri‐miRNA folds into a stem‐loop structure via intramolecular base pairing 5 and is then cleaved by the microprocessor complex, which consists of Drosha and DGCR8. The product is a 60–120 nucleotide long hairpin miRNA precursor, called pre‐miRNA 6, 7. Pre‐miRNA is actively transported from the nucleus by Exportin‐5 8. In the cytoplasm, the RNase III enzyme Dicer, makes the final cleavage, producing a ~22‐nucleotide miRNA duplex. Association of Dicer with RNA‐binding domain factors, PACT or TRBP, can also produce slightly different‐sized miRNAs, termed isomiRs, which have altered target‐binding specificities 9. After the mature miRNA is formed, the duplex is unwound. While one strand is normally degraded, the other is incorporated into the RNA‐induced silencing complex (RISC). Within RISC, the miRNA serves as a template for recognizing its complementary mRNA molecule 10, most frequently within the 3′ untranslated region. This interaction results in degradation and/or translational repression of target genes.
Figure 1.
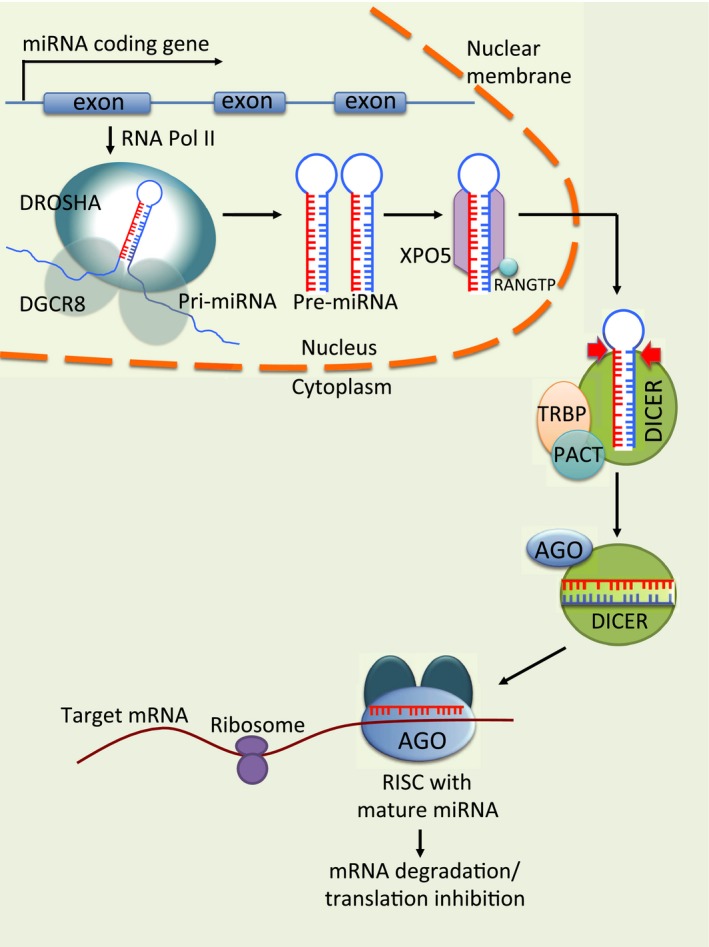
Biogenesis of miRNAs. miRNAs are encoded by nuclear DNA and are transcribed by RNA polymerase II to generate pri‐miRNAs. Pri‐miRNAs then fold into stem‐loop structures via intramolecular base pairing and are cleaved by the microprocessor complex. The resulting long hairpin miRNA precursors, called pre‐miRNAs, are then transported from the nucleus by Exportin‐5. In the cytoplasm, pre‐miRNAs are cleaved by the RNase III enzyme Dicer, to produce miRNA duplexes. The duplexes are then unwound and the guide strands are selected by Argonaute for integration into the RNA‐induced silencing complex (RISC). Within RISC, miRNAs serve as templates for recognizing complementary mRNA molecules, resulting in degradation and/or translational repression of target genes.
There are currently 2588 mature human sequences recorded in miRBase, a public miRNA repository (www.mirbase.org, accessed January 2016). Each is predicted to individually regulate hundreds to thousands of mRNAs, depending on cell type, context, and binding of cofactors 11, 12. Further, specific mRNAs can also be modulated by numerous different miRNAs, thus allowing for widespread regulation of gene expression. Indeed, it is estimated that miRNAs regulate up to one‐third of human genes 13, participating in various processes including cell proliferation, differentiation, cell cycle regulation, and apoptosis.
Importantly, miRNAs have also been implicated in various pathological conditions, including cancer. Virtually all tumors display globally abnormal miRNA expression patterns 14, and more than 50% of the human miRNA‐encoding genes are found in fragile chromosomal sites associated with cancer 15. These alterations in miRNA expression can occur through a variety of mechanisms including altered transcriptional regulation, abnormalities in miRNA processing, defects in localization of miRNAs, gene mutations, chromosomal changes, epigenetic aberrations, and alterations in the machinery involved in miRNA biogenesis 16. This frequently results in upregulation of oncogenes and/or downregulation of tumor suppressors, thereby functioning to support carcinogenesis.
miRNAs in GBM
Many studies have begun to map out the expression profiles and functions of miRNAs in GBM, aiming to garner new insight that can be used to combat this insidious disease. A recent systematic review by Møller et al. 17 found that the most common aberration in miRNA expression with GBM is upregulation. Indeed, 256 miRNAs were found to be significantly overexpressed (notably miR‐10b, miR‐17–92 cluster, miR‐21, and miR‐93) and 95 miRNAs were significantly underexpressed (including miR‐7, miR‐34a, miR‐128, and miR‐137) in GBM as compared to normal brain tissue 17. Moreover, individual miRNAs have been correlated with different glioma stages. Investigating the miRNA expression profiles of WHO grade II gliomas that spontaneously progressed to WHO grade IV GBM, Malkorn et al. 18 identified 12 miRNAs (miR‐9, miR‐15a, miR‐16, miR‐17, miR‐19a, miR‐20a, miR‐21, miR‐25, miR‐28, miR‐130b, miR‐140, and miR‐210) that were upregulated and two (miR‐184 and miR‐328) that were downregulated during progression 18. Other studies have additionally found miRNAs that are differentially expressed in advanced clinical stages of GBM, including miR‐182, which is upregulated and miR‐137, which is downregulated in late stages 19, 20.
Importantly, miRNAs seem to have a predilection for targeting developmental genes, making them fundamental in the regulation of proliferation, differentiation, and apoptosis. In relation to cancer pathobiology, this means that miRNAs impact many of the hallmarks of cancer 21, 22. Indeed, miRNAs have been found to contribute to sustained proliferative signaling and evasion of growth suppressors, resistance to cell death, enabling of replicative immortality, induction of angiogenesis, and activation of invasion and metastasis. Further, miRNAs are also important regulators of drug resistance in GBM. Table 1 provides an overview of miRNAs that are known to participate in GBM, including which pathways they have been found to interact with. In the following sections, we briefly discuss the impact of various miRNAs on these pathways.
Table 1.
miRNAs associated with Glioblastoma multiforme
| miRNA | mRNA targets | Expression change with GBM/ poor prognosis | Role(s) in cancer progression | References |
|---|---|---|---|---|
| let‐7 | NRAS, KRAS, CCND1 | Decrease | Proliferation, apoptosis, migration, invasion, chemoresistance | 35, 143, 144 |
| miR‐7 | EGFR, RAF1, PI3K, FAK, IRS2 | Decrease | Survival, proliferation, apoptosis, invasion, angiogenesis | 23, 24, 75, 145, 146, 147, 148, 149 |
| miR‐9/miR‐9* | SOX2, PTCH1, FOXP1, CAMTA1 | Increase;decrease | Proliferation, stemness, chemoresistance | 18, 138, 150, 151, 152, 153, 154 |
| miR‐10a/b | BCL2L11, TFAP2C, CDKN2A, CDKN1A, CSMD, HOXD10, E2F1 | Increase | Proliferation, apoptosis, migration, invasion, stemness | 72, 148, 149, 155, 156, 157, 158 |
| miR‐15a | Increase | 18 | ||
| miR‐15b | NRP2, CCND1 | Decrease | Proliferation, apoptosis, invasion, angiogenesis | 159, 160, 161 |
| miR‐16 | BMI1, NFKB1, BCL2, ZYX | Decrease;increase 18 | Proliferation, apoptosis, migration, invasion, angiogenesis | 18, 162, 163, 164 |
| miR‐17 | CAMTA1, PTEN, MDM2 | Increase | Survival, proliferation, migration, invasion, stemness, chemoresistance, stress response | 18, 84, 154, 165, 166 |
| miR 17–92 cluster | CTFG | Increase | Proliferation, apoptosis, stemness | 167 |
| miR‐18a/18a* | NEO1, DLL3, CTGF, SMAD3 | Increase | Proliferation, apoptosis, migration, invasion, stemness | 168, 169, 170 |
| miR‐19a/b | PTEN | Increase | Survival, proliferation | 18, 171 |
| miR‐20a | TIMP2 | Increase | Invasion | 18, 84, 172 |
| miR‐21 | PDCD4, IGFB3, FBXO11, ANP32A, SMARCA4, LRRFIP1, HNRPK, TP63, RECK, TIMP3, TPM1, FASLG, SPRY2 | Increase | Survival, proliferation, apoptosis, migration, invasion, chemoresistance | 18, 37, 38, 39, 40, 46, 47, 69, 70, 90, 149, 173, 174, 175, 176, 177, 178 |
| miR‐23b | PTK2B | Decrease | Migration, invasion | 179 |
| miR‐24 | ST7L | Increase | Proliferation, apoptosis, invasion | 180 |
| miR‐25 | CDKN1C, NEFL, MDM2, TSC1 | Increase; decrease 181 | Viability, proliferation, invasion | 18, 181, 182, 183 |
| miR‐26a | PTEN, ATM | Increase; decrease 184 | Radioresistance | 184, 185 |
| miR‐26b | EPHA2 | Decrease | Proliferation, migration, invasion, vasculogenic mimicry | 186 |
| miR‐27a | FOXO3a | Increase; decrease with higher grade 187 | Proliferation, apoptosis, invasion | 187, 188 |
| miR‐27b | Increase | Proliferation, apoptosis, invasion | 189 | |
| miR‐28 | Increase | 18 | ||
| miR‐29a/b | MCL1, PDPN | Increase; decrease 190 | Proliferation, apoptosis, invasion | 190, 191 |
| miR‐30a | SOCS3, SEPT7 | Increase | Proliferation, apoptosis, invasion, stemness | 192, 193, 194 |
| miR‐31 | FIH1, RDX, TRADD | Decrease; increase 66 | Proliferation, apoptosis, migration, invasion, stemness, angiogenesis, chemoresistance | 66, 84, 149, 195, 196, 197 |
| miR‐32 | MDM2, TSC1 | Decrease | Survival, proliferation | 198 |
| miR‐34a | MET, NOTCH1, NOTCH2, CCND1, CDK6, RICTOR, SIRT1 | Decrease | Survival, proliferation, apoptosis, migration, invasion, stemness | 31, 199, 200, 201, 202 |
| miR‐92a | BCL2L11 | Increase | Proliferation, apoptosis | 203 |
| miR‐92b | DKK3, NLK | Increase | Proliferation, apoptosis, invasion | 204, 205 |
| miR‐93 | ITGB3 | Increase | Survival, proliferation, angiogenesis, stemness | 83, 148 |
| miR‐95 | Decrease | 148 | ||
| miR‐100 | SMRT/NCOR2, ATM | Decrease; increase 206 | Proliferation, apoptosis, radioresistance | 206, 207 |
| miR‐101 | KLF6 | Decrease | Proliferation, apoptosis, invasion, migration | 149, 208 |
| miR‐106a | SLC2A3, TIMP2 | Decrease; increase in GSCs 172 | Proliferation, invasion, metabolism | 172, 209 |
| miR‐107 | SALL4, NOTCH2, CDK6 | Decrease | Proliferation, apoptosis, invasion | 210, 211, 212, 213 |
| miR‐124 | PIM3, NRAS, SOS1, PPP1R3L, RRAS, NRAS, SNAI2, MAPK14, TEAD1, SERP1, LAMB1, CDK4, IQGAP1 | Decrease | Proliferation, apoptosis, migration, invasion, stemness, angiogenesis, chemoresistance, radioresistance, stress response | 29, 148, 156, 214, 215, 216, 217, 218, 219, 220, 221, 222 |
| miR‐125a | NRG1, PDPN | Decrease | Proliferation, apoptosis, invasion, migration | 190, 223 |
| miR‐125b | LIN28, BAK1, MAPK14, CDK6, CDC25A, BMF, MAZ, E2F2 | Decrease80, 224, 225, 226, 227; increase 228, 229, 230 | Survival, proliferation, apoptosis, invasion, stemness, angiogenesis, chemoresistance | 80, 224, 225, 226, 227, 228, 229, 230, 231, 232 |
| miR‐126 | Decrease | 233 | ||
| miR‐128 | P70S6K1, SUZ12, BMI1, PDGFRα, EGFR, E2F3a, WEE1, MSI1 | Decrease | Proliferation, apoptosis, angiogenesis, stemness, radioresistance | 25, 26, 27, 28, 82, 90, 148, 178, 234, 235 |
| miR‐130a | Decrease | Chemoresistance | 236, 237 | |
| miR‐130b | MST1, SAV1 | Increase | Stemness | 18, 238 |
| miR‐132 | Increase; decrease 148 | 148, 239, 240 | ||
| miR‐135b | ADAM12, SMAD5, GSK3β | Decrease | Proliferation, migration, stemness, radioresistance | 241, 242 |
| miR‐136 | AEG1, BCL2 | Decrease | Apoptosis, chemoresistance | 243, 244 |
| miR‐137 | RTVP1, PTGS2, MSI1 | Decrease | Proliferation, apoptosis, migration, invasion, stemness | 29, 245 27, 149, 246, 247, 248 |
| miR‐139 | ELTD1, MCL1 | Decrease | Proliferation, apoptosis, chemoresistance | 148, 249 250 |
| miR‐140 | Increase | 18, 156 | ||
| miR‐143 | HK2, RAS | Decrease; increase 251 | Migration, invasion, angiogenesis, chemoresistance, stemness, glycolysis | 251, 252, 253 |
| miR‐145 | CTGF, ABCG2 | Decrease; increase 251 | Proliferation, migration, invasion | 251, 254, 255 |
| miR‐146 | NOTCH1 | Decrease; increase 84 | Proliferation | 84, 256 |
| miR‐148a | FIH1 | Increase | Proliferation, stemness, angiogenesis | 27, 84 |
| miR‐152 | KLF4, MMP3, XIST | Decrease | Proliferation, apoptosis, migration, invasion, stemness | 67, 159, 257 |
| miR‐153 | BCL2, MCL1, IRS1 | Decrease | Viability, proliferation, apoptosis, stemness | 258, 259, 260 |
| miR‐155 | GABRA 1, EAG1, MAPK13, MAPK14, FOXO3a, MXI1 | Increase | Proliferation, apoptosis, invasion, chemoresistance | 261, 262, 263 264, 265 237, 266 |
| miR‐181 | FOS, KPNA4, MGMT, RAP1B, BCL2, NOTCH2, MGMT, KRAS, BCL2, MDM2 | Decrease | Proliferation, apoptosis, invasion, stemness, chemoresistance, radioresistance | 88, 111, 178, 234, 267, 268, 269, 270, 271, 272, 273, 274, 275 |
| miR‐182 | BCL2L12, HIF2A, MET, CYLD, LRRC4 | Increase | Proliferation, apoptosis, invasion, angiogenesis, chemoresistance, stemness | 19, 135, 276, 277 |
| miR‐184 | FOXO3, SND1 | Decrease | Proliferation, invasion, chemoresistance | 18, 278 279 |
| miR‐193 | SMAD3 | Increase | Proliferation | 84, 280 |
| miR‐195 | CCND1, CCNE1 | Decrease; increase in TMZ‐resistant GBM 87 | Proliferation, chemoresistance | 281 178, 87 |
| miR‐196 | NFKBIA | Increase | Proliferation, apoptosis | 85, 178, 248, 282, 283 |
| miR‐200a/b | MGMT | Decrease; increase | Chemoresistance | 84, 284 |
| miR‐203 | SNAI2, PLD2 | Decrease | Chemoresistance | 285, 286, 287 |
| miR‐205 | VEGFA, LRP1 | Decrease | Proliferation, apoptosis, invasion, migration | 102, 288, 289 |
| miR‐210 | HIF3A | Increase | Survival, chemoresistance | 18, 81, 237 |
| miR‐218 | LEF1, IKBKB, BIM1, ECOP, CDK6 | Decrease | Survival, proliferation, apoptosis, migration, invasion, stemness | 54, 55, 290, 291, 292, 293 |
| miR‐221/222 | PTEN, PUMA, CDKN1B, CDKN1C, PTPRM, MGMT, SEMA3B, TIMP3, GJA1 | Increase; decrease 84, 178 | Viability, proliferation, apoptosis, migration, invasion, chemoresistance, radioresistance | 234, 294 48, 149, 178 52, 84, 295, 296, 297, 298 |
| miR‐296‐3p | EAG1 | Decrease | Proliferation, chemoresistance | 299 |
| miR‐302/367 cluster | CXCR4 | Invasion, chemoresistance, stemness | 300, 301 | |
| miR‐320 | E2F1 | Decrease | Proliferation, migration | 302 |
| miR‐323 | IGFR1 | Decrease; increase 237 | Proliferation, apoptosis, migration | 237, 303 |
| miR‐326 | NOB1 | Decrease | Proliferation, apoptosis | 304, 305 237 |
| miR‐328 | SRFP1 | Decrease; increased in low grade GBM 306 | Proliferation, invasion | 18, 96, 306 |
| miR‐329 | E2F1 | Decrease; increase 237 | Survival, proliferation, apoptosis | 237, 307 |
| miR‐330 | SH3GL2 | Increase; decrease 149 | Proliferation, apoptosis, migration, invasion | 149, 308, 309 |
| miR‐331‐3p | NRP2, HER2 | Decrease | Proliferation, apoptosis, migration, invasion, angiogenesis | 145, 310 |
| miR‐335 | DAAM1, PAX6 | Increase | Survival, proliferation, apoptosis, invasion, stemness | 53, 311, 312, 313 |
| miR‐340 | PLAT, ROCK1, CDK6, CCND1, CCND2 | Decrease | Proliferation, apoptosis, migration, invasion, stemness | 314, 315, 316 |
| miR‐363 | BIM, CASP3 | Increase | Survival | 317 |
| miR‐372 | PHLPP2 | Increase | Proliferation, apoptosis invasion | 318 |
| miR‐377 | SP1 | Decrease | Proliferation, invasion | 319 |
| miR‐378 | Decrease | Migration, invasion | 320 | |
| miR‐381 | LLRC4 | Increase | Proliferation | 277, 321 |
| miR‐410 | MET | Decrease | Proliferation, invasion | 322 |
| miR‐451 | CAB39 | Decrease | Proliferation, invasion, apoptosis, stemness | 56, 323, 324 |
| miR‐455‐3p | SMAD2 | Increase | Chemoresistance | 87, 325 |
| miR‐483‐5p | ERK1 | Decrease | Proliferation | 326 |
| miR‐487b | Decrease | Proliferation, apoptosis | 148 | |
| miR‐491‐3p | IGFBP2, CDK6 | Decrease | Proliferation, invasion, stemness | 327 |
| miR‐491‐5p | BCL2L1, EGFR, CDK6, MMP9 | Decrease | Proliferation, invasion, stemness | 327, 328 |
| miR‐513 | LRP6 | Decrease | Proliferation | 329 |
| miR‐582‐5p | CASP3, CASP9 | Increase | Proliferation, apoptosis | 317 |
| miR‐603 | MGMT, WIF1, CTNNBIP1 | Increase | Proliferation, chemoresistance | 268, 330 |
| miR‐655 | SENP6 | Increase | Invasion | 331 |
| miR‐663 | PIK3CD | Decrease | Proliferation, invasion | 332 |
| miR‐873 | IGF2BP1, BCL2 | Decrease; increase 260 | Proliferation, apoptosis, migration, invasion, chemoresistance | 148, 333, 334 |
| miR‐874 | Decrease | 156 |
ECOP, epidermal growth factor receptor coamplified and overexpressed protein; KLF4, Krüppel‐like factor 4; MAZ, myc‐associated zinc finger protein; MMP, matrix metalloproteinase; PUMA, p53‐upregulated modulator of apoptosis;TMZ, temozolomide.
