SUMMARY
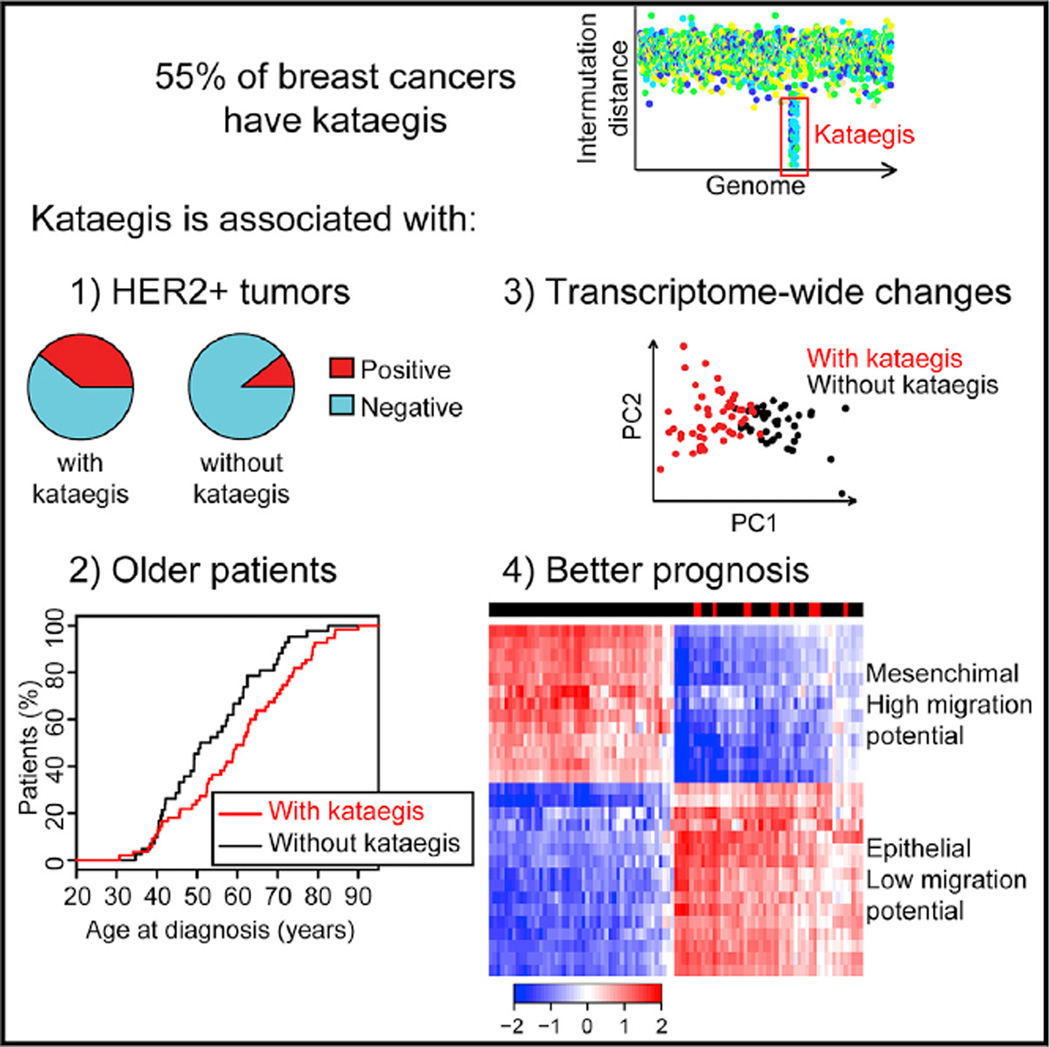
Kataegis is a mutational process observed in ~55% of breast tumors that results in hypermutation in localized genomic regions. Using whole-genome sequence data of 97 tumors, we examined the distribution of kataegis loci, showing that these somatic mutations are over-represented on chromosomes 8,17, and 22 and enriched in genic regions and active chromatin elements. We show that tumors harboring kataegis are associated with transcriptome-wide expression changes consistent with low invasive potential. We exploit the kataegis expression signature to predict kataegis status in 412 breast cancers with transcriptome but not whole-genome sequence data and show that kataegis loci are enriched in high-grade, HER2+ tumors in patients diagnosed with breast cancer at an older age and who have a later age at death. Our study demonstrates that kataegis loci are associated with important clinical features in breast cancer and may serve as a marker of good prognosis.
Graphical abstract
INTRODUCTION
Kataegis is a mutational process that has been observed in several cancer types (Alexandrov et al., 2013) and results in hypermutation (a few to several hundred C > T and C > G substitutions enriched at TpCpN trinucleotides) on the same DNA strand in small localized genomic regions (Taylor et al., 2013). Alexandrov et al. (2013) computationally modeled a variety of mutational signatures and defined kataegis as six or more consecutive mutations with average intermutation distances of ≤1 kb. Kataegis was first studied in breast cancer, in which >50% of tumors contain one or more kataegis loci (Nik-Zainal et al., 2012b), often in the vicinity of structural rearrangements (Nik-Zainal et al., 2012a). A subfamily of the APOBEC (apolipo-protein B mRNA editing enzyme, catalytic polypeptide-like) cytidine deaminases has been implicated as a source of kataegis mutations in part because aberrant expression in yeast generates a similar C > T substitution mutational signature (Roberts et al., 2013; Taylor et al., 2013). Whether kataegis plays a role in breast cancer etiology and is associated with clinical features or is simply a byproduct of aberrant APOBEC activity is unknown.
Here, we investigated the functional and clinical impact of kataegis on breast cancer by studying: (1) associations between the chromosomal positions of kataegis loci and those of functional elements, (2) gene expression differences between tumors that do and those that do not harbor kataegis loci, and (3) associations between the occurrence of kataegis and the occurrence of clinical features. We show that kataegis loci are not randomly distributed across the genome but are enriched in regions containing genes and functional regulatory elements, in addition to being over-represented on chromosomes 8, 17, and 22 and depleted on chromosomes 2, 9, and 16. Our study also shows that genes in the vicinity of kataegis loci (within 500 kb) are less likely to be aberrantly expressed than distal genes. We determined that breast cancers harboring kataegis have a transcriptome-wide expression signature that is consistent with low invasive potential and enables the kataegis status of a tumor to be predicted using RNA sequencing (RNA-seq) data. Furthermore, breast cancers that harbor kataegis loci are enriched in patients with high-grade, HER2+ tumors who are diagnosed at an older age and have a higher age at death.
RESULTS
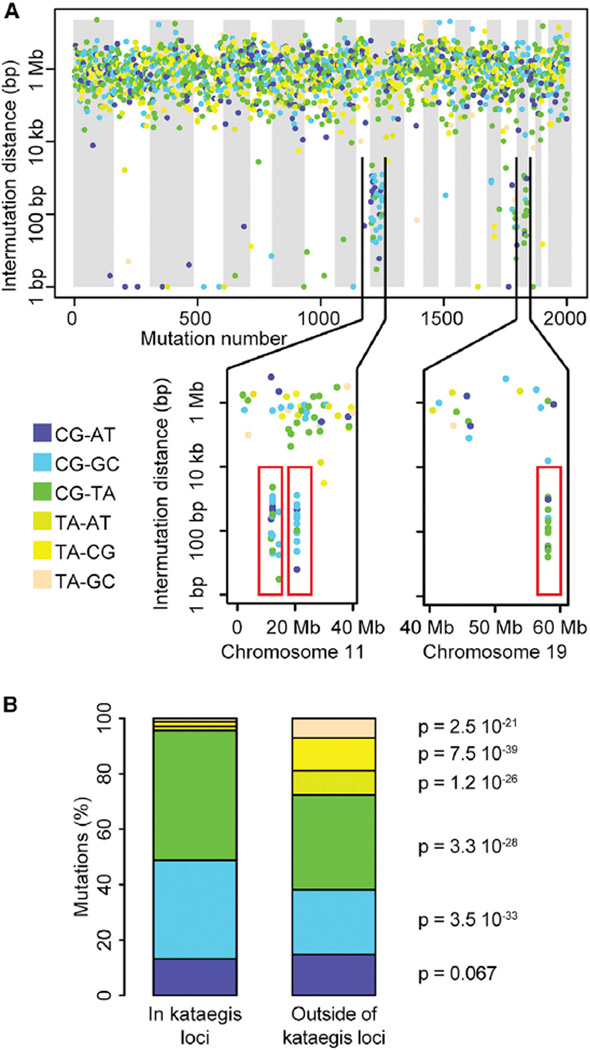
We analyzed the whole-genome sequences of 97 breast tumors and their associated normal DNA, obtained from the Cancer Genome Atlas (TCGA), and detected 387,289 high-confidence somatic mutations (Table S1A). For each tumor sample, we calculated distances between somatic substitutions and found a total of 132 kataegis loci (1–10 per sample) distributed across 55 samples (56.7%) (Figures 1A and S1; Table S1B). These 132 kataegis loci are significantly enriched for C > T and C > G substitutions, as expected (Figure 1B) (Taylor et al., 2013).
