Abstract
Mercury is a widespread environmental contaminant with exposures eliciting a well-documented catalog of adverse effects. Yet, knowledge regarding the underlying mechanisms by which mercury exposures are translated into biological effects remains incomplete. DNA methylation is an epigenetic modification that is sensitive to environmental cues, and alterations in DNA methylation at the global level are associated with a variety of diseases. Using a liquid chromatography tandem mass spectrometry-based (LC-MS/MS) approach, global DNA methylation levels were measured in red blood cells of 144 wild American alligators (Alligator mississippiensis) from 6 sites with variable levels of mercury contamination across Florida’s north-south axis. Variation in mercury concentrations measured in whole blood was highly associated with location, allowing the comparison of global DNA methylation levels across different “treatments” of mercury. Global DNA methylation in alligators across all locations was weakly associated with increased mercury exposure. However, a much more robust relationship was observed in those animals sampled from locations more highly contaminated with mercury. Also, similar to other vertebrates, global DNA methylation appears to decline with age in alligators. The relationship between age-associated loss of global DNA methylation and varying mercury exposures was examined to reveal a potential interaction. These findings demonstrate that global DNA methylation levels are associated with mercury exposure, and give insights into interactions between contaminants, aging, and epigenetics.
Keywords: DNA methylation, mercury, American alligator, environmental contaminants, liquid chromatography, mass spectrometry, deoxyribonucleoside
1. Introduction
Epigenetic modifications (alteration of gene expression rather than genetic code) can mediate a variety of environmental influences on organisms. Advances in understanding the molecular nature of epigenetic modifications provides opportunities for studies aimed at elucidating the interplay between specific modifications and environmental factors. However, due to computational and/or economic constraints, many experimental approaches for examining epigenetic modifications are prohibitive at the population level. Although some studies have begun to examine epigenome-by-environment interactions in human populations, the aforementioned impediments have restricted the majority of wildlife studies in terms of both the species and the number of individuals (Alvarado et al., 2014; Calvanese et al., 2009; Christensen et al., 2009). Over the past decade, direct measurement of changes in global DNA methylation using liquid chromatography-tandem mass spectrometry (LC-MS/MS) has become a viable approach, and the inherent advantage of this approach (cost, throughput, simplicity) make it suitable for both environmental applications and studies at the population level (Kok et al., 2007; Le et al., 2011; Liu et al., 2009; Ma et al., 2009; Parrott et al., 2014; Quinlivan and Gregory, 2008b; Song et al., 2005). In this study, LC-MS/MS was used to characterize changes in global DNA methylation in a long-lived environmental sentinel to investigate the relationship between a fundamental epigenetic modification and whole blood mercury concentrations.
The American alligator (Alligator mississippiensis) is a long-lived (~80 years) apex predator that displays high site fidelity (Elsey et al., 2008; Lance, 2003). These characteristics provide an ideal model in which to study the long-term effects of chronic environmental contaminant exposures on the epigenome. Alligators are also an economically important natural resource across the southeastern United States as they are both commercially harvested for meat and leather, as well as recreationally harvested for personal consumption. Mercury (Hg) has been a contaminant of growing concern across the state of Florida since the 1980s, when concentrations in higher trophic level species such as large-mouth bass (Micropterus salmoides) and alligators were found to be above the United States Environmental Protection Agency’s Human Health Advisory level of 0.30 μg/g at several locations (Delany et al., 1988; Lange et al., 1994; Ware et al., 1990). In particular, the Florida Everglades has been identified as a ‘hotspot’ region for mercury accumulation, due to the unique environmental conditions of that ecosystem (e.g., hydroperiod, warm temperatures, shallow water depths) that promote the methylation of mercury (Julian, 2013). Elevated concentrations of mercury found in the Everglades are reflected throughout the trophic levels of the biota, from apple snails (Pomacea paludosa) to alligators (Eisemann et al., 1997; Heaton-Jones et al., 1997; Jagoe et al., 1998; Kannan et al., 1998; Rumbold et al., 2002; Ugarte et al., 2005; Yanochko et al., 1997). Mercury has been shown to cause reproductive and neurological impairment as a function of direct exposure and consumption of contaminated prey (Crump and Trudeau, 2009; Frederick and Jayasena, 2010; Heath et al., 2005; Khan and Tansel, 2000; Kurland et al., 1960). This is problematic for all trophic levels in the Everglades, as the bioaccumulation of mercury in this ecosystem begins at the base of the food web (Julian, 2013; Khan and Tansel, 2000). Other effects, such as inflammation and tissue necrosis, have been observed in internal organs of adult spotted sea trout (Cynoscion nebulosus) from areas highly contaminated with mercury (Adams et al., 2010). However, the mechanisms by which mercury affects various organs and systems need to be further elucidated, particularly for long-lived species undergoing chronic exposure.
DNA methylation, consisting of the covalent addition of a methyl-group to the 5′ carbon of cytosine, is an epigenetic modification functioning in roles that include regulating gene expression, promoting chromosome stability, and silencing the transcription of transposons (Di Giacomo et al., 2013; Parrott et al., 2014; Rodriguez et al., 2006). Alterations to the methylome can occur at specific loci or more broadly across the genome. While methods of resolving the methylome at base pair resolution allow for the identification of loci-specific alterations, analyses of the comprehensive methylome at base pair are not currently feasible at the population level. However, measures of global DNA methylation can be made across a large number of samples and DNA methylation at the global level is intimately linked to one carbon metabolism as this process provides key methyl donors for both maintenance and de novo DNA methylation (Anderson et al., 2012; Parrott et al., 2014). Alterations to one-carbon metabolism, either through environmental or genetic factors, have been shown to influence measures of global DNA methylation (Drake et al., 2015; Kruman and Fowler, 2014). Further, while targeted approaches can provide understanding of the influence of DNA methylation on transcriptional activity at a specific locus, they are not able to provide insights into the other aforementioned broader roles of DNA methylation.
Studies in humans and other lab models have demonstrated the highly dynamic nature of DNA methylation patterning across the vertebrate lifespan. Shortly after fertilization, the maternal and paternal pronuclei undergo genome-wide demethylation and subsequently begin to acquire tissue- and cell-type specific methylomes during development and differentiation (Brandeis et al., 1993; MacDonald and Mann, 2014). Later in life, variation in the DNA methylome becomes tightly associated with age, with global measures of genomic DNA methylation consistently found to decline as a function of increasing age (Bollati et al., 2009; Fuke et al., 2004; Hannum et al., 2013; Heyn et al., 2012). However, the drivers of these age-associated changes in DNA methylation are not known. In humans, lifestyle and environmental factors such as diet, smoking, alcohol consumption, traffic pollution, and exposures to endocrine disrupting compounds have all been associated with altered levels of global DNA methylation (Bromer et al., 2010; Christensen et al., 2009; Dolinoy et al., 2007; Guo et al., 2014; Rusiecki et al., 2008). Previous reports have shown that exposure to certain metals (e.g., arsenic, cadmium, copper, mercury) can negatively affect global DNA methylation (Arita and Costa, 2009; Argos et al., 2015; Niedzwiecki et al., 2013; Tellez-Plaza et al., 2014). Moreover, it is known that specific metals can interact to produce varying biological responses (Raymond and Ralston, 2004; Yang et al., 2010). However, the molecular mechanisms by which environmental factors, such as metals, influence DNA methylation in natural populations remain unclear.
