Abstract
Cyclooxygenase-2 (COX-2) is an important contributor to ischemic brain injury. Identification of the downstream mediators of COX-2 toxicity may allow the development of targeted therapies. Of particular interest is the cyclopentenone family of prostaglandin metabolites. Cyclopentenone prostaglandins (CyPGs) are highly reactive molecules that form covalent bonds with cellular thiols. Protein disulfide isomerase (PDI) is an important molecule for the restoration of denatured proteins following ischemia. Because PDI has several thiols, including thiols within the active thioredoxin-like domain, we hypothesized that PDI is a target of CyPGs and that CyPG binding of PDI is detrimental. CyPG–PDI binding was detected in vitro via immunoprecipitation and MS. CyPG–PDI binding decreased PDI enzymatic activity in recombinant PDI treated with CyPG, and PDI immunoprecipitated from neuronal culture treated with CyPG or anoxia. Toxic effects of binding were demonstrated in experiments showing that: (a) pharmacologic inhibition of PDI increased cell death in anoxic neurons, (b) PDI overexpression protected neurons exposed to anoxia and SH-SY5Y cells exposed to CyPG, and (c) PDI overexpression in SH-SY5Y cells attenuated ubiquitination of proteins and decreased activation of pro-apoptotic caspases. In conclusion, CyPG production and subsequent binding of PDI is a novel and potentially important mechanism of ischemic brain injury. We show that CyPGs bind to PDI, cyclopentenones inhibit PDI activity, and CyPG–PDI binding is associated with increased neuronal susceptibility to anoxia. Additional studies are necessary to determine the relative role of CyPG-dependent inhibition of PDI activity in ischemia and other neurodegenerative disorders.
Keywords: brain, cyclooxygenase, cyclopentenone, ischemia, protein disulfide isomerase
Introduction
Cyclooxygenase (COX) converts arachidonic acid into the intermediate prostaglandin H2 (PGH2), which is subsequently converted via specific prostaglandin synthases into one of the biologically active prostaglandins (PGI2, PGA2, PGD2, and PGE2). The inducible isoform of COX, cyclooxygenase-2 (COX-2), is the most prominent isoform in brain. COX-2 is induced in response to neuronal stress and has been identified as an important contributor to brain damage following hypoxia–ischemia. COX-2 inhibition is neuroprotective and COX-2 knockout mice are protected from hypoxic–ischemic injury, suggesting a possible therapeutic role for COX-2 inhibitors [1]. Unfortunately, COX-2 inhibitors are prothrombotic, limiting their use as neuroprotective agents [2]. Identification of the specific downstream mediators of COX-2 toxicity may allow the development of targeted therapies without prothrombotic side effects [3]. Potential mediators include specific prostaglandins, as well as prostaglandin metabolites [4,5]. Of particular interest is the cyclopentenone family of prostaglandin metabolites. Cyclopentenone prostaglandins (CyPGs) are highly reactive molecules containing an electrophilic carbonyl capable of forming covalent bonds with nucleophiles, including proteins with free cysteine groups. Our group has recently shown that CyPGs are increased in a COX-2-dependent manner in ischemic brain [6,7] and CyPGs exacerbate cell death in hypoxic primary neuronal culture [8,9]. A likely mechanism for CyPG neurotoxicity is via covalent adduction of CyPGs with accessible, free-cysteine nucleophiles on proteins with subsequent alteration of structure or function in the target proteins [10,11]. For example, we have recently shown that CyPGs can form a covalent bond with specific cysteines within UCH-L1, an enzyme within the ubiquitin–proteasome pathway, and this binding causes a conformational change within UCH-L1 that inhibits its activity and leads to accumulation of ubiquitinated proteins [12].
There are currently more than 50 protein targets of CyPGs identified [13,14] including several proteins that regulate cell death and survival [11,15–17]. One of the earliest identified targets was the redox protein thioredoxin [18]. This prompted us to examine protein disulfide isomerase (PDI), which contains four thioredoxin-like domains, as a potential target of CyPGs. PDI is an abundant and important housekeeping protein that resides in the endoplasmic reticulum (ER) constituting ~ 0.8% of total cellular protein (approaching mM concentration). PDI facilitates the proper folding of nascent proteins and refolding of partially denatured proteins by catalyzing disulfide bond formation and rearrangement via oxidation/reduction within two thioredoxin-like domains containing the cysteine–X–X–cysteine (CXXC) motif [19,20]. PDI has special relevance as a potential target in ischemic brain injury because altered PDI activity has been linked to neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis [21]. Furthermore, PDI is upregulated in rat forebrain ischemia and PDI overexpression is protective against ischemic injury [22]. Accordingly, we hypothesized that: (a) CyPGs can bind to cysteines within the active thioredoxin-like sites of PDI, (b) covalent binding of CyPG and PDI will inhibit PDI function, and (c) covalent binding of PDI by CyPG will exacerbate hypoxic neuronal death.
Results
15d-PGJ2 modifies PDI at CXXC motif in the catalytic thioredoxin-like domain
Our and others’ previous work has demonstrated that CyPGs, such as 15-deoxy Δ12,14-PGJ2 (15d-PGJ2), can modify a series of proteins on their cysteine residues through Michael addition and therefore profoundly change the protein’s functions [8,11–15]. PDI, an abundant protein located in the ER, facilitates isomerization of the disulfide bond in nascent and denatured proteins and therefore plays an important role in ER stress [19,20]. The disulfide oxidoreductase activity of PDI is dependent on its catalytic thioredoxin-like domain with the CXXC motif. Because PDI contains seven cysteine residues, including four in two critical CXXC motifs, it is possible that 15d-PGJ2 may directly adduct PDI and interfere with its function.
To test this hypothesis, an in vitro binding assay was performed using recombinant PDI protein and biotinylated (b-) 15d-PGJ2. PDI protein (1 μg) was incubated with 5 μM b-15d-PGJ2 or methyl acetate as vehicle control (Veh) for 90 min. The resultant b-15d-PGJ2–PDI adduct was detected by immunoblotting with horseradish peroxidase-conjugated streptavidin (streptavidin–HRP). To confirm assay specificity, PDI was also preincubated with 500 μM 15d-PGJ2 (100-fold excess) for 30 min prior to b-15d-PGJ2 treatment. As shown in Fig. 1 (A), b-15d-PGJ2–PDI adducts were detected when PDI was incubated with b-15d-PGJ2; adduct formation was decreased when PDI was pretreated with unlabeled 15d-PGJ2. Next, to assess 15d-PGJ2 modification of endogenous PDI in intact neurons, we incubated primary neurons with either 15d-PGJ2 or b-15d-PGJ2 for 2 h. Intracellular b-15d-PGJ2–PDI adducts were then detected by avidin pull-down assay (Fig. 1B, upper). After 15d-PGJ2 or b-15d-PGJ2 incubation, primary neuronal cell lysates were incubated with NeutrAvidin beads and the bound biotinylated proteins were then eluted for immunoblotting. PDI was detected in the eluent of the b-15d-PGJ2-treated group but not the 15d-PGJ2-treated group, confirming that modification of 15d-PGJ2 by endogenous PDI occurs within the intact neuron. In addition, b-15d-PGJ2–PDI adduct was also detected in b-15d-PGJ2-treated primary neuron cell lysates using immunoprecipitation (IP, Fig. 1B, lower) with a Direct IP kit from Pierce. Rat primary neurons were incubated with 10 μM b-15d-PGJ2 (+) or vehicle (−) for 2 h before being harvested. Cell lysates were then incubated with PDI antibody conjugated resin (R) overnight before wash and elution. A nonreactive control resin was included as a negative control. The b-15d-PGJ2–PDI adduct and PDI in the IP eluent were detected by immunoblotting with streptavidin-HRP (Str-H, upper right) and PDI antibody (lower right), respectively. Because both PDI and PDI-associated proteins may be present in the IP eluent, multiple bands in the Str-H blot may indicate that multiple PDI-associated proteins together with PDI are modified by 15d-PGJ2 in primary neurons. The band representing the b-15d-PGJ2–PDI adduct was determined by protein size (indicated with an arrow).