Enhancing cell proliferation
The ability to continuously proliferate is a fundamental characteristic of all cancers that is achieved through deregulation of cell signaling pathways. Importantly, miRNAs can influence the promotion of sustained signaling for proliferation as well as the ability to evade growth suppressors, enhancing the expansion capacities of cancer cells.
This is exemplified by survival and proliferation pathways modulated by EGFR and Akt. In GBM, where amplification of EGFR is a characteristic trait of primary tumors, miRNAs involved in EGFR regulation display corresponding disruptions in expression with disease progression. miR‐7, which acts as a tumor suppressor, directly targets EGFR and can independently repress the Akt pathway through targeting of its upstream regulators. In GBM, miR‐7 is frequently downregulated, allowing for enhanced activation of the Akt pathway, and thus increased viability and invasiveness of tumor cells 23, 24. These protumorigenic effects can be corrected with transfection of mimic miR‐7 oligonucleotides, which results in decreased invasiveness and increased apoptosis of GBM cell lines 23, 24.
miR‐128 also exerts an antiproliferative effect and is frequently downregulated with GBM 25. Its growth‐suppressive functions are mediated through numerous pathways via targeting of (1) EGFR and growth factor receptor PDGFRA 25, (2) WEE1, a regulator of cell cycle progression that can block entry into mitosis through inhibition of CDK1 26, (3) MSI1, which regulates the expression of the NOTCH1 antagonist NUMB, influencing proliferation and the maintenance of stem cells 27, and (4) the transcription factor E2F3A, which is a cell cycle activator 28. Thus, repression of miR‐128 in cancer cells can be an advantageous mechanism for overcoming normal proliferative signaling.
However, these pathways can also be targeted by other miRNAs. The E2F transcription factor family plays a pivotal role in the cell cycle. In response to mitogenic signaling, RB1 is phosphorylated by CDK/cyclin complexes leading to activation of E2F‐responsive genes, thereby promoting cell cycle progression. This pathway can be suppressed by miR‐137 and miR‐124a, which inhibit CDK6 expression as well as phosphorylation of RB1. As both miR‐137 and miR‐124a are consistently downregulated in GBM 29, transfection of either miRNA can prevent proliferation of GBM cells. Further, forced miR‐137 overexpression can also mediate MSI1 27 and PTGS2 30, which additionally function to reduce proliferation and invasion.
miR‐34a is also persistently downregulated in GBM and has similar targets, repressing CDK6, CCND1, MET, NOTCH1, NOTCH2, and SIRT1 31, 32, 33, 34. Of particular importance to proliferative signaling, targeting of CDK6 and CCND1 prevents the downstream prosurvival signaling of the cyclin/CDK pathway. Thus, restoration of miR‐34a can reduce CDK6 protein expression, inhibiting cell survival, proliferation, and invasion as well as inducing apoptosis 31, 32.
RAS proteins, which process signals downstream of growth receptors, are also targeted by miRNAs and play a key role in the deregulation of proliferation pathways in many cancers, including GBM. let‐7 is an miRNA which is decreased in gliomas and inversely correlates with the presence of RAS proteins. Restoration of let‐7 reduces the expression of RAS, resulting in decreased proliferation and migration of tumor cells in vitro and inhibition of tumor growth in vivo. Importantly, transfection of let‐7 has no effect on normal human astrocytes, indicating that normal cells are able to more effectively regulate miRNA activity 35.
In addition to miRNA modulation of RAS proteins, RAS proteins can conversely influence miRNA expression. Indeed, miR‐21 is a transcriptional target of the AP‐1 complex and RAS oncogenes are well‐known inducers of AP‐1 activity. AP‐1 is a transcription factor that regulates a variety of target genes, leading to an increase in cell proliferation, invasion, and angiogenesis during tumor development. AP‐1 induces miR‐21, which downregulates tumor suppressors PDCD4 and PTEN. Inhibition of PDCD4 then contributes to an increase in AP‐1 activity, revealing an AP‐1 autoregulatory mechanism in RAS transformation 36. In addition to PDCD4 and PTEN, miR‐21 also exerts its prooncogenic effect by downregulating numerous targets including ANP32A, SMARCA4, SPRY2, IGFBP3, LRRFIP1, and RECK. Through these targets, miR‐21 can influence numerous biological processes in addition to the promotion of cell cycle progression, including promotion of invasion and metastasis and resistance to chemotherapeutics 37, 38, 39, 40, 41, 42, 43, 44. Notably, inhibition of miR‐21 expression can repress tumor growth 37, 43, 45.
Resisting cell death
In addition to sustained proliferation, the ability to evade apoptosis is an essential characteristic of cancer. Dysregulation of miRNA expression is one mechanism for allowing cancer cells to bypass signals for programmed cell death. Further, miRNA mediation of apoptosis is strongly linked to drug resistance, as many treatments aim to initiate apoptotic pathways by damaging cancer cells. Thus, the ability to override signals for cell death is critical for not only sustained proliferation but also for acquiring drug immunity.
miRNAs can have either pro‐ or antiapoptotic functions and are therefore differentially regulated during cancer progression. Antiapoptotic miRNAs target proapoptotic genes and are frequently upregulated with GBM. miR‐21 is antiapoptotic and enhances tumor formation by targeting the signaling pathways of P53 and TGF‐β as well as the mitochondrial apoptotic pathway 46. Inhibition of miR‐21 results in activation of caspases, suspension of cell growth, reduced invasion, increased apoptosis, and enhanced chemosensitivity. These effects are mediated in part by decreased repression of targets including HNRPK, TAP63, and PDCD4 37, 44, 46. Further, miR‐21 can modulate the extrinsic apoptotic pathway through downregulation of FASL, an ability that has been particularly evident in cancer stem cells 47. Thus, miR‐21 has widespread effects on cell death pathways, making it a critical player in GBM pathogenesis and a promising target for therapeutic interventions.
miR‐221 and miR‐222 are also overexpressed in GBM and have numerous targets involved in gliomagenesis, including in apoptotic pathways. miR‐221/222 can regulate cell death through targeting of p53‐upregulated modulator of apoptosis (PUMA). Under normal conditions, PUMA binds Bcl‐2 and Bcl‐x, rapidly inducing apoptosis. Thus, forced expression of miR‐221/222 and the subsequent downregulation of PUMA, promotes cell survival. Therefore, the knockdown of miR‐221/222 is able to induce cell death and decrease tumor growth 48, 49. miR‐221/222 can also target the cell growth‐suppressive CDK inhibitors P27 and P57 50 and thus are tightly linked to cell cycle checkpoints for initiation of S phase. When human glioblastoma U251 cells are treated with antisense miR‐221/222, the cell cycle is arrested in G0 or G1 phase 51. Moreover, treatment with antisense oligonucleotides for miR‐221/222 enhances the effects of both temozolomide (TMZ) and radiation 49, 52.
miR‐335 is also antiapoptotic and is upregulated with GBM. miR‐335 targets potential tumor suppressor, disheveled‐associated activator of morphogenesis 1 (DAAM1), thereby promoting growth and invasion of astrocytoma cells. Thus, inhibition of miR‐335 is able to effectively suppress growth and induce apoptosis of astrocytoma cells both in vitro and in vivo. Indeed, delivery of a miR‐335 antagonist to rat glioma C6 cells resulted in growth arrest, activation of apoptosis, repression of invasion, and marked regression of astrocytoma xenografts 53. Contributing to its widespread influence on cellular function, miR‐335 also regulates RB1 and controls cell proliferation in a p53‐dependent manner 53.
On the opposing side of the spectrum are proapoptotic miRNAs, which target antiapoptotic genes and are often downregulated during GBM progression. miR‐218 is proapoptotic and exerts its influence through targeting of CDK6. Thus, its downregulation corresponds with an increase in cellular proliferation with GBM. Exogenous administration of miR‐218 can effectively repress expression of CDK6, inhibiting cell proliferation and inducing apoptosis of malignant glioma cells 54. Further, miR‐218 was found to sensitize glioma cells to apoptosis by regulating epidermal growth factor receptor coamplified and overexpressed protein (ECOP), which can suppress the transcriptional activity of NFκB, thereby modulating the cellular apoptotic response 55.
miR‐451 also acts as a tumor suppressor and is subject to downregulation in GBM tissue. Correcting the loss of miR‐451 via administration of miR‐451 mimics has been shown to inhibit cell growth, decrease invasive capacity, induce G0/G1 phase arrest, and increase apoptosis of GBM cells. This activity is predicted to result from modulation of the PI3K/Akt pathway as administration of miR‐451 mimics results in decreased protein expression of Akt1, Cyclin D1, MMP‐2, MMP‐9, and Bcl‐2 as well as increased expression of p27 56.
Autophagy
An additional means of miRNA regulation of cell death is through modulation of autophagy, an evolutionarily conserved, multistep lysosomal degradation process that functions to catabolize unnecessary or dysfunctional cellular components. It is unclear whether autophagic activity in dying cells is the cause of death or a survival mechanism, and thus, the role of autophagy in cancer has not been completely elucidated. Autophagy may protect against cancer by isolating damaged organelles, promoting cell differentiation, increasing protein catabolism, or potentially by prompting apoptosis of cancerous cells. Alternatively, autophagy can contribute to cancer progression by degrading apoptotic mediators and by providing a means of surviving nutrient depletion or absence of growth factors 57.
Various miRNAs contribute to both autophagy and progression of GBM. miR‐17 is frequently overexpressed in gliomas and targets ATG7, one of the master regulators of autophagy. Downregulation of miR‐17 promotes ATG7 protein expression and induction of autophagy, leading to a subsequent decrease in cell viability and proliferation. Moreover, anti‐miR‐17 treatment can enhance the effectiveness of radiation and TMZ, indicating that modulation of autophagy is an important aspect of GBM survival and progression 58.
miR‐21 also inhibits autophagy and enhances resistance of glioma cells to radiation therapy 59. A study by Gwak et al. 59 found that miR‐21 expression levels positively correlated with resistance to radiation in glioma cells. Anti‐miR‐21 treatment of U373 and U87 malignant glioma cell lines resulted in an increased sensitivity to radiation. Further, administration of anti‐miR‐21 was correlated with an increased expression of molecular factors associated with autophagosome formation as well as autophagic activity. This increase in autophagy resulted in an expansion of the apoptotic population following irradiation 59. Thus, these studies indicate that autophagy may play a protective role against cancer, which could potentially be exploited using miRNA‐based therapies.
Enabling replicative immortality
While normal cells have a limited number of potential divisions, cancer cells frequently display infinite replicative capacity. This is particularly evident in cancer stem cells, which serve as potent drivers of tumor growth and development. Indeed, CD133+ GBM cancer stem cells (GSCs) are able to initiate tumors with as few as 100 cells when injected into the brains of NOD‐SCID mice 60. Due to their unique characteristics and extremely aggressive nature, including resistance to radio‐ and chemotherapy 61, it has been proposed that GBM cells, and GSCs in particular, may originate from adult neural stem cells or neural precursors that have undergone transformation, allowing them to retain stem cell characteristics 62.
In support of stem cell origins, Lavon et al. 63 demonstrated that gliomas display neural precursor cell (NPC)‐like miRNA signatures. In a human glioma panel, 71 of the 180 miRNAs investigated had a distinct expression pattern which matched that of the embryonic stem cells and neural precursor cells. This included upregulation of miR‐21 and downregulation of miR‐124 and miR‐137, which have previously been implicated in gliomagenesis. Interestingly, about half of the miRNAs expressed in the shared profile clustered in seven genomic regions which are prone to genetic or epigenetic aberrations with various cancers. Further, this expression signature was consistent across cell lines as well as unsorted GBM human tumors, indicating that a majority of GBM tumor cells may share this profile.
Silber et al. 29 likewise confirmed that miR‐124 and miR‐137 were significantly decreased in GBM as compared to normal brain tissue. They further found that miR‐124 and miR‐137 induced differentiation of adult mouse neural stem cells and human GBM‐derived stem cells, suggesting that repression of these miRNAs is indeed critical for maintenance of stem cell properties.
miRNAs have also been associated with various facets of the selection and promotion process for GSCs. For example, miR‐128 targets the oncogene BMI1 and is downregulated in GBM, allowing for enhanced self‐renewal of GSCs 64. miR‐326 is also decreased in gliomas and its expression is inversely correlated with expression of smoothened (SMO), a component of the Hedgehog pathway. The Hedgehog pathway is a signaling cascade that is important for the modulation of embryonic stem cells as well as for the proliferation of adult stems cells. Overexpression of miR‐326 was able to repress self‐renewal and partially prompted differentiation of U251 tumor stem cells. Further, the transfection of miR‐326 reduced intracranial tumorigenicity 65.
A study by Wong et al. 66 found that miRNAs can also regulate GBM growth through maintenance of tumor stem cells and stem cell niches. Inhibition of miR‐148a and miR‐31 using antisense oligonucleotides was able to reduce cancer cell proliferation, deplete stem cells, and normalize tumor vasculature. These effects were mediated in part through miR‐148a and miR‐31 repression of factor‐inhibiting hypoxia‐inducible factor 1 (FIH1), which can influence angiogenesis and tumor stemness through HIF1α and Notch signaling 66. Thus, their inhibition was able to effectively suppress tumor growth and prolong survival time in orthotopic xenograft GBM mouse models.
Other miRNAs, such as miR‐152, serve as tumor suppressors and are downregulated in GSCs. Restoration of miR‐152 is able to reduce proliferation, migration, and invasion, as well as induce apoptosis of GSCs. These activities are mediated through targeting of Krüppel‐like factor 4 (KLF4), which is associated with downregulation of LGALS3 and reduced activation of the MEK1/2 and PI3K signaling pathways. Importantly, miR‐152 overexpression is able to decrease tumor volume and enhance survival time in mouse models 67.
Activating invasion and metastasis
While metastasis to other parts of the body is rare, GBM is characterized by extensive, diffuse infiltration throughout the brain. This capacity for pervasive invasion and metastasis is enabled through various mechanisms including modulation of cell‐to‐cell and cell‐to‐matrix interactions, degradation and remodeling of the extracellular matrix, cytoskeletal reorganization, and gain of migratory behavior. Epithelial to mesenchymal transition (EMT) is an important part of this process. EMT is characterized by the loss of an epithelial phenotype, including expression of E‐cadherin, and the acquisition of mesenchymal markers such as fibronectin, vimentin, and N‐cadherin. Many distinct molecular processes occur during EMT including activation of transcription factors, expression of specific cell‐surface proteins, reorganization of cytoskeletal proteins, and production of extracellular matrix‐degrading enzymes 68. As a result, tumor cells develop increasing invasive and migratory potential. Importantly, these processes are accompanied by changes in expression of many miRNAs, which could serve as potential targets for inhibiting invasion and metastasis.
As previously noted, miR‐21 is frequently overexpressed during gliomagenesis and contributes to a variety of protumorigenic pathways. Through the targeting of matrix metalloproteinase (MMP) inhibitors, such as RECK and TIMP3, as well as tumor suppressors including ANP32A and SPRY2, miR‐21 can increase the expression and activity of various MMPs, facilitate Ras/Raf binding, and induce ERK phosphorylation, thereby enhancing the invasive potential of GBM cells 40, 69, 70. Administration of antisense oligonucleotides to miR‐21 is able to ameliorate these effects, resulting in elevated levels of RECK and TIMP3, a reduction in MMP activity, and decreased migration and invasion of GBM cells 70.