In this study, we aimed to investigate the presence of trace metals in an environmental model and interrogate potential associations with global DNA methylation, with a particular focus on mercury, as its impact throughout the southeastern United States has been well-documented (Facemire et al., 1995). Very few studies to date have examined how mercury exposures might affect the epigenome, especially in free-living animals (Basu et al., 2013; Hanna et al., 2012; Pilsner et al., 2010). In one study, indirect measures of global DNA methylation weakly trended negative with increasing mercury levels in brain tissues from polar bears (Ursus maritimus). However, a clear correlation was not observed (Pilsner et al., 2010). Studies using captive animals have shown that there is a relationship between DNA hypomethylation in brain tissue and environmentally relevant concentrations of mercury for mink (Neovison vison), but not for fish (Perca flavescens) or chickens (Gallus gallus) (Basu et al., 2013). However, current literature does not provide a clear consensus regarding the relationship between DNA methylation and Hg exposures. Here, we report our findings examining the relationship between direct measures of global DNA methylation and mercury concentrations, as well as sixteen other metals, in blood samples collected from adult and sub-adult alligators from 6 different sites distributed along Florida’s north-south axis. Whereas the cellular heterogeneity associated with mammalian blood samples has resulted in controversy and provides challenges for interpreting epigenetic data, red blood cells (RBCs) in non-mammalian vertebrates are nucleated and provide a relatively homogenous cellular composition ideal for epigenetic studies (Reinius et al., 2012). Further, collection of RBCs in alligators is non-lethal, minimally invasive, and allows for sampling a large population of animals. Here, we take advantage of these aspects to explore the hypothesis that long-term mercury exposure is linked to measures of DNA methylation.
2. Materials and Methods
2.1 Sample Collection
Animals were collected by researchers with the Florida Fish and Wildlife Conservation Commission and the Medical University of South Carolina using guidelines provided by the American Society of Ichthyologists and Herpetologists (ASIH, 2004). Animals were selected at random from sites known to have either a low, moderate, or high concentrations of mercury in the upper trophic levels (Delany et al., 1988; Hord et al., 1990). In this study, we use measures of body length (snout-vent length (SVL)) as a proxy for age. Two size classes were noted in this study, sub-adult (45 cm ≤ SVL < 90 cm) and adult (SVL ≥ 90 cm), based on a previous report in Florida pertaining to reproductive maturity (Woodward et al., 1992). Twenty-four alligators, 12 sub-adults and 12 adults of both sexes included (Tables A1 and A2), were sampled from each location during the spring of 2012 (Fig 1A). Whole blood was collected from the post-occipital venous sinus with a sterile needle and syringe immediately following capture as described by Myburgh et al. (2014). Whole blood samples were then transferred to 8 mL lithium-heparin Vacutainer blood collection tubes (BD, Franklin Lakes, NJ), and kept on wet ice for no longer than 5 h. For mercury measures, whole blood was frozen at −80 °C until analysis. Red blood cell samples were collected from a separate tube by centrifugation, fixed in RNA Later (Sigma-Aldrich, St. Louis, MO), and frozen at −80 °C until DNA extraction.
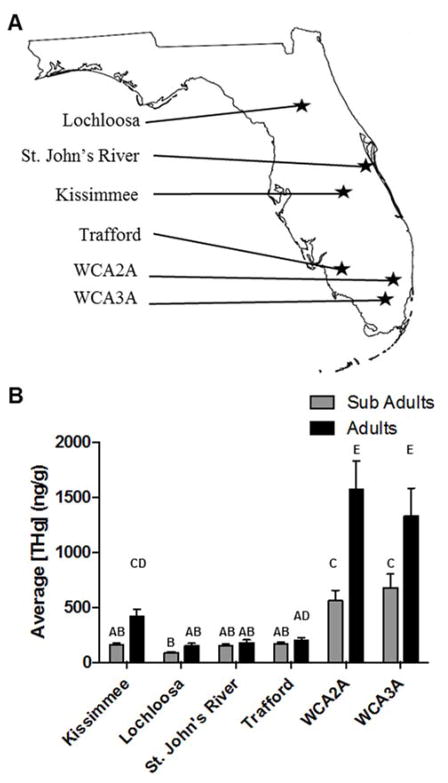
Figure 1.
Map of Florida showing the six sites from which American alligators (Alligator mississippiensis) were sampled in this study (A). The mean (± SD) total mercury concentrations (ng/g, wet mass) in alligator whole blood partitioned as a function of size class and sampling location in Florida (B). Significantly different mean mercury concentrations are denoted by different letters (significance noted by p < 0.05).
2.2 Trace Metal Analysis
For analysis of mercury and sixteen other trace elements (aluminum (Al), vanadium (V), chromium (Cr), manganese (Mn), cobalt (Co), nickel (Ni), copper (Cu), zinc (Zn), arsenic (As), selenium (Se), rubidium (Rb), strontium (Sr), molybdenum (Mo), cadmium (Cd), tin (Sn) and lead (Pb)) whole blood samples were thawed, gently rocked for homogenization and divided into subsamples for analysis. The mass fraction of total mercury was determined in one aliquot (100 μL) of alligator whole blood with a direct mercury analyzer (DMA-80, Milestone, Shelton, CT), utilizing National Institute of Standards and Technology (NIST) Standard Reference Material (SRM) 3133, Mercury Standard Solution, for external calibration and SRM 955c Level 3, Toxic Metals in Caprine Blood, as a control material, certified for total mercury at 17.8 ± 1.6 ng/g. The replicated measurements of the control material had an average value of 18.7 ng/g ± 1.45 ng/g, falling within the confidence interval (16.2 – 19.4 ng/g). Replicated unknown samples were analyzed throughout the sample queue, with a relative standard deviation (RSD) of 0.7%. Procedural and field blanks were analyzed concurrently, by use of an empty sample vessel or Milli-Q water, respectively. If blanks were found to be above the detection limit, the samples were blank corrected. Total mercury (THg) was measured as previous studies of reptiles and birds have shown that methylmercury comprises greater than 70% of the total mercury content in whole blood, and in the species most closely related to the American alligator, the ratio is 100% when mercury concentrations are above 1,000 ng/g and drop to 70% at concentrations below 1000 ng/g (Bergeron et al., 2007; Rimmer et al., 2005). This measurement is used as a proxy for the most biologically relevant form of mercury, methylmercury, until species specific information regarding the ratio of methylmercury to total mercury in crocodilian whole blood becomes available.