Fig. 1.
15d-PGJ2 binds to PDI in vitro and in primary neurons. (A) PDI recombinant protein was preincubated with or without 500 μM 15d-PGJ2 for 30 min before being incubated with vehicle or 5 μM b-15d-PGJ2 for another 90 min. The b-15d-PGJ2–PDI adducts were detected by immunoblotting with streptavidin–HRP. (B) 15d-PGJ2 binds to PDI in primary neurons. (Upper) Rat primary neurons were incubated with 10 μM 15d-PGJ2 or b-15d-PGJ2 for 2 h prior to harvest. The avidin pull-down assay was performed with cell lysates, and b-15d-PGJ2–PDI adducts were detected with PDI antibody. (Lower) Rat primary neurons were incubated with 10 μM b-15d-PGJ2 (+) or vehicle (−) for 2 h before harvest. Cell lysates were either subjected to immunoblotting to detect biotinylated proteins with streptavidin–HRP (Str-H, upper left) and PDI levels with an PDI antibody (lower left) or subjected to IP to detect the b-15d-PGJ2–PDI adduct (right). For IP, cell lysates were incubated with PDI antibody-conjugated resin (R) overnight before elution. A nonreactive control resin was included as a negative control. The b-15d-PGJ2–PDI adduct and PDI in the eluent were detected by immunoblotting with streptavidin–HRP (upper right) and PDI antibody (lower right), respectively. The arrow indicates the band representing b-15d-PGJ2–PDI adduct. (C) Avidin pull-down assay detecting arachidonic acid (AA) metabolite-modified PDI in primary neurons. Neurons were incubated with b-arachidonic acid (b-AA) then underwent hypoxia (+) or normoxia (−) before being harvested at the indicated time points. (Upper) Avidin-bead-bound PDI was detected by immunoblotting with PDI antibody. (Lower) Biotinylated proteins and endogenous PDI in cell lysates were detected by immunoblot with streptavidin–HRP and PDI antibody. (D) Fragmentation spectra of 15d-PGJ2-modified PDI (Uniprot Accession: P07237). The tryptic peptides indicate cysteine (C*) modifications at C-57 or C-60 (upper) and C-401 or C-404 (lower). Cysteine carbamidomethylation from sample processing for mass spectrometry is denoted by C#. MS/MS has an abundance of y- and b-ions N terminal to proline (underlined) with mass shifts corresponding to 316.204 Da indicating 15d-PGJ2 adduction.
Our previous work has shown that 15d-PGJ2 generation is significantly increased in the post-ischemia rat brain via COX-2 activation, and the conversion of 15d-PGJ2 from arachidonic acid is also increased in post-hypoxic primary neuronal culture [6–8]. Consistent with these findings, biotinylated PDI adduct was detected in post-hypoxic neurons when pretreated with biotinylated arachidonic acid in the NeutrAvidin pull-down assay (Fig. 1C). The time-dependent nature of this finding (increasing from 3 to 6 h of hypoxia) is consistent with conversion of the biotinylated arachidonic acid into biotinylated CyPG followed by adduction to proteins, including PDI. Taken together, the above data support the hypothesis that 15d-PGJ2 adducts PDI in primary neuronal cells and that this intracellular modification may be increased under pathological conditions such as hypoxia.
Next, to explore which cysteine residues in the PDI molecule are targets for 15d-PGJ2 adduction, MS/MS was performed on trypsin-digested peptides of PDI incubated with 5 μM 15d-PGJ2. Database searches of MS/MS data allowed for 15d-PGJ2-modified cysteine residues with a mass shift of 316.204 Da (15d-PGJ2 reduced). Modified peptides with characteristic mass shifts for a single 15d-PGJ2 modification were detected on the peptides carrying the two CXXC motifs, but not other cysteine residues outside the motif. Fragment ions indicating cysteine adduction with 15d-PGJ2 were evidenced by a continuous series of b- and y-type ions with mass shifts corresponding to 316.2 Da (Fig. 1D). The site localization of 15d-PGJ2 adduction to cysteine residues in the motif was ambiguous. This is explained by the presence of proline residues in the identified peptides. The presence and position of proline affects peptide dissociation. Termed the ‘proline effect’, peptides carrying prolines have fragmentation spectra dominated by an abundance of fragment ions N terminus to the prolines [23]. The position of the 15d-PGJ2-modified cysteines in both the identified peptides is C terminus to prolines. High-intensity fragment ions were observed N terminus to the proline residue, and lack of diagnostic ions around the CXXC motif did not allow for unambiguous site localization of 15d-PGJ2 adduction. However, high-resolution MS data and diagnostic b- and y-ions with mass shifts corresponding to 15d-PGJ2 adduction, provide strong evidence for 15d-PGJ2 adduction within the two CXXC motifs in PDI. This interesting finding suggests that the modification by 15d-PGJ2 to the CXXC motif may significantly interfere with PDI protein function.
15d-PGJ2 modification inhibits PDI thiol reductase activity
To examine whether 15d-PGJ2 modification interferes with PDI activity, a thiol reductase activity assay was performed with dieosin glutathione disulfide (Di-E-GSSG), a pseudo substrate for PDI. Di-E-GSSG, a fluorescence self-quenching molecule, exhibits a rapid 70-fold increase in fluorescence upon reduction of its disulfide bond by PDI [24]. In this experiment, 5 μM recombinant PDI was incubated with 1.65–33.3 μM 15d-PGJ2 for 3 h before Di-E-GSSG was added. The initial rate of disulfide bond reduction (dependent on PDI activity) was measured by spectrofluorometry, and data were normalized to untreated PDI (100% relative activity). As shown in Fig. 2 (A), preincubation with 15d-PGJ2 significantly decreases the PDI thiol reductase activity in a dose-dependent manner. Next, primary neurons were incubated with 10 μM 15d-PGJ2 or vehicle for 24 h. Endogenous PDI protein was then extracted by IP and PDI thiol reductase activity was measured. Compared with vehicle control, 15d-PGJ2 treatment significantly decreased neuronal PDI reductase activity (Fig. 2B). Taken together, these data indicate that covalent modification by 15d-PGJ2 inhibits PDI activity both in vitro and in neuronal cells.
Fig. 2.
PDI thiol reductase activity is inhibited by 15d-PGJ2 modification. (A) 15d-PGJ2 reduces recombinant PDI protein thiol reductase activity. Recombinant PDI (5 μM) was incubated with 1.65–33.3 μM of 15d-PGJ2 for 3 h followed by Di-E-GSSG substrate. Thiol reduction was measured by fluorescence plate reader and normalized to untreated recombinant PDI control. Data are means ± SD. n = 2–3 per group. (B) PDI activity is decreased in rat primary neuronal culture after treatment with 15d-PGJ2. Rat primary neuronal cultures were treated with 10 μM vehicle (Veh, methyl acetate) or 10 μM 15d-PGJ2 for 24 h prior to harvest. Neuronal PDI proteins were precipitated with anti-PDI antibody and a thiol reductase activity assay was performed. n = 3 per group. Data are means ± SD. **P < 0.001.