MMPs are also targeted by other miRNAs, including miR‐146b. miR‐146b inhibits expression of MMP16, an enzyme that functions in proteolysis of extracellular matrix components and is therefore critical for the migration and invasion properties of tumor cells. Thus, miR‐146b is often decreased during gliomagenesis, allowing for upregulation of MMP16 71.
miR‐10b can also modulate MMPs, indirectly targeting MMP14 as well as urokinase receptor (uPAR) and RhoC through direct suppression of their upstream target HOXD10. Augmented miR‐10b expression levels are correlated with higher grade gliomas, 72, 73 and indeed, GBM cells display reduced growth, invasion, and angiogenesis, as well as enhanced cell death when treated with antisense oligonucleotides to miR‐10b 73. Further, in an orthotopic human glioma mouse model, inhibition of miR‐10b diminished the growth, invasiveness, and angiogenicity of glioma cells in the brain, significantly prolonging the survival of glioma‐bearing mice 74.
miR‐7 also has many targets involved in metastasis including FAK, PI3K, EGFR, and RAF1 23, 24, 75, 76. As indicated by its targets, miR‐7 functions as a tumor suppressor and is downregulated in GBM. Its overexpression can inhibit metastasis and invasion of GBM cells by directly repressing FAK, a mediator of cell‐extracellular matrix signaling, as well as by reducing the expression of MMP2 and MMP9, thereby decreasing the ability of GBM cells to move through extracellular matrix 75. Further, by suppressing EGFR expression and inhibiting the Akt pathway, miR‐7 can decrease the viability and invasiveness of GBM cells 23.
Inducing angiogenesis
GBM is distinguished from lower grade gliomas by excessive microvascular proliferation, which results in highly vascularized tumors. Enhanced vascularization allows for increased proliferative and invasive capacity of cancer cells due to the greater availability of oxygen and nutrients. Additionally, blood vessels can aid in cell migration, supporting diffuse infiltrative migration of single cells into the brain parenchyma as well as perivascular migration of cells along the microvasculature 77.
A group of miRNAs now termed angiomiRs, have recently been identified as important contributors to neovascularization in GBM 78. These molecules can act on either tumor cells or neighboring tumor‐associated cells. miR‐296 is an angiomiR that has been found to be upregulated in endothelial cells with the presence of glioma cells or angiogenic growth factors such as VEGF. Augmented expression of miR‐296 is associated with increased endothelial cell tube formation and enhanced vascularization of tumors, while knockdown of miR‐296 results in reduced tumor angiogenesis 79.
miR‐125b is an angiomiR that has conversely been found to be repressed in GBM‐associated endothelial cells. This downregulation results in increased expression of its target, myc‐associated zinc finger protein (MAZ), a transcription factor that regulates VEGF. Decreased expression of miR‐125b in target cells promotes vascularization of tumors 80.
Within GBM tumor cells, hypoxia can also influence miRNA expression as a mechanism for augmenting angiogenesis. Exemplifying this, hypoxia induces expression of miR‐210‐3p, which directly targets HIF3A, a negative regulator of hypoxic response that acts through downregulation of VEGF. Thus, miR‐210‐3p overexpression induces HIF, VEGF, and CA9 transcriptional activity, enhancing vasculogenesis. However, inhibition of miR‐210‐3p under hypoxic conditions prevents HIF‐mediated induction of VEGF and CA9, reducing vascular density, and abating growth of tumors in vivo 81.
miR‐128 is also able to modulate angiogenesis, functioning through suppression of P70S6K1, a kinase upstream of HIF‐1α and VEGF. miR‐128 is decreased in gliomas, which promotes angiogenesis, cell proliferation, and tumor growth. Restoration of miR‐128 is able to attenuate these effects, while forced expression of P70S6K1 can partly rescue the inhibitory function of miR‐128 on cancer growth 82.
miR‐93, a member of the miR‐17 family and part of the miR‐106b‐25 cluster, is upregulated in GBM and enhances cell survival, sphere formation, and tumor growth, in part through promotion of angiogenesis. miR‐93‐expressing cells induce blood vessel formation, potentially through suppression of integrin‐β8, a protein involved in cell–cell and cell–matrix interactions. Fang et al. 83 found that vasculogenesis could be enhanced by overexpressing miR‐93 in the human glioblastoma cell line U87 and then coculturing the GBM cells with endothelial cells. This resulted in an increase in endothelial cell proliferation and tube formation in vitro and highly increased blood vessel formation in GBM xenograft tumors in mice in vivo 83. These studies illustrate the important role that miRNAs play in the cross‐talk between cells, serving as critical mediators in tumor cell modulation of their microenvironment.
miRNAs in GBM drug resistance
Currently, the standard treatment for newly diagnosed GBM is cytoreductive surgery followed by concurrent radiation and chemotherapy with TMZ 2. However, even the most successful treatment plans are only palliative, aiming to delay relapse for as long as possible. A major challenge is that the response to treatments is highly variable across patients. This is largely due to the heterogeneity of tumors, including differences in genotypes and miRNA profiles between each tumor. However, an advantage is that the correlation between miRNA expression levels and GBM progression can be exploited and miRNAs can be used as predictors of treatment response or overall survival. Indeed, Srinivasan et al. 84 found a 10‐miRNA signature of GBM tumors that was associated with patient survival. The signature included three miRNAs (miR‐20a, miR‐106a, and miR‐17‐5p) that were protective and seven (miR‐31, miR‐222, miR‐148a, miR‐221, miR‐146b, miR‐200b, and miR‐193a) that were risky with respect to the association between their expression and patient survival. Additionally, other studies have found that high expression of miR‐21, miR‐182, and miR‐196 as well as low expression of miR‐181b and miR‐106a are associated with poor patient outcomes 19, 85, 86.
As the standard GBM treatment utilizes radiation and TMZ, many studies have focused on the potential role of miRNAs in resistance to these therapeutics. To identify miRNAs involved in TMZ resistance in GBM, Ujifuku et al. 87 used miRNA microarrays to perform a comprehensive analysis of miRNA expression in a TMZ‐sensitive GBM cell line. They found that miR‐195, miR‐455‐3p, and miR‐10a∗ were the three most upregulated miRNAs in resistant cells. Of the three, knockdown of miR‐195 had the greatest effect on initiating tumor cell death, significantly enhancing the effectiveness of TMZ. Slaby et al. 88 also examined the correlation between expression levels of selected miRNAs and TMZ resistance in GBM, but used 22 primary GBM tumors instead of cell lines. They found that miR‐221, miR‐222, miR‐181b, miR‐181c, and miR‐128 were significantly downregulated with GBM, while miR‐21 was overexpressed. Downregulation of miR‐181b and miR‐181c had the strongest correlation with responsiveness to TMZ treatment, indicating that their presence could serve as a predictive marker for response to TMZ therapy.
As miR‐21 is consistently upregulated in GBM and targets numerous pathways including those involved in survival, proliferation, invasion, and apoptosis, it is unsurprising that it has also been found to play a role in drug resistance. Shi et al. 89 determined that overexpression of miR‐21 significantly inhibited the effect of TMZ on apoptosis, which was mediated through downregulation of proapoptotic proteins Bax and caspase‐3 as well as upregulation of antiapoptotic protein Bcl‐2. Moreover, numerous other studies have investigated the impact of miR‐21 on drug resistance in GBM, finding that inhibiting miR‐21 can enhance the chemosensitivity of human GBM cells to TMZ, paclitaxel, sunitinib, doxorubicin, and VM‐26 39, 90, 91, 92, 93, 94.
miRNAs can also influence drug resistance by targeting the multidrug resistance protein ABCG2 (ATP‐binding cassette subfamily G member 2), a transporter which regulates shuttling of substrates across the cellular membrane. miR‐328 directly targets ABCG2 and is underexpressed in many cancers, including GBM. This allows for elevated expression of ABCG2, enhancing chemoresistance 95. Moreover, high expression of miR‐328 is a protective factor in GBM, while low levels are associated with shorter survival times 96. Thus, restoration of miR‐328 may be an effective option for combination therapy with radiation or chemotherapeutics.
Extracellular miRNAs and GBM
miRNAs are present and stable at high levels in body fluid, including serum, plasma, saliva, urine, and milk. Additionally, miRNA profiles and concentrations in body fluids are correlated with pathological conditions such as cancer, suggesting that miRNAs could serve as promising noninvasive prognostic and diagnostic biomarkers 97, 98. Thus, in tumors such as GBM, where it is often difficult to obtain tissue samples, miRNA profiling via liquid biopsies could provide a promising alternative.
Wang et al. 99 characterized plasma‐derived miRNA expression profiles, finding that with GBM, miR‐21 was upregulated while miR‐128 and miR‐342‐3p were downregulated. Further, expression levels correlated with glioma stage and could be used to distinguish gliomas from other brain tumors such as pituitary adenomas and meningiomas. Roth et al. 100 also explored the blood‐derived miRNA profiles of GBM patients, substantiating a decrease in miR‐342‐3p, but conversely finding that expression of miR‐128 was significantly increased with disease. However, a more recent study investigating miR‐128 supported Wang et al., finding that it was significantly decreased in the serum of glioma patients prior to surgery, and was elevated post surgery (although still not reaching normal levels) 101.
miR‐205 has also been identified as a potential biomarker due to its significantly lower levels in the serum of GBM patients as compared to controls. In a study of 64 glioma patients, serum miR‐205 levels displayed a stepwise decrease with ascending tumor grade and additionally increased following surgery and decreased again during tumor recurrence. Further, patients with advanced pathological grade demonstrated longer overall survival times when serum miR‐205 levels were elevated 101, 102. Thus, serum miRNAs could serve as critical indicators of disease progression and outcome.
The mechanisms for release of miRNAs from cancer cells as well as for crossing the blood–brain barrier to enter systemic circulation are not completely understood. Potential means of release and stabilization include binding of miRNAs to protein complexes or packaging within extracellular vesicles. Studies investigating the presence of extracellular miRNAs have found that a vast majority of those present in blood or cell culture media are independent of exosomes and are bound to Ago2, which enhances their stability 103, 104. However, the selection mechanism for release of miRNAs into extracellular space has yet to be elucidated. Turchinovich et al. 103 proposed that instead of being actively released, large portions of circulating miRNAs could be derived from dead or dying cells.
In addition to protein attachment, horizontal transfer of miRNAs from cancer cells could be facilitated by extracellular vesicles (EV), which are able to protect proteins and nucleotides from degradation 105. Although it has been suggested that a minority of miRNAs in circulation are found in EVs, these vesicles may represent a potent means of paracrine signaling to nearby cells. Cancer cells are able to use EVs to export selected molecules as a mechanism for modulating the tumor microenvironment and establishing premetastatic niches, thereby facilitating dissemination and distant engraftment. The extracellular presence of miRNAs suggests that they may be involved in this process, contributing to the metastatic potential of cancer cells and the mediation of communication between cancer cells and their environment.
Extracellular vesicles such as exosomes and microvesicles, are among the most well‐studied mechanisms of lateral transfer. While microvesicles bud directly from the plasma membrane, exosomes are formed from intraluminal vesicles, which bud into early endosomes and then form multivesicular endosomes (MVE). MVEs are then released by either fusion with the plasma membrane or direct release from the plasma membrane 106. Importantly, the release of miRNAs via exosomes is selective, based on factors such as cell of origin and malignancy. For example, while EVs derived from dendritic cells contain costimulatory proteins necessary for T‐cell activation, the content of vesicles from tumor cells more frequently aid in tumor growth and invasiveness 107. Additionally, Pigati et al. 108 found that approximately 30% of the miRNAs released into exosomes did not reflect the intracellular profile. In particular, a majority of the miR‐451 and miR‐1246 produced by malignant mammary epithelial cells was released, while the majority of these same miRNAs produced by nonmalignant mammary epithelial cells were retained, indicating that there is a selection mechanism for miRNA release 108. Moreover, selectively exported miRNAs from malignant cells may be packaged in structures that differ from normal cells and further, some miRNAs exclusively associate with certain vesicles 109.
The formation of EVs to transport cellular information has been confirmed in GBM. Cultured primary cells from GBM tumors were found to be covered with microvesicles of various sizes, containing both RNA and proteins 107. Moreover, the exosomes from GBM tumors were internalized by endosome‐like structures of brain endothelial cells, where they were able to stimulate tubule formation. This confirmed that exosomes can serve as a means of intercellular communication and thus are a critical tool for tumor cells to use in influencing proximal cells 107. GBM microvesicles were also able to stimulate the proliferation of human glioma cell lines, indicating a potential self‐promoting function 107.
miRNAs released from tumor cells have also been shown to modulate the tumor microenvironment. Tominaga et al. 110 discovered that miR‐181c was significantly upregulated in EVs from breast cancer cells, particularly in brain metastatic breast cancer. Further, miR‐181c was able to trigger the breakdown of an in vitro model of the blood–brain barrier. EVs from the cancer cells were selectively incorporated into endothelial cells (although not into pericytes or astrocytes), where miR‐181c suppressed PDPK1, initiating downregulation of phosphorylated cofilin. This led to abnormal localization of actin and thus weakening of the blood–brain barrier 110. However, miR‐181c has also been shown to act as a tumor suppressor, undergoing downregulation in GBM 111. This suggests that miRNAs may have different functions depending on the cancer type and microenvironment.
An additional example of miRNA regulation of the tumor microenvironment is demonstrated by miR‐105. miR‐105 released from cancer cells is able to destroy tight junctions through inhibition of tight junction protein ZO‐1. Downregulation or loss of tight junctions contributes to cancer progression by altering cell migration, proliferation, polarity, and differentiation. Further, overexpression of miR‐105 in nonmetastatic cancer cells induces metastasis as well as vascular permeability in distant organs. Fittingly, miR‐105 can be detected in circulation at the premetastatic stage. Moreover, its levels in the blood and tumor are associated with ZO‐1 expression and metastatic progression in early‐stage breast cancer 112.
It is important to note that exosomal transfer of miRNAs is not exclusively involved in cancer promotion and can also serve as a physiological mechanism for suppressing cancer proliferation. Kosaka et al. 113 demonstrated that noncancerous cells can secrete and transfer antiproliferative miR‐143 exclusively to cancer cells where miR‐143 is deregulated, thereby suppressing their proliferation. As cells with normal levels of miR‐143 were not affected by its overexpression, it appears that there may be a threshold for miRNA activity or that normal cells are more effectively able to regulate miRNAs.
miRNA‐based therapeutics
Increasing evidence demonstrates that aberrant miRNA expression profiles and signaling pathways are present in many cancers. As these molecules have the ability to target several genes within the same pathway or even multiple oncogenic pathways, miRNAs represent promising therapeutic targets with potential for more comprehensive benefits than other targets with more limited activities. Importantly, miRNAs can function as either oncogenes or tumor suppressors, which then correspond to two different approaches to miRNA‐targeted therapy. These methods aim to either (1) reduce the expression of target miRNAs via antisense technologies or miRNA sponges or (2) restore miRNA expression using synthetic mimics or gene replacement therapy. There are currently a multitude of different strategies being investigated under each category.
Antisense oligonucleotides
Overexpressed intracellular miRNAs can be inhibited via administration of synthetic antisense, single‐stranded RNA‐based oligonucleotides, termed antagomirs or antimiRs. Antagomirs are complementary to mature endogenous miRNAs, allowing for binding and silencing of their targets (Fig. 2A). Efficient inhibition by antagomirs requires optimization of the oligonucleotides for high binding affinity, high resistance to nuclease degradation, low toxicity, and efficient in vivo delivery. These factors can be modified through alterations of the sugar, the nucleobase, the internucleotide linkages, or with the addition of nonnucleotide modifiers 114.
Figure 2.
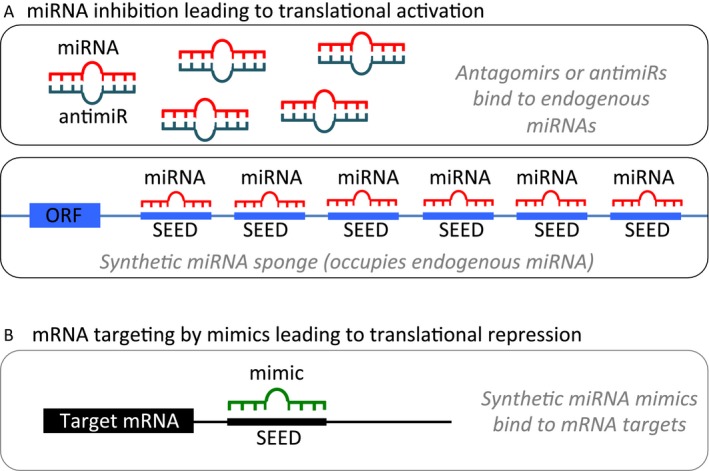
miRNA modulation strategies for therapeutic intervention. (A). miRNA inhibition. (1) Antagomirs are synthetic, single‐stranded RNA‐based oligonucleotides that are complementary to mature endogenous miRNAs, allowing for binding and silencing of their targets. (2) miRNA sponges contain multiple binding sites to an miRNA of interest, competitively inhibiting it from binding to its target mRNA. As the binding sites are specific to an miRNA's seed region, sponges can inhibit an entire family of related miRNAs. (B) miRNA mimics are synthetic, double‐stranded RNA molecules that have identical sequences to their naturally occurring equivalents, allowing for restoration or amplification of the activity of a target miRNA.
Phosphorothioate oligodeoxynucleotides were the most common type of first‐generation antisense oligonucleotides, having high resistance to nucleases but low binding stability and high toxicity. These were followed by 2′‐O‐methyl‐modified oligonucleotides, which displayed increased binding stability and reduced nonspecific effects, but suboptimal efficiency 115. 2′‐O‐methyl‐modified oligonucleotides were further improved on by the development of locked nucleic acid (LNA)‐modified oligonucleotides, which have higher stability, efficacy, and specificity as well as lower toxicity 116, 117.
