Abstract
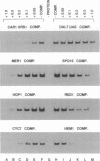
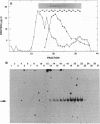
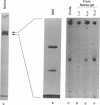
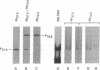
The protein that binds to the URS1 site situated upstream of many genes in Saccharomyces cerevisiae is a central element responsible for global negative control of transcription in this organism. Among the genes whose expression is regulated by this protein are those that participate in nitrogen metabolism, carbon metabolism, electron transport, inositol metabolism, heat shock response, meiosis, and sporulation. This factor, binding URS1 factor (BUF), has been purified and shown to be a heteromeric protein composed of 37.5- and 73.5-kDa monomers. The heteromeric form of BUF is stably maintained both in solution and bound to its DNA target site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beacham I. R., Schweitzer B. W., Warrick H. M., Carbon J. The nucleotide sequence of the yeast ARG4 gene. Gene. 1984 Sep;29(3):271–279. doi: 10.1016/0378-1119(84)90056-8. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Cohen G., Rapatz W., Ruis H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988 Sep 1;176(1):159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- Cohen R., Yokoi T., Holland J. P., Pepper A. E., Holland M. J. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol. 1987 Aug;7(8):2753–2761. doi: 10.1128/mcb.7.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covitz P. A., Herskowitz I., Mitchell A. P. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-alpha 2. Genes Dev. 1991 Nov;5(11):1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Roeder G. S. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol. 1990 May;10(5):2379–2389. doi: 10.1128/mcb.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Dumas K. S., Livak K. J. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 1985 Jun 11;13(11):4011–4027. doi: 10.1093/nar/13.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig A., Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986 Nov 3;160(3):487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Roeder R. G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985 Jul;41(3):955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Labieniec L., Swimmer C., Holland M. J. Homologous nucleotide sequences at the 5' termini of messenger RNAs synthesized from the yeast enolase and glyceraldehyde-3-phosphate dehydrogenase gene families. The primary structure of a third yeast glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1983 Apr 25;258(8):5291–5299. [PubMed] [Google Scholar]
- Kovari L. Z., Cooper T. G. Participation of ABF-1 protein in expression of the Saccharomyces cerevisiae CAR1 gene. J Bacteriol. 1991 Oct;173(20):6332–6338. doi: 10.1128/jb.173.20.6332-6338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari L., Sumrada R., Kovari I., Cooper T. G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Oct;10(10):5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavasic M. J., Elder R. T. Complementary transcripts from two genes necessary for normal meiosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jun;10(6):2809–2819. doi: 10.1128/mcb.10.6.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Dec;11(12):6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. D., Luche R. M., Cooper T. G. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992 Apr 25;20(8):1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. O., Craig E. A. Transcriptional regulation of a yeast HSP70 gene by heat shock factor and an upstream repression site-binding factor. Genes Dev. 1991 Jul;5(7):1299–1308. doi: 10.1101/gad.5.7.1299. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc Natl Acad Sci U S A. 1985 Feb;82(3):643–647. doi: 10.1073/pnas.82.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J. M. Regulatory DNA-binding proteins in yeast: an overview. Yeast. 1990 Jul-Aug;6(4):271–297. doi: 10.1002/yea.320060402. [DOI] [PubMed] [Google Scholar]
- Walter P., Gangloff J., Bonnet J., Boulanger Y., Ebel J. P., Fasiolo F. Primary structure of the Saccharomyces cerevisiae gene for methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 May;80(9):2437–2441. doi: 10.1073/pnas.80.9.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren H. J., Daugherty J. R., Rai R., Cooper T. G. Upstream induction sequence, the cis-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J Bacteriol. 1991 Nov;173(22):7186–7195. doi: 10.1128/jb.173.22.7186-7195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]