PDI overexpression attenuates 15d-PGJ2-induced cell death in SH-SY5Y cells
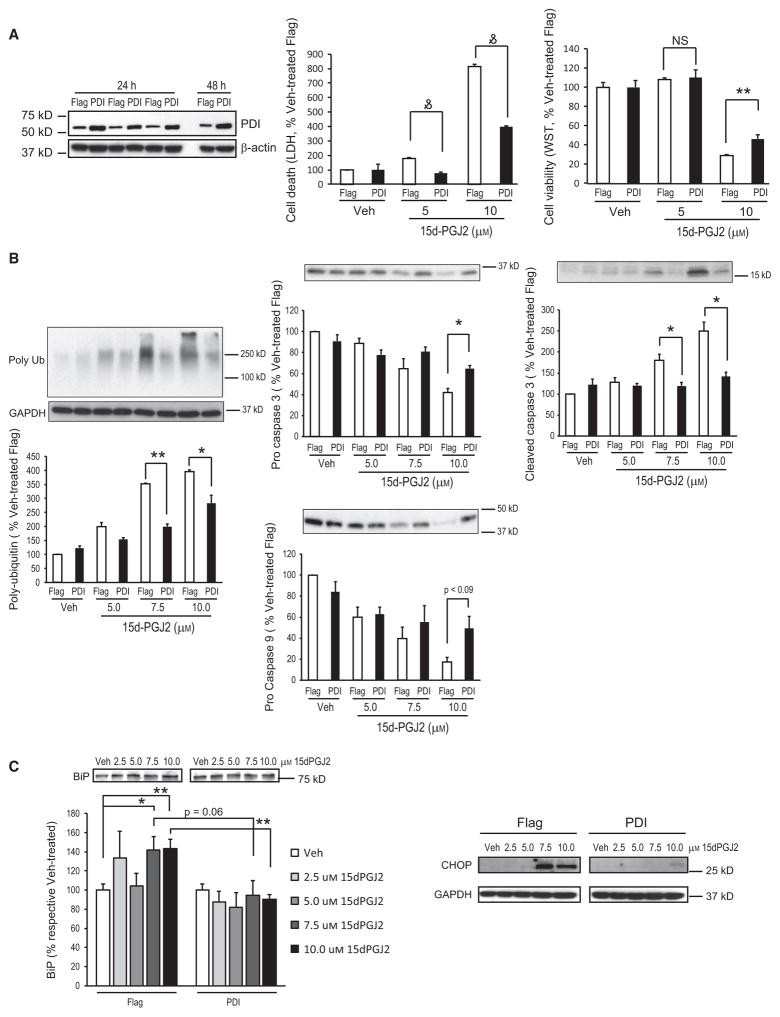
Previous reports regarding 15d-PGJ2 toxicity have suggested that cell apoptosis is induced via caspase pathway activation and that this effect is related to interference of the ubiquitin proteasome system and intracellular ubiquitinated protein accumulation [7,8]. To explore the role of PDI in 15d-PGJ2-induced cell death, a plasmid carrying sequence of Flag-tagged PDI, Flag–PDI/pcDNA3.1 (PDI vector), was constructed and an empty vector containing the Flag tag sequence (Flag vector) was applied as a negative control in the following experiments. Overexpression of PDI in transfected cells was confirmed with an PDI antibody using western blotting (Fig. 3A). Twenty-four hours after transfection, cells were treated with 15d-PGJ2 at 5 or 10 μM, or vehicle control for 24 h. Cell viability and cell death were measured and cell lysates were immunoblotted. As shown in Fig. 3(A), 15d-PGJ2 induced cell death in SH-SY5Y cells and this toxic effect was attenuated by the overexpression of PDI. Consistent with other reports [7,8,25], 15d-PGJ2-induced SH-SY5Y cell death is accompanied with ubiquitinated protein accumulation and caspase pathway activation (Fig. 3B). Polyubiquitinated protein accumulation was observed in Flag vector-transfected cells treated with 15d-PGJ2 compared with those treated with vehicle; overexpression of PDI significantly attenuated intracellular ubiquitinated protein build-up compared with Flag vector-transfected cells. Pro-caspase 3 was decreased in Flag vector-transfected cells treated with 15d-PGJ2 accompanied by an increase in cleaved caspase 3. Compared with the Flag vector-transfected group, overexpression of PDI significantly inhibited 15d-PGJ2-induced caspase 3 cleavage. A similar effect was also observed for pro-caspase 9 in 15d-PGJ2-treated SH-SY5Y cells; PDI vector transfection conferred resistance to caspase 9 cleavage. Additionally, we also found that the protein levels of the ER stress markers c/EBP homologous Protein (CHOP) and Binding of Immunoglobulin Protein (BiP), were significantly increased after 16 h of 7.5 or 10 μM 15d-PGJ2 treatment in Flag vector-transfected cells, indicating that 15d-PGJ2 induces ER stress in SH-SY5Y cells. Increased CHOP and BiP levels were attenuated in PDI overexpressing cells compared with vector control. Therefore, PDI function is protective against 15d-PGJ2-induced ubiquitinated protein accumulation, ER stress and apoptosis.
Fig. 3.
Overexpression of PDI decreases 15d-PGJ2-induced cell death in SH-SY5Y cells. SH-SY5Y cells were transfected with Flag–PDI/pcDNA3.1 (PDI, black bar) or control empty vector (Flag, white bar) for 24 h before treatment with 2.5–10 μM 15d-PGJ2 or vehicle (Veh) for an additional 16 h (C) or 24 h (A,B). (A) (Left) Overexpression of PDI in transfected SH-SY5Y cells. SH-SY5Y cells were transfected with Flag–PDI/pcDNA3.1 (PDI) or control empty vector (Flag) then harvested at 24 h (n = 3) and 48 h. Cell lysates were subjected to immunoblotting using anti-PDI antibody, and β-actin was used as loading control. (Center and right) Cell death was measured by LDH assay and cell viability by WST-1 assay. n = 6–12 per group. (B) Representative immunoblots using caspase 9, caspase 3, and poly-ubiquitinated protein antibodies. Densitometric analysis is shown below representative immunoblots (n = 3 per group). GAPDH was used to verify equal protein loading. (C) Representative immunoblots using BiP and CHOP antibodies. Densitometric analysis is shown below for Bip (n = 4 per group). GAPDH was used as a loading control. Data are means ± SE and are normalized to vehicle-treated control (Flag). *P < 0.05; **P < 0.01; and P < 0.001.
PDI thiol reductase activity is inhibited in post-hypoxic neurons and post-ischemic brain tissue
The above data have shown that 15d-PGJ2 adducts PDI, inhibits its reductase activity, and this modification may be increased after hypoxia. To assess the role of PDI and its modification in neuronal post-hypoxia/ischemia injury, primary neurons were subjected to hypoxia or normoxia. Twenty-four hours after reperfusion, cells were harvested and endogenous PDI reductase activity was measured with immunoprecipitated PDI as described above. As shown in Fig. 4 (A), PDI activity was significantly inhibited in vehicle-treated samples from the hypoxia group compared with normoxia control. In a second cohort of cells, the selective COX-2 inhibitor, SC65872, was applied to neuronal cells 2 h prior to hypoxia/normoxia. SC65872 had no effect on PDI activity in the normoxia group, but significantly attenuated hypoxia-induced PDI inhibition compared with vehicle control. This result indicates that PDI activity is compromised in post-hypoxia neurons at least in part, by a COX-2-mediated mechanism. Furthermore, PDI activity was also measured in post-ischemic rat hippocampus. Male juvenile rats underwent asphyxia cardiac arrest or sham surgery and were sacrificed 24 h after resuscitation. Hippocampal tissue was dissected and PDI protein was extracted by IP. As shown in Fig. 4(B), PDI activity significantly decreased after asphyxia cardiac arrest compared with the sham surgery group. Interestingly, the PDI protein levels in these two groups did not change significantly, suggesting the modification of PDI rather than protein degradation is more likely to be responsible for inhibition of PDI activity.