LNAs are modified, conformationally locked nucleotide analogs with a high level of affinity to DNA and RNA nucleotides and high nuclease resistance 118. LNA‐linked oligonucleotides are currently the most widely used class of antagomirs and have shown promise for treating various conditions. Indeed, a PBS‐formulated LNA antagomir was found to be effective at reducing serum cholesterol levels in African green monkeys. As miR‐122 plays a critical role in fat and lipid metabolism, the study utilized an LNA‐antagomir against liver‐expressed miR‐122. Upon administration, the antagomir was taken into the cytoplasm of hepatocytes, where it was able to successfully deplete miR‐122. This was accompanied by effective lowering of plasma cholesterol levels and no evidence of toxicity or histopathological changes 119.
LNA‐based antagomirs have also been utilized in studies targeting hepatitis C virus (HCV), which requires miR‐122 for replication. Treatment with LNA‐based anti‐miR‐122 oligonucleotides was able to suppress viremia and improve HCV‐induced liver pathology in chimpanzees, also without negative side effects 120. Subsequent studies resulted in the development of miravirsen, an LNA‐modified antisense oligonucleotide against miR‐122, which was the first miRNA‐targeting drug in clinical use. Phase I and II clinical trials evaluated the safety and efficacy of miravirsen, finding that patients had a dose‐dependent reduction in HCV levels and no adverse side effects 121. However, as miRNAs have widespread impact and can function in various pathways, the long‐term effects of antagomirs must be more thoroughly investigated. Notably, in other contexts, miR‐122 has been shown to function as a tumor suppressor, including in GBM, where decreased expression of miR‐122 is associated with decreased patient survival 122. Thus, antagonizing miRNAs likely has different consequences depending on the disease context.
Beyond determining long‐term effects, one of the greatest challenges with miRNA‐based therapeutics is finding effective delivery systems, particularly to the brain. There are various obstacles to efficient delivery including cellular resistance to the uptake of oligonucleotides, sequestration of treatments in the liver, and the blood–brain barrier, which prevents the delivery of most drugs to the brain. A recent study by Song et al. 123 addressed the first issue, finding that R3V6 peptides were able to protect anti‐miR‐21 oligonucleotides from nucleases, while delivering the treatment into the cells more efficiently than other modifications, including polyethylenimine. The peptide was also effective at reducing miR‐21 levels and inducing apoptosis in GBM cells, suggesting that the R3V6 peptide may be a useful carrier for antisense oligonucleotides 123. Other modifications have also been tested, including cholesterol, which allows for injection of antagomirs. However, cholesterol‐conjugated miRNAs are not able to access all tissues, primarily accumulating in the liver 124. Thus, much work remains to be done on optimization of delivery strategies.
miRNA sponges
An alternative to antisense oligonucleotides is the miRNA sponge, which contains multiple binding sites for the miRNA of interest, competitively inhibiting it from binding to its target mRNA. Importantly, the sponge's binding sites are specific to the miRNA's seed region, allowing a single sponge to block all miRNA family members containing the same seed sequence (Fig. 2A) 125.
The efficacy of miRNA sponges depends on the affinity and avidity of binding sites as well as the concentration of sponge RNA relative to miRNAs. miRNA sponges can be modified by manipulation of binding sites and their separating spacers. Binding sites can be either perfectly antisense or can contain mismatches in the middle positions, decreasing vulnerability to Ago2‐mediated endonuclease cleavage. The number of binding sites can also be altered, although increasing the number can expedite sponge RNA degradation. Further, variations in the mismatches and spacers can be introduced to reduce the risk of recombination during cloning or of introducing unintended binding motifs for other factors 125.
Chen et al. 126 demonstrated the utility of miRNA sponges in a glioma cell line and orthotopic mouse model, using an miRNA sponge to inhibit miR‐23b, which functions as an oncogene in GBM. Knockdown of miR‐23b led to a significant reduction in tumor malignancy accompanied by downregulation of HIF1α, β‐catenin, MMP2, MMP9, VEGF, and ZEB1, as well as increased expression of VHL and E‐cadherin. This was able to reduce angiogenesis, migration, and invasion of the GBM cells, thereby inhibiting cancer progression 126.
Further improvements have been made on first‐generation sponges, resulting in optimized, more potent miRNA sponges such as “tough decoy RNAs” (TuD RNA). TuD RNAs place the miRNA binding site(s) in the single‐stranded region of short stem‐loops, precisely presenting them for binding to miRNA complexes. This modification allows for more specific and efficient biological effects, including long‐term suppression of specific miRNAs 127.
There are also naturally occurring miRNA sponges, such as circular RNAs (circRNA). These RNAs are highly abundant, with thousands found in mammals 128. CircRNAs are generated via backsplicing or through partial degradation of intron lariat RNAs, forming stable closed loops that are resistant to debranching enzymes and RNA exonucleases. A circRNA sponge for miR‐7 (ciRS‐7) has been of particular interest after Hansen et al. 129 found that ciRS‐7 and miR‐7 were coexpressed in the mouse brain. ciRS‐7 contains more than 70 conserved miRNA target sites and strongly suppresses miR‐7 activity, resulting in increased levels of miR‐7 targets. As circRNAs are more thoroughly investigated, they may provide further insight for designing more effective strategies for regulating miRNAs and miRNA target genes.
Mimics
miRNA replacement therapy aims to restore or amplify a loss of function, particularly of tumor suppressor activity. This can be achieved through administration of synthetic miRNA mimics, which have identical sequences to their naturally occurring equivalents (Fig. 2B). For example, Chen et al. found that miR‐203 was significantly decreased in high WHO grade gliomas as compared to low WHO grade gliomas and normal brain tissue. Transfection of miR‐203 mimics into U251 human GBM cells markedly downregulated expression of phospholipase D2, a target of miR‐213 which is thought to be oncogenic in GBM. This suppressed the proliferation and invasion of U251 cells, demonstrating the utility of mimics for correcting miRNA depletion.
Mimics can also be delivered systemically with technologies used for delivering short interfering RNAs. Indeed, Ibrahim et al. 130 was able to use polyethylenimine (PEI)‐mediated delivery of chemically synthesized, unmodified miR‐145 and miR‐33a to preclinically validate the delivery method in a mouse model of colon carcinoma. Intraperitoneal injection of PEI‐complexed miR‐145 substantially reduced tumor proliferation and increased apoptosis of tumor cells, while miR‐33a decreased oncogenic kinase Pim‐1 and dampened tumor cell proliferation. Administration of both miRNAs resulted in significant reductions in tumor size, illustrating efficacy of both the mimics and the delivery mechanism.
Investigations of miRNA mimics in clinical trials have also begun. MRX34, a liposome‐formulated mimic of miR‐34 is currently undergoing a Phase I study conducted by Mirna Therapeutics. This study will examine its effects on unresectable primary liver cancer or advanced metastatic cancer with liver involvement (ClinicalTrials.gov Identifier: NCT01829971). While not targeting GBM, the expansion of miRNA use in treatment strategies represents promising advancement.
Additional delivery methods
As previously noted, one of the greatest challenges to efficacy of miRNA‐based therapies is the absence of effective delivery mechanisms. Currently, there are a myriad of strategies being investigated, focusing on delivery to various locations. One approach that has been gaining traction for use in gene therapy is the utilization of vectors based on adeno‐associated viruses (AAV). AAVs provide a long‐term treatment option that displays high transduction efficacy and tissue‐specific tropism 131. There are currently 12 AAV serotypes and more than 100 variants for the transfer of foreign genes into the liver, pancreas, heart, lung, skeletal muscle, and central nervous system 132. A major barrier to using AAVs in humans is the low but persistent host immune response seen in preclinical and clinical trials 133. However, there are now modified, or recombinant AAVs (rAAV), which are even more effective than the wild‐type, with lower risk and greater predictability. Currently, rAAVs are being used in clinical trials for various diseases including muscular dystrophy, Parkinson's, cystic fibrosis, and hemophilia B 133.
As gene transfer represents a long‐term strategy, the most useful miRNAs for AAV‐mediated delivery are those that are highly expressed and well tolerated in normal tissues but are lost in tumor cells. Kota et al. 134 used this approach with miR‐26a, an miRNA that is expressed at high levels in normal tissue but is lost in hepatocellular carcinoma. Systemic administration of miR‐26a via AAVs resulted in increased levels of miR‐26a in the tumors and subsequent inhibition of cancer cell proliferation, induction of tumor‐specific apoptosis, and dramatic protection from disease progression without toxicity. Thus, AAVs may represent one efficacious approach for miRNA delivery, although not appropriate for all contexts.
Nanoparticles provide another mechanism for delivery of miRNAs. Nanoparticles are highly modifiable and can be optimized for different uses by modulating the material, size, charge, and presence of surface proteins. Kouri et al. 135 used gold nanoparticles to develop a novel construct of RNA‐based spherical nucleic acids (SNA) functionalized with mature miR‐182 sequences (182‐SNA). SNAs exhibit high cellular uptake, high stability, high resistance to nuclease degradation, low activation of the innate immune system, and no significant acute or long‐term toxicity in animal models. SNAs also display preferential accumulation within intracerebral gliomas due to the sequestration of nanomaterials in tumors because of the abnormal architecture of tumor blood vessels. 182‐SNAs were able to effectively cross the blood–brain and blood–tumor barrier following systemic intravenous administration in glioma‐bearing mice. The constructs disseminated throughout the glioma parenchyma where glioma cells displayed robust cellular uptake of the 182‐SNAs. This was accompanied by potent downregulation of target proteins as well as enhanced sensitivity toward chemotherapy‐induced apoptosis, resulting in significantly reduced tumor burden and increased survival time with no significant adverse side effects 135, 136.
Mesenchymal stem cells are also predicted to be effective delivery vehicles due to their ability to preferentially home to tumor tissue. Lee et al. 137 found that MSCs derived from bone marrow, adipose tissue, placenta, and umbilical cords could deliver synthetic miRNA mimics to glioma cells through both gap junction‐dependent and ‐independent mechanisms. MSCs transfected with Cy‐3‐labeled miR‐124 and miR‐145 efficiently delivered the miRNAs to adjacent cocultured glioma cells. This resulted in downregulation of miR‐124 and miR‐145 target genes and decreased the migration and self‐renewal capacities of glioma cells 137. Munoz et al. 138 likewise confirmed that MSCs could transfer miRNA‐based therapies to GBM cells. They found that anti‐miR‐9 was transferred from MSCs to GBM cells via gap junctional intercellular communication as well as through the release of microvesicles. The delivery of anti‐miR‐9 sensitized GBM cells to TMZ, increasing caspase activity and cell death 138. Further, studies have demonstrated that MSCs are capable of localizing to endogenous high‐grade gliomas after intraarterial delivery in mice 139.
Challenges
Despite advances in understanding of both miRNAs and GBM, there are still major difficulties to overcome before widespread application of miRNA‐based treatments becomes feasible. The lack of an effective delivery system with enough specificity and efficacy is one of the greatest challenges in utilizing miRNAs for therapeutics. This is particularly relevant for GBM, where both the blood–brain and blood–tumor barriers prevent the accumulation of therapeutic drug concentrations in brain tumors. However, with the development of more accurate in vitro models of the blood–brain barrier, we have the opportunity to more rapidly and easily screen new delivery mechanisms for potential in penetrating the blood–brain barrier and specifically targeting tumor sites 140. Alternatively, emphasis on the development of more effective local delivery techniques that can bypass the blood–brain barrier could also offer more efficacious solutions 141.
Additional challenges could arise from the ability of miRNAs to engage with multiple targets. While offering many advantages, this ability is a double‐edged sword. The potential for widespread effects increases the risk of deleterious repercussions from impact on unintended targets. Additionally, as mRNAs can be targeted by numerous different miRNAs, compensation mechanisms through alternative pathways could compromise treatment outcomes.
Finally, tumor heterogeneity is a major challenge as there are substantial variations across individuals as well as between different cells within tumors. This includes the presence of GSCs, which can dramatically impact tumor aggressiveness and resistance to treatments. Thus, each tumor may respond differently to the same therapeutic strategies, necessitating a need for more individualized treatment options. With the increasing ease in obtaining genomic information for each patient, miRNA profiling could be used as a critical factor for evaluating response to treatment. Thus, more comprehensive characterization of potential predictive and prognostic miRNA markers is essential to optimizing personalized treatments for GBM.
Conclusion
miRNAs have been called “small RNA molecules with a huge impact” due to their wide range of targets and functions in carcinogenesis 142. The understanding of miRNAs is rapidly developing with potential to drastically change the conceptualization of developmental biology, including the development of pathological conditions such as cancer. In particular, miRNA expression profiling has enhanced our understanding of GBM disease progression, providing valuable information on pathogenesis and potential targets. Detecting and quantifying miRNAs in tissue and serum will likely become a standard diagnostic and prognostic tool for GBM, potentially serving as a mechanism for creating personalized treatment strategies. As the role of miRNAs in GBM become more well‐understood and novel delivery methods are developed and optimized, miRNA‐based targeted therapies could provide a promising advancement in cancer therapeutics. Thus, mapping of expression profiles for different disease states as well as identification of pathways targeted by miRNAs could be a critical step forward in determining more effective treatment mechanisms for various cancers, including GBM.
Conflict of Interest
The authors declare that they have no conflict of interest.