Figure 1. Mutational Profile of Kataegis Loci.
(A) Mutational profile (rainfall plot) in sample TCGA-B6-A0I2. The x axis shows mutations ordered by mutation number (from the first mutated position on chromosome 1 to the last mutated position on chromosome X), and the y axis represents intermutation distance in log scale. The lower section shows three kataegis loci (red rectangles): two on chromosome 11 and one on chromosome 19. Rainfall plots for all 97 breast tumors are in Figure S1.
(B) Distributions of substitution types of 1,732 mutations that compose all 132 kataegis loci and 385,557 mutations outside of kataegis loci in the 55 tumors that harbor kataegis. The p values were calculated using a chi-square test. See also Figure S1 and Table S1.
Distribution of Kataegis Loci in the Genome
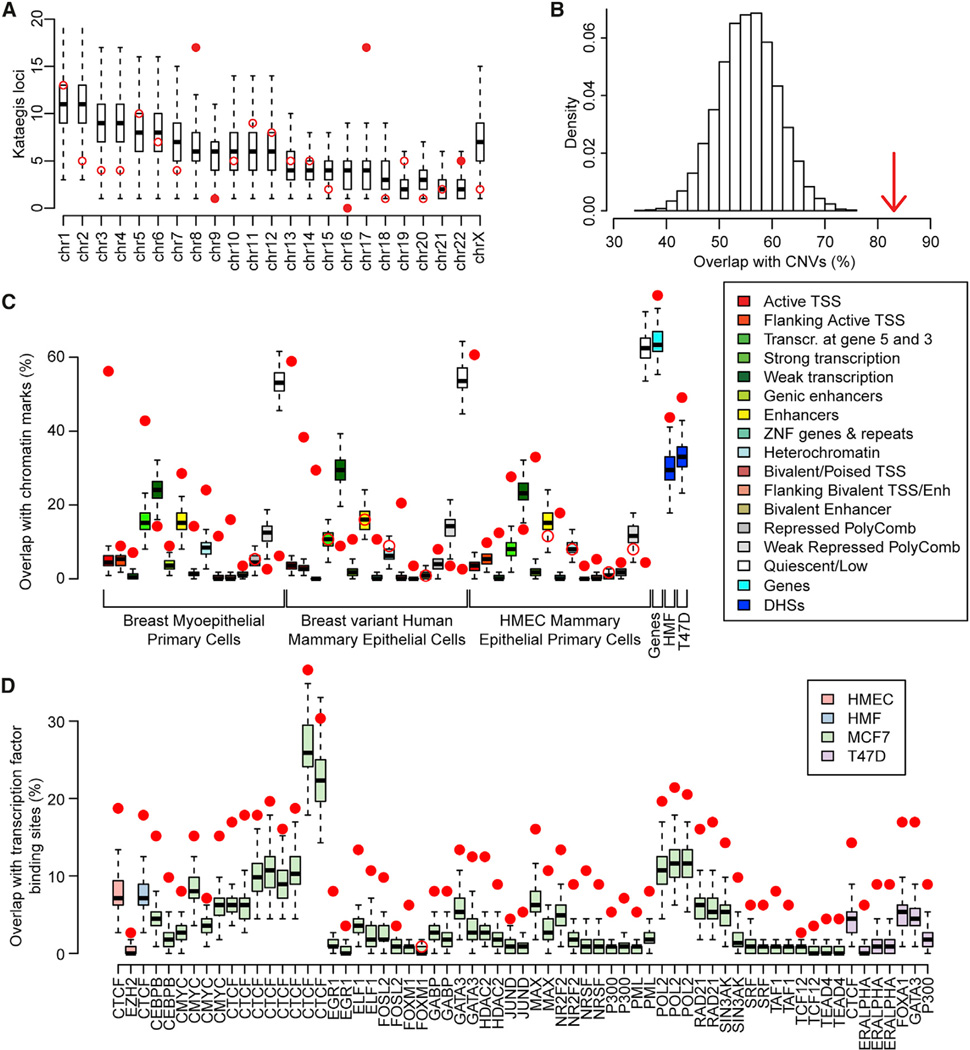
We examined the distribution of kataegis loci across the genome with respect to chromosomal positions and functional elements. We observed that the 132 kataegis loci are preferentially located on chromosomes 8, 17, and 22 and depleted on chromosomes 2, 9, and 16 (Figure 2A) as determined by permutation testing (10,000 permutations). The coordinates of kataegis loci overlap with copy number variations (CNVs = 83, corresponding to 62.8% of all loci) at a higher rate than expected by chance (Figure 2B), consistent with the findings of a previous study (Nik-Zainal et al., 2012a). Next, we intersected the coordinates of each kataegis locus with functional elements including (1) 15 chromatin states defined in three Roadmap Epigenomics breast cell lines (Ernst et al., 2011; Roadmap Epigenomics Consortium et al., 2015); (2) gene coordinates derived from Gencode; (3) DNase I hypersensitive sites (DHSs), which are marks of active chromatin, defined in two Encyclopedia of DNA Elements (ENCODE) breast cell lines, human mammary fibroblast (HMF), and T47D (Neph et al., 2012); and (4) binding sites for 25 transcription factors derived from 66 different chromatin immunoprecipitation sequencing (ChIP-seq) experiments in four ENCODE breast cell lines: human mammary epithelial primary cell (HMEC), T47D, HMF, and MCF7. After correcting for local sequence characteristics (see Experimental Procedures), we determined that kataegis loci occur at a significantly higher rate than expected within marks of active chromatin, coding sequences, DHSs, and transcription factor binding sites (65 of the 66 ChIP-seq experiments) (Figures 2C and 2D; Table S2), which is consistent with previous findings (Kazanov et al., 2015). All three Roadmap breast cell lines demonstrate strong enrichment (~15 times higher than the expected random distribution) in regions corresponding to active transcription start sites (TSSs) and high depletion (on average 14 times lower than the expected random distribution) in quiescent regions (genomic loci that are not transcribed and do not overlap ChIP-seq peaks associated with transcription factor binding sites or histone modifications) (Lawrence et al., 2013). These data demonstrate that kataegis loci are not found at random positions in the genome but are preferentially located in functional sequences that correspond to active chromatin in normal breast tissue and breast cancer.
Figure 2. Associations between Kataegis Loci and Chromatin Features.
(A and B) Distributions of kataegis loci (A) in each chromosome and (B) overlapping CNVs. Red circles represent the observed number of kataegis loci, while the distributions of kataegis loci based on 10,000 permutations are represented by boxplots in (A) and as a histogram in (B). Filled circles in (A) show the chromosomes with a Z score >2. The arrow in (B) shows the observed percentage of kataegis loci that overlap CNVs (Z score = 5.28).
(C and D) Boxplots showing the expected overlap between kataegis loci and (C) chromatin states from three Roadmap breast cell lines, Gencode genes, and DHSs in two ENCODE breast cell lines and (D) transcription factor binding sites derived from 66 ChIP-seq experiments in four ENCODE breast cell lines. Filled circles show a Z score >2 or a Z score <-2.
See also Table S2.