The second aliquot of whole blood was used to measure total trace element concentrations. Approximately 0.25 g of sample and 0.25 g of internal standard (Ru, Y, Sc, High-Purity Standards, Charleston, SC) were weighed into a pre-cleaned Teflon digestion vessel (CEM, Matthews, NC). After the addition of 3.5 mL of high purity nitric acid (Optima, Fisher, Pittsburg, PA), samples were digested in a CEM MARS Xpress microwave (Microwave Accelerate Reaction System, CEM, Matthews, NC), set at a maximum temperature of 210 °C holding for 15 min. Samples were diluted to 50 g in 3% high purity hydrochloric acid prior to analysis. The samples were measured on a Thermo X2 quadrupole inductively coupled plasma-mass spectrometer (ICP-MS) system (software build, 2.6.0.334; Thermo, Waltham, MA), equipped with a standard introduction system and an ESI SC4 auto-sampler (Elemental Scientific, Omaha, NE). The procedural blanks, field blanks and control materials were treated in a similar manner to the samples.
The ICP-MS was tuned daily using a standard 1 ng/g multi-element tuning solution (Solution “F”, Thermo, Wilmington, DE). The ICP-MS was operated in two collision cell modes utilizing 8 % H2 in 92 % He or 1 % NH3 in 99 % He as the collision gas to help eliminate isobaric interferences. Quadrupole MS routine methods utilize peak jumping, with each of five replicate runs consisting of 50 sweeps, and a dwell time of 25 ms. Seronorm Trace Metals in Whole Blood L-3 standard solution (Sero, Bilingstad, Norway Lot# 1112691) was used as a control material for ICP-MS analysis. The main trace element of interest, Selenium, measured an average value of 204.5 ng/g with a standard deviation of 9.7 ng/g across all control material replicates; falling within the acceptable range of 256.6 ± 51.9 ng/g. All other trace metals were within acceptable range for sample analysis (Al, Cu, Zn, As, Rb, Sr, Pb) or were found to be below the limits of detection (V, Cr, Mn, Co, Ni, Mo, Cd, Sn). Replicated unknown samples were analyzed throughout the sample queue, with a relative standard deviation (RSD) of 0.1 %, 4 %, 4 %, 7 %, 1 %, 5 %, 4 %, and 1 % for Al, Cu, Zn, As, Se, Rb, Sr and Pb, respectively. Data were blank corrected based on the procedural blanks analyzed within the same day. Working external calibration standards were prepared by gravimetric dilution of primary standard NIST SRM 3100 series solutions. First order linear fits were applied to each isotope analyzed and the slope and intercept resulting from the curve was used to calculate the mass fraction of trace elements in the blood samples.
2.3 DNA Isolation
DNA was extracted following the protocol from RBCs preserved with RNA Later using the Promega total DNA Isolation kit (Madison, WI). DNA concentration and purity was assessed by measuring optical density using a NanoDrop UV-Vis Spectrophotometer (Thermo, Wilmington, DE).
2.4 Preparation of Deoxyribonucleoside Solutions and Calibration
2′-deoxyguanosine monohydrate (dG, Sigma Aldrich, St. Louis, MO) and 5-methyl-2′-deoxycytidine (5mdC, Santa Cruz Biotechnology Inc., Dallas, TX) were used as the deoxyribonucleoside standards. Initial stock solutions of each deoxyribonucleoside standard were made by gravimetric addition of ≈10 mg neat standard into 10 mL of Milli-Q water to produce ≈1000 ng standard/mg water solutions. To enhance solubility, sodium hydroxide pellets (402.24 mg, Sigma Aldrich, 97 % ACS reagent) were gravimetrically added to the dG stock solution. Using the initial stock solutions, 20 ng standard/mg water working solutions were prepared by gravimetric addition. The final calibration solutions included the gravimetric addition of 200 mg of the 20 ng/mg dG solution and varying amounts of the 20 ng/mg 5mdC solution to produce twelve increments from 0.1 % to 10 % solutions of 5mdC to dG (each calibration solution was analyzed in quadruplicate).
2.5 Hydrolysis of Final Calibration Solutions and Whole Blood DNA Extracts
To hydrolyze genomic DNA into individual nucleosides, the method described by Quinlivan and Gregory (2008) was used and modified as follows (Quinlivan and Gregory, 2008a). The genomic DNA extracted from whole blood (18 μL at [20 μg/μL]) was added to hydrolysis buffer in a 1:1 ratio (final volume = 36 μL). Both the genomic DNA samples and the final calibration curve solutions (18 μL of each final calibration solution and 18 μL of hydrolysis buffer) were incubated at 37 °C for 11 h in an Innova 4200 Incubator (New Brunswick Scientific, Edison, NJ). The hydrolysis buffer was made as previously reported by Quinlivan and Gregory (2008), with the Tris-HCl Buffer at a pH = 7.75 (Quinlivan and Gregory, 2008a). To confirm hydrolysis, aliquots of 5 samples were assessed with their non-hydrolyzed counterparts via 1% agarose gel electrophoresis (Figure A1). Post-hydrolysis, 30 μL of each hydrolyzed sample/calibration solution and 40 μL of Milli-Q water were added to autosampler vials, for a final volume of 70 μL. Using calculated amounts (ng) in each calibration vial, gravimetrically derived weight ratios were calculated for 5mdC to dG. The weight ratios (x-axis) were used to construct calibration lines for the % 5mdC to dG.
2.6 LC-MS/MS Method
LC-MS/MS was employed to directly calculate the proportion of methylated deoxycytosine (5mdC) to deoxyguanosine (dG) within genomic DNA extracted from RBCs. Deoxyribonucleosides, (5mdC and dG) present in the DNA extracts, calibration solutions, and blanks were chromatographically separated using an Agilent 1100 LC and Autosampler. The deoxyribonucleosides were separated on a temperature-controlled (20 °C) Kinetex C18 column (100 × 3.0 mm, 2.6 μm, Phenomenex, Torrance, CA). After each injection (7.5 μL), separation of the nucleosides was achieved using the solvent mixtures of (A) Optima LC-MS grade acetonitrile (ACN, Fisher Scientific, Fair Lawn, NJ) with 0.1 % formic acid ((98 %, EMD, Germany) and (B) water with 0.1 % formic acid in a gradient as follows: 0 to 1 min (100 % B), 1 to 14 min (92 % B), 14 to 15 min (100 % B), and continued from 15 to 20 min for re-equilibration, with a flow rate of 250 μL/min. Other deoxyribonucleosides, such as 2′-deoxycytidine (dC, Sigma-Aldrich, St. Louis, MO), thymidine (T, Sigma-Aldrich, St. Louis, MO), 2′-deoxyadenosine monohydrate (dA, Sigma Aldrich), and 5-hydroxymethyl-2′-deoxycytidine (5hmdC, Berry and Associates, Dexter, MI), were also separated using the described method (Figure A2). To assess the reproducibility of the LC-MS/MS assay and serve as a Quality Control (QC) measure, three separate aliquots of a pooled DNA sample were prepared and each was analyzed in triplicate (RSD < 10 %). The alligator whole blood DNA extracts (n = 122), calibration solutions (n = 13), QC samples (n = 3) and blanks (n = 3) were queued in randomized sets and analyzed in triplicate. Blank replicates were randomly analyzed every 30 samples during the sample queue (in triplicate).