Cancer Medicine 2016; 5(8):1917–1946
References
- 1. Ostrom, Q. T. , Gittleman H., Liao P., Rouse C., Chen Y., Dowling J., et al. 2014. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007‐2011. Neuro. Oncol. 16(Suppl. 4):iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp, R. , Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J., Janzer R. C., et al. 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5‐year analysis of the EORTC‐NCIC trial. Lancet Oncol. 10:459–466. [DOI] [PubMed] [Google Scholar]
- 3. Vella, M. C. , Choi E. Y., Lin S. Y., Reinert K., and Slack F. J.. 2004. The C. elegans microRNA let‐7 binds to imperfect let‐7 complementary sites from the lin‐41 3′UTR. Genes Dev. 18:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee, Y. , Kim M., Han J., Yeom K. H., Lee S., Baek S. H., et al. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winter, J. , Jung S., Keller S., Gregory R. I., and Diederichs S.. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11:228–234. [DOI] [PubMed] [Google Scholar]
- 6. Lee, Y. , Ahn C., Han J., Choi H., Kim J., Yim J., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. [DOI] [PubMed] [Google Scholar]
- 7. Han, J. , Lee Y., Yeom K. H., Kim Y. K., Jin H., and Kim V. N.. 2004. The Drosha‐DGCR8 complex in primary microRNA processing. Genes Dev. 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yi, R. , Qin Y., Macara I. G., and Cullen B. R.. 2003. Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev. 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee, H. Y. , Zhou K., Smith A. M., Noland C. L., and Doudna J. A.. 2013. Differential roles of human Dicer‐binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 41:6568–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawamata, T. , Yoda M., and Tomari Y.. 2011. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 12:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacobsen, A. , Silber J., Harinath G., Huse J. T., Schultz N., and Sander C.. 2013. Analysis of microRNA‐target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 20:1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin, H. C. , Wani S., Steptoe A. L., Krishnan K., Nones K., Nourbakhsh E., et al. 2014. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 15:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis, B. P. , Burge C. B., and Bartel D. P.. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. [DOI] [PubMed] [Google Scholar]
- 14. Calin, G. A. , and Croce C. M.. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6:857–866. [DOI] [PubMed] [Google Scholar]
- 15. Calin, G. A. , Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., et al. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Visone, R. , and Croce C. M.. 2009. MiRNAs and cancer. Am. J. Pathol. 174:1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moller, H. G. , Rasmussen A. P., Andersen H. H., Johnsen K. B., Henriksen M., and Duroux M.. 2013. A systematic review of microRNA in glioblastoma multiforme: micro‐modulators in the mesenchymal mode of migration and invasion. Mol. Neurobiol. 47:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malzkorn, B. , Wolter M., Liesenberg F., Grzendowski M., Stuhler K., Meyer H. E., et al. 2010. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 20:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang, L. , Mao P., Song L., Wu J., Huang J., Lin C., et al. 2010. miR‐182 as a prognostic marker for glioma progression and patient survival. Am. J. Pathol. 177:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun, G. , Cao Y., Shi L., Sun L., Wang Y., Chen C., et al. 2013. Overexpressed miRNA‐137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother. Radiopharm. 28:327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanahan, D. , and Weinberg R. A.. 2000. The hallmarks of cancer. Cell 100:57–70. [DOI] [PubMed] [Google Scholar]
- 22. Hanahan, D. , and Weinberg R. A.. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- 23. Kefas, B. , Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., et al. 2008. microRNA‐7 inhibits the epidermal growth factor receptor and the Akt pathway and is down‐regulated in glioblastoma. Cancer Res. 68:3566–3572. [DOI] [PubMed] [Google Scholar]
- 24. Liu, Z. , Liu Y., Li L., Xu Z., Bi B., Wang Y., et al. 2014. MiR‐7‐5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumour Biol. 35:10177–10184. [DOI] [PubMed] [Google Scholar]
- 25. Papagiannakopoulos, T. , Friedmann‐Morvinski D., Neveu P., Dugas J. C., Gill R. M., Huillard E., et al. 2012. Pro‐neural miR‐128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 31:1884–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wuchty, S. , Arjona D., Li A., Kotliarov Y., Walling J., Ahn S., et al. 2011. Prediction of associations between microRNAs and Gene expression in glioma biology. PLoS ONE 6:e14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vo, D. T. , Qiao M., Smith A. D., Burns S. C., Brenner A. J., and Penalva L. O.. 2011. The oncogenic RNA‐binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol. 8:817–828. [DOI] [PubMed] [Google Scholar]
- 28. Zhang, Y. , Chao T., Li R., Liu W., Chen Y., Yan X., et al. 2009. MicroRNA‐128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. (Berl) 87:43–51. [DOI] [PubMed] [Google Scholar]
- 29. Silber, J. , Lim D. A., Petritsch C., Persson A. I., Maunakea A. K., Yu M., et al. 2008. miR‐124 and miR‐137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pratheeshkumar, P. , Budhraja A., Son Y. O., Wang X., Zhang Z., Ding S., et al. 2012. Quercetin inhibits angiogenesis mediated human prostate tumor growth by targeting VEGFR‐ 2 regulated AKT/mTOR/P70S6K signaling pathways. PLoS ONE 7:e47516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lukiw, W. J. , Cui J. G., Li Y. Y., and Culicchia F.. 2009. Up‐regulation of micro‐RNA‐221 (miRNA‐221; chr Xp11.3) and caspase‐3 accompanies down‐regulation of the survivin‐1 homolog BIRC1 (NAIP) in glioblastoma multiforme (GBM). J. Neurooncol. 91:27–32. [DOI] [PubMed] [Google Scholar]
- 32. Sun, F. , Fu H., Liu Q., Tie Y., Zhu J., Xing R., et al. 2008. Downregulation of CCND1 and CDK6 by miR‐34a induces cell cycle arrest. FEBS Lett. 582:1564–1568. [DOI] [PubMed] [Google Scholar]
- 33. Bueno, M. J. , and Malumbres M.. 2011. MicroRNAs and the cell cycle. Biochim. Biophys. Acta 1812:592–601. [DOI] [PubMed] [Google Scholar]
- 34. Luan, S. , Sun L., and Huang F.. 2010. MicroRNA‐34a: a novel tumor suppressor in p53‐mutant glioma cell line U251. Arch. Med. Res. 41:67–74. [DOI] [PubMed] [Google Scholar]
- 35. Lee, S. T. , Chu K., Oh H. J., Im W. S., Lim J. Y., Kim S. K., et al. 2011. Let‐7 microRNA inhibits the proliferation of human glioblastoma cells. J. Neurooncol. 102:19–24. [DOI] [PubMed] [Google Scholar]
- 36. Talotta, F. , Cimmino A., Matarazzo M. R., Casalino L., De Vita G., D'Esposito M., et al. 2009. An autoregulatory loop mediated by miR‐21 and PDCD4 controls the AP‐1 activity in RAS transformation. Oncogene 28:73–84. [DOI] [PubMed] [Google Scholar]
- 37. Gaur, A. B. , Holbeck S. L., Colburn N. H., and Israel M. A.. 2011. Downregulation of Pdcd4 by mir‐21 facilitates glioblastoma proliferation in vivo. Neuro. Oncol. 13:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang, C. H. , Yue J., Pfeffer S. R., Fan M., Paulus E., Hosni‐Ahmed A., et al. 2014. MicroRNA‐21 promotes glioblastoma tumorigenesis by down‐regulating insulin‐like growth factor‐binding protein‐3 (IGFBP3). J. Biol. Chem. 289:25079–25087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li, Y. , Li W., Yang Y., Lu Y., He C., Hu G., et al. 2009. MicroRNA‐21 targets LRRFIP1 and contributes to VM‐26 resistance in glioblastoma multiforme. Brain Res. 1286:13–18. [DOI] [PubMed] [Google Scholar]
- 40. Kwak, H. J. , Kim Y. J., Chun K. R., Woo Y. M., Park S. J., Jeong J. A., et al. 2011. Downregulation of Spry2 by miR‐21 triggers malignancy in human gliomas. Oncogene 30:2433–2442. [DOI] [PubMed] [Google Scholar]
- 41. Chen, Y. , Liu W., Chao T., Zhang Y., Yan X., Gong Y., et al. 2008. MicroRNA‐21 down‐regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 272:197–205. [DOI] [PubMed] [Google Scholar]
- 42. Abba, M. , Patil N., and Allgayer H.. 2014. MicroRNAs in the regulation of MMPs and metastasis. Cancers (Basel) 6:625–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou, X. , Ren Y., Moore L., Mei M., You Y., Xu P., et al. 2010. Downregulation of miR‐21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest. 90:144–155. [DOI] [PubMed] [Google Scholar]
- 44. Zhou, X. , Zhang J., Jia Q., Ren Y., Wang Y., Shi L., et al. 2010. Reduction of miR‐21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol. Rep. 24:195–201. [DOI] [PubMed] [Google Scholar]
- 45. Corsten, M. F. , Miranda R., Kasmieh R., Krichevsky A. M., Weissleder R., and Shah K.. 2007. MicroRNA‐21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S‐TRAIL in human gliomas. Cancer Res. 67:8994–9000. [DOI] [PubMed] [Google Scholar]
- 46. Papagiannakopoulos, T. , Shapiro A., and Kosik K. S.. 2008. MicroRNA‐21 targets a network of key tumor‐suppressive pathways in glioblastoma cells. Cancer Res. 68:8164–8172. [DOI] [PubMed] [Google Scholar]
- 47. Shang, C. , Guo Y., Hong Y., Liu Y., and Xue Y.. 2015. MiR‐21 up‐regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting FASLG. Mol. Biol. Rep. 42:721–727. [DOI] [PubMed] [Google Scholar]
- 48. Zhang, C. Z. , Zhang J. X., Zhang A. L., Shi Z. D., Han L., Jia Z. F., et al. 2010. MiR‐221 and miR‐222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer. 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen, L. , Zhang J., Han L., Zhang A., Zhang C., Zheng Y., et al. 2012. Downregulation of miR‐221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol. Rep. 27:854–860. [DOI] [PubMed] [Google Scholar]
- 50. Medina, R. , Zaidi S. K., Liu C. G., Stein J. L., van Wijnen A. J., Croce C. M., et al. 2008. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 68:2773–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang, C. , Kang C., You Y., Pu P., Yang W., Zhao P., et al. 2009. Co‐suppression of miR‐221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int. J. Oncol. 34:1653–1660. [DOI] [PubMed] [Google Scholar]
- 52. Li, W. , Guo F., Wang P., Hong S., and Zhang C.. 2014. miR‐221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr. Mol. Med. 14:185–195. [DOI] [PubMed] [Google Scholar]
- 53. Shu, M. , Zheng X., Wu S., Lu H., Leng T., Zhu W., et al. 2011. Targeting oncogenic miR‐335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer. 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang, J. M. , Sun C. Y., Yu S. Z., Wang Q., An T. L., Li Y. Y., et al. 2011. Relationship between miR‐218 and CDK6 expression and their biological impact on glioma cell proliferation and apoptosis. Zhonghua Bing Li Xue Za Zhi 40:454–459. [PubMed] [Google Scholar]
- 55. Xia, H. , Yan Y., Hu M., Wang Y., Wang Y., Dai Y., et al. 2013. MiR‐218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP‐mediated suppression of NF‐kappaB activity. Neuro. Oncol. 15:413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nan, Y. , Han L., Zhang A., Wang G., Jia Z., Yang Y., et al. 2010. MiRNA‐451 plays a role as tumor suppressor in human glioma cells. Brain Res. 1359:14–21. [DOI] [PubMed] [Google Scholar]
- 57. Mathew, R. , Karantza‐Wadsworth V., and White E.. 2007. Role of autophagy in cancer. Nat. Rev. Cancer 7:961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Comincini, S. , Allavena G., Palumbo S., Morini M., Durando F., Angeletti F., et al. 2013. microRNA‐17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low‐dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 14:574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gwak, H. S. , Kim T. H., Jo G. H., Kim Y. J., Kwak H. J., Kim J. H., et al. 2012. Silencing of microRNA‐21 confers radio‐sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS ONE 7:e47449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Singh, S. K. , Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., et al. 2004. Identification of human brain tumour initiating cells. Nature 432:396–401. [DOI] [PubMed] [Google Scholar]
- 61. Bao, S. , Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A. B., et al. 2006. Stem cell‐like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66:7843–7848. [DOI] [PubMed] [Google Scholar]
- 62. Vescovi, A. L. , Galli R., and Reynolds B. A.. 2006. Brain tumour stem cells. Nat. Rev. Cancer 6:425–436. [DOI] [PubMed] [Google Scholar]
- 63. Lavon, I. , Zrihan D., Granit A., Einstein O., Fainstein N., Cohen M. A., et al. 2010. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro. Oncol. 12:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Godlewski, J. , Nowicki M. O., Bronisz A., Williams S., Otsuki A., Nuovo G., et al. 2008. Targeting of the Bmi‐1 oncogene/stem cell renewal factor by microRNA‐128 inhibits glioma proliferation and self‐renewal. Cancer Res. 68:9125–9130. [DOI] [PubMed] [Google Scholar]
- 65. Du, W. , Liu X., Chen L., Dou Z., Lei X., Chang L., et al. 2015. Targeting the SMO oncogene by miR‐326 inhibits glioma biological behaviors and stemness. Neuro. Oncol. 17:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wong, H. K. , Fatimy R. E., Onodera C., Wei Z., Yi M., Mohan A., et al. 2015. The cancer genome atlas analysis predicts microrna for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 23:1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ma, J. , Yao Y., Wang P., Liu Y., Zhao L., Li Z., et al. 2014. MiR‐152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel‐like factor 4. Cancer Lett. 355:85–95. [DOI] [PubMed] [Google Scholar]
- 68. Kalluri, R. , and Weinberg R. A.. 2009. The basics of epithelial‐mesenchymal transition. J. Clin. Invest. 119:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schramedei, K. , Morbt N., Pfeifer G., Lauter J., Rosolowski M., Tomm J. M., et al. 2011. MicroRNA‐21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene 30:2975–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gabriely, G. , Wurdinger T., Kesari S., Esau C. C., Burchard J., Linsley P. S., et al. 2008. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 28:5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li, Y. , Wang Y., Yu L., Sun C., Cheng D., Yu S., et al. 2013. miR‐146b‐5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 339:260–269. [DOI] [PubMed] [Google Scholar]
- 72. Sasayama, T. , Nishihara M., Kondoh T., Hosoda K., and Kohmura E.. 2009. MicroRNA‐10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer 125:1407–1413. [DOI] [PubMed] [Google Scholar]
- 73. Sun, L. , Yan W., Wang Y., Sun G., Luo H., Zhang J., et al. 2011. MicroRNA‐10b induces glioma cell invasion by modulating MMP‐14 and uPAR expression via HOXD10. Brain Res. 1389:9–18. [DOI] [PubMed] [Google Scholar]
- 74. Lin, J. , Teo S., Lam D. H., Jeyaseelan K., and Wang S.. 2012. MicroRNA‐10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis. 3:e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu, D. G. , Wang Y. Y., Fan L. G., Luo H., Han B., Sun L. H., et al. 2011. MicroRNA‐7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin. Med. J. (Engl.) 124:2616–2621. [PubMed] [Google Scholar]
- 76. Lu, Z. J. , Liu S. Y., Yao Y. Q., Zhou Y. J., Zhang S., Dai L., et al. 2011. The effect of miR‐7 on behavior and global protein expression in glioma cell lines. Electrophoresis 32:3612–3620. [DOI] [PubMed] [Google Scholar]
- 77. Wurdinger, T. , and Tannous B. A.. 2009. Glioma angiogenesis: towards novel RNA therapeutics. Cell Adh. Migr. 3:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang, S. , and Olson E. N.. 2009. AngiomiRs–key regulators of angiogenesis. Curr. Opin. Genet. Dev. 19:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wurdinger, T. , Tannous B. A., Saydam O., Skog J., Grau S., Soutschek J., et al. 2008. miR‐296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14:382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Smits, M. , Wurdinger T., van het Hof B., Drexhage J. A., Geerts D., Wesseling P., et al. 2012. Myc‐associated zinc finger protein (MAZ) is regulated by miR‐125b and mediates VEGF‐induced angiogenesis in glioblastoma. FASEB J. 26:2639–2647. [DOI] [PubMed] [Google Scholar]
- 81. Agrawal, R. , Pandey P., Jha P., Dwivedi V., Sarkar C., and Kulshreshtha R.. 2014. Hypoxic signature of microRNAs in glioblastoma: insights from small RNA deep sequencing. BMC Genom. 15:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shi, Z. M. , Wang J., Yan Z., You Y. P., Li C. Y., Qian X., et al. 2012. MiR‐128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 7:e32709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fang, L. , Deng Z., Shatseva T., Yang J., Peng C., Du W. W., et al. 2011. MicroRNA miR‐93 promotes tumor growth and angiogenesis by targeting integrin‐beta8. Oncogene 30:806–821. [DOI] [PubMed] [Google Scholar]
- 84. Srinivasan, S. , Patric I. R., and Somasundaram K.. 2011. A ten‐microRNA expression signature predicts survival in glioblastoma. PLoS ONE 6:e17438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guan, Y. , Mizoguchi M., Yoshimoto K., Hata N., Shono T., Suzuki S. O., et al. 2010. MiRNA‐196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin. Cancer Res. 16:4289–4297. [DOI] [PubMed] [Google Scholar]
- 86. Zhi, F. , Chen X., Wang S., Xia X., Shi Y., Guan W., et al. 2010. The use of hsa‐miR‐21, hsa‐miR‐181b and hsa‐miR‐106a as prognostic indicators of astrocytoma. Eur. J. Cancer 46:1640–1649. [DOI] [PubMed] [Google Scholar]
- 87. Ujifuku, K. , Mitsutake N., Takakura S., Matsuse M., Saenko V., Suzuki K., et al. 2010. miR‐195, miR‐455‐3p and miR‐10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 296:241–248. [DOI] [PubMed] [Google Scholar]
- 88. Slaby, O. , Lakomy R., Fadrus P., Hrstka R., Kren L., Lzicarova E., et al. 2010. MicroRNA‐181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma 57:264–269. [DOI] [PubMed] [Google Scholar]
- 89. Shi, L. , Chen J., Yang J., Pan T., Zhang S., and Wang Z.. 2010. MiR‐21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl‐2 ratio and caspase‐3 activity. Brain Res. 1352:255–264. [DOI] [PubMed] [Google Scholar]