Fig. 4.
PDI thiol reductase activity is inhibited in post-hypoxic neurons and post-ischemic brain. (A) The COX-2 inhibitor SC65872 attenuates decrease in PDI activity after hypoxia. Rat primary neurons were treated with dimethylsulfoxide (Veh) or SC65872 (1 μM) 2 h prior to normoxia or hypoxia. Cells were harvested 24 h later, PDI was immunoprecipitated, and PDI thiol reductase activity measured. Data are means ± SE. n = 9 per group (three independent experiments combined, n = 3 each). *P < 0.05; **P < 0.001 using one way ANOVA with Dunnett’s post hoc analysis. (B) PDI thiol reductase activity is reduced after asphyxial cardiac arrest. Male juvenile rats underwent asphyxial cardiac arrest or sham surgery (n = 8 per group) and were killed 24 h after resuscitation. Hippocampal PDI was immunoprecipitated followed by PDI thiol reductase activity assay (upper). Immunoblot of hippocampal cell lysate (lower) indicates PDI protein expression is unchanged after asphyxial cardiac arrest. Data normalized to sham and are presented as means ± SE. **P < 0.001.
PDI activity protects against hypoxia-induced primary neuronal cell death
To assess the effects of PDI inhibition on hypoxia-induced primary neuronal cell death, two PDI inhibitors [26,27], methyl 2-(2-chloroacetyl)-1-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-1-carboxylate (16F16; 0.5–8 μM) or nitazoxanide (at 1 or 5 μM), were incubated with primary neurons for 48 h after mild hypoxia. Cell viability and cell death were determined with WST-1 and CytoTox-Fluor cytotoxicity assays, respectively. Prior to hypoxia, additional cells treated with 1 μM MK801 (as 100% live control) or 20 μM staurosporin (as 100% death control). As shown in Fig. 5 (A,B), incubation with 16F16 or nitazoxanide both significantly decreased cell viability and exacerbated post-hypoxia neuronal cell death compared with vehicle control in a dose-dependent manner.
Fig. 5.
PDI inhibition exacerbates hypoxia-induced neuronal cell death, whereas PDI overexpression attenuates cell death. Rat primary neurons were treated with the PDI inhibitors 16F16 (A) or nitazoxanide (NTZ) (B) for 48 h then analyzed for cell death (LDH) and cell viability (WST). n = 6–12 per group. Data are means ± SE. *P < 0.05. (C,D) Rat primary neurons were infected with lentivirus-carrying PDI or empty vector (EV, as control). (C) Representative fluorescent images of lentivirus infection (red, tdTomato) in rat primary neurons. Blue is Hoechst nuclear stain. Photos taken at ×20 with an Olympus BX51 microscope with appropriate filters. (D) Cell viability 24 h after hypoxia (left) or treatment with 25 μM 15d-PGJ2 (right). n = 6 per group. *P < 0.05; **P < 0.01. MK, MK801 (1 μM) as control for 100% live cells; SP, staurosporin (20 μM) as control for 100% cell death; UN, untreated.
To confirm that PDI activity protects against hypoxic neuronal injury, we constructed a lentiviral expression vector to overexpress PDI in primary neuronal cells. Lenti-PDI vector expresses PDI and red fluorescent tdTomato protein from a bicistronic mRNA transcript, allowing tdTomato to be used as an indicator of transduction. After viral particle generation and purification, primary neurons were seeded on 96-well plates and infected with lentivirus carrying PDI sequence (PDI) or empty vector (EV) at 1.2 × 108 particles·well−1. Infection was performed at DIV2 overnight, then medium was changed and hypoxia was performed at DIV 10. Fluoromicroscopy indicates that ~ 30% of neuronal cells were infected with lentivirus as determined by red fluorescence and Hoechst staining (Fig. 5C). Cell viability was determined 24 h after hypoxia. PDI overexpression significantly increased cell viability (Fig. 5D left) compared with the EV-infected control group. These data further support the hypothesis that PDI activity plays a significant role in protecting neuronal cells against hypoxic insults. Consistent with our above data using SH-SY5Y cells, overexpression of PDI in primary neuronal cells affords protection from 25 μM 15d-PGJ2-induced cell death, as shown in Fig. 5 (D, right).
Discussion
This is the first study to identify PDI as a target of CyPGs causing increased susceptibility of neurons to anoxia. Evidence of CyPG–PDI binding includes: (a) recombinant PDI binds to biotinylated CyPG, (b) biotinylated arachidonic acid (upstream of CyPG production) is incorporated into PDI in primary neuronal culture exposed to anoxia, (c) PDI and biotinylated CyPG is detected by antibody pull-down of PDI in primary neuronal culture, and (d) MS of recombinant PDI incubated with CyPG identified CyPG-adducted fragments at the predicted cysteine-containing peptides.
CyPG covalent binding of cysteine residues within the thioredoxin domains of PDI would be expected to inhibit the enzymatic activity of PDI. Consistent with this hypothesis, we found decreased enzymatic activity in recombinant PDI treated with CyPG. We also found decreased activity in immunoprecipitated PDI from primary neuronal culture treated with CyPG or anoxia.
We have previously shown that neuronal death in primary neuronal culture exposed to anoxia is partially mediated by COX-2-dependent products including CyPGs [28,29]. Here, we show that anoxia causes decreased PDI activity in primary neuronal culture and this activity may be partially protected by preincubation with a COX-2 inhibitor; thus identifying PDI inhibition as a potential mechanism for CyPG-dependent anoxic injury. We complemented the in vitro work with our in vivo model of asphyxial cardiac arrest in rats [30]. This model results in robust induction of COX-2 in hippocampus following resuscitation accompanied by increased production of prostaglandins and CyPGs [6]. Similar to the in vitro experiments, immunoprecipitated PDI from hippocampus in asphyxiated rats showed decreased PDI activity. Taken together, this evidence supports that CyPG production following neuronal ischemia inhibits PDI activity. Because PDI plays a critical role in refolding denatured proteins, inhibition of PDI activity following an ischemic injury would be expected to worsen injury. Consistent with this hypothesis, we show that (a) pharmacologic inhibition of PDI increases cell death in primary neuronal culture exposed to anoxia, (b) overexpression of PDI protects neurons exposed to anoxia and SH-SY5Y cells exposed to CyPG, and (c) overexpression of PDI in SH-SY5Y cells decreases activation of the pro-apoptotic effectors caspase 3 and caspase 9 as well as attenuates ER stress as measured by BiP, CHOP, and protein ubiquitination.
CyPGs are highly reactive molecules with many potential protein targets but PDI is likely to be a particularly important target in ischemic brain because of its known association with other neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and amyotrophic lateral sclerosis [21,31]. Its protective role in ischemic disease is demonstrated by the work of Tanaka et al. using a PDI vector injected into the hippocampus of rats exposed to forebrain ischemia [22]. The common element of many neurodegenerative diseases is disordered protein folding or aggregation. Thus, PDI’s folding/refolding activities, as well as its chaperone activity, are central to the injury and repair paradigm of many neurodegenerative diseases including hypoxic–ischemic injury. Hypoxic–ischemic injury results in denatured proteins, as well as selected upregulation of newly formed proteins requiring proper folding. The cellular response to denatured and aggregating proteins, named the unfolded protein response, involves a series of adaptive mechanisms to inhibit unnecessary protein production while facilitating production and proper folding of pro-survival proteins. The role of the unfolded protein response and PDI in ischemic injury is reviewed in DeGracia & Montie [32]. Importantly, PDI is upregulated in response to hypoxic–ischemic injury [22,33–35] and is neuroprotective [22,36]. To date, the focus on PDI in neurodegenerative disorders has been on oxidative-stress-mediated nitrosylation of PDI as a contributing factor to impaired PDI activity in neurodegenerative disorders [37–39]. Here, we describe another potential mechanism, CyPG adduction, which deserves investigation in these same diseases.