Chromatographically separated deoxyribonucleosides were detected by multiple reaction monitoring (MRM) using an AB Sciex API 4000 triple quadrupole mass spectrometer (equipped with a TurboV electrospray ionization source). Operation of the LC and MS was controlled using Analyst software (v.1.52, SCIEX, Framingham, MA). The method employed scheduled MRM, which was set to scan using a 180 s scan window from the retention times noted in Table A3. The target scan time for each MRM scan was 2 s. The MRM transition for each deoxyribonucleoside (Q1 mass (Da) → Q3 mass (Da)), and the tune-optimized compound-specific MS/MS parameters are also shown in Table A3. The tune-optimized source parameters were: collisionally activated dissociation (8.0); curtain gas (15 psi); gas 1 (50 psi); gas 2 (30 psi); source temperature (500 °C); interface heater (on); and ion spray voltage (5000 V).
2.7 Global DNA Methylation Quantitation
Peaks for each nucleoside were integrated using the Analyst quantitation software. The percent global methylation (% 5mdC, or 5mdC/dG) was calculated using a calibration curve constructed by relating the calibration solution peak area ratios to the gravimetrically determined weight ratios of 5mdC and dG, as previously described, with some modification (contribution of deoxyribonucleoside adducts was negligible and were not factored into this analysis) (Parrott et al., 2014; Quinlivan and Gregory, 2008b). To normalize differences in ionization efficiency between 5mdC and dG, the dG peak area was multiplied by a response factor (0.8971) before the peak area ratio was calculated, as previously described using an equimolar solution (Parrott et al., 2014). The QC and calibration solutions were run throughout the sample queue and the resultant calibration line is shown in Figure A3 (r2 = 0.9994). Peak areas for each deoxyribonucleoside in each sample (n = 3) were corrected (dG, as previously described), and averages as well as the % of 5mdC were calculated using the calibration line.
2.8 Statistical Analysis
All trace element data failed to meet the assumptions of normality and homoscedasticity based on the Shapiro-Wilk Goodness of Fit Test and Levene’s Test for Unequal Variances. After the trace element data were log10 transformed, THg measurements met the parametric assumptions; the remaining trace metal measurements did not. Linear Regression, Spearman Correlation, and Pearson Product Moment Correlations were used to compare the trace element data to the DNA methylation data, where appropriate. Student’s T-tests were used to assess differences in THg concentration and DNA methylation pattern due to sex. No significant differences were found and both sexes were grouped for the remaining analyses. The Two-Way Factorial ANOVA was used to compare mercury concentrations found at each of the six sites, with the Tukeys’ HSD Multiple Comparison post-hoc test for comparisons among sites and age classes. An ANCOVA analysis was not used as all the parameters did not meet the assumptions, particularly the homogeneity of regression. All statistical analyses were conducted using GraphPad (Prism 6, La Jolla, CA) and JMP 11 (SAS, Cary, NC). Statistical significance was determined by p < 0.05 for all tests.
3. Results and Discussion
3.1 Variation in total mercury levels in alligators is site and age-class dependent
Total mercury (THg) concentrations in whole blood of adult American alligators from 6 sites in Florida (Fig 1A) were measured. Values are expressed on a wet mass basis. At each site, adults were found to have greater concentrations of mercury than sub-adults. Statistically significant differences between size classes were only seen at the three locations with the greatest mercury concentrations (WCA2A, WCA3A, and Kissimmee) (Fig 1B, Tables A1 and A2). Comparisons of mercury concentrations across the sites revealed significant differences among many of the groups (Fig 1B, Table A4). With exception of one site, Lake Trafford, increasing concentrations of mercury were observed towards the southern part of the state (Fig 1B). Animals sampled at the two sites within the Everglades, WCA2A and WCA3A, were observed to have the greatest concentrations of mercury. These findings are consistent with previous studies identifying the Everglades region as a whole but, particularly WCA3A, as a ‘hotspot’ for mercury accumulation, with animals from this region demonstrating elevated mercury concentrations in comparison to animals from other regions of the state (Delany et al., 1988; Heaton-Jones et al., 1997; Julian, 2013).
Interestingly, there appeared to be an increasing gradient of mercury concentrations along the north-to-south axis, through the central Florida drainage system, from Lake Kissimmee to the Everglades (Figure A5, A), although it should be noted that for adults, mercury concentrations in WCA2 were slightly higher than in WCA3 (Fig 1B). The trend in mean mercury concentration for the “high mercury” sites (Kissimmee, WCA2, and WCA3) was found to be 417, 1570, 1330 ng/g, in adults and 160, 560, and 680 ng/g, in sub-adults, respectively. In comparison, mercury in the northern-central “low mercury” sites (Lochloosa, St. John’s River, Trafford) were over three times lower than those in the Everglades for sub-adults (over five times lower for adults) (Table A5). Previous localized atmospheric sources, such as medical waste incineration, have been thought to contribute significantly to the deposition of mercury in the Everglades in the 1980’s and 1990’s (Frederick et al., 2005). Reductions in these local sources of mercury emissions in the early 1990’s resulted in a concomitant reduction in mercury concentration in resident Everglades fish and wildlife; however, atmospheric deposition of mercury, dominated by global sources, have persisted with relatively stable amounts of deposition since that time (Florida Department of Environmental Protection, 2013; Julian et al., 2015). The unique hydrology of the Everglades continues to enable the rapid conversion of mercury to the biologically available form which accumulates to concentrations in biota that are potentially unsafe for human consumption (Frederick et al., 2005; Julian, 2013). The increasing concentrations of mercury observed in animals from Lake Kissimmee to the Everglades locations suggests that anthropogenic influence could be an additional source of mercury in this drainage system. This series of connected watersheds begins north of Lake Kissimmee, drains through part of urban Orlando and nearby tourist attractions, which both add to the water effluent being filtered through the more southern watersheds. This drainage system continues through central Florida down through the Everglades to Florida Bay (Figure A5, A) (Florida Department of Environmental Protection and Restoration, 2015). Mercury concentrations in animals from the three sites that are not part of this drainage system, Lochloosa Lake, St. John’s River and Lake Trafford (Fig A5, B), were below those concentrations observed in the Everglades, despite Lake Trafford’s close proximity to the WCAs. The combined effects of the anthropogenic influence on the central Florida drainage system and the unique biogeochemical characteristics of the Everglades that control production, transport, binding, and bioaccumulation of methylmercury, provides a unique location to study a chronic environmental “treatment” of mercury in the organisms that reside there.