Kataegis Loci Stabilize the Expression of Neighboring Genes
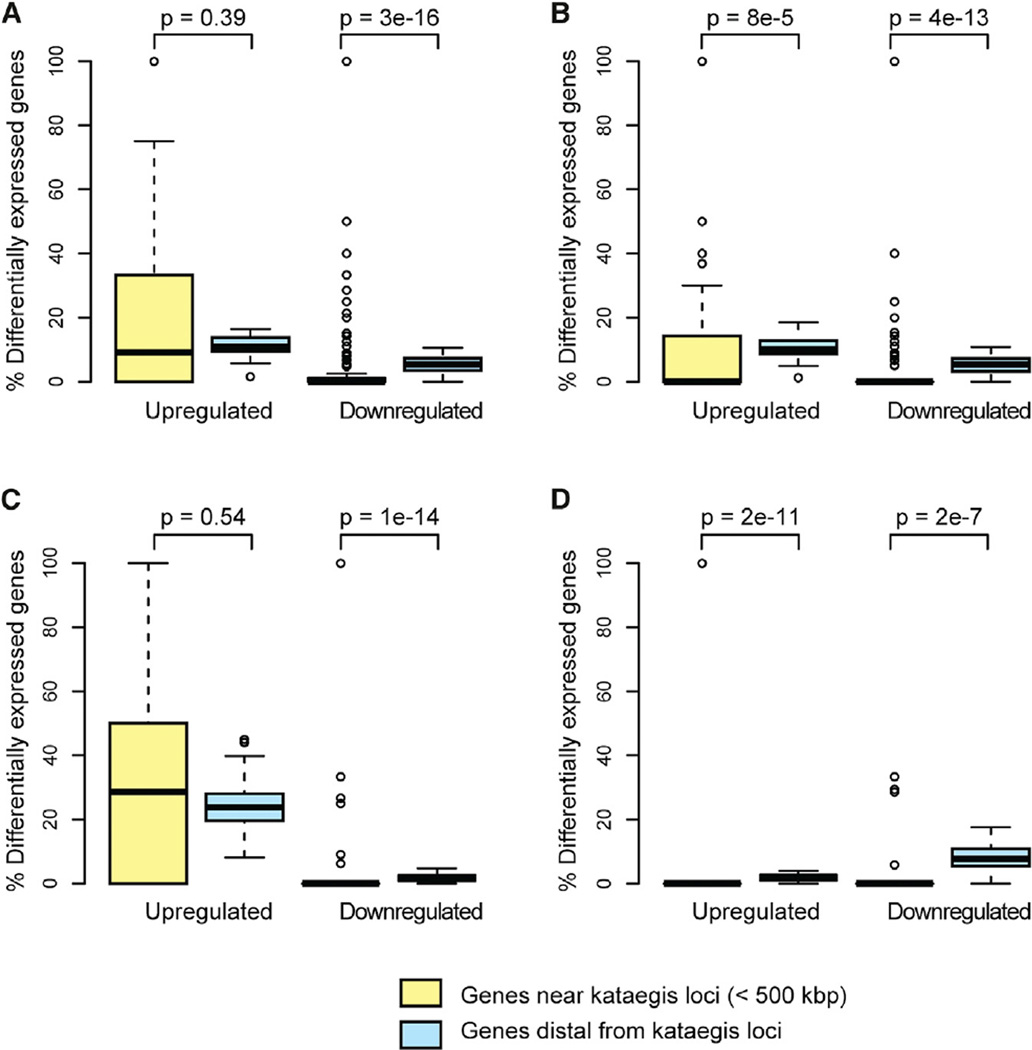
Because kataegis loci are enriched in regulatory elements (in particular, TSS) and coding sequences, we investigated whether they influence the expression of neighboring genes. For each tumor harboring kataegis, we compared the expression level of each gene (20,502 total genes) to the expression levels observed in 106 unrelated normal breasts derived from TCGA. Considering all 132 kataegis loci, nearby genes (within 500 kb) are less likely to be downregulated than the transcriptome-wide average (p = 3.0 × 10−16) (Figure 3A). Because somatic CNVs can affect gene expression, we investigated kataegis loci that do not overlap CNVs, that overlap CNV amplifications, and that overlap CNV deletions as separate groups. We confirmed that neighboring genes are less likely to be downregulated than expected in all three groups of kataegis loci (Figures 3B-3D). In addition, neighboring genes of kataegis loci that do not overlap CNVs or overlap CNV deletions are less likely to be upregulated compared to the transcriptome-wide average. These data show that genes neighboring kataegis loci are less likely to be aberrantly expressed, suggesting that the kataegis loci are somehow stabilizing their expression.
Figure 3. Influence of Kataegis Loci on Local Gene Expression.
Boxplots showing the percentage of upregulated and downregulated genes near to (<500 kbp) and distal from kataegis loci for (A) all kataegis loci (n = 132), (B) kataegis loci that do not overlap CNVs (n = 49), (C) kataegis loci that overlap CNV amplifications (n = 39), and (D) kataegis loci that overlap CNV deletions (n = 44). The p values were calculated with a Wilcoxon test.
Gene Expression Signature Can Be Used to Predict Kataegis Status in Tumors
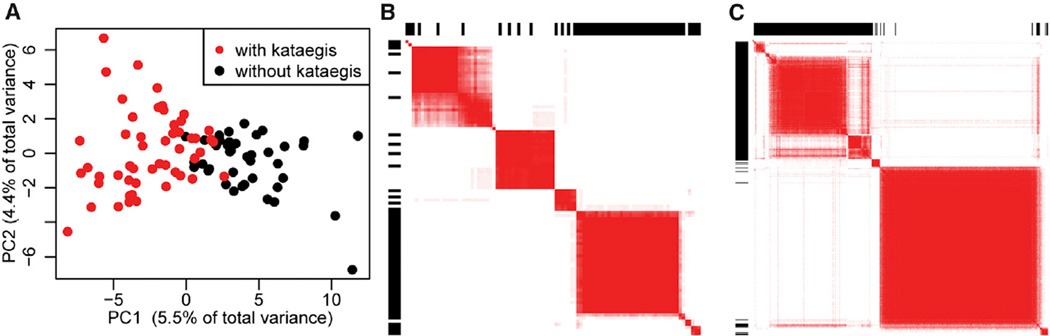
We investigated whether kataegis mutations are associated with transcriptome-wide expression differences. Using RNA-seq data, we examined 20,502 human genes (Table S3) for differential expression between the 55 tumors with and the 42 tumors without kataegis and detected significant differences in 628 genes. A principal-component analysis (PCA) using the expression levels of these genes showed distinct gene expression signatures for breast cancer samples with and without kataegis (Figures 4A and S2). To confirm the presence of these distinct expression signatures, we performed consensus clustering (Monti et al., 2003) using the first five principal components and showed that samples with and without kataegis clearly separate into distinct clusters (Figure 4B). Overall, the results from PCA and consensus clustering indicate that gene expression data can be used to discriminate between tumors with and those without kataegis.
Figure 4. Transcriptome-wide Kataegis Expression Signature.
(A) PCA analysis of the levels of 628 differentially expressed genes in the 97 tumors separates samples with (red) and without (black) kataegis into two classes.
(B) Heatmap showing results of consensus clustering using the first five principal components in the 97 tumors. Samples with kataegis are indicated by a black bar in the outer rectangle area. The intensity of the heatmap represents the number of times (over a total of 20 runs) each pair of samples is clustered together (white = 0, red = 20).
(C) Heatmap showing consensus clusters using the first five principal components in 412 tumors. Samples predicted to harbor kataegis loci by the GLM (95% CI) are indicated by a black bar in the outer rectangle area.
We explored whether the gene expression signature could be exploited to predict the presence of kataegis in 998 TCGA breast cancer samples that have RNA-seq but not whole-genome sequence data. We built a generalized linear model (GLM) to predict the presence of kataegis loci using the first five principal components and kataegis status of the 97 TCGA discovery tumors as input. We performed a 10-fold cross-validation on the 97 TCGA discovery tumors and using the 95% confidence interval (CI) of the GLM observed a receiver operating characteristic (ROC) area under the curve (AUC) of 0.94 (Figure S2C; Table S4A). We applied this model to the 998 TCGA tumors with only RNA-seq data, of which 412 had values at the 95% confidence level and could be assigned to one of the two categories (180 breast tumors with kataegis and 232 without kataegis) (Figure 4C; Table S4B), while 586 samples could not be assigned and were excluded from further analyses. These findings demonstrate that our GLM (95% CI) is able, with high confidence, to separate samples with predicted kataegis loci from those without predicted kataegis loci.