A limitation to studying PDI is that it is a family of 20 proteins, all sharing the thioredoxin active domain, with both specificity and redundancy for a large number of cysteine-containing protein substrates [20,40,41]. Thus, knocking down or overexpressing a single member of the family will have limited impact. Indeed, we were unable to show an effect from knocking down one PDI family member (data not shown), whereas pharmacologic inhibition of all PDI family members increased susceptibility to anoxia. For the same reason, our overexpression experiments using a single PDI vector in anoxia and CyPG exposure had only modest improvements in survival. Another limitation is that PDI can be modified by nitrosylation [38,39,42], glutathionylation [43,44], lipid peroxidation products [45], and CyPG adduction; thus, the effect of inhibiting one pathway may be masked by compensatory damage from the remaining mechanisms. The relative contribution of these different mechanisms was beyond the scope of our experiments but deserves additional study. Of importance, there are small-molecule PDI mimics in development that may provide cross-mechanistic protection and may be of therapeutic benefit in the future [46–51]. Another challenge to understanding the role of PDI is that there is evidence it may contribute to apoptotic death [27], thus like many housekeeping proteins it can contribute to either death or survival depending upon the conditions within the cell. Accordingly, experiments might yield conflicting results with slightly different experimental conditions. But if apoptosis is an adaptive mechanism, the role of PDI in both survival and death is important and inhibition of its activity by CyPG adduction would be detrimental. Also, we investigated only one of many documented CyPG interactions: both neuroprotective and neurodegenerative effects have been described in various models of CNS injury [52]. Lastly, neuroprotection from overexpression of PDI might occur via: (a) providing additional functional PDI to overwhelm CyPG adduction and augment enzymatic capacity; or (b) providing additional PDI to act as a ‘decoy’ to quench CyPGs and protect other, more important, proteins. Although it is not possible to be certain which of these two mechanisms is relevant, the data showing alteration of the unfolded protein response (ubiquitination, CHOP, BiP) with PDI overexpression favor a direct role for PDI.
In summary, PDI is a novel and potentially important target for post-ischemic COX-2-dependent neuronal injury. We show that CyPGs bind to PDI, cyclopentenone binding inhibits PDI activity, and cyclopentenone binding of PDI increases neuronal susceptibility to anoxia and CyPGs in vitro. Additional studies are necessary to determine the relative role of CyPG-dependent inhibition of PDI activity in ischemia and other neurodegenerative disorders. Future therapeutic directions might include inhibition of CyPG production or adduction, restoration of PDI function, and administration of PDI mimics.
Experimental procedures
Animal studies were performed with the approval of the University of Pittsburgh Institutional Animal Care and Use Committee and conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animals were housed in a temperature and humidity controlled environment with 12 h light cycles and free access to food and water.
Asphyxial cardiac arrest
Male juvenile rats (16–18 days old) underwent asphyxia cardiac arrest or sham surgery as described previously [30], n = 8 per group. All animals received 1 mL warmed 0.45% saline/5% dextrose subcutaneously prior to extubation. Rats were killed 24 h after resuscitation and brain hippocampi removed for PDI thiol reductase activity assay.
Reagents and antibodies
Human PDI recombinant protein was purchased from Assay Designs (Ann Arbor, MI, USA) and anti-PDI antibody was from Abcam (Cambridge, MA, USA) and Cell Signaling Technology (Danvers, MA, USA). Free or biotinylated arachidonic acid and b-15d-PGJ2 were from Cayman Chemical (Ann Arbor, MI, USA); monoclonal caspase 3, CHOP, BiP and caspase 9 antibodies were from Cell Signaling; GAPDH antibody was from Covance (Berkeley, CA, USA); Cy3-conjugated monoclonal mouse biotin and Alexafluor 488-conjugated secondary antibodies were from Jackson Immunoresearch Lab (West Grove, PA, USA). Mouse monoclonal mono- and polyubiquitinated proteins antibody (clone FK2) were from Enzo Life Sciences (Plymouth Meeting, PA, USA). NeutrAvidin beads and streptavidin–HRP, and immunoprecipitation kits were from Pierce (Rockford, IL, USA). UPLC organic solvents and water were from VWR (West Chester, PA, USA). β-actin antibody and all other chemicals were from Sigma-Aldrich unless otherwise noted. The lentiviral expression vector, pLVX-IRES-tdTomato vector, and Lenti-X HTX concentrator and packaging system were purchased from Clontech Laboratories (Mountain View, CA, USA). WST-1 cell proliferation assay was also from Clontech. The selective COX-2 inhibitor SC65872 was generously donated by Peter Isakson (Monsanto Searle). PDI inhibitor 16F16 was a gift from Brent Stockwell (Columbia University) and was also purchased from Enzo Life Sciences; the PDI inhibitor nitazoxanide was from Sigma.
Plasmid constructs
The DNA sequence encoding full-length rat PDI was amplified by PCR, and cloned into a modified pcDNA3.1 vector (Novagen, San Diego, CA, USA) with a N terminus Flag tag sequence. The resulting construct was confirmed by sequencing. For lentiviral expression vector construction, PDI cDNA was inserted into pLVX-IRES–tdTomato vector with XhoI and BamHI restrictive sites to generate lenti-PDI. This vector expresses PDI and the red fluorescent protein tdTomato from a bicistronic mRNA transcript, allowing tdTomato to be used as an indicator of transduction. To generate infectious lentiviral particles, Lenti-X 293T cells (Clontech) were transfected with the lenti-PDI vector or empty lenti vector together with Lenti-X packaging mix using XFect (Clontech) following manufacturer’s instructions. Lentiviral particles were then collected, concentrated, and purified. Both lenti-PDI and lenti-empty virus titration was determined with a Lenti-X qRT-PCR titration kit (Clontech).
Primary neuronal cell culture and lenti-virus infection
Rat cortical primary neuronal cultures were prepared from E17 fetal rats (Sprague–Dawley, Charles River, Wilmington, MA, USA) as previously described [28] and used for experiments after 9 DIV. Cells were grown in serum-free Neurobasal medium (Invitrogen, Carlsbad, CA, USA) supplemented with B27 and GlutaMAX (Invitrogen). To overexpress PDI in primary neurons, cells were seeded in 96-well plates and lenti-PDI particles or empty lentiviral particles were added to the culture medium at 1.2 × 108 particles·well−1 for 18 h. Viral infection was performed at DIV2, and cells underwent hypoxia or 25 μM 15d-PGJ2 treatment at DIV10. Cell viability and cell death were detected 24 h after hypoxia or 15d-PGJ2 treatment.
SH-SY5Y cell culture and transfection
Human neuroblast cell line SH-SY5Y cells were maintained in Dulbecco’s modified Eagle’s medium plus 10% FBS. Cells were transfected with Flag–PDI/pcDNA3.1 or empty vector using JetPrime transfection reagent (Polyplus transfection, New York, NY, USA). Twenty-four hours post transfection, cells were treated with 15d-PGJ2 for 24 h in Dulbecco’s modified Eagle’s medium without FBS. Cell death and cell viability assays were then performed or cell lysates were harvested for western blotting.