3.2 DNA hypomethylation is associated with mercury exposure
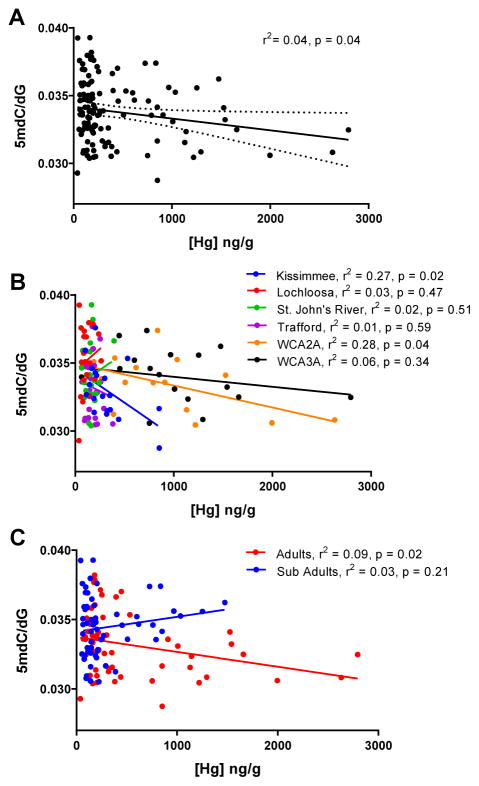
The relationship between global measures of DNA methylation and concentrations of all trace elements quantified (Al, Cu, Zn, As, Se, Rb, Sr, Pb and THg) was investigated. Decreased measures of global DNA methylation (5mdC/dG) were weakly, but significantly correlated to increasing mercury concentrations across all individuals by linear regression (p = 0.04; r2 = 0.04) (Fig 2A). However, correlations between global DNA methylation and other trace elements were not observed (Table A5). To test if the negative relationship between global DNA methylation and THg concentrations was different across sampling sites, each site was analyzed independently. The relationships between global DNA methylation and THg were found to be significantly different and varied widely across sites (Fig 2B; F5, 107 = 3.05, p = 0.01). The relationship appeared more pronounced in alligators from Kissimmee (p = 0.02; R2 = 0.27), and WCA2A (p = 0.04; R2 = 0.28), two of the sites with the greatest concentrations of THg (Fig 2B). A significant relationship was not observed at WCA3A. There was no discernible relationship between global DNA methylation and THg at the three sites with the lowest measured Hg concentrations. Taken together, these data suggest that higher Hg exposures are linked to decreases in global DNA methylation in alligators living in areas with higher, but not lower, levels of Hg contamination.
Figure 2.
Global measures of DNA methylation in whole blood samples from American alligators (Alligator mississippiensis) from Florida. All individuals (n = 119) are plotted either (A) all together, (B) according to site, or (C) according to size class. Results of linear regression analyses are reported. Top and bottom lines in (A) represent 95% confidence intervals.
Alligators from the sites with the least THg also appear to have greater selenium concentrations when compared to other locations; however, the relationship was not statistically significant (Table A5). Selenium has been shown to have a protective role in ameliorating deleterious effects associated with mercury exposure, as it facilitates the demethylation process (Burger and Gochfeld, 2011; Ralston and Raymond, 2010; Raymond and Ralston, 2004; Yang et al., 2008). Ralston and Raymond (2010) propose that a selenium to mercury (Se: Hg) molar ratio above 1:1 may afford protection from the effects of mercury exposure in rodents. There are many factors to consider apart from the singular molar ratio, as not all selenium measured is biologically available for demethylation of mercury, and therefore not available to aid in mercury detoxification. Still, reporting molar ratios has been deemed an acceptable basis of comparison (Burger and Gochfeld, 2011). Alligators examined in this study demonstrated a wide range of Se:Hg molar ratios, with the animals from the locations with greater mercury concentrations, having lower Se:Hg ratios (Table A5). The adult animals from the Everglades locations demonstrated Se:Hg molar ratios below 1:1, suggesting that these individuals might be more susceptible to the effects of mercury exposure (Table A5) (Ralston and Raymond, 2010). In this study, lower Se:Hg ratios are associated with decreased methylation, possibly due to the inability of selenium to counteract mercury efficiently when mercury is present in much greater concentrations. However, the variation in Se:Hg ratios reported in this study appear to be driven more by changes in Hg rather than Se, and therefore prevent strong conclusions about the protective effect of Se in these animals.
The relationship between global DNA methylation and THg concentrations in different size classes was also examined. This relationship was different across the two size classes (F1,115 = 7.00, p < 0.01), with a significant inverse relationship observed between the larger and sexually mature adult animals (p = 0.02), but not for sub-adults (Fig 2C), or between the sexes (data not shown). Because adults were found to have greater THg levels than sub-adults, these findings further suggest that only at the higher ranges of exposure does mercury contamination begin to affect the methylome. Pilsner et al. (2010) examined global DNA methylation and THg in the brains of polar bears (Ursus maritimus), and found no correlation between the two. In addition, mean THg concentrations in bear brains were lower those observed in the studies of other wildlife (Bastos et al., 2015; Fortin et al., 2001; Heaton-Jones et al., 1997). However, the size class of bears sampled was heavily skewed toward sub-adults. If the relationship between global DNA methylation and mercury exposure is size class-dependent in brain tissue from polar bears as it appears to be in whole blood from alligators, our findings in this study might explain why no correlation was observed by Pilsner et al. (2010). Diet is also known to affect DNA methylation patterns and maternal ingestion of endocrine disrupting contaminants has been shown to cause hypomethylation in the offspring of female mice (Dolinoy et al., 2007). Thus, while maternal effects rather than chronic exposure cannot be ruled out, the observed relationship in adults but not sub-adults supports a model in which chronic and/or bioaccumulation negatively impacts global DNA methylation in these animals.
3.3 Age-associated DNA methylation loss and relationship to THg levels
The effect of site and size class on global measures of DNA methylation was then examined. A two-way ANOVA revealed that global DNA methylation is significantly associated with both site (F5,107 = 2.80, p = 0.02) and size class (F1,107 = 12.33, p < 0.001), with adults categorically displaying decreased global DNA methylation when compared to sub-adults (Fig 3).