- 90. Costa, P. M. , Cardoso A. L., Nobrega C., Pereira de Almeida L. F., Bruce J. N., Canoll P., et al. 2013. MicroRNA‐21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum. Mol. Genet. 22:904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wong, S. T. , Zhang X. Q., Zhuang J. T., Chan H. L., Li C. H., and Leung G. K.. 2012. MicroRNA‐21 inhibition enhances in vitro chemosensitivity of temozolomide‐resistant glioblastoma cells. Anticancer Res. 32:2835–2841. [PubMed] [Google Scholar]
- 92. Barker, C. A. , Chang M., Chou J. F., Zhang Z., Beal K., Gutin P. H., et al. 2012. Radiotherapy and concomitant temozolomide may improve survival of elderly patients with glioblastoma. J. Neurooncol. 109:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang, S. , Han L., Wei J., Shi Z., Pu P., Zhang J., et al. 2015. Combination treatment with doxorubicin and microRNA‐21 inhibitor synergistically augments anticancer activity through upregulation of tumor suppressing genes. Int. J. Oncol. 46:1589–1600. [DOI] [PubMed] [Google Scholar]
- 94. Ren, Y. , Zhou X., Mei M., Yuan X. B., Han L., Wang G. X., et al. 2010. MicroRNA‐21 inhibitor sensitizes human glioblastoma cells U251 (PTEN‐mutant) and LN229 (PTEN‐wild type) to taxol. BMC Cancer 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li, W. Q. , Li Y. M., Tao B. B., Lu Y. C., Hu G. H., Liu H. M., et al. 2010. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA‐328 may decrease their chemoresistance. Med. Sci. Monit. 16:HY27–HY30. [PubMed] [Google Scholar]
- 96. Wu, Z. , Sun L., Wang H., Yao J., Jiang C., Xu W., et al. 2012. MiR‐328 expression is decreased in high‐grade gliomas and is associated with worse survival in primary glioblastoma. PLoS ONE 7:e47270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Challagundla, K. B. , Fanini F., Vannini I., Wise P., Murtadha M., Malinconico L., et al. 2014. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev. Mol. Diagn. 14:565–574. [DOI] [PubMed] [Google Scholar]
- 98. Wang, W. T. , and Chen Y. Q.. 2014. Circulating miRNAs in cancer: from detection to therapy. J. Hematol. Oncol. 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang, Q. , Li P., Li A., Jiang W., Wang H., Wang J., et al. 2012. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. 31:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Roth, P. , Wischhusen J., Happold C., Chandran P. A., Hofer S., Eisele G., et al. 2011. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 118:449–457. [DOI] [PubMed] [Google Scholar]
- 101. Sun, J. , Liao K., Wu X., Huang J., Zhang S., and Lu X.. 2015. Serum microRNA‐128 as a biomarker for diagnosis of glioma. Int. J. Clin. Exp. Med. 8:456–463. [PMC free article] [PubMed] [Google Scholar]
- 102. Yue, X. , Lan F., Hu M., Pan Q., Wang Q., and Wang J.. 2015. Downregulation of serum microRNA‐205 as a potential diagnostic and prognostic biomarker for human glioma. J. Neurosurg. 124:122–128. [DOI] [PubMed] [Google Scholar]
- 103. Turchinovich, A. , Weiz L., Langheinz A., and Burwinkel B.. 2011. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Arroyo, J. D. , Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., et al. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chen, X. , Liang H., Zhang J., Zen K., and Zhang C. Y.. 2012. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell 3:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Raposo, G. , and Stoorvogel W.. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Skog, J. , Wurdinger T., van Rijn S., Meijer D. H., Gainche L., Sena‐Esteves M., et al. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pigati, L. , Yaddanapudi S. C., Iyengar R., Kim D. J., Hearn S. A., Danforth D., et al. 2010. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 5:e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Palma, J. , Yaddanapudi S. C., Pigati L., Havens M. A., Jeong S., Weiner G. A., et al. 2012. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 40:9125–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tominaga, N. , Kosaka N., Ono M., Katsuda T., Yoshioka Y., Tamura K., et al. 2015. Brain metastatic cancer cells release microRNA‐181c‐containing extracellular vesicles capable of destructing blood‐brain barrier. Nat. Commun. 6:6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ruan, J. , Lou S., Dai Q., Mao D., Ji J., and Sun X.. 2015. Tumor suppressor miR‐181c attenuates proliferation, invasion, and self‐renewal abilities in glioblastoma. NeuroReport 26:66–73. [DOI] [PubMed] [Google Scholar]
- 112. Zhou, W. , Fong M. Y., Min Y., Somlo G., Liu L., Palomares M. R., et al. 2014. Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25:501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kosaka, N. , Iguchi H., Yoshioka Y., Hagiwara K., Takeshita F., and Ochiya T.. 2012. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J. Biol. Chem. 287:1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lennox, K. A. , Owczarzy R., Thomas D. M., Walder J. A., and Behlke M. A.. 2013. Improved performance of anti‐miRNA oligonucleotides Using a Novel Non‐Nucleotide Modifier. Mol. Ther. Nucleic Acids 2:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yoo, B. H. , Bochkareva E., Bochkarev A., Mou T. C., and Gray D. M.. 2004. 2′‐O‐methyl‐modified phosphorothioate antisense oligonucleotides have reduced non‐specific effects in vitro. Nucleic Acids Res. 32:2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nana‐Sinkam, S. P. , and Croce C. M.. 2013. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 93:98–104. [DOI] [PubMed] [Google Scholar]
- 117. Shibata, C. , Otsuka M., Kishikawa T., Yoshikawa T., Ohno M., Takata A., et al. 2013. Current status of miRNA‐targeting therapeutics and preclinical studies against gastroenterological carcinoma. Mol. Cell. Ther. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Elmen, J. , Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., et al. 2005. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 33:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Elmen, J. , Lindow M., Schutz S., Lawrence M., Petri A., Obad S., et al. 2008. LNA‐mediated microRNA silencing in non‐human primates. Nature 452:896–899. [DOI] [PubMed] [Google Scholar]
- 120. Lanford, R. E. , Hildebrandt‐Eriksen E. S., Petri A., Persson R., Lindow M., Munk M. E., et al. 2010. Therapeutic silencing of microRNA‐122 in primates with chronic hepatitis C virus infection. Science 327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Janssen, H. L. , Reesink H. W., Lawitz E. J., Zeuzem S., Rodriguez‐Torres M., Patel K., et al. 2013. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- 122. Wang, G. , Zhao Y., and Zheng Y.. 2014. MiR‐122/Wnt/beta‐catenin regulatory circuitry sustains glioma progression. Tumour Biol. 35:8565–8572. [DOI] [PubMed] [Google Scholar]
- 123. Song, H. , Oh B., Choi M., Oh J., and Lee M.. 2015. Delivery of anti‐microRNA‐21 antisense‐oligodeoxynucleotide using amphiphilic peptides for glioblastoma gene therapy. J. Drug Target. 23:360–370. [DOI] [PubMed] [Google Scholar]
- 124. Krutzfeldt, J. , Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., et al. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689. [DOI] [PubMed] [Google Scholar]
- 125. Ebert, M. S. , and Sharp P. A.. 2010. MicroRNA sponges: progress and possibilities. RNA 16:2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chen, L. , Zhang K., Shi Z., Zhang A., Jia Z., Wang G., et al. 2014. A lentivirus‐mediated miR‐23b sponge diminishes the malignant phenotype of glioma cells in vitro and in vivo. Oncol. Rep. 31:1573–1580. [DOI] [PubMed] [Google Scholar]
- 127. Haraguchi, T. , Ozaki Y., and Iba H.. 2009. Vectors expressing efficient RNA decoys achieve the long‐term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Song, D. P. , Lin Y., Gai Y., Colella N. S., Li C., Liu X. H., et al. 2015. Controlled supramolecular self‐assembly of large nanoparticles in amphiphilic brush block copolymers. J. Am. Chem. Soc. 137:3771–3774. [DOI] [PubMed] [Google Scholar]
- 129. Hansen, T. B. , Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. [DOI] [PubMed] [Google Scholar]
- 130. Ibrahim, A. F. , Weirauch U., Thomas M., Grunweller A., Hartmann R. K., and Aigner A.. 2011. MicroRNA replacement therapy for miR‐145 and miR‐33a is efficacious in a model of colon carcinoma. Cancer Res. 71:5214–5224. [DOI] [PubMed] [Google Scholar]
- 131. Xie, J. , Burt D., and Gao G.. 2015. AAV‐mediated miRNA delivery and therapeutics. Semin. Liver Dis. 35:081–088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Asokan, A. , Schaffer D. V., and Samulski R. J.. 2012. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 20:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mingozzi, F. , and High K. A.. 2011. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12:341–355. [DOI] [PubMed] [Google Scholar]
- 134. Kota, J. , Chivukula R. R., O'Donnell K. A., Wentzel E. A., Montgomery C. L., Hwang H. W., et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kouri, F. M. , Hurley L. A., Daniel W. L., Day E. S., Hua Y., Hao L., et al. 2015. miR‐182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 29:732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kouri, F. M. , Ritner C., and Stegh A. H.. 2015. miRNA‐182 and the regulation of the glioblastoma phenotype ‐ toward miRNA‐based precision therapeutics. Cell Cycle 14:3794–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lee, H. K. , Finniss S., Cazacu S., Bucris E., Ziv‐Av A., Xiang C., et al. 2013. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self‐renewal. Oncotarget 4:346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Munoz, J. L. , Bliss S. A., Greco S. J., Ramkissoon S. H., Ligon K. L., and Rameshwar P.. 2013. Delivery of functional anti‐miR‐9 by mesenchymal stem cell‐derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Doucette, T. , Rao G., Yang Y., Gumin J., Shinojima N., Bekele B. N., et al. 2011. Mesenchymal stem cells display tumor‐specific tropism in an RCAS/Ntv‐a glioma model. Neoplasia 13:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wilhelm, I. , and Krizbai I. A.. 2014. In vitro models of the blood‐brain barrier for the study of drug delivery to the brain. Mol. Pharm. 11:1949–1963. [DOI] [PubMed] [Google Scholar]
- 141. Purow, B. 2011. The elephant in the room: do microRNA‐based therapies have a realistic chance of succeeding for brain tumors such as glioblastoma? J. Neurooncol. 103:429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Iorio, M. V. , and Croce C. M.. 2009. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 27:5848–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Guo, Y. , Yan K., Fang J., Qu Q., Zhou M., and Chen F.. 2013. Let‐7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J. Exp. Clin. Cancer Res. 32:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang, X. R. , Luo H., Li H. L., Cao L., Wang X. F., Yan W., et al. 2013. Overexpressed let‐7a inhibits glioma cell malignancy by directly targeting K‐ras, independently of PTEN. Neuro. Oncol. 15:1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Giles, K. M. , Barker A., Zhang P. M., Epis M. R., and Leedman P. J.. 2011. MicroRNA regulation of growth factor receptor signaling in human cancer cells. Methods Mol. Biol. 676:147–163. [DOI] [PubMed] [Google Scholar]
- 146. Babae, N. , Bourajjaj M., Liu Y., Van Beijnum J. R., Cerisoli F., Scaria P. V., et al. 2014. Systemic miRNA‐7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 5:6687–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Liu, Z. , Jiang Z., Huang J., Huang S., Li Y., Yu S., et al. 2014. miR‐7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 44:1571–1580. [DOI] [PubMed] [Google Scholar]
- 148. Skalsky, R. L. , and Cullen B. R.. 2011. Reduced expression of brain‐enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS ONE 6:e24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Visani, M. , de Biase D., Marucci G., Cerasoli S., Nigrisoli E., Bacchi Reggiani M. L., et al.: Group Ps , 2014. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I‐III. Mol. Oncol. 8:417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jeon, H. M. , Sohn Y. W., Oh S. Y., Kim S. H., Beck S., Kim S., et al. 2011. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR‐9*‐mediated suppression of SOX2. Cancer Res. 71:3410–3421. [DOI] [PubMed] [Google Scholar]
- 151. Wu, Z. , Wang L., Li G., Liu H., Fan F., Li Z., et al. 2013. Increased expression of microRNA‐9 predicts an unfavorable prognosis in human glioma. Mol. Cell. Biochem. 384(1–2):263–268. [DOI] [PubMed] [Google Scholar]
- 152. Munoz, J. L. , Rodriguez‐Cruz V., Ramkissoon S. H., Ligon K. L., Greco S. J., and Rameshwar P.. 2015. Temozolomide resistance in glioblastoma occurs by miRNA‐9‐targeted PTCH1, independent of sonic hedgehog level. Oncotarget 6:1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gomez, G. G. , Volinia S., Croce C. M., Zanca C., Li M., Emnett R., et al. 2014. Suppression of microRNA‐9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. 74:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Schraivogel, D. , Weinmann L., Beier D., Tabatabai G., Eichner A., Zhu J. Y., et al. 2011. CAMTA1 is a novel tumour suppressor regulated by miR‐9/9* in glioblastoma stem cells. EMBO J. 30:4309–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gabriely, G. , Yi M., Narayan R. S., Niers J. M., Wurdinger T., Imitola J., et al. 2011. Human glioma growth is controlled by microRNA‐10b. Cancer Res. 71:3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lang, M. F. , Yang S., Zhao C., Sun G., Murai K., Wu X., et al. 2012. Genome‐wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS ONE 7:e36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Guessous, F. , Alvarado‐Velez M., Marcinkiewicz L., Zhang Y., Kim J., Heister S., et al. 2013. Oncogenic effects of miR‐10b in glioblastoma stem cells. J. Neurooncol. 112:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Teplyuk, N. M. , Uhlmann E. J., Wong A. H., Karmali P., Basu M., Gabriely G., et al. 2015. MicroRNA‐10b inhibition reduces E2F1‐mediated transcription and miR‐15/16 activity in glioblastoma. Oncotarget 6:3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zheng, X. , Chopp M., Lu Y., Buller B., and Jiang F.. 2013. MiR‐15b and miR‐152 reduce glioma cell invasion and angiogenesis via NRP‐2 and MMP‐3. Cancer Lett. 329:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Sun, G. , Shi L., Yan S., Wan Z., Jiang N., Fu L., et al. 2014. MiR‐15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. BioMed. Res. Int. 2014:687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Sun, G. , Yan S., Shi L., Wan Z., Jiang N., Li M., et al. 2015. Decreased Expression of miR‐15b in human gliomas is associated with poor prognosis. Cancer Biother. Radiopharm. 30:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chen, F. , Chen L., He H., Huang W., Zhang R., Li P., et al. 2015. Up‐regulation of microRNA‐16 in glioblastoma inhibits the function of endothelial cells and tumor angiogenesis by targeting Bmi‐1. Anticancer Agents Med. Chem. 16:609–620. [DOI] [PubMed] [Google Scholar]
- 163. Yang, T. Q. , Lu X. J., Wu T. F., Ding D. D., Zhao Z. H., Chen G. L., et al. 2014. MicroRNA‐16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor‐kappaB1/MMP9 signaling pathway. Cancer Sci. 105:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Li, X. , Ling N., Bai Y., Dong W., Hui G. Z., Liu D., et al. 2013. MiR‐16‐1 plays a role in reducing migration and invasion of glioma cells. Anat. Rec. (Hoboken) 296:427–432. [DOI] [PubMed] [Google Scholar]
- 165. Lu, S. , Wang S., Geng S., Ma S., Liang Z., and Jiao B.. 2012. Increased expression of microRNA‐17 predicts poor prognosis in human glioma. J. Biomed. Biotechnol. 2012:970761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Li, H. , and Yang B. B.. 2012. Stress response of glioblastoma cells mediated by miR‐17‐5p targeting PTEN and the passenger strand miR‐17‐3p targeting MDM2. Oncotarget 3:1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ernst, A. , Campos B., Meier J., Devens F., Liesenberg F., Wolter M., et al. 2010. De‐repression of CTGF via the miR‐17‐92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29:3411–3422. [DOI] [PubMed] [Google Scholar]
- 168. Song, Y. , Wang P., Zhao W., Yao Y., Liu X., Ma J., et al. 2014. MiR‐18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 324:54–64. [DOI] [PubMed] [Google Scholar]
- 169. Turchi, L. , Debruyne D. N., Almairac F., Virolle V., Fareh M., Neirijnck Y., et al. 2013. Tumorigenic potential of miR‐18A* in glioma initiating cells requires NOTCH‐1 signaling. Stem Cells 31:1252–1265. [DOI] [PubMed] [Google Scholar]
- 170. Fox, J. L. , Dews M., Minn A. J., and Thomas‐Tikhonenko A.. 2013. Targeting of TGFbeta signature and its essential component CTGF by miR‐18 correlates with improved survival in glioblastoma. RNA 19:177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Jia, Z. , Wang K., Zhang A., Wang G., Kang C., Han L., et al. 2013. miR‐19a and miR‐19b overexpression in gliomas. Pathol. Oncol. Res. 19:847–853. [DOI] [PubMed] [Google Scholar]
- 172. Wang, Z. , Wang B., Shi Y., Xu C., Xiao H. L., Ma L. N., et al. 2015. Oncogenic miR‐20a and miR‐106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP‐2. Oncogene 34:1407–1419. [DOI] [PubMed] [Google Scholar]
- 173. Giunti, L. , da Ros M., Vinci S., Gelmini S., Iorio A. L., Buccoliero A. M., et al. 2015. Anti‐miR21 oligonucleotide enhances chemosensitivity of T98G cell line to doxorubicin by inducing apoptosis. Am. J. Cancer Res. 5:231–242. [PMC free article] [PubMed] [Google Scholar]
- 174. Costa, P. M. , Cardoso A. L., Custodia C., Cunha P., Pereira de Almeida L., andPedroso de Lima M. C.. 2015. MiRNA‐21 silencing mediated by tumor‐targeted nanoparticles combined with sunitinib: a new multimodal gene therapy approach for glioblastoma. J Control Release 207:31–39. [DOI] [PubMed] [Google Scholar]
- 175. Lu, Z. , Liu M., Stribinskis V., Klinge C. M., Ramos K. S., Colburn N. H., et al. 2008. MicroRNA‐21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27:4373–4379. [DOI] [PubMed] [Google Scholar]