In vitro hypoxia and cell death measurements
Hypoxia was performed using a hypoxic glove box (Coy Laboratories, Grass Lake, MI, USA) flushed with 92% argon, 5% CO2 and 3% H2 for 2–3 h as described previously [28] resulting in ~ 40% cell death after 24 h reperfusion. Staurosporin (20 μM, a 100% cell death internal standard) and 1 μM dizocilpine (MK801, a 100% cell survival internal standard) treatments were included in each cell death assay experiment. Cell death was quantitatively assessed by measuring lactate dehydrogenase (LDH) release into the culture medium 24 h after hypoxia or using CytoTox-Fluor cytotoxicity assay (Promega). Cell viability was assessed with WST-1 assay. Rat primary neurons were treated with PDI inhibitors 16F16 (0.5–8 μM) or nitazoxanide (1 or 5 mM) for 48 h or vehicle (Veh) after hypoxia then harvested for cell death and cell viability measurements (n = 6–12 wells per group).
In vitro binding assay
Purified recombinant PDI protein (1 μg) was incubated with 1 or 5 μM b-15d-PGJ2 or vehicle for 1 h in 100 μL of binding buffer (20 mM Tris, pH 7.0; 45 mM NaCl; 5 mM MgCl2; 0.1 mM dithiothreitol, 1% glycerol) before immunoblotting with streptavidin–HRP. For the competition-binding assay, PDI protein was pre-incubated with 500 μM of 15d-PGJ2 for 30 min at room temperature before adding b-15d-PGJ2.
Avidin pull-down assay
The avidin pull-down assay was performed as previously described [8]. Primary neurons were incubated with 20 μM biotinylated arachidonic acid and subjected to hypoxia or normoxia treatment. Cells were then harvested at 0, 3 and 6 h post hypoxia. Equal amounts of protein were incubated with NeutrAvidin beads for 4 h at 4 °C. Bound proteins were washed three times with lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100) before elution and detection by immunoblot with β-actin and PDI antibodies, or streptavidin–HRP for biotin-incorporated proteins.
Immunoblotting
Immunoblotting was performed as previously described [8]. For pro-caspase 3, cleaved caspase 3, pro-caspase 9, BiP and CHOP detection, cell lysates were resolved on 10% or 12% SDS/PAGE. After blocking with 5% non-fat milk in TBS/Tween-20, membranes were incubated with pro-caspase 3, cleaved caspase 3, pro-caspase 9, BiP or CHOP (1 : 1000, all), antibodies at 4 °C overnight. For ubiquitinated protein detection, cell lysates were resolved on a 4–20% linear gradient polyacrylamide gel (BioRad, Hercules, CA, USA) before detection with poly-ubiquitinated conjugates antibody (1 : 1000). Blots were washed and the appropriate secondary antibodies applied. Protein signal was visualized with ECL reagents (Pierce). Blots were then stripped and reprobed using GAPDH antibody as a loading control.
Protein in-gel digestion and tandem MS
Recombinant PDI protein (1 μg) was incubated with 15d-PGJ2 (5 μM) at room temperature for 2 h before being subjected to SDS/PAGE. The gel was then stained with Coomassie Brilliant Blue and protein bands were excised and digested with porcine Trypsin Gold (Promega, Madison, WI, USA), as previously described [53] with minor modifications.
Digests were analyzed by nano LC MS/MS on a Thermo Fisher LTQ Orbitrap Velos (Thermo Fisher, Pittsburgh, PA, USA). The LTQ Orbitrap Velos was connected to a Waters Acquity UPLC system (Waters Corp., Milford, MA, USA) consisting of sample trap (nanoAcuity UPLC trap column, 180 μm × 20 mm, 5 μm particle size, Symmetry C18, Waters Corp.) and a C18 column (nanoAcquity UPLC column, 75 μm × 250 mm, 1.7 μm particle size, BEH300 C18, Waters Corp.) interfaced with a nanospray ionization source. Solvent A was 0.1% formic acid in water. Solvent B was 0.1% formic acid in acetonitrile. Samples were loaded on the sample trap with 1% solvent B at a flow rate of 15 μL·min−1 for 1 min. Peptides were then eluted off the column with a 90-min gradient running at 300 nL·min−1: 5% B for 3 min, 5–55% B in 60 min, 30– 95% B in 1 min, 95% B for 5 min, 95–5% B in 1 min, 5% B for 20 min. All MS spectra were acquired in the Orbitrap detector at 60 000 resolution in profile mode while tope nine data-dependent MS/MS spectra were fragmented by collision-induced dissociation in the ion trap of the LTQ mass spectrometer but acquired in the high resolution Orbitrap at 7500 resolution.
Database searches were carried out with Proteome Discoverer 1.4 against the complete Uniprot human database appended to a contaminant database (88 575 sequences, 35 106 826 residues) with the Sequest search engine, for a trypsin digest, with two missed cleavages, and three dynamic modifications (oxidation of methionines, carbamidomethylation of cysteines, and 15d-PGJ2 adduction of cysteines). The mass tolerance was set at 10 ppm for precursor mass and 0.8 Da for collision-induced dissociation fragment ion masses. Peptide identifications were validated using the Percolator algorithm with a 5% global false discovery rate. The spectra of 15d-PGJ2 adducted peptides were manually inspected.
Immunoprecipitation
Endogenous PDI was extracted from brain tissue or primary neuron lysates using the Pierce Seize® X Protein A immunoprecipitation kit per manufacturer’s instructions (Pierce). Briefly, anti-PDI antibody was immobilized to protein A beads using the cross-linker disuccinimidyl subsrate. Protein lysates were then incubated with antibody-linked beads at 4 °C overnight then washed and antibody-bound protein eluted with a glycine elution buffer. Following elution, the eluate was neutralized with 1 M Tris, pH 9.5. To detect b-15d-PGJ2–PDI adducts in primary neuronal cells, a Direct IP Kit (Pierce) was used following the manufacturer’s instructions. The antibody becomes covalently attached to aldehyde-activated beaded agarose resin. By contrast with traditional IP methods, the Direct IP Kit enables elution of antigen without antibody contamination, which favors the detection of PDI (~ 50 kDa). A nonreactive control resin was included in the IP as a negative control.
PDI thiol reductase activity measurement
Recombinant PDI
Thiol reductase activity was measured as previously described [24,54]. Briefly, 5 μM recombinant PDI (Affinity Bioreagents) was incubated with 1.65 to 33.3 μM 15d-PGJ2 (Cayman Chemical) at 37 °C for 3 h then di-eosin-labeled oxidized glutathione (Di-E-GSSG) substrate was added to the mixture. Di-E-GSSG emits fluorescence upon reduction of its disulfide bond. The rate of reduction (dependent upon PDI thiol reductase activity) was measured using a fluorescent plate reader over 2 min. Data are normalized to control recombinant PDI (without 15d-PGJ2 addition). The x-axis, representing the concentration of 15d-PGJ2, is scaled to indicate the number of molecules of 15d-PGJ2 relative to the number of thiols available from PDI in each experiment.
Primary neuronal culture treated with 15d-PGJ2
Cultures were treated with vehicle (methyl acetate) or 10 μM 15d-PGJ2 for 24 h prior to harvest (three per group). Primary neuronal culture with hypoxia: Cells were treated with the COX-2 inhibitor SC65872 (1 μM) or vehicle (dimethylsulfoxide) 2 h prior to normoxia or hypoxia. Cells were harvested 24 h later; n = 9 per group (data combined from three experiments run on separate days, n = 3 each). Data are means ± SD.