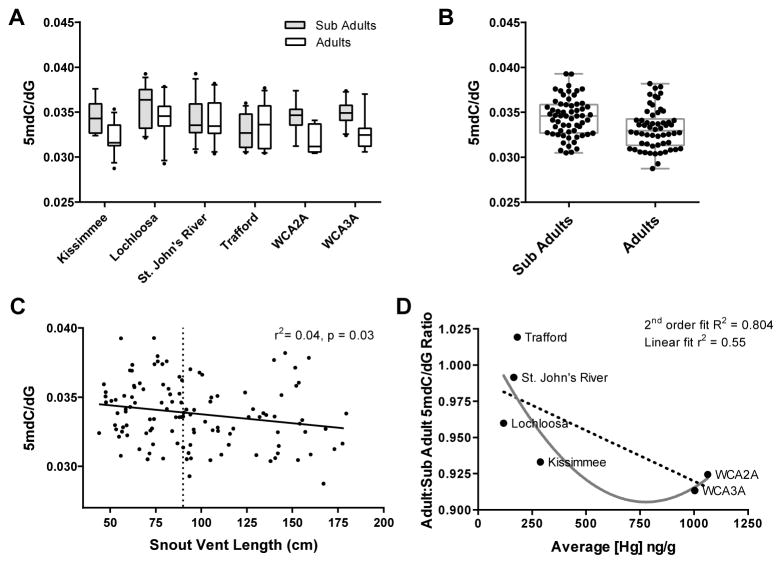
Figure 3.
Influence of site and size class (age) on global measures of DNA methylation in the American alligators (Alligator mississippiensis) sampled in Florida using whole blood (THg analysis) and red blood cells (DNA methylation analysis). (A) Box-and-whisker plots of global DNA methylation measures across site and age class. (B) Across all individuals, sub adults have greater measures of global DNA methylation when compared to adults. (C) Snout-vent length is plotted against 5mdC/dG for each individual. Results of a linear regression analysis is reported, dotted line demarcates the sub-adults and adults. (D) The proportion of global DNA methylation in adults relative to sub-adults from the same site is plotted against the mean concentrations of THg measured for each site. r2 values are reported for linear regression analyses; R2 values are reported for non-linear regression analyses.
The differences in global DNA methylation for sub-adults compared to adults is consistent with previous studies in which captive juvenile alligators were found to have increased measures of global methylation when compared to wild adults (Parrott et al., 2014). Alligators undergo prolonged growth and in many cases, measures of animal length serve as the closest proxy to age available. In an effort to further explore the relationship between global DNA methylation and alligator size (and age) class, a linear regression analysis was performed incorporating all captured animals and a weak, yet significant inverse relationship was found between global DNA methylation and snout-vent length (Fig 3C; r2 = 0.04, p = 0.03). Taken together, both categorical and continuous analyses suggest that similar to humans, alligators undergo age-associated loss of global DNA methylation. However, when linear regression analyses were performed on either adults or sub-adults alone (Figure A4), there was no apparent link between length and DNA methylation, suggesting that instead of a continuous loss over time, age-associated global methylation loss might be more punctuated and occur over the course of sexual maturation.
In Figure 3A, the differences between sub-adult and adult global DNA methylation appear more pronounced in alligators captured at sites observed to have the greatest concentrations of THg (Kissimmee, WCA2A, and WCA3A). In light of the finding that increased THg concentrations are associated with decreased global DNA methylation, it is hypothesized that alligators living in sites with the greatest concentrations of THg could undergo a more pronounced age class-associated reduction in global DNA methylation. We tested for an interaction between THg exposure and size-class on measures of global DNA methylation. We categorized the three sites with the highest THg (WCA2A, WCA3A, and Kissimmee) as “high mercury” and three sites with the lowest THg (Lochloosa, Trafford, and St. Johns River) as “low mercury.” We performed a two-way ANOVA with mercury level and size class set as independent variables and observed a significant interaction between them (F1,117 = 6.23, p = 0.014). To investigate this interaction more closely, the average THg concentration for each site was plotted against the ratio of adult to sub-adult global DNA methylation (Fig. 3D). Interestingly, the three sites with the greatest concentrations of THg showed the most dramatic size class-associated loss of DNA methylation, with WCA3A, WCA2A, and Kissimmee adults retaining only 91%, 92%, and 93%, respectively, of the global DNA methylation levels measured in their sub-adult counterparts. In contrast, adult alligators living in the three sites with the lowest THg concentrations retained greater than 95% of the DNA methylation observed in their sub-adult counterparts. Because the vast majority of DNA methylation takes place only within the context of CpG dinucleotides, it is important to note that small percentage changes in global methylation (relative to all cytosines) represents a substantially larger change in the proportion of methylated CpG dinucleotides. Further, both linear (r2 = 0.55, p = 0.09) and 2nd order polynomial regressions (R2 = 0.80, p = 0.27) yielded models suggestive of a trend in which those alligators living in sites with the greatest THg concentrations undergo the most age-associated decrease in global DNA methylation.
Conclusions
By utilizing a long-lived sentinel species, we demonstrate that global measures of genomic DNA methylation are negatively associated with high mercury exposure, aging, and also identify a possible link connecting environmental exposures to the age-associated loss of global DNA methylation. While global measures provide a feasible approach to examine DNA methylation across large numbers of animals, it is still unclear what regions of the genome undergo epigenetic remodeling in response to mercury exposures and aging. Christensen et al. (2009), found that DNA methylation in humans increased in the context of CpG islands and decreased outside of islands as a function of aging. A similar study found that centenarians displayed hypomethylation in all regions of the genome except CpG islands when compared to newborns (Heyn et al., 2012). Experiments aimed at elucidating the alligator methylome at the base pair level are needed in order to better understand how the epigenome changes as a function of aging and if age-associated changes in humans are broadly conserved in non-mammalian vertebrates. Future studies examining if both mercury exposure and aging affect specific and overlapping regions of the methylome will reveal insights into mechanisms by which environmental factors impact the aging process. Interactions between environmental factors and fundamental aging processes could potentially shape aging trajectories and affect the downstream development of age-associated health outcomes.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported in part by the CoEE Center for Marine Genomics, National Institute of Standards and Technology (NIST), and NIST Grant #60NANB12D225.
We would like to thank the members of the Guillette laboratory group and the Florida Fish and Wildlife Conservation Commission that aided in sample collection. This work is dedicated to the memory of Louis J. Guillette Jr., a mentor, colleague, friend, and full-fledged supporter of interdisciplinary research.
Footnotes
Disclaimer
Certain commercial equipment or instruments are identified in the paper to specify adequately the experimental procedures. Such identification does not imply recommendations or endorsement by the NIST nor does it imply that the equipment or instruments are the best available for the purpose.