Kataegis Status Is Associated with Clinical Features
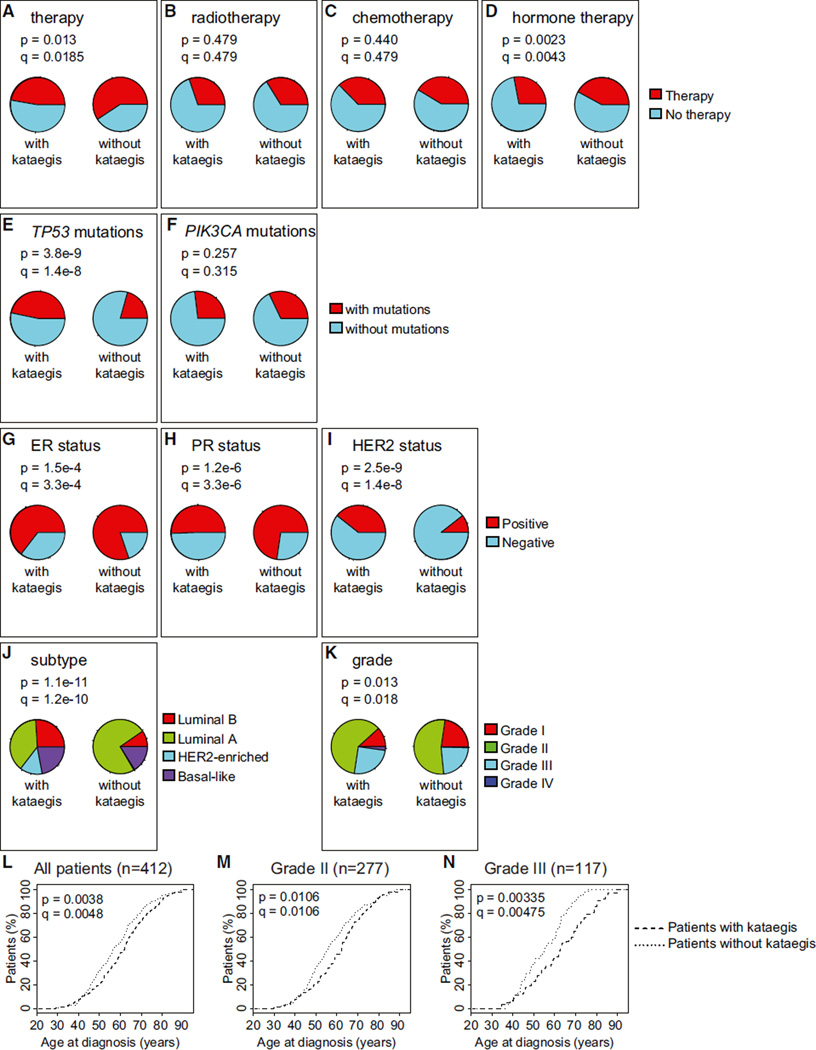
Kataegis is associated with a specific expression signature, which leads us to hypothesize that tumors carrying these mutations may also be correlated with particular clinical profiles. Therefore, we studied the association between presence of kataegis and therapeutic treatment, presence of known driver mutations, breast cancer subtypes, tumor grade, and patient age at diagnosis and at death. The treating physicians of these patients did not know the kataegis status of their tumors. We initially examined the 412 patients with predicted kataegis status for differences in clinical properties and then investigated whether the same associations hold in the 97 patients with known kataegis status. Comparison of the 180 patients and the 232 patients whose tumors are predicted to harbor and not harbor kataegis loci, respectively, shows that the former are less likely to undergo therapy (q = 0.0185, Fisher’s exact test with Benjamini-Hochberg correction for multiple testing hypothesis) and, in particular, hormone therapy (q = 0.0043) (Figures 5A-5D), more likely to have mutations in TP53 (q = 1.4 10−8), and more likely to be estrogen receptor-negative (ER−) (q = 3.3 10−4), progesterone receptor-negative (PR−) (q = 3.3 10−6), and HER2+ (q = 1.4 10−8) (Figures 5E-5I). We did not detect significant associations between predication of kataegis and chemotherapy, radiotherapy, or presence of mutations in PIK3CA. The set of breast cancers with predicted kataegis loci are composed of fewer luminal A (ER+ and/or PR+ and HER2−) and a greater number of luminal B (ER+ and/or PR+ and HER2+) and HER2-enriched tumors (ER−, PR−, and HER2+, q = 1.2 10−10) (Figure 5J), and a lower proportion are grade I tumors (q = 0.018) (Figure 5K). We also examined 1,992 breast cancer samples in the METABRIC data-set (Curtis et al., 2012) and observed significant associations between kataegis expression signature and ER−, PR−, and HER2+ status and mutations in TP53 (Figure S3). Tumors with predicted kataegis are enriched in the HER2+ subtype, which is consistent with the finding that they tend to be higher grade (Slamon et al., 2006) and carry TP53 mutations (Cancer Genome Atlas Network, 2012). Likewise, tumors with predicted kataegis tend to be ER−and PR−, which is consistent with the finding that the patients are less likely to undergo hormone therapy, which targets these receptors. Overall, these data show that kataegis tends to occur in higher-grade breast tumors that are HER2+.
Figure 5. Association between Predicted Kataegis Status and Clinical Features, Genomic Features, and Age at Diagnosis.
(A–J) The distribution of samples with and without kataegis were examined for differences in (A) any type of therapy, (B) radiotherapy, (C) chemotherapy, (D) hormone therapy, (E) mutations in TP53, (F) mutations in PIK3CA, (G) ER status, (H) PR status, (I) HER2 status, (J) tumor subtype, and (K) grade. The Benjamini-Hochberg method was used to adjust p values for multiple testing hypotheses (q values).
(L–N) Empirical distributions showing age at diagnosis (L) for all patients (n = 412), (M) for grade II patients (n = 277), and (N) for grade III patients (n = 117) with and without predicted kataegis. The p values were calculated using the Wald test from the Cox proportional hazard model. The p values were adjusted with the Benjamini-Hochberg method (q values).
See also Figures S3, S4, S5, and S6.
We also found that patients with predicted kataegis loci tend to present with breast cancer at a later age (q = 0.0048, Cox proportional hazard test adjusted with the Benjamini-Hochberg method for a multiple testing hypothesis) (Figures 5L and S4A–S4C), which is observed when restricting the analysis to only patients with grade II tumors (Figure 5M) and more prominent when considering only grade III tumors (Figures 5N and S4C). The age at death for patients with predicted kataegis is higher (q = 0.097, Wilcoxon test adjusted with the Benjamini-Hochberg method for a multiple testing hypothesis) (Figures S4D–S4F). We observed that there is significantly shorter survival after diagnosis for patients with predicted kataegis (Figures S4G–S4I) but propose that this is due to a significantly higher age at diagnosis and consequently a shorter interval before death due to non-cancer-related causes. Considering all 412 TCGA patients, we observe a significant negative correlation (−0.09, p = 0.039) between age at diagnosis and survival after diagnosis. To determine whether the difference in survival after diagnosis is due to differences in age at diagnosis, we divided the patients into four age groups, assessed the differences in survival after diagnosis between patients with and those without predicted kataegis in each age group, and found no significant differences (Figures S4J–S4M), confirming that the shorter survival after diagnosis for patients with predicted kataegis is likely due to their older age. These data show that tumors with kataegis are enriched in patients diagnosed at later ages who tend to have a higher age at death.
Analyses of the 97 TCGA patients with known kataegis status shows similar trends with regards to clinical features, as observed in 412 TCGA patients with predicted kataegis status (Figures S5A–S5K). In particular, patients with kataegis loci are enriched for being HER2+ (p = 0.066) (Figure S5I) and grade III (p = 0.015, Fisher’s exact test) (Figure S5K). These patients also tend to present with breast cancer at a later age (age at diagnosis is 60.3 years compared to 54.1 years in patients without kataegis, p = 0.0303, q = 0.0909, Cox proportional hazard test adjusted with the Benjamini-Hochberg method for a multiple testing hypothesis) (Figures S5L–S5N) and the age at death is higher (median age at death is 78 years compared to 47 years in patients without kataegis, p = 0.008, q = 0.059) (Figures S5O–S5Q). There is no observed difference in the survival after diagnosis when considering all patients with known kataegis status (q = 0.809, Cox proportional hazard test) (Figures S5R–S5T), but patients with grade II tumors containing kataegis tend to have improved survival (q = 0.0599) (Figure S5S). Because whole-genome sequence data are available for these 97 TCGA tumors, we were able to examine whether the association between CNVs and kataegis loci confounds the relationship between kataegis and clinical features. We investigated whether patients with tumors harboring kataegis loci that overlap (n = 42) or do not overlap with CNVs (n = 13) vary in age at diagnosis and determined that no differences exist (p = 0.631, Wilcoxon test) (Figure S5U). This analysis suggests that the associations we observe between kataegis loci and clinical variables are not confounded by the association between kataegis and CNVs. These analyses confirm that tumors with kataegis loci are enriched in patients who are diagnosed at an older age, die later in life, and have tumors that are higher grade and HER2+.