Statistical analysis
Data are expressed as means ± SE and were analyzed using one-way ANOVA with Bonferroni or Dunnett’s post-hoc testing where appropriate. Results were considered to be significant when P < 0.05.
Acknowledgments
This work was supported by National Institutes of Health, National Institute of Child Health and Human Development and National Institute of Neurological Disorders and Stroke: R21HD058846 (RWH), R01NS37549 (SHG) and the VA Merit Review program (SHG). The work done by the Biomedical Mass Spectrometry Center was supported in part by the Cancer Center Support Grant P30CA047904, National Cancer Institute. The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Abbreviations
- 16F16
methyl 2-(2-chloroacetyl)-1-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-1-carboxylate
- BiP
Binding of Immunoglobulin Protein
- CHOP
c/EBP homologous protein
- COX-2
cyclooxygenase-2
- CXXC
cysteine-X-X-cysteine
- CyPG
cyclopentenone prostaglandin
- Di-E-GSSG
dieosin glutathione disulfide
- ER
endoplasmic reticulum
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- PDI
protein disulfide isomerase
- PG
prostaglandin
Footnotes
Author contributions
Hao Liu - experimental design, performed experiments, wrote the manuscript, analyzed data. Jie Chen - performed cell culture experiments. Wenjin Li - performed cell culture experiments. Marie E. Rose - performed animal experiments, analyzed data, wrote the manuscript. Sunita N. Shinde - performed biochemical assays. Manimalha Balasubramani - performed MS/MS assays. Guy T. Uechi - performed MS/MS assays. Bulent Mutus - thiol reductase activity assays. Steven H. Graham - experimental design. Robert W. Hickey - experimental design, analyzed data, wrote the manuscript.
References
- 1.Candelario-Jalil E, Fiebich BL. Cyclooxygenase inhibition in ischemic brain injury. Curr Pharm Des. 2008;14:1401–1418. doi: 10.2174/138161208784480216. [DOI] [PubMed] [Google Scholar]
- 2.Amer M, Bead VR, Bathon J, Blumenthal RS, Edwards DN. Use of nonsteroidal anti-inflammatory drugs in patients with cardiovascular disease: a cautionary tale. Cardiol Rev. 2010;18:204–212. doi: 10.1097/CRD.0b013e3181ce1521. [DOI] [PubMed] [Google Scholar]
- 3.Iadecola C, Gorelick PB. The Janus face of cyclooxygenase-2 in ischemic stroke: shifting toward downstream targets. Stroke. 2005;36:182–185. doi: 10.1161/01.STR.0000153797.33611.d8. [DOI] [PubMed] [Google Scholar]
- 4.Andreasson K. Prostaglandin signalling in cerebral ischaemia. Br J Pharmacol. 2010;160:844–846. doi: 10.1111/j.1476-5381.2010.00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewett SJ, Bell SC, Hewett JA. Contributions of cyclooxygenase-2 to neuroplasticity and neuropathology of the central nervous system. Pharmacol Ther. 2006;112:335–357. doi: 10.1016/j.pharmthera.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Rose ME, Miller TM, Li W, Shinde SN, Pickrell AM, Poloyac SM, Graham SH, Hickey RW. COX2-derived primary and cyclopentenone prostaglandins are increased after asphyxial cardiac arrest. Brain Res. 2013;1519:71–77. doi: 10.1016/j.brainres.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Li W, Ahmad M, Rose ME, Miller TM, Yu M, Chen J, Pascoe JL, Poloyac SM, Hickey RW, et al. Increased generation of cyclopentenone prostaglandins after brain ischemia and their role in aggregation of ubiquitinated proteins in neurons. Neurotox Res. 2013;24:191–204. doi: 10.1007/s12640-013-9377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Li W, Ahmad M, Miller TM, Rose ME, Poloyac SM, Uechi G, Balasubramani M, Hickey RW, Graham SH. Modification of ubiquitin-C-terminal hydrolase-L1 by cyclopentenone prostaglandins exacerbates hypoxic injury. Neurobiol Dis. 2011;41:318–328. doi: 10.1016/j.nbd.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Li W, Rose ME, Pascoe JL, Miller TM, Ahmad M, Poloyac SM, Hickey RW, Graham SH. Prostaglandin D toxicity in primary neurons is mediated through its bioactive cyclopentenone metabolites. Neurotoxicology. 2013;39C:35–44. doi: 10.1016/j.neuro.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oeste CL, Perez-Sala D. Modification of cysteine residues by cyclopentenone prostaglandins: interplay with redox regulation of protein function. Mass Spectrom Rev. 2014;33:110–125. doi: 10.1002/mas.21383. [DOI] [PubMed] [Google Scholar]
- 11.Garzon B, Oeste CL, Diez-Dacal B, Perez-Sala D. Proteomic studies on protein modification by cyclopentenone prostaglandins: expanding our view on electrophile actions. J Proteomics. 2011;74:2243–2263. doi: 10.1016/j.jprot.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 12.Koharudin LM, Liu H, Di Maio R, Kodali RB, Graham SH, Gronenborn AM. Cyclopentenone prostaglandin-induced unfolding and aggregation of the Parkinson disease-associated UCH-L1. Proc Natl Acad Sci USA. 2010;107:6835–6840. doi: 10.1073/pnas.1002295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Gomez FJ, Cernuda-Morollon E, Stamatakis K, Perez-Sala D. Protein thiol modification by 15-deoxy-Delta12,14-prostaglandin J2 addition in mesangial cells: role in the inhibition of proinflammatory genes. Mol Pharmacol. 2004;66:1349–1358. doi: 10.1124/mol.104.002824. [DOI] [PubMed] [Google Scholar]
- 14.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo M, Shibata T, Kumagai T, Osawa T, Shibata N, Kobayashi M, Sasaki S, Iwata M, Noguchi N, Uchida K. 15-Deoxy-Delta(12,14)-prostaglandin J(2): the endogenous electrophile that induces neuronal apoptosis. Proc Natl Acad Sci USA. 2002;99:7367–7372. doi: 10.1073/pnas.112212599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh T, Lipton SA. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci. 2007;30:37–45. doi: 10.1016/j.tins.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Uchida K, Shibata T. 15-Deoxy-Delta(12,14)-prostaglandin J2: an electrophilic trigger of cellular responses. Chem Res Toxicol. 2008;21:138–144. doi: 10.1021/tx700177j. [DOI] [PubMed] [Google Scholar]
- 18.Shibata T, Yamada T, Ishii T, Kumazawa S, Nakamura H, Masutani H, Yodoi J, Uchida K. Thioredoxin as a molecular target of cyclopentenone prostaglandins. J Biol Chem. 2003;278:26046–26054. doi: 10.1074/jbc.M303690200. [DOI] [PubMed] [Google Scholar]
- 19.Hatahet F, Ruddock LW, Ahn K, Benham A, Craik D, Ellgaard L, Ferrari D, Ventura S. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 20.Kozlov G, Maattanen P, Thomas DY, Gehring K. A structural overview of the PDI family of proteins. FEBS J. 2010;277:3924–3936. doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- 21.Andreu CI, Woehlbier U, Torres M, Hetz C. Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett. 2012;586:2826–2834. doi: 10.1016/j.febslet.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka S, Uehara T, Nomura Y. Upregulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J Biol Chem. 2000;275:10388–10393. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- 23.Bleiholder C, Suhai S, Harrison AG, Paizs B. Towards understanding the tandem mass spectra of protonated oligopeptides. 2: the proline effect in collision-induced dissociation of protonated Ala-Ala-Xxx-Pro-Ala (Xxx = Ala, Ser, Leu, Val, Phe, and Trp) J Am Soc Mass Spectrom. 2011;22:1032–1039. doi: 10.1007/s13361-011-0092-1. [DOI] [PubMed] [Google Scholar]