Information detailing the sample collection sites, analytical method employed and associated analysis conducted is provided in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- Adams DH, Sonne C, Basu N, Dietz R, Nam D-H, Leifsson PS, et al. Mercury contamination in spotted seatrout, Cynoscion nebulosus: An assessment of liver, kidney, blood, and nervous system health. Sci Total Environ. 2010;408:5808–5816. doi: 10.1016/j.scitotenv.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Alvarado S, Fernald RD, Storey KB, Szyf M. The Dynamic Nature of DNA Methylation: A Role in Response to Social and Seasonal Variation. Integr Comp Biol. 2014;54:68–76. doi: 10.1093/icb/icu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–9. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, et al. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect. 2015;123:64–71. doi: 10.1289/ehp.1307884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASIH ASoIaH. Guidelines for use of live amphibians and reptiles in field and laboratory research. Herpetological Animal Care and Use Committee of the ASIH; Washington, DC: 2004. [Google Scholar]
- Bastos WR, Dórea JG, Bernardi JVE, Lauthartte LC, Mussy MH, Hauser M, et al. Mercury in muscle and brain of catfish from the Madeira river, Amazon, Brazil. Ecotoxicol Environ Safety. 2015;118:90–97. doi: 10.1016/j.ecoenv.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Basu N, Head J, Nam D-H, Pilsner JR, Carvan MJ, Chan HM, et al. Effects of methylmercury on epigenetic markers in three model species: mink, chicken and yellow perch. Comp Biochem Physiol C Toxicol Pharmacol. 2013;157:322–327. doi: 10.1016/j.cbpc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron CM, Husak JF, Unrine JM, Romanek CS, Hopkins WA. Influence of feeding ecology on blood mercury concentrations in four species of turtles. Environ Toxicol Chem. 2007;26:1733–1741. doi: 10.1897/06-594r.1. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–9. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Ariel M, Cedar H. Dynamics of DNA methylation during development. Bioessays. 1993;15:709–713. doi: 10.1002/bies.950151103. [DOI] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J: Fed Am Soc Exp Biol. 2010;24:2273–80. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, Gochfeld M. Mercury and selenium levels in 19 species of saltwater fish from New Jersey as a function of species, size, and season. Sci Total Environ. 2011;409:1418–1429. doi: 10.1016/j.scitotenv.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLOS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump KL, Trudeau VL. Mercury-induced reproductive impairment in fish. Environ Toxicol Chem. 2009;28:895–907. doi: 10.1897/08-151.1. [DOI] [PubMed] [Google Scholar]
- Delany MF, Bell JU, Sundlof SF. Concentrations of contaminants in muscle of the American alligator in Florida. J Wildl Dis. 1988;24:62–66. doi: 10.7589/0090-3558-24.1.62. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–8. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AJ, O’Shaughnessy PJ, Bhattacharya S, Monterio A, Kerrigan D, Goetz S, et al. In utero exposure to cigarette chemicals induces sex-specific disruption of one-carbone metabolism and DNA methylation in the human fetal liver. BMC Med. 2015;13:18. doi: 10.1186/s12916-014-0251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisemann J, Beyer W, Bennetts R, Morton A. Mercury residues in south Florida apple snails (Pomacea paludosa) Bull Environ Contam Toxicol. 1997;58:739–743. doi: 10.1007/s001289900395. [DOI] [PubMed] [Google Scholar]
- Elsey RM, Trosclair PL, III, Glenn TC. Nest-site fidelity in American alligators in a Louisiana coastal marsh. Southeast Nat. 2008;7:737–743. [Google Scholar]
- Facemire C, Augspurger T, Bateman D, Brim M, Conzelmann P, Delchamps S, et al. Impacts of mercury contamination in the southeastern United States. Water Air Soil Poll. 1995;80:923–926. [Google Scholar]
- Florida Department of Environmental Protection, Restoration DoWRMaEAa. Florida Watersheds and River Basins Map. 2015. [Google Scholar]
- Florida Department of Environmental Protection F. Final REPORT Mercury TMDL for the State of Florida; Watershed Evaluation and TMDL Section. 2013 http://www.dep.state.fl.us/water/tmdl/final_tmdl.htm.
- Fortin C, Beauchamp G, Dansereau M, Lariviere N, Belanger D. Spatial variation in mercury concentrations in wild mink and river otter carcasses from the James Bay Territory, Quebec, Canada. Arch Environ Contam Toxicol. 2001;40:121–127. doi: 10.1007/s002440010154. [DOI] [PubMed] [Google Scholar]
- Frederick P, Axelrad D, Atkson T, Pollman C. Contaminants research and policy: the Everglades mercury story. Natl Wetlands Newsl. 2005;27:3–6. [Google Scholar]
- Frederick P, Jayasena N. Altered pairing behaviour and reproductive success in white ibises exposed to environmentally relevant concentrations of methylmercury. Proc R Soc B Biol Sci. 2010:rspb20102189. doi: 10.1098/rspb.2010.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Guo L, Byun HM, Zhong J, Motta V, Barupal J, Zheng Y, et al. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ Mol Mutagen. 2014;55:322–335. doi: 10.1002/em.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Bloom MS, Robinson WP, Kim D, Parsons PJ, vom Saal FS, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod. 2012;27:1401–1410. doi: 10.1093/humrep/des038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JA, Frederick PC, Karasov W. Relationships among mercury concentrations, hormones, and nesting effort of white ibises (Eudocimus albus) in the Florida Everglades. The Auk. 2005;122:255–267. [Google Scholar]
- Heaton-Jones TG, Homer BL, Heaton-Jones D, Sundlof SF. Mercury distribution in American alligators (Alligator mississippiensis) in Florida. J Zoo Wildl Med. 1997:62–70. [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord L, Jennings M, Brunell A. Mercury contamination of Florida alligators. Crocodiles: Proceedings of the 10th Working Meeting of the Crocodile Specialist Group, IUCN-The World Conservation Union; Gland Switzerland. 1990. pp. 1–15. [Google Scholar]
- Jagoe C, Arnold-Hill B, Yanochko G, Winger P, Brisbin I., Jr Mercury in alligators (Alligator mississippiensis) in the southeastern United States. Sci Total Environ. 1998;213:255–262. doi: 10.1016/s0048-9697(98)00098-9. [DOI] [PubMed] [Google Scholar]
- Julian P. Mercury hotspot identification in Water Conservation Area 3, Florida, USA. Ann GIS. 2013;19:79–88. [Google Scholar]