Kataegis Status Is a Marker of Good Prognosis
Given the high degree of covariance between kataegis and several well-established clinical variables, we investigated whether kataegis status could be predicted by one or more clinical variables. We fit a linear model to predict the kataegis status in the 412 TCGA tumors based on the following variables: HER2 status, PR status, ER status, tumor grade, and presence of mutations in TP53. Of all the clinical features examined, only tumor grade was a weak predictor of kataegis (p = 0.048, t test) (Figure S6A), suggesting that the observed associations between kataegis and age of diagnosis and at death are not confounded.
To examine kataegis as a prognostic marker, we used two independent methods. We initially defined a bad prognosis as death before 55 years of age, while living patients and those who died when they were older than 55 years were considered to have a good prognosis. We fit a linear model to predict prognosis using clinical variables (HER2 status, PR status, ER status, tumor grade, and presence of mutations in TP53) and kataegis status in the 412 TCGA patients. Kataegis is the only variable that significantly contributes to prognosis (p = 0.020, t test) (Figure S6B). In addition, by analyzing the coefficients of the linear model, we determined that, among these variables, kataegis has the highest impact on prognosis (Figures S6C–S6F). We next examined whether kataegis can predict age of death using a Cox proportional hazard model. We used HER2 status, PR status, ER status, presence of mutations in TP53, tumor grade, and kataegis status as input variables, with the age at death or age at last contact as the response variable. We found kataegis to be the only significant predictor of age at death (Figure S6G). These analyses demonstrate that kataegis is a better marker for good prognosis than established clinical variables.
Functional Characterization of Tumors with Kataegis
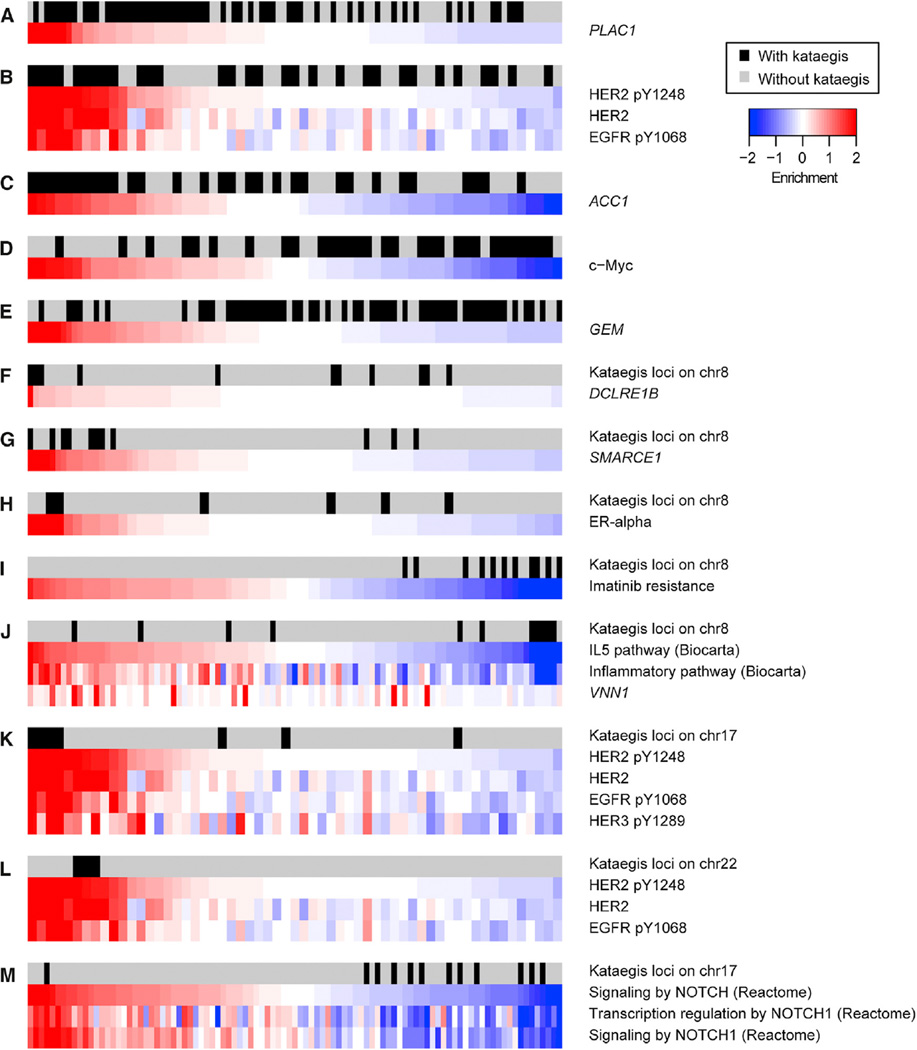
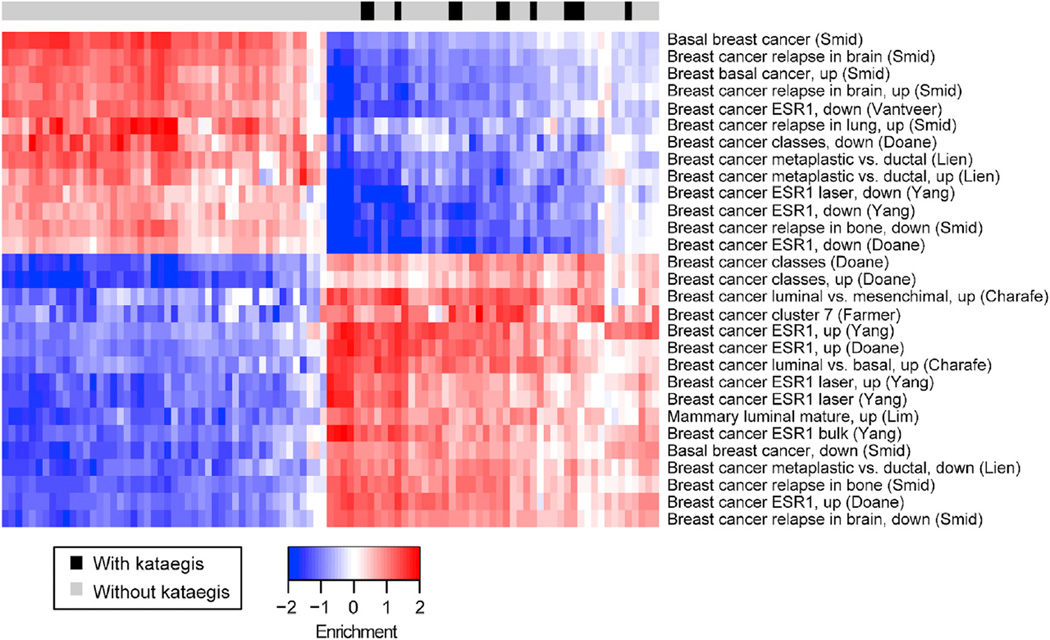
To investigate the molecular underpinnings of the clinical differences between tumors with and those without kataegis loci, we used the information coefficient (IC) (Kim et al., 2016) to examine the 97 TCGA tumors with known kataegis status. IC is an information-theoretic association metric that allows the discovery of linear or non-linear correlations between genomic alterations (here the presence or absence of kataegis loci) and functional phenotypes, such as gene expression levels, protein expression data, and pathway enrichment (Table S5). Using this approach, we found that samples with kataegis are significantly associated with high mRNA levels of PLAC1 (an immunotherapeutic target for gastric adenocarcinoma) (Figure 6A) (Liu et al., 2015), high HER2 (native and phosphorylated), EGFR (Figure 6B), and ACC1 (involved in regulation of hypoxia-induced apoptosis) protein levels (Figure 6C) (Keenan et al., 2015). In addition, these samples display low c-Myc protein (Figure 6D) and Ras-like protein GEM levels (Figure 6E). We next used the IC to investigate molecular differences between tumors harboring kataegis loci on chromosomes 8 and all other breast tumors. Kataegis loci on chromosome 8 are associated with high expression of DCLRE1B (a gene involved in DNA cross-link repair whose expression is influenced by SNP rs11552449, a risk factor for breast cancer (Caswell et al., 2015) (Figure 6F; Table S5). In addition, they are associated with high expression of SMARCE1 (a suppressor of EGFR) (Figure 6G) (Papadakis et al., 2015), high protein levels of ER-alpha (Figure 6H), and low inflammation levels, as shown by the downregulation of the interleukin-5 (IL-5) pathway and the expression levels of VNN1, a gene involved in lymphocyte migration (Figure 6J). The IC analysis was also used to identify molecular differences between tumors with kataegis loci on chromosome 17 and all other tumors and between tumors with kataegis on chromosome 22 and all other tumors. Tumors with kataegis on chromosome 17 or 22 are associated with upregulation of HER2 and EGFR (Figures 6K and 6L; Table S5). Of the 16 tumors with kataegis loci on one of these two chromosomes, 11 tumors are HER2+(68.8%) (Table S1A), whereas only 8 tumors of the remaining 73 tumors are HER2+ (11.0%, p = 2.1 10−6, Fisher’s exact test). Kataegis loci on chromosome 17 are also associated with inactivation of the NOTCH1 pathway (Figure 6M). Furthermore, kataegis loci on chromosome 17 are strongly associated with the downregulation of several pathways associated with aggressive metastatic tumor behavior and HER2− tumors (in particular, basal-like breast cancers, which are ER−, PR−, and HER2−) and the upregulation of pathways associated with HER2+ tumors (in particular, luminal breast cancer) (Figure 7; Table S5). Overall, the IC analyses conducted showed that kataegis occurs in tumors that may have better response to treatment, given the overexpression of the immunotherapeutic target PLAC1 and the downregulation of genes involved in imatinib resistance. In addition, kataegis loci across the genome are associated with higher levels of HER2 and EGFR, while those on chromosome 17 are specifically associated with low levels of NOTCH1 and gene sets predictive of low invasiveness.