- 24.Raturi A, Vacratsis PO, Seslija D, Lee L, Mutus B. A direct, continuous, sensitive assay for protein disulphide-isomerase based on fluorescence self-quenching. Biochem J. 2005;391:351–357. doi: 10.1042/BJ20050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Melandri F, Berdo I, Jansen M, Hunter L, Wright S, Valbrun D, Figueiredo-Pereira ME. Delta12-Prostaglandin J2 inhibits the ubiquitin hydrolase UCH-L1 and elicits ubiquitin-protein aggregation without proteasome inhibition. Biochem Biophys Res Commun. 2004;319:1171–1180. doi: 10.1016/j.bbrc.2004.05.098. [DOI] [PubMed] [Google Scholar]
- 26.Dickerhof N, Kleffmann T, Jack R, McCormick S. Bacitracin inhibits the reductive activity of protein disulfide isomerase by disulfide bond formation with free cysteines in the substrate-binding domain. FEBS J. 2011;278:2034–2043. doi: 10.1111/j.1742-4658.2011.08119.x. [DOI] [PubMed] [Google Scholar]
- 27.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Wu S, Hickey RW, Rose ME, Chen J, Graham SH. Neuronal cyclooxygenase-2 activity and prostaglandins PGE2, PGD2, and PGF2 alpha exacerbate hypoxic neuronal injury in neuron-enriched primary culture. Neurochem Res. 2008;33:490–499. doi: 10.1007/s11064-007-9462-2. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Wu S, Ahmad M, Jiang J, Liu H, Nagayama T, Rose ME, Tyurin VA, Tyurina YY, Borisenko GG, et al. The cyclooxygenase site, but not the peroxidase site of cyclooxygenase-2 is required for neurotoxicity in hypoxic and ischemic injury. J Neurochem. 2010;113:965–977. doi: 10.1111/j.1471-4159.2010.06674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fink EL, Alexander H, Marco CD, Dixon CE, Kochanek PM, Jenkins LW, Lai Y, Donovan HA, Hickey RW, Clark RS. Experimental model of pediatric asphyxial cardiopulmonary arrest in rats. Pediatr Crit Care Med. 2004;5:139–144. doi: 10.1097/01.pcc.0000112376.29903.8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T, Lipton SA. S-nitrosylation of critical protein thiols mediates protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Antioxid Redox Signal. 2011;14:1479–1492. doi: 10.1089/ars.2010.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeGracia DJ, Montie HL. Cerebral ischemia and the unfolded protein response. J Neurochem. 2004;91:1–8. doi: 10.1111/j.1471-4159.2004.02703.x. [DOI] [PubMed] [Google Scholar]
- 33.Truettner JS, Hu K, Liu CL, Dietrich WD, Hu B. Subcellular stress response and induction of molecular chaperones and folding proteins after transient global ischemia in rats. Brain Res. 2009;1249:9–18. doi: 10.1016/j.brainres.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura Y. Neuronal apoptosis and protection: effects of nitric oxide and endoplasmic reticulum-related proteins. Biol Pharm Bull. 2004;27:961–963. doi: 10.1248/bpb.27.961. [DOI] [PubMed] [Google Scholar]
- 35.Hwang IK, Yoo KY, Kim DW, Han BH, Kang TC, Choi SY, Kim JS, Won MH. Protein disulfide isomerase immunoreactivity and protein level changes in neurons and astrocytes in the gerbil hippocampal CA1 region following transient ischemia. Neurosci Lett. 2005;375:117–122. doi: 10.1016/j.neulet.2004.10.079. [DOI] [PubMed] [Google Scholar]
- 36.Kam KY, Yu SJ, Jeong N, Hong JH, Jalin AM, Lee S, Choi YW, Lee CK, Kang SG. p-Hydroxybenzyl alcohol prevents brain injury and behavioral impairment by activating Nrf2, PDI, and neurotrophic factor genes in a rat model of brain ischemia. Mol Cells. 2011;31:209–215. doi: 10.1007/s10059-011-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal R, Miranda M, Narayan M. Nitrosative stress-induced Parkinsonian Lewy-like aggregates prevented through polyphenolic phytochemical analog intervention. Biochem Biophys Res Commun. 2011;404:324–329. doi: 10.1016/j.bbrc.2010.11.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. Snitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 39.Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD. Protein disulphide isomerase protects against protein aggregation and is Snitrosylated in amyotrophic lateral sclerosis. Brain. 2010;133:105–116. doi: 10.1093/brain/awp267. [DOI] [PubMed] [Google Scholar]
- 40.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274:5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Guan T, Li C, Shang H, Cui L, Li XM, Kong J. SOD1 aggregation in astrocytes following ischemia/reperfusion injury: a role of NO-mediated S-nitrosylation of protein disulfide isomerase (PDI) J Neuroinflammation. 2012;9:237. doi: 10.1186/1742-2094-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uys JD, Xiong Y, Townsend DM. Nitrosative stress-induced S-glutathionylation of protein disulfide isomerase. Methods Enzymol. 2011;490:321–332. doi: 10.1016/B978-0-12-385114-7.00018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Townsend DM, Manevich Y, He L, Xiong Y, Bowers RR, Jr, Hutchens S, Tew KD. Nitrosative stress-induced s-glutathionylation of protein disulfide isomerase leads to activation of the unfolded protein response. Cancer Res. 2009;69:7626–7634. doi: 10.1158/0008-5472.CAN-09-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 46.Descamps E, Petrault-Laprais M, Maurois P, Pages N, Bac P, Bordet R, Vamecq J. Experimental stroke protection induced by 4-hydroxybenzyl alcohol is cancelled by bacitracin. Neurosci Res. 2009;64:137–142. doi: 10.1016/j.neures.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Woycechowsky KJ, Wittrup KD, Raines RT. A small-molecule catalyst of protein folding in vitro and in vivo. Chem Biol. 1999;6:871–879. doi: 10.1016/s1074-5521(00)80006-x. [DOI] [PubMed] [Google Scholar]
- 48.Kersteen EA, Raines RT. Catalysis of protein folding by protein disulfide isomerase and smallmolecule mimics. Antioxid Redox Signal. 2003;5:413–424. doi: 10.1089/152308603768295159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang GZ, Dong XY, Sun Y. Acyl cystamine: small-molecular foldase mimics accelerating oxidative refolding of disulfide-containing proteins. Biotechnol Prog. 2011;27:377–385. doi: 10.1002/btpr.517. [DOI] [PubMed] [Google Scholar]
- 50.Lees WJ. Small-molecule catalysts of oxidative protein folding. Curr Opin Chem Biol. 2008;12:740–745. doi: 10.1016/j.cbpa.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 51.Beld J, Woycechowsky KJ, Hilvert D. Small-molecule diselenides catalyze oxidative protein folding in vivo. ACS Chem Biol. 2010;5:177–182. doi: 10.1021/cb9002688. [DOI] [PubMed] [Google Scholar]
- 52.Musiek ES, Milne GL, McLaughlin B, Morrow JD. Cyclopentenone eicosanoids as mediators of neurodegeneration: a pathogenic mechanism of oxidative stress-mediated and cyclooxygenase-mediated neurotoxicity. Brain Pathol. 2005;15:149–158. doi: 10.1111/j.1750-3639.2005.tb00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 54.Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]