- Julian P, Gu B, Redfield G, Weaver K, Lange T, Frederick P, et al. South Florida Environmental Report. South Florida Water Management District; West Palm Beach, FL: 2015. Chapter 3B: Mercury and sulfur environmental assessment for the everglades. [Google Scholar]
- Kannan K, Smith R, Jr, Lee R, Windom H, Heitmuller P, Macauley J, et al. Distribution of total mercury and methyl mercury in water, sediment, and fish from south Florida estuaries. Arch Environ Contam Toxicol. 1998;34:109–118. doi: 10.1007/s002449900294. [DOI] [PubMed] [Google Scholar]
- Khan B, Tansel B. Mercury Bioconcentration Factors in American Alligators (Alligator mississippiensis) in the Florida Everglades. Ecotoxicol Environ Safety. 2000;47:54–58. doi: 10.1006/eesa.2000.1923. [DOI] [PubMed] [Google Scholar]
- Kok RM, Smith DE, Barto R, Spijkerman AM, Teerlink T, Gellekink HJ, et al. Global DNA methylation measured by liquid chromatography-tandem mass spectrometry: analytical technique, reference values and determinants in healthy subjects. Clin Chem Lab Med. 2007;45:903–911. doi: 10.1515/CCLM.2007.137. [DOI] [PubMed] [Google Scholar]
- Kruman II, Fowler AK. Impaired one carbon metabolism and DNA methylation in alcohol toxicity. J Neurochem. 2014;129:770–80. doi: 10.1111/jnc.12677. [DOI] [PubMed] [Google Scholar]
- Kurland T, Faro SN, Siedler H. Minamata disease. The outbreak of a neurologic disorder in Minamata, Japan, and its relationship to the ingestion of seafood contaminated by mercuric compounds. World Neurol. 1960;1:370–95. [PubMed] [Google Scholar]
- Lance VA. Alligator physiology and life history: the importance of temperature. Exp Gerontol. 2003;38:801–805. doi: 10.1016/s0531-5565(03)00112-8. [DOI] [PubMed] [Google Scholar]
- Lange T, Royals H, Connor L. Mercury accumulation in largemouth bass (Micropterus salmoides) in a Florida lake. Arch Environ Contam Toxicol. 1994;27:466–471. doi: 10.1007/BF00214837. [DOI] [PubMed] [Google Scholar]
- Le T, Kim K-P, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Anal Biochem. 2011;412:203–209. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu J, Xie Z, Liu S, Fan-Havard P, Huang TH-M, et al. Quantification of regional DNA methylation by liquid chromatography/tandem mass spectrometry. Anal Biochem. 2009;391:106–113. doi: 10.1016/j.ab.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhang W, Hu J, Yu Z, Chen Y, Luo Q, et al. Analysis of global DNA methylation levels in human blood using high-performance liquid chromatography/tandem electrospray ionization mass spectrometry. Eur J Mass Spectrom. 2009;15:555. doi: 10.1255/ejms.1007. [DOI] [PubMed] [Google Scholar]
- MacDonald WA, Mann MR. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev. 2014;81:126–40. doi: 10.1002/mrd.22220. [DOI] [PubMed] [Google Scholar]
- Myburgh JG, Kirberger RM, Steyl JC, Soley JT, Booyse DG, Huchzermeyer FW, et al. The post-occipital spinal venous sinus of the Nile crocodile (Crocodylus niloticus): its anatomy and use for blood sample collection and intravenous infusions: original research. J S Afr Vet Assoc. 2014;85:1–10. doi: 10.4102/jsava.v85i1.965. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki MM, Hall MN, Liu X, Oka J, Harper KN, Slavkovich V, et al. A dose-response study of arsenic exposure and global DNA methylation of peripheral blood mononuclear cell DNA in Bangladeshi adults. Environ Health Perspect. 2013;121:1306–1312. doi: 10.1289/ehp.1206421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott BB, Bowden JA, Kohno S, Cloy-McCoy JA, Hale MD, Bangma JT, et al. Influence of tissue, age, and environmental quality on DNA methylation in Alligator mississippiensis. Reproduction. 2014 doi: 10.1530/REP-13-0498. REP-13-0498. [DOI] [PubMed] [Google Scholar]
- Pilsner RJ, Lazarus AL, Nam DH, Letcher RJ, Sonne C, Dietz R, et al. Mercury-associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol. 2010;19:307–314. doi: 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- Quinlivan EP, Gregory JF. DNA digestion to deoxyribonucleoside: a simplified one-step procedure. Anal Biochem. 2008a;373:383–385. doi: 10.1016/j.ab.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan EP, Gregory JF. DNA methylation determination by liquid chromatography–tandem mass spectrometry using novel biosynthetic [U-15N] deoxycytidine and [U-15N] methyldeoxycytidine internal standards. Nucleic Acids Res. 2008b;36:e119–e119. doi: 10.1093/nar/gkn534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston NV, Raymond LJ. Dietary selenium’s protective effects against methylmercury toxicity. Toxicol. 2010;278:112–123. doi: 10.1016/j.tox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Raymond LJ, Ralston NV. Mercury: selenium interactions and health implications. SMDJ. 2004;7:72–77. doi: 10.1016/j.neuro.2020.09.020. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer CC, McFarland KP, Evers DC, Miller EK, Aubry Y, Busby D, et al. Mercury concentrations in Bicknell’s thrush and other insectivorous passerines in montane forests of northeastern North America. Ecotoxicol. 2005;14:223–240. doi: 10.1007/s10646-004-6270-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- Rumbold D, Fink L, Laine K, Niemczyk S, Chandrasekhar T, Wankel S, et al. Levels of mercury in alligators (Alligator mississippiensis) collected along a transect through the Florida Everglades. Sci Total Environ. 2002;297:239–252. doi: 10.1016/s0048-9697(02)00132-8. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, et al. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ Health Perspect. 2014;122:946–954. doi: 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte CA, Rice KG, Donnelly MA. Variation of total mercury concentrations in pig frogs (Rana grylio) across the Florida Everglades, USA. Sci Total Environ. 2005;345:51–59. doi: 10.1016/j.scitotenv.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Ware FJ, Royals H, Lange T. Mercury contamination in Florida largemouth bass. Proceedings of the Annual Conference of the Southeastern Association of Fish Wildlife Agencies. 1990;44:5–12. [Google Scholar]
- Woodward AR, Moore CT, Delaney MF. Experimental alligator harvest. Final Report; Florida Game and Fresh Water Fish Commission; Gainesville, FL. 1992. p. 118. Study Number 7567. [Google Scholar]
- Yang D-Y, Chen Y-W, Gunn JM, Belzile N. Selenium and mercury in organisms: interactions and mechanisms. Environ Rev. 2008;16:71–92. [Google Scholar]
- Yanochko G, Jagoe C, Brisbin I., Jr Tissue mercury concentrations in alligators (Alligator mississippiensis) from the Florida Everglades and the Savannah River site, South Carolina. Arch Environ Contam Toxicol. 1997;32:323–328. doi: 10.1007/s002449900192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.