Figure 6. Association among Kataegis, Gene Levels, Protein Levels, and Pathway Enrichment.
(A–M) Heatmaps showing gene levels, protein levels, and pathway enrichment for the most significant terms derived from the IC analysis (p values shown in Table S5). Data associated with each feature were normalized to have mean = 0 and SD = 1. The first row of each heatmap shows samples with kataegis (black) and samples without kataegis (gray). Enrichments are shown for (A) PLAC1 expression levels; (B) phosphorylated HER2, native HER2, and EGFR protein levels; (C) ACC1 gene expression levels; (D) c-Myc protein levels; (E) GEM gene expression levels; (F) DCLRE1B expression levels (for kataegis loci on chromosome 8); (G) SMARCE1 gene expression levels (chromosome 8); (H) ER-alpha protein levels (chromosome 8); (I) genes involved in imatinib resistance (chromosome 8); (J) IL-5 pathway, inflammatory pathway, and VNN1 gene expression levels (chromosome 8); (K) phosphorylated HER2, native HER2, EGFR, and phosphorylated HER3 protein levels (chromosome 17); (L) phosphorylated HER2, native HER2, and EGFR protein levels (chromosome 22); and (M) NOTCH signaling pathway, transcription regulation by NOTCH1, and signaling by NOTCH1 pathways (chromosome 17).
Figure 7. Association between Kataegis Loci on Chromosome 17 and Gene Sets Predictive of Low Invasiveness.
The heatmap shows the pathway enrichment for the significant terms associated with breast cancer in the “Chemical and genetic perturbations” gene set class derived from the IC analysis(p values shown in Table S5). The first row shows samples with kataegis (black) and samples without kataegis (gray). Data associated with each feature were normalized to have mean = 0 and SD = 1.
See also Table S5.
To further characterize the functional differences between tumors with and without kataegis, we performed single-sample gene set enrichment analysis (ssGSEA) (Barbie et al., 2009; Subramanian et al., 2005) using the 50 hallmark Molecular Signatures Database (MSigDB) gene sets derived from Liberzon et al. (2015). We discovered that tumors harboring kataegis loci significantly downregulate genes involved in cell-cell interactions, response to hypoxia, epithelial to mesenchymal transition, angiogenesis, and hematopoietic lineage, as well as KRAS, necrosis factor κB (NF-κB), and transforming growth factor β (TGF-β) signaling pathways (Figure S7; Table S6). These expression differences suggest that tumors with kataegis are more epithelial-like and have less organized extra-cellular matrix and vasculature, with consequent lower migration potential. These findings suggest that tumors harboring kataegis loci may be less invasive.
DISCUSSION
Kataegis loci have previously been associated with several tumor types, including breast cancer, pancreatic cancer, lymphoma, leukemia, and lung adenocarcinoma (Alexandrov et al., 2013). Although this phenomenon is present in more than 50% of breast tumors (Nik-Zainal et al., 2012b), its role in tumorigenesis has not been thoroughly investigated. We show that kataegis loci are strongly associated with a transcriptome-wide expression signature, which we exploited to predict the presence of kataegis in an independent set of 412 breast cancers from TCGA. Our findings suggest that kataegis loci may mitigate the aberrant expression of neighboring genes and thus do not affect tumorigenesis in a manner similar to known regulatory driver mutations (Killela et al., 2013; Weinhold et al., 2014). We demonstrate that kataegis is enriched in patients whose tumors are high grade and/or HER2+ and in patients who present cancer at a later age and have a higher age at death. Although we do not have information about the cause of death for breast cancer patients included in TCGA, patients without kataegis tend to die younger (median age at death is 47 years) than patients with kataegis (median age is 78 years), which leads us to hypothesize that they are more likely to die from breast cancer than are patients with kataegis.
It was previously shown that kataegis loci are often associated with genomic rearrangements, although most rearrangements do not exhibit kataegis (Alexandrov et al., 2013). We show that although there is a significant difference in the age at diagnosis between breast cancer patients with and those without kataegis, in the 55% of patients with tumors that have kataegis, there is no difference in age at diagnosis between those that overlap (76%) and those that do not overlap (~24%) with CNVs. In addition to being co-localized with CNVs, kataegis loci are enriched in genomic intervals harboring genes and regulatory elements (DHSs, promoters, and transcription factor binding sites). These data suggest that the previously observed association between kataegis loci and CNVs may be confounded by both types of mutations occurring preferentially in regions of open chromatin (Lu et al., 2014). The observation that mutations driven by APOBEC localize in open chromatin regions (Kazanov et al., 2015) supports this hypothesis.
The differences in pathway enrichment, gene expression, and protein levels between breast cancers with and those without kataegis, as determined by IC analysis, are consistent with the associations found with clinical features (i.e., tumors with kataegis are associated with higher age at diagnosis and later age at death, high HER2 levels, and higher grade). In particular, IC analysis confirms that tumors with kataegis express high levels of HER2, which are known to have good response to treatment and better prognosis (Voduc et al., 2010; Yang et al., 2011). In addition, tumors with kataegis upregulate a known immunotherapeutic target (PLAC1) (Liu et al., 2015), and tumors with kataegis loci on chromosome 8 are correlated with downregulation of genes involved in imatinib resistance (Mahadevan et al., 2007). Thus, tumors containing kataegis loci may have better response to treatment and kataegis may potentially serve as a predictive marker for therapy outcome. We found that kataegis is associated with the inhibition of oncogenic pathways, such as KRAS and NF-κB, and reduced expression of genes involved in migration, which is predictive of less invasive tumors (Eser et al., 2014). Kataegis loci on chromosome 17 are strongly associated with the downregulation of several pathways involved in aggressive metastatic tumor behavior and HER2− tumors. Furthermore, tumors with kataegis display a downregulation of genes and pathways involved in epithelial-to-mesenchymal transition, extracellular matrix organization, and vasculature, suggesting that they likely have a higher proliferation rate and lower invasion potential (Mallini et al., 2014). Thus, pathway enrichment and expression analysis suggest that tumors with kataegis likely have low invasive potential, which is consistent with enrichment in patients with a higher age at diagnosis and later age at death. In conclusion, our results provide important insights into the role kataegis plays in cancer etiology and suggest that the kataegis expression signature may serve as a marker for better prognosis in breast cancer.