- 176. Yang, C. H. , Pfeffer S. R., Sims M., Yue J., Wang Y., Linga V. G., et al. 2015. The oncogenic microRNA‐21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J. Biol. Chem. 290:6037–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zhu, S. , Si M. L., Wu H., and Mo Y. Y.. 2007. MicroRNA‐21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282:14328–14336. [DOI] [PubMed] [Google Scholar]
- 178. Lakomy, R. , Sana J., Hankeova S., Fadrus P., Kren L., Lzicarova E., et al. 2011. MiR‐195, miR‐196b, miR‐181c, miR‐21 expression levels and O‐6‐methylguanine‐DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 102:2186–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Loftus, J. C. , Ross J. T., Paquette K. M., Paulino V. M., Nasser S., Yang Z., et al. 2012. miRNA expression profiling in migrating glioblastoma cells: regulation of cell migration and invasion by miR‐23b via targeting of Pyk2. PLoS ONE 7:e39818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Chen, L. , Zhang A., Li Y., Zhang K., Han L., Du W., et al. 2013. MiR‐24 regulates the proliferation and invasion of glioma by ST7L via beta‐catenin/Tcf‐4 signaling. Cancer Lett. 329:174–180. [DOI] [PubMed] [Google Scholar]
- 181. Suh, S. S. , Yoo J. Y., Nuovo G. J., Jeon Y. J., Kim S., Lee T. J., et al. 2012. MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc. Natl Acad. Sci. USA 109:5316–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zhang, J. , Gong X., Tian K., Chen D., Sun J., Wang G., et al. 2015. miR‐25 promotes glioma cell proliferation by targeting CDKN1C. Biomed. Pharmacother. 71:7–14. [DOI] [PubMed] [Google Scholar]
- 183. Peng, G. , Yuan X., Yuan J., Liu Q., Dai M., Shen C., et al. 2015. miR‐25 promotes glioblastoma cell proliferation and invasion by directly targeting NEFL. Mol. Cell. Biochem. 409(1–2):103–111. [DOI] [PubMed] [Google Scholar]
- 184. Guo, P. , Lan J., Ge J., Nie Q., Guo L., Qiu Y., et al. 2014. MiR‐26a enhances the radiosensitivity of glioblastoma multiforme cells through targeting of ataxia‐telangiectasia mutated. Exp. Cell Res. 320:200–208. [DOI] [PubMed] [Google Scholar]
- 185. Huse, J. T. , Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S. H., et al. 2009. The PTEN‐regulating microRNA miR‐26a is amplified in high‐grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 23:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wu, N. , Zhao X., Liu M., Liu H., Yao W., Zhang Y., et al. 2011. Role of microRNA‐26b in glioma development and its mediated regulation on EphA2. PLoS ONE 6:e16264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Rivera‐Diaz, M. , Miranda‐Roman M. A., Soto D., Quintero‐Aguilo M., Ortiz‐Zuazaga H., Marcos‐Martinez M. J., et al. 2015. MicroRNA‐27a distinguishes glioblastoma multiforme from diffuse and anaplastic astrocytomas and has prognostic value. Am. J. Cancer Res. 5:201–218. [PMC free article] [PubMed] [Google Scholar]
- 188. Ge, Y. F. , Sun J., Jin C. J., Cao B. Q., Jiang Z. F., and Shao J. F.. 2013. AntagomiR‐27a targets FOXO3a in glioblastoma and suppresses U87 cell growth in vitro and in vivo. Asian Pac. J. Cancer Prev. 14:963–968. [DOI] [PubMed] [Google Scholar]
- 189. Chen, L. , Li H., Han L., Zhang K., Wang G., Wang Y., et al. 2011. Expression and function of miR‐27b in human glioma. Oncol. Rep. 26:1617–1621. [DOI] [PubMed] [Google Scholar]
- 190. Cortez, M. A. , Nicoloso M. S., Shimizu M., Rossi S., Gopisetty G., Molina J. R., et al. 2010. miR‐29b and miR‐125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosom. Cancer 49:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Aldaz, B. , Sagardoy A., Nogueira L., Guruceaga E., Grande L., Huse J. T., et al. 2013. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem‐like cells. PLoS ONE 8:e77098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Che, S. , Sun T., Wang J., Jiao Y., Wang C., Meng Q., et al. 2015. miR‐30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 36:6805–6811. [DOI] [PubMed] [Google Scholar]
- 193. Wang, K. , Jia Z., Zou J., Zhang A., Wang G., Hao J., et al. 2013. Analysis of hsa‐miR‐30a‐5p expression in human gliomas. Pathol. Oncol. Res. 19:405–411. [DOI] [PubMed] [Google Scholar]
- 194. Jia, Z. , Wang K., Wang G., Zhang A., and Pu P.. 2013. MiR‐30a‐5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS ONE 8:e55008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 195. Hua, D. , Ding D., Han X., Zhang W., Zhao N., Foltz G., et al. 2012. Human miR‐31 targets radixin and inhibits migration and invasion of glioma cells. Oncol. Rep. 27:700–706. [DOI] [PubMed] [Google Scholar]
- 196. Zhou, R. J. , Xu X. Y., Liu B. X., Dai W. Z., Cai M. Q., Bai C. F., et al. 2015. Growth‐inhibitory and chemosensitizing effects of microRNA‐31 in human glioblastoma multiforme cells. Int. J. Mol. Med. 36:1159–1164. [DOI] [PubMed] [Google Scholar]
- 197. Rajbhandari, R. , McFarland B. C., Patel A., Gerigk M., Gray G. K., Fehling S. C., et al. 2015. Loss of tumor suppressive microRNA‐31 enhances TRADD/NF‐kappaB signaling in glioblastoma. Oncotarget 6:17805–17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kim, Y. S. , Kim S. H., Cho J., Kim J. W., Chang J. H., Kim D. S., et al. 2012. MGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide‐based chemoradiotherapy: a single‐institution study. Int. J. Radiat. Oncol. Biol. Phys. 84:661–667. [DOI] [PubMed] [Google Scholar]
- 199. Gao, H. , Zhao H., and Xiang W.. 2013. Expression level of human miR‐34a correlates with glioma grade and prognosis. J. Neurooncol. 113:221–228. [DOI] [PubMed] [Google Scholar]
- 200. Guessous, F. , Zhang Y., Kofman A., Catania A., Li Y., Schiff D., et al. 2010. microRNA‐34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 9:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Rathod, S. S. , Rani S. B., Khan M., Muzumdar D., and Shiras A.. 2014. Tumor suppressive miRNA‐34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio. 4:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Fan, Y. N. , Meley D., Pizer B., and See V.. 2014. Mir‐34a mimics are potential therapeutic agents for p53‐mutated and chemo‐resistant brain tumour cells. PLoS ONE 9:e108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Niu, H. , Wang K., Zhang A., Yang S., Song Z., Wang W., et al. 2012. miR‐92a is a critical regulator of the apoptosis pathway in glioblastoma with inverse expression of BCL2L11. Oncol. Rep. 28:1771–1777. [DOI] [PubMed] [Google Scholar]
- 204. Li, Q. , Shen K., Zhao Y., Ma C., Liu J., and Ma J.. 2013. MiR‐92b inhibitor promoted glioma cell apoptosis via targeting DKK3 and blocking the Wnt/beta‐catenin signaling pathway. J. Transl. Med. 11:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Wang, K. , Wang X., Zou J., Zhang A., Wan Y., Pu P., et al. 2013. miR‐92b controls glioma proliferation and invasion through regulating Wnt/beta‐catenin signaling via nemo‐like kinase. Neuro. Oncol. 15:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Ng, W. L. , Yan D., Zhang X., Mo Y. Y., and Wang Y.. 2010. Over‐expression of miR‐100 is responsible for the low‐expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst.) 9:1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 207. Alrfaei, B. M. , Vemuganti R., and Kuo J. S.. 2013. microRNA‐100 targets SMRT/NCOR2, reduces proliferation, and improves survival in glioblastoma animal models. PLoS ONE 8:e80865.24244722 [Google Scholar]
- 208. Yao, Y. L. , Ma J., Wang P., Xue Y. X., Li Z., Zhao L. N., et al. 2015. miR‐101 acts as a tumor suppressor by targeting Kruppel‐like factor 6 in glioblastoma stem cells. CNS Neurosci. Ther. 21:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Dai, D. W. , Lu Q., Wang L. X., Zhao W. Y., Cao Y. Q., Li Y. N., et al. 2013. Decreased miR‐106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer 13:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. He, J. , Zhang W., Zhou Q., Zhao T., Song Y., Chai L., et al. 2013. Low‐expression of microRNA‐107 inhibits cell apoptosis in glioma by upregulation of SALL4. Int. J. Biochem. Cell Biol. 45:1962–1973. [DOI] [PubMed] [Google Scholar]
- 211. Chen, L. , Zhang R., Li P., Liu Y., Qin K., Fa Z. Q., et al. 2013. P53‐induced microRNA‐107 inhibits proliferation of glioma cells and down‐regulates the expression of CDK6 and Notch‐2. Neurosci. Lett. 534:327–332. [DOI] [PubMed] [Google Scholar]
- 212. Chen, L. , Chen X. R., Zhang R., Li P., Liu Y., Yan K., et al. 2013. MicroRNA‐107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J. Neurooncol. 112:59–66. [DOI] [PubMed] [Google Scholar]
- 213. Chen, L. , Chen X. R., Chen F. F., Liu Y., Li P., Zhang R., et al. 2013. MicroRNA‐107 inhibits U87 glioma stem cells growth and invasion. Cell. Mol. Neurobiol. 33:651–657. [DOI] [PubMed] [Google Scholar]
- 214. Lv, Z. , and Yang L.. 2013. MiR‐124 inhibits the growth of glioblastoma through the downregulation of SOS1. Mol. Med. Rep. 8:345–349. [DOI] [PubMed] [Google Scholar]
- 215. Zhao, W. H. , Wu S. Q., and Zhang Y. D.. 2013. Downregulation of miR‐124 promotes the growth and invasiveness of glioblastoma cells involving upregulation of PPP1R13L. Int. J. Mol. Med. 32:101–107. [DOI] [PubMed] [Google Scholar]
- 216. Shi, Z. , Chen Q., Li C., Wang L., Qian X., Jiang C., et al. 2014. MiR‐124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R‐Ras and N‐Ras. Neuro. Oncol. 16:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Xia, H. , Cheung W. K., Ng S. S., Jiang X., Jiang S., Sze J., et al. 2012. Loss of brain‐enriched miR‐124 microRNA enhances stem‐like traits and invasiveness of glioma cells. J. Biol. Chem. 287:9962–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Fowler, A. , Thomson D., Giles K., Maleki S., Mreich E., Wheeler H., et al. 2011. miR‐124a is frequently down‐regulated in glioblastoma and is involved in migration and invasion. Eur. J. Cancer 47:953–963. [DOI] [PubMed] [Google Scholar]
- 219. Mucaj, V. , Lee S. S., Skuli N., Giannoukos D. N., Qiu B., Eisinger‐Mathason T. S., et al. 2015. MicroRNA‐124 expression counteracts pro‐survival stress responses in glioblastoma. Oncogene 34:2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Chen, J. , Chen H., Zhan Y., Yang X., and Yu M.. 2014. Lentivirus‐mediated interference of E3 ubiquitin ligase RNF31 inhibits tumor‐necrosis factor‐alpha‐induced activation of nuclear factor‐kappaB pathway. Nan Fang Yi Ke Da Xue Xue Bao 34:1713–1720. [PubMed] [Google Scholar]
- 221. Deng, X. , Ma L., Wu M., Zhang G., Jin C., Guo Y., et al. 2013. miR‐124 radiosensitizes human glioma cells by targeting CDK4. J. Neurooncol. 114:263–274. [DOI] [PubMed] [Google Scholar]
- 222. Lu, S. H. , Jiang X. J., Xiao G. L., Liu D. Y., and Yuan X. R.. 2014. miR‐124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and beta‐catenin. Oncol. Rep. 32:2104–2110. [DOI] [PubMed] [Google Scholar]
- 223. Yin, F. , Zhang J. N., Wang S. W., Zhou C. H., Zhao M. M., Fan W. H., et al. 2015. MiR‐125a‐3p regulates glioma apoptosis and invasion by regulating Nrg1. PLoS ONE 10:e0116759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Shi, L. , Zhang S., Feng K., Wu F., Wan Y., Wang Z., et al. 2012. MicroRNA‐125b‐2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int. J. Oncol. 40:119–129. [DOI] [PubMed] [Google Scholar]
- 225. Wan, Y. , Sun G., Wang Z., Guo J., and Shi L.. 2014. miR‐125b promotes cell proliferation by directly targeting Lin28 in glioblastoma stem cells with low expression levels of miR‐125b. NeuroReport 25:289–296. [DOI] [PubMed] [Google Scholar]
- 226. Shi, L. , Zhang J., Pan T., Zhou J., Gong W., Liu N., et al. 2010. MiR‐125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 1312:120–126. [DOI] [PubMed] [Google Scholar]
- 227. Wu, N. , Xiao L., Zhao X., Zhao J., Wang J., Wang F., et al. 2012. miR‐125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS Lett. 586:3831–3839. [DOI] [PubMed] [Google Scholar]
- 228. Xia, H. F. , He T. Z., Liu C. M., Cui Y., Song P. P., Jin X. H., et al. 2009. MiR‐125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell. Physiol. Biochem. 23(4–6):347–358. [DOI] [PubMed] [Google Scholar]
- 229. Chen, J. , Fu X., Wan Y., Wang Z., Jiang D., and Shi L.. 2014. miR‐125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting Bak1. Tumour Biol. 35:6293–6302. [DOI] [PubMed] [Google Scholar]
- 230. Wu, N. , Lin X., Zhao X., Zheng L., Xiao L., Liu J., et al. 2013. MiR‐125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38MAPK‐independent pathways. Br. J. Cancer 109:2853–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Shi, L. , Wan Y., Sun G., Gu X., Qian C., Yan W., et al. 2012. Functional differences of miR‐125b on the invasion of primary glioblastoma CD133‐negative cells and CD133‐positive cells. Neuromolecular Med. 14:303–316. [DOI] [PubMed] [Google Scholar]
- 232. Shi, L. , Fei X., Wang Z., and You Y.. 2015. PI3K inhibitor combined with miR‐125b inhibitor sensitize TMZ‐induced anti‐glioma stem cancer effects through inactivation of Wnt/beta‐catenin signaling pathway. In Vitro Cell. Dev. Biol. Anim. 51:1047–1055. [DOI] [PubMed] [Google Scholar]
- 233. Cui, H. , Mu Y., Yu L., Xi Y. G., Matthiesen R., Su X., et al. 2015. Methylation of the miR‐126 gene associated with glioma progression. Fam. Cancer 15:317–324. [DOI] [PubMed] [Google Scholar]
- 234. Ciafre, S. A. , Galardi S., Mangiola A., Ferracin M., Liu C. G., Sabatino G., et al. 2005. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 334:1351–1358. [DOI] [PubMed] [Google Scholar]
- 235. Peruzzi, P. , Bronisz A., Nowicki M. O., Wang Y., Ogawa D., Price R., et al. 2013. MicroRNA‐128 coordinately targets polycomb repressor Complexes in glioma stem cells. Neuro. Oncol. 15:1212–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Chen, H. , Li X., Li W., and Zheng H.. 2015. miR‐130a can predict response to temozolomide in patients with glioblastoma multiforme, independently of O6‐methylguanine‐DNA methyltransferase. J. Transl. Med. 13:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Qiu, S. , Lin S., Hu D., Feng Y., Tan Y., and Peng Y.. 2013. Interactions of miR‐323/miR‐326/miR‐329 and miR‐130a/miR‐155/miR‐210 as prognostic indicators for clinical outcome of glioblastoma patients. J. Transl. Med. 11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Zhu, G. , Wang Y., Mijiti M., Wang Z., Wu P. F., and Jiafu D.. 2015. Upregulation of miR‐130b enhances stem cell‐like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem. Biophys. Res. Commun. 465:194–199. [DOI] [PubMed] [Google Scholar]
- 239. Parker, N. R. , Correia N., Crossley B., Buckland M. E., Howell V. M., and Wheeler H. R.. 2013. Correlation of MicroRNA 132 Up‐regulation with an unfavorable clinical outcome in patients with primary glioblastoma multiforme treated with radiotherapy plus concomitant and adjuvant temozolomide chemotherapy. Transl. Oncol. 6:742–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Liu, Q. , Liao F., Wu H., Cai T., Yang L., Wang Z. F., et al. 2014. Upregulation of miR‐132 expression in glioma and its clinical significance. Tumour Biol. 35:12299–12304. [DOI] [PubMed] [Google Scholar]
- 241. Lulli, V. , Buccarelli M., Martini M., Signore M., Biffoni M., Giannetti S., et al. 2015. miR‐135b suppresses tumorigenesis in glioblastoma stem‐like cells impairing proliferation, migration and self‐renewal. Oncotarget 6:37241–37256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Xiao, S. , Yang Z., Lv R., Zhao J., Wu M., Liao Y., et al. 2014. miR‐135b contributes to the radioresistance by targeting GSK3beta in human glioblastoma multiforme cells. PLoS ONE 9:e108810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Wu, H. , Liu Q., Cai T., Chen Y. D., Liao F., and Wang Z. F.. 2014. MiR‐136 modulates glioma cell sensitivity to temozolomide by targeting astrocyte elevated gene‐1. Diagn. Pathol. 9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Yang, Y. , Wu J., Guan H., Cai J., Fang L., Li J., et al. 2012. MiR‐136 promotes apoptosis of glioma cells by targeting AEG‐1 and Bcl‐2. FEBS Lett. 586:3608–3612. [DOI] [PubMed] [Google Scholar]
- 245. Bier, A. , Giladi N., Kronfeld N., Lee H. K., Cazacu S., Finniss S., et al. 2013. MicroRNA‐137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP‐1. Oncotarget 4:665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Tamim, S. , Vo D. T., Uren P. J., Qiao M., Bindewald E., Kasprzak W. K., et al. 2014. Genomic analyses reveal broad impact of miR‐137 on genes associated with malignant transformation and neuronal differentiation in glioblastoma cells. PLoS ONE 9:e85591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Chen, L. , Wang X., Wang H., Li Y., Yan W., Han L., et al. 2012. miR‐137 is frequently down‐regulated in glioblastoma and is a negative regulator of Cox‐2. Eur. J. Cancer 48:3104–3111. [DOI] [PubMed] [Google Scholar]
- 248. Guan, Y. , Chen L., Bao Y., Qiu B., Pang C., Cui R., et al. 2015. High miR‐196a and low miR‐367 cooperatively correlate with unfavorable prognosis of high‐grade glioma. Int. J. Clin. Exp. Pathol. 8:6576–6588. [PMC free article] [PubMed] [Google Scholar]
- 249. Dai, S. , Wang X., Li X., and Cao Y.. 2015. MicroRNA‐139‐5p acts as a tumor suppressor by targeting ELTD1 and regulating cell cycle in glioblastoma multiforme. Biochem. Biophys. Res. Commun. 467:204–210. [DOI] [PubMed] [Google Scholar]
- 250. Li, R. Y. , Chen L. C., Zhang H. Y., Du W. Z., Feng Y., Wang H. B., et al. 2013. MiR‐139 inhibits Mcl‐1 expression and potentiates TMZ‐induced apoptosis in glioma. CNS Neurosci. Ther. 19:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Koo, S. , Martin G. S., Schulz K. J., Ronck M., and Toussaint L. G.. 2012. Serial selection for invasiveness increases expression of miR‐143/miR‐145 in glioblastoma cell lines. BMC Cancer 12:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Zhao, S. , Liu H., Liu Y., Wu J., Wang C., Hou X., et al. 2013. miR‐143 inhibits glycolysis and depletes stemness of glioblastoma stem‐like cells. Cancer Lett. 333:253–260. [DOI] [PubMed] [Google Scholar]
- 253. Liu, X. , Yu H., Cai H., and Wang Y.. 2014. The expression and clinical significance of miR‐132 in gastric cancer patients. Diagn. Pathol. 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Lee, H. K. , Bier A., Cazacu S., Finniss S., Xiang C., Twito H., et al. 2013. MicroRNA‐145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS ONE 8:e54652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Shi, L. , Wang Z., Sun G., Wan Y., Guo J., and Fu X.. 2014. miR‐145 inhibits migration and invasion of glioma stem cells by targeting ABCG2. Neuromolecular Med. 16:517–528. [DOI] [PubMed] [Google Scholar]