EXPERIMENTAL PROCEDURES
Genome Analysis
The BAM files of whole-genome sequences of 97 matched breast tumor and blood samples were downloaded from the Cancer Genomics Hub (CGHub, https://browser.cghub.ucsc.edu/, frozen December 19, 2013) at the University of California, Santa Cruz (UCSC) (Wilks et al., 2014). Duplicated reads were removed using Samtools 0.1.18 (Li et al., 2009), and then base-quality recali-bration and local realignment around insertions or deletions were performed using Genome Analysis Toolkit (GATK) v.1.6-5-g557da77 (DePristo et al., 2011). MuTect (Cibulskis et al., 2013) was used to call somatic point substitutions in the 97 tumor genomes. Only substitutions with at least 14 reads coverage in the tumor, 8 reads in the matched normal, and an allelic fraction of >10% in the tumor were retained (Lawrence et al., 2013), resulting in 986,526 somatic mutations. All mutations within repetitive elements (Repeat-Masker track from the UCSC Genome Browser) (Smit, 1999) were removed to decrease the number of potential false positives, resulting in 387,289 high-confidence somatic mutations.
Identification of Breast Functional Elements
Functional elements active in breast were derived from four sources. First, 15 ChromHMM states (Ernst et al., 2011) associated with three breast cell lines (E027, breast myoepithelial primary cells; E028, breast variant human mammary epithelial cells [vHMECs]; and E119, HMECs) were retrieved from Road-map Epigenomics (http://egg2.wustl.edu/roadmap/web_portal/) (Roadmap Epigenomics Consortium et al., 2015). Next, the coordinates of 34,020 genes were derived from Gencode v.19 (Mudge and Harrow, 2015). Then, 266,757 DHSs from a HMF line (Wang et al., 2010) and 278,680 from the T47D breast cancer cell line (Judge and Chatterton, 1983) were retrieved from the UCSC Genome Browser track wgEncodeUwDgf by ENCODE (Neph et al., 2012). Finally, ChIP-seq data for transcription factor binding sites was retrieved from the ENCODE Transcription Factor Binding super-track on the UCSC Genome Browser. To analyze transcription factor binding sites, we downloaded 66 tracks from four cell lines (HMEC, T47D, HMF, and MCF7), including 11 NarrowPeak files from the Uniform transcription factor binding site (TFBS) track (wgEncodeAwgTfbsUniform),42 BroadPeak files from the Hudson Alpha Institute for Biotechnology (HAIB) TFBS Track (wgEncodeHaibTfbs), 11 NarrowPeak files from the University of Texas, Austin (UTA) TFBS Track (wgEnco-deOpenChromChip), and 2 BroadPeak files from the University of Washington (UW) CTCF Binding (wgEncodeUwTfbs).
Permutation Tests
The position of each kataegis locus was permutated 10,000 times on the genome using the runif function in R. The number of kataegis loci per sample was maintained constant for each randomization, which was important for the CNV analysis, because the distributions of CNVs are different across the 97 breast cancer genomes. Positions of each kataegis locus and all randomized loci were intersected with (1) the 22 autosomal chromosomes and the X chromosome, and (2) CNVs obtained from TCGA. Z scores were calculated as the difference between the number of kataegis loci observed to overlap each class of elements and the mean value in the 10,000 randomizations, divided by the SD in the 10,000 randomizations.
Well-defined sequence characteristics are known to influence local mutation probability. To determine whether kataegis loci are enriched, particularly chromatin features, analyses were conducted by comparing each kataegis locus to genomic intervals with similar characteristics. We excluded centromeric regions and divided the rest of the genome into 3 kb intervals (corresponding to the average kataegis locus length) (M.D.A., D. Weghorn, A. D’Antonio-Chronowska, C. DeBoever, F. Drees, A. Arias, F. Coulet, R.B. Schwab, S.R. Sunyaev, and K.A.F., unpublished data). We performed k-means clustering on the basis of the values of (1) DNA replication timing (Koren et al., 2012), (2) open and closed chromatin status measured by Hi-C mapping (GEO: GSE35156) (Dixon et al., 2012), (3) GC content (Chapman et al., 2011), and (4) local gene density (Lawrence et al., 2013). Local gene density was calculated as the fraction of base pairs within 250 kb overlapping RefSeq genes, while the values for the other covariates were derived from the corresponding publication. Genomic intervals were binned into 637 clusters. We determined the percentage of kataegis loci that overlap each chromatin state or transcription factor binding site and compared it with the null distribution derived from selecting 100 random regions from the same clusters as the kataegis loci. Z scores were calculated as the number of SDs between the mean percentage across the 100 random sets and the observed value for kataegis loci.
Expression Analysis of Genes within 500 kb of Kataegis Loci
For 106 normal breast samples (unrelated to the tumor samples), normalized expression levels derived from RNA-seq data were downloaded from TCGA. The expression levels for each gene in each of the 55 breast tumors with kataegis were compared with the 106 normal breast samples using edgeR with default parameters (Robinson and Smyth, 2008). We considered a gene in a tumor upregulated or downregulated if it had p < 0.05 after adjustment for multiple testing using the Benjamini-Hochberg method. The number of upregulated and downregulated genes near to (<500 kb distance) and distal from each kataegis locus were calculated in each of the 55 tumors with kataegis, and the Wilcoxon method was used to test for significance.
Identifying Upregulated and Downregulated Genes in Tumors with Kataegis
Normalized expression levels of 20,502 human genes derived from RNA-seq data for the 97 breast cancers were downloaded from TCGA. EdgeR with default parameters (Robinson and Smyth, 2008) was run to determine the differences in expression levels between samples with and those without kataegis for all 20,502 genes. The p values derived from edgeR were adjusted for multiple testing hypotheses with the Benjamini-Hochberg method. Adjusted p < 0.05 was considered significant.
Model to Predict the Presence or Absence of Kataegis Loci in Breast Cancer Samples
PCA was performed on the 628 genes with significant expression differences between the 55 tumors with kataegis and the 42 tumors without kataegis. The first five principal components, as well as kataegis status (presence of kataegis = 1, absence of kataegis = 0), were used as input to train a GLM using the function glm in R with parameter family = binomial (link = logit). We also tested a random forest method (using the function RandomForest from the Random-Forest package in R)and a more stringent method that used the 95% CI values of the GLM prediction (samples with a lower boundary >0.75 were considered to be harboring kataegis loci, and samples with an upper CI boundary <0.25 were considered to not be harboring kataegis). To assess the performance of these three models, we conducted 10-fold cross-validation; the 97 tumors were divided into ten bins, and the presence of kataegis was predicted on one bin (the testing set) using the other nine as training sets. By repeating this analysis using each of the ten bins as the testing set, the accuracy of predicting kataegis status in all 97 tumors was determined (Figure S2C; Table S4). The GLM (95% CI) performed the best, with a ROC AUC of 0.94.
Predicting the Kataegis Status in 998 TCGA Tumors
The normalized expression levels of the 20,502 human genes in 998 breast cancers derived from RNA-seq data were downloaded from TCGA. Although the GLM predictor and random forest methods were able to predict kataegis status in all samples, we decided to employ the GLM (95% CI) method for greater accuracy. Principal-component data were determined for the 998 breast cancer samples and used to predict the presence of kataegis, using the function predict.glm in R with parameters type = link, se.fit = TRUE. This parameter allows retrieval of the standard error of the prediction, which was used to determine the prediction’s 95% CI.
Clinical Features of Breast Cancers
Level 2 Biotab files containing clinical information for the 1,095 breast cancer patients included in this study were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). Values marked as Equivocal, Indeterminate, [Not available], and [Not evaluated] were not used in the statistical tests shown in Figures 5 and S6.
Assessing the Impact of Kataegis on Prognosis in Breast Cancer
A linear model (function lm in R) was used to show that kataegis status could not be predicted based on clinical variables (HER2 status, PR status, ER status, tumor grade, and presence of mutations in TP53) and to show that kataegis is the only significant predictor of prognosis. To determine the relative impact of each of these variables on prognosis, the function calc.relimp (included in the relaimpo package in R) (Grömping, 2006) was applied to the linear model using four distinct methods (Figure S6B): (1) R2 contribution averaged over orderings among regressors (Lindeman, Merenda, and Gold’s method [LMG]), (2) usefulness, (3) squared covariance between prognosis and each variable, and (4) product of the standardized coefficient and the correlation (Darlington, 1968; Grömping, 2007).
The survival Cox proportional hazard linear regression model was performed using the coxph function in the Survival package in R. Input variables were HER2 status, PR status, ER status, tumor grade, kataegis status, and presence of mutations in TP53, while age at death or age at last contact was used as output.
Functional Analysis of Tumors with and without Kataegis with IC and ssGSEA
The IC analysis (Kim et al., 2016) was performed using 11 distinct datasets as input: (1) RNA-seq gene expression levels derived from TCGA (19,921 genes), (2) reverse-phase protein array (RPPA) levels for 142 proteins derived from TCGA, (3) ssGSEA pathway enrichment levels for 4,496 chemical and genetic perturbation gene sets derived from MSigDB (Liberzon et al., 2011, 2015), (4) ssGSEA pathway enrichment levels for 217 Biocarta pathways, (5) ssGSEA pathway enrichment levels for 674 Reactome pathways, (6) ssGSEA enrichment levels for the targets of 221 microRNA, (7) ssGSEA enrichment levels for the targets of 608 transcription factors, (8) ssGSEA enrichment levels for 426 cancer gene neighborhoods, (9) ssGSEA enrichment levels for 431 cancer modules, (10) ssGSEA enrichment levels for 279 oncogenic signatures, and (11) ssGSEA enrichment levels for 52 hallmark gene sets derived from MSigDB (Liberzon et al., 2011, 2015). The IC analysis was run separately for (1) all tumors with kataegis, (2) tumors with kataegis loci on chromosome 8, (3) tumors with kataegis loci on chromosome 17, and (4) tumors with kataegis loci on chromosome 22. The IC analysis is based on the use of a normalized differential mutual information function, which is a non-linear correlation coefficient (Kim et al., 2016) with values in the range [−1,1], with 1 representing a perfect match with kataegis and −1 representing a perfect anti-match. For the cases of binary phenotypes (e.g., the kataegis status) and continuous genomic features (e.g., gene expression), the IC becomes the generalized Jensen-Shannon divergence (Kim et al., 2016). An ssGSEA (Barbie et al., 2009) was used to project the breast cancer samples into the space of the 50 hallmarks and the larger collection of 9,092 gene sets divided into 11 distinct datasets as input mentioned earlier. The significance of the associations between the Kataegis phenotype and the genomic features was obtained by an empirical permutation test on the Kataegis phenotype. The permutation IC values are then used to create a null distribution from which nominal, family-wise error rate (FWER), and Bonferroni p values and false discovery rates (FDRs) are computed.
Supplementary Material
Highlights.
Kataegis results in a strong expression signature that can be used to predict status
Kataegis occurs in higher-grade breast tumors that are HER2+
Kataegis occurs in patients diagnosed at an older age and with a higher age at death
Kataegis is a novel marker of good prognosis and low invasiveness in breast cancer
In Brief.
D’Antonio et al. show kataegis is associated with breast cancer patients diagnosed at an older age and with a higher age at death and in HER2+ tumors. Tumors harboring kataegis are associated with transcriptome-wide expression changes consistent with low invasive potential. Kataegis is a marker for good prognosis in breast cancer.
Acknowledgments
This work was supported by NIH grants UL1TR000100, P30CA023100, and R01CA154480. We thank Shamil Sunyaev, Richard Schwab, Erin Smith, Agnieszka D’Antonio-Chronowska, Mike Lawrence, Chip Steward, and Julian Hess for comments and discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.06.026.
AUTHOR CONTRIBUTIONS
K.A.F. and M.D. conceived the study. M.D. performed data processing and computational analyses. P.T. performed the IC analysis. P.T. and J.P.M. contributed to the functional analysis. M.D. and K.A.F. prepared the manuscript.
REFERENCES
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell JL, Camarda R, Zhou AY, Huntsman S, Hu D, Brenner SE, Zaitlen N, Goga A, Ziv E. Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum. Mol. Genet. 2015;24:7421–7431. doi: 10.1093/hmg/ddv432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. METABRIC Group. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington RB. Multiple regression in psychological research and practice. Psychol. Bull. 1968;69:161–182. doi: 10.1037/h0025471. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grömping U. Relative importance for linear regression in R: the package relaimpo. J. Stat. Softw. 2006;17:1–27. [Google Scholar]
- Grömping U. Estimators of relative importance in linear regression based on variance decomposition. Am. Stat. 2007;61:139–147. [Google Scholar]
- Judge SM, Chatterton RT., Jr Progesterone-specific stimulation of triglyceride biosynthesis in a breast cancer cell line (T-47D) Cancer Res. 1983;43:4407–4412. [PubMed] [Google Scholar]
- Kazanov MD, Roberts SA, Polak P, Stamatoyannopoulos J, Klimczak LJ, Gordenin DA, Sunyaev SR. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, genedense, and active chromatin regions. Cell Rep. 2015;13:1103–1109. doi: 10.1016/j.celrep.2015.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan MM, Liu B, Tang X, Wu J, Cyr D, Stevens RD, Ilkayeva O, Huang Z, Tollini LA, Murphy SK, et al. ACLY and ACC1 regulate hypoxia-induced apoptosis by modulating ETV4 via a-ketoglutarate. PLoS Genet. 2015;11:e1005599. doi: 10.1371/journal.pgen.1005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Botvinnik OB, Abudayyeh O, Birger C, Rosenbluh J, Shrestha Y, Abazeed ME, Hammerman PS, DiCara D, Konieczkowski DJ, et al. Characterizing genomic alterations in cancer by complementary functional associations. Nat. Biotechnol. 2016;34:539–546. doi: 10.1038/nbt.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Siva-chenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Shen D, Kang X, Zhang C, Song Q. New tumour antigen PLAC1/CP1, a potentially useful prognostic marker and immunotherapy target for gastric adenocarcinoma. J. Clin. Pathol. 2015;68:913–916. doi: 10.1136/jclinpath-2015-202978. [DOI] [PubMed] [Google Scholar]
- Lu J, Li H, Hu M, Sasaki T, Baccei A, Gilbert DM, Liu JS, Collins JJ, Lerou PH. The distribution of genomic variations in human iPSCs is related to replication-timing reorganization during reprogramming. Cell Rep. 2014;7:70–78. doi: 10.1016/j.celrep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Cooke L, Riley C, Swart R, Simons B, Della Croce K, Wisner L, Iorio M, Shakalya K, Garewal H, et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26:3909–3919. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- Mallini P, Lennard T, Kirby J, Meeson A. Epithelial-to-mesen-chymal transition: what is the impact on breast cancer stem cells and drug resistance. Cancer Treat. Rev. 2014;40:341–348. doi: 10.1016/j.ctrv.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov JP, Golub TR. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003;52:91–118. [Google Scholar]
- Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. Anexpan-sive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Breast Cancer Working Group of the International Cancer Genome Consortium. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012a;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. Breast Cancer Working Group of the International Cancer Genome Consortium. The life history of 21 breast cancers. Cell. 2012b;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis AI, Sun C, Knijnenburg TA, Xue Y, Grernrum W, Hölzel M, Nijkamp W, Wessels LF, Beijersbergen RL, Bernards R, Huang S. SMARCE1 suppresses EGFR expression and controls responses to MET and ALK inhibitors in lung cancer. Cell Res. 2015;25:445–458. doi: 10.1038/cr.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bi-lenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stoja-nov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Romond EH, Perez EA CME Consultants, Inc. Advances in adjuvant therapy for breast cancer. Clin. Adv. Hematol. Oncol. 2006;4:4–9. [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Me-sirov JP. Gene set enrichment analysis:aknowledge-basedapproach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Ken-necke H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- Wang X, Sun L, Maffini MV, Soto A, Sonnenschein C, Kaplan DL. A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials. 2010;31:3920–3929. doi: 10.1016/j.biomaterials.2010.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks C, Cline MS, Weiler E, Diehkans M, Craft B, Martin C, Murphy D, Pierce H, Black J, Nelson D, et al. The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database (Oxford) 2014;2014:1–10. doi: 10.1093/database/bau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J. Natl. Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.