- 256. Mei, J. , Bachoo R., and Zhang C. L.. 2011. MicroRNA‐146a inhibits glioma development by targeting Notch1. Mol. Cell. Biol. 31:3584–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Yao, Y. , Ma J., Xue Y., Wang P., Li Z., Liu J., et al. 2015. Knockdown of long non‐coding RNA XIST exerts tumor‐suppressive functions in human glioblastoma stem cells by up‐regulating miR‐152. Cancer Lett. 359:75–86. [DOI] [PubMed] [Google Scholar]
- 258. Zhao, S. , Deng Y., Liu Y., Chen X., Yang G., Mu Y., et al. 2013. MicroRNA‐153 is tumor suppressive in glioblastoma stem cells. Mol. Biol. Rep. 40:2789–2798. [DOI] [PubMed] [Google Scholar]
- 259. Xu, J. , Liao X., and Wong C.. 2010. Downregulations of B‐cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG‐05MG. Int. J. Cancer 126:1029–1035. [DOI] [PubMed] [Google Scholar]
- 260. Xu, J. , Liao X., Lu N., Liu W., and Wong C. W.. 2011. Chromatin‐modifying drugs induce miRNA‐153 expression to suppress Irs‐2 in glioblastoma cell lines. Int. J. Cancer 129:2527–2531. [DOI] [PubMed] [Google Scholar]
- 261. Liu, Q. , Zou R., Zhou R., Gong C., Wang Z., Cai T., et al. 2015. miR‐155 Regulates glioma cells invasion and chemosensitivity by p38 Isforms in vitro. J. Cell. Biochem. 116:1213–1221. [DOI] [PubMed] [Google Scholar]
- 262. Ling, N. , Gu J., Lei Z., Li M., Zhao J., Zhang H. T., et al. 2013. microRNA‐155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol. Rep. 30:2111–2118. [DOI] [PubMed] [Google Scholar]
- 263. Zhou, J. , Wang W., Gao Z., Peng X., Chen X., Chen W., et al. 2013. MicroRNA‐155 promotes glioma cell proliferation via the regulation of MXI1. PLoS ONE 8:e83055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Sun, J. , Shi H., Lai N., Liao K., Zhang S., and Lu X.. 2014. Overexpression of microRNA‐155 predicts poor prognosis in glioma patients. Med. Oncol. 31:911. [DOI] [PubMed] [Google Scholar]
- 265. Meng, W. , Jiang L., Lu L., Hu H., Yu H., Ding D., et al. 2012. Anti‐miR‐155 oligonucleotide enhances chemosensitivity of U251 cell to taxol by inducing apoptosis. Cell Biol. Int. 36:653–659. [DOI] [PubMed] [Google Scholar]
- 266. D'Urso, P. I. , D'Urso O. F., Storelli C., Mallardo M., Gianfreda C. D., Montinaro A., et al. 2012. miR‐155 is up‐regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int. J. Oncol. 41:228–234. [DOI] [PubMed] [Google Scholar]
- 267. Wang, H. , Tao T., Yan W., Feng Y., Wang Y., Cai J., et al. 2015. Upregulation of miR‐181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci. Rep. 5:13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Kushwaha, D. , Ramakrishnan V., Ng K., Steed T., Nguyen T., Futalan D., et al. 2014. A genome‐wide miRNA screen revealed miR‐603 as a MGMT‐regulating miRNA in glioblastomas. Oncotarget 5:4026–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. She, X. , Yu Z., Cui Y., Lei Q., Wang Z., Xu G., et al. 2014. miR‐181 subunits enhance the chemosensitivity of temozolomide by Rap1B‐mediated cytoskeleton remodeling in glioblastoma cells. Med. Oncol. 31:892. [DOI] [PubMed] [Google Scholar]
- 270. Chen, G. , Zhu W., Shi D., Lv L., Zhang C., Liu P., et al. 2010. MicroRNA‐181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl‐2. Oncol. Rep. 23:997–1003. [DOI] [PubMed] [Google Scholar]
- 271. Conti, A. , Aguennouz M., La Torre D., Tomasello C., Cardali S., Angileri F. F., et al. 2009. miR‐21 and 221 upregulation and miR‐181b downregulation in human grade II‐IV astrocytic tumors. J. Neurooncol. 93:325–332. [DOI] [PubMed] [Google Scholar]
- 272. Shi, L. , Cheng Z., Zhang J., Li R., Zhao P., Fu Z., et al. 2008. hsa‐mir‐181a and hsa‐miR‐181b function as tumor suppressors in human glioma cells. Brain Res. 1236:185–193. [DOI] [PubMed] [Google Scholar]
- 273. Zhang, W. , Zhang J., Hoadley K., Kushwaha D., Ramakrishnan V., Li S., et al. 2012. miR‐181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro. Oncol. 14:712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Wang, X. F. , Shi Z. M., Wang X. R., Cao L., Wang Y. Y., Zhang J. X., et al. 2012. MiR‐181d acts as a tumor suppressor in glioma by targeting K‐ras and Bcl‐2. J. Cancer Res. Clin. Oncol. 138:573–584. [DOI] [PubMed] [Google Scholar]
- 275. Sun, Y. C. , Wang J., Guo C. C., Sai K., Wang J., Chen F. R., et al. 2014. MiR‐181b sensitizes glioma cells to teniposide by targeting MDM2. BMC Cancer 14:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Song, L. , Liu L., Wu Z., Li Y., Ying Z., Lin C., et al. 2012. TGF‐beta induces miR‐182 to sustain NF‐kappaB activation in glioma subsets. J. Clin. Investig. 122:3563–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Tang, H. , Wang Z., Liu Q., Liu X., Wu M., and Li G.. 2014. Disturbing miR‐182 and ‐381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PLoS ONE 9:e84146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Cui, Q. K. , Liu W. D., Zhu J. X., Wang Y. H., and Wang Z. G.. 2014. MicroRNA‐184 promotes proliferation ability of glioma cells by regulating FOXO3. Asian Pac. J. Trop. Med. 7:776–779. [DOI] [PubMed] [Google Scholar]
- 279. Emdad, L. , Janjic A., Alzubi M. A., Hu B., Santhekadur P. K., Menezes M. E., et al. 2015. Suppression of miR‐184 in malignant gliomas upregulates SND1 and promotes tumor aggressiveness. Neuro. Oncol. 17:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Zhong, Q. , Wang T., Lu P., Zhang R., Zou J., and Yuan S.. 2014. miR‐193b promotes cell proliferation by targeting Smad3 in human glioma. J. Neurosci. Res. 92:619–626. [DOI] [PubMed] [Google Scholar]
- 281. Hui, W. , Yuntao L., Lun L., WenSheng L., ChaoFeng L., HaiYong H., et al. 2013. MicroRNA‐195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS ONE 8:e54932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Yang, G. , Han D., Chen X., Zhang D., Wang L., Shi C., et al. 2014. MiR‐196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IkappaBalpha both in vitro and in vivo. Neuro. Oncol. 16:652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Ma, R. , Yan W., Zhang G., Lv H., Liu Z., Fang F., et al. 2012. Upregulation of miR‐196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS ONE 7:e38096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Berthois, Y. , Delfino C., Metellus P., Fina F., Nanni‐Metellus I., Al Aswy H., et al. 2014. Differential expression of miR200a‐3p and miR21 in grade II‐III and grade IV gliomas: evidence that miR200a‐3p is regulated by O(6)‐methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol. Ther. 15:938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. He, J. , Deng Y., Yang G., and Xie W.. 2013. MicroRNA‐203 down‐regulation is associated with unfavorable prognosis in human glioma. J. Surg. Oncol. 108:121–125. [DOI] [PubMed] [Google Scholar]
- 286. Liao, H. , Bai Y., Qiu S., Zheng L., Huang L., Liu T., et al. 2015. MiR‐203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial‐mesenchymal transition via SNAI2. Oncotarget 6:8914–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Chen, Z. , Li D., Cheng Q., Ma Z., Jiang B., Peng R., et al. 2014. MicroRNA‐203 inhibits the proliferation and invasion of U251 glioblastoma cells by directly targeting PLD2. Mol. Med. Rep. 9:503–508. [DOI] [PubMed] [Google Scholar]
- 288. Hou, S. X. , Ding B. J., Li H. Z., Wang L., Xia F., Du F., et al. 2013. Identification of microRNA‐205 as a potential prognostic indicator for human glioma. J. Clin. Neurosci. 20:933–937. [DOI] [PubMed] [Google Scholar]
- 289. Song, H. , and Bu G.. 2009. MicroRNA‐205 inhibits tumor cell migration through down‐regulating the expression of the LDL receptor‐related protein 1. Biochem. Biophys. Res. Commun. 388:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Liu, Y. , Yan W., Zhang W., Chen L., You G., Bao Z., et al. 2012. MiR‐218 reverses high invasiveness of glioblastoma cells by targeting the oncogenic transcription factor LEF1. Oncol. Rep. 28:1013–1021. [DOI] [PubMed] [Google Scholar]
- 291. Song, L. , Huang Q., Chen K., Liu L., Lin C., Dai T., et al. 2010. miR‐218 inhibits the invasive ability of glioma cells by direct downregulation of IKK‐beta. Biochem. Biophys. Res. Commun. 402:135–140. [DOI] [PubMed] [Google Scholar]
- 292. Mathew, L. K. , Skuli N., Mucaj V., Lee S. S., Zinn P. O., Sathyan P., et al. 2014. miR‐218 opposes a critical RTK‐HIF pathway in mesenchymal glioblastoma. Proc. Natl Acad. Sci. USA 111:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Tu, Y. , Gao X., Li G., Fu H., Cui D., Liu H., et al. 2013. MicroRNA‐218 inhibits glioma invasion, migration, proliferation, and cancer stem‐like cell self‐renewal by targeting the polycomb group gene Bmi1. Cancer Res. 73:6046–6055. [DOI] [PubMed] [Google Scholar]
- 294. Xie, Q. , Yan Y., Huang Z., Zhong X., and Huang L.. 2014. MicroRNA‐221 targeting PI3‐K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology 34:455–464. [DOI] [PubMed] [Google Scholar]
- 295. Quintavalle, C. , Garofalo M., Zanca C., Romano G., Iaboni M., del Basso De Caro M., et al. 2012. miR‐221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 31:858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Cai, G. , Qiao S., and Chen K.. 2015. Suppression of miR‐221 inhibits glioma cells proliferation and invasion via targeting SEMA3B. Biol. Res. 48:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Zhang, C. , Zhang J., Hao J., Shi Z., Wang Y., Han L., et al. 2012. High level of miR‐221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Hao, J. , Zhang C., Zhang A., Wang K., Jia Z., Wang G., et al. 2012. miR‐221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol. Rep. 27:1504–1510. [DOI] [PubMed] [Google Scholar]
- 299. Bai, Y. , Liao H., Liu T., Zeng X., Xiao F., Luo L., et al. 2013. MiR‐296‐3p regulates cell growth and multi‐drug resistance of human glioblastoma by targeting ether‐a‐go‐go (EAG1). Eur. J. Cancer 49:710–724. [DOI] [PubMed] [Google Scholar]
- 300. Yang, C. M. , Chiba T., Brill B., Delis N., von Manstein V., Vafaizadeh V., et al. 2015. Expression of the miR‐302/367 cluster in glioblastoma cells suppresses tumorigenic gene expression patterns and abolishes transformation related phenotypes. Int. J. Cancer Suppl. 137:2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Fareh, M. , Turchi L., Virolle V., Debruyne D., Almairac F., de‐la‐Forest Divonne S. , et al. 2012. The miR 302‐367 cluster drastically affects self‐renewal and infiltration properties of glioma‐initiating cells through CXCR4 repression and consequent disruption of the SHH‐GLI‐NANOG network. Cell Death Differ. 19:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Sun, J. Y. , Xiao W. Z., Wang F., Wang Y. Q., Zhu Y. H., Wu Y. F., et al. 2015. MicroRNA‐320 inhibits cell proliferation in glioma by targeting E2F1. Mol. Med. Rep. 12:2355–2359. [DOI] [PubMed] [Google Scholar]
- 303. Lian, H. W. , Zhou Y., Jian Z. H., and Liu R. Z.. 2014. MiR‐323‐5p acts as a tumor suppressor by targeting the insulin‐like growth factor 1 receptor in human glioma cells. Asian Pac. J. Cancer Prev. 15:10181–10185. [DOI] [PubMed] [Google Scholar]
- 304. Wang, S. , Lu S., Geng S., Ma S., Liang Z., and Jiao B.. 2013. Expression and clinical significance of microRNA‐326 in human glioma miR‐326 expression in glioma. Med. Oncol. 30:373. [DOI] [PubMed] [Google Scholar]
- 305. Zhou, J. , Xu T., Yan Y., Qin R., Wang H., Zhang X., et al. 2013. MicroRNA‐326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1). PLoS ONE 8:e68469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Delic, S. , Lottmann N., Stelzl A., Liesenberg F., Wolter M., Gotze S., et al. 2014. MiR‐328 promotes glioma cell invasion via SFRP1‐dependent Wnt‐signaling activation. Neuro. Oncol. 16:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Xiao, B. , Tan L., He B., Liu Z., and Xu R.. 2013. MiRNA‐329 targeting E2F1 inhibits cell proliferation in glioma cells. J. Transl. Med. 11:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Yao, Y. , Xue Y., Ma J., Shang C., Wang P., Liu L., et al. 2014. MiR‐330‐mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS ONE 9:e95060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Qu, S. , Yao Y., Shang C., Xue Y., Ma J., Li Z., et al. 2012. MicroRNA‐330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS ONE 7:e46010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 310. Epis, M. R. , Giles K. M., Candy P. A., Webster R. J., and Leedman P. J.. 2014. miR‐331‐3p regulates expression of neuropilin‐2 in glioblastoma. J. Neurooncol. 116:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Cheng, Q. , Cao H., Chen Z., Ma Z., Wan X., Peng R., et al. 2014. PAX6, a novel target of miR‐335, inhibits cell proliferation and invasion in glioma cells. Mol. Med. Rep. 10:399–404. [DOI] [PubMed] [Google Scholar]
- 312. Jiang, J. , Sun X., Wang W., Jin X., Bo X., Li Z., et al. 2012. Tumor microRNA‐335 expression is associated with poor prognosis in human glioma. Med. Oncol. 29:3472–3477. [DOI] [PubMed] [Google Scholar]
- 313. Shu, M. , Zhou Y., Zhu W., Zhang H., Wu S., Chen J., et al. 2012. MicroRNA 335 is required for differentiation of malignant glioma cells induced by activation of cAMP/protein kinase A pathway. Mol. Pharmacol. 81:292–298. [DOI] [PubMed] [Google Scholar]
- 314. Yamashita, D. , Kondo T., Ohue S., Takahashi H., Ishikawa M., Matoba R., et al. 2015. miR340 suppresses the stem‐like cell function of glioma‐initiating cells by targeting tissue plasminogen activator. Cancer Res. 75:1123–1133. [DOI] [PubMed] [Google Scholar]
- 315. Huang, D. , Qiu S., Ge R., He L., Li M., Li Y., et al. 2015. miR‐340 suppresses glioblastoma multiforme. Oncotarget 6:9257–9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Li, X. , Gong X., Chen J., Zhang J., Sun J., and Guo M.. 2015. miR‐340 inhibits glioblastoma cell proliferation by suppressing CDK6, cyclin‐D1 and cyclin‐D2. Biochem. Biophys. Res. Commun. 460:670–677. [DOI] [PubMed] [Google Scholar]
- 317. Floyd, D. H. , Zhang Y., Dey B. K., Kefas B., Breit H., Marks K., et al. 2014. Novel anti‐apoptotic microRNAs 582‐5p and 363 promote human glioblastoma stem cell survival via direct inhibition of caspase 3, caspase 9, and Bim. PLoS ONE 9:e96239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Chen, X. , Hao B., Han G., Liu Y., Dai D., Li Y., et al. 2015. miR‐372 regulates glioma cell proliferation and invasion by directly targeting PHLPP2. J. Cell. Biochem. 116:225–232. [DOI] [PubMed] [Google Scholar]
- 319. Zhang, R. , Luo H., Wang S., Chen W., Chen Z., Wang H. W., et al. 2014. MicroRNA‐377 inhibited proliferation and invasion of human glioblastoma cells by directly targeting specificity protein 1. Neuro. Oncol. 16:1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Li, B. , Wang Y., Li S., He H., Sun F., Wang C., et al. 2015. Decreased expression of miR‐378 correlates with tumor invasiveness and poor prognosis of patients with glioma. Int. J. Clin. Exp. Pathol. 8:7016–7021. [PMC free article] [PubMed] [Google Scholar]
- 321. Tang, H. , Liu X., Wang Z., She X., Zeng X., Deng M., et al. 2011. Interaction of hsa‐miR‐381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 1390:21–32. [DOI] [PubMed] [Google Scholar]
- 322. Chen, L. , Zhang J., Feng Y., Li R., Sun X., Du W., et al. 2012. MiR‐410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem. Cell Biol. 44:1711–1717. [DOI] [PubMed] [Google Scholar]
- 323. Tian, Y. , Nan Y., Han L., Zhang A., Wang G., Jia Z., et al. 2012. MicroRNA miR‐451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int. J. Oncol. 40:1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Gal, H. , Pandi G., Kanner A. A., Ram Z., Lithwick‐Yanai G., Amariglio N., et al. 2008. MIR‐451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 376:86–90. [DOI] [PubMed] [Google Scholar]
- 325. Tezcan, G. , Tunca B., Bekar A., Preusser M., Berghoff A. S., Egeli U., et al. 2014. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell. Mol. Neurobiol. 34:679–692. [DOI] [PubMed] [Google Scholar]
- 326. Wang, L. , Shi M., Hou S., Ding B., Liu L., Ji X., et al. 2012. MiR‐483‐5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett. 586:1312–1317. [DOI] [PubMed] [Google Scholar]
- 327. Li, X. , Liu Y., Granberg K. J., Wang Q., Moore L. M., Ji P., et al. 2015. Two mature products of MIR‐491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene 34:1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Yan, W. , Zhang W., Sun L., Liu Y., You G., Wang Y., et al. 2011. Identification of MMP‐9 specific microRNA expression profile as potential targets of anti‐invasion therapy in glioblastoma multiforme. Brain Res. 1411:108–115. [DOI] [PubMed] [Google Scholar]
- 329. Xu, J. , Sun T., and Hu X.. 2015. microRNA‐513c suppresses the proliferation of human glioblastoma cells by repressing low‐density lipoprotein receptor‐related protein 6. Mol. Med. Rep. 12:4403–4409. [DOI] [PubMed] [Google Scholar]
- 330. Guo, M. , Zhang X., Wang G., Sun J., Jiang Z., Khadarian K., et al. 2015. miR‐603 promotes glioma cell growth via Wnt/beta‐catenin pathway by inhibiting WIF1 and CTNNBIP1. Cancer Lett. 360:76–86. [DOI] [PubMed] [Google Scholar]
- 331. Zhang, Z. , Song X., Feng X., Miao Y., Wang H., Li Y., et al. 2015. Norcantharidin modulates miR‐655‐regulated SENP6 protein translation to suppresses invasion of glioblastoma cells. Tumour Biol. doi: 10.3892/mmr.2014.2150. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 332. Shi, Y. , Chen C., Zhang X., Liu Q., Xu J. L., Zhang H. R., et al. 2014. Primate‐specific miR‐663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin. Cancer Res. 20:1803–1813. [DOI] [PubMed] [Google Scholar]
- 333. Wang, R. J. , Li J. W., Bao B. H., Wu H. C., Du Z. H., Su J. L., et al. 2015. MicroRNA‐873 (miRNA‐873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J. Biol. Chem. 290:8938–8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Chen, X. , Zhang Y., Shi Y., Lian H., Tu H., Han S., et al. 2015. miR‐873 acts as a novel sensitizer of glioma cells to cisplatin by targeting Bcl‐2. Int. J. Oncol. 47:1603–1611. [DOI] [PubMed] [Google Scholar]


