ABSTRACT
NBR2 (neighbor of BRCA1 gene 2) is a non–protein coding gene that resides adjacent to tumor suppressor gene BRCA1, but its role in cancer biology has remained unknown. Our recent study showed that NBR2 encodes a long non-coding RNA and suppresses tumor development through regulation of adenosine monophosphate–activated protein kinase (AMPK) activation.
KEYWORDS: AMPK, BRCA1, energy stress response, lncRNA, NBR2, tumor suppression
The BRCA1 tumor suppressor gene encodes a nuclear protein that plays critical roles in the maintenance of genome integrity, and germline mutations of this gene account for most familial cases of breast and ovarian cancer. BRCA1 mutations are relatively rare in sporadic human cancers; instead, its expression is often downregulated, which has prompted detailed analysis of its gene promoter and transcriptional control. Previous genomic studies1-3 revealed that the mouse Brca1 gene lies head-to-head with the Nbr1 (neighbor of BRCA1 gene 1) gene, with only several hundred base pairs (bp) between the transcription start sites of these 2 genes (Fig. 1A). Interestingly, during the evolution from mouse to primate, genomic duplication and subsequent genomic alterations resulted in the BRCA1-NBR2-BRCA1P1 (BRCA1 pseudogene 1)-NBR1 genomic structure on human chromosome 17q21, in which the BRCA1 gene lies head-to-head with the NBR2 gene, while the BRCA1P1 gene resides right next to the NBR1 gene3 (Fig. 1A).
Figure 1.
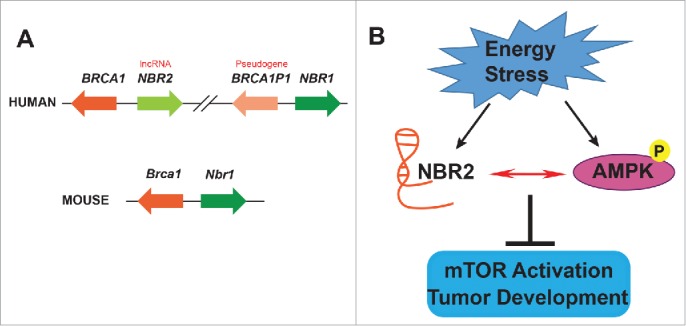
Genomic structure of the NBR2 gene and its proposed role in the regulation of adenosine monophosphate–activated protein kinase (AMPK). (A) Human and mouse BRCA1 loci, with arrows indicating the direction of transcription. Genes are not drawn to scale. Orthologous genes are shown in identical colors. (B) NBR2 and AMPK reciprocally regulate each other in response to energy stress and function in mTORC1 (mammalian target of rapamycin complex 1, also known as mechanistic target of rapamycin complex 1) inactivation and tumor suppression.
Although the NBR1 and NBR2 genes share high sequence homology at their 5′ ends, the remaining sequences share no significant homology. NBR1 encodes a protein of 966 amino acids that functions as an autophagy receptor.4 In contrast, current evidence suggests that the NBR2 gene encodes a long non-coding RNA (lncRNA), a class of non-coding RNAs that are longer than 200 nucleotides.5 NBR2 transcripts ranging from 1 to 3 kb were expressed in most of the tissues examined in a previous study.2 Although the NBR2 cDNA contains an open reading frame (ORF) of 112 amino acids, a strong Kozak signal has not been identified within this ORF. In addition, the stop codon is more than 55 bp upstream of the last splicing site for the putative NBR2 ORF, suggesting that even if the NBR2 transcript were subjected to protein synthesis it would have been degraded by nonsense-mediated mRNA decay.2 There is only 218 bp between the transcription start sites of the human BRCA1 and NBR2 genes, and the 2 genes share a bi-directional promoter.2 Given the close proximity of the human BRCA1 and NBR2 genes, it has been suggested that these 2 genes may be coordinately expressed. Indeed, BRCA1 and NBR2 genes are often co-deleted in familial breast and ovarian cancers.6,7 These observations suggest that, like BRCA1, NBR2 may play a role in tumor suppression. For many years, however, NBR2 has been regarded as a “junk gene” as it does not appear to encode a protein, and its potential function in tumor biology has remained unexplored.
Our recent study revealed, for the first time, the function of NBR2 in tumor suppression.8 The discovery stemmed from our analysis and characterization of energy stress-induced lncRNAs. RNA sequencing and computational analyses of non-coding RNA profiles of cells cultured in glucose-containing or glucose-free medium identified NBR2 as one of several lncRNAs that were induced upon glucose starvation. Subsequent analyses revealed that NBR2 functions to promote energy stress–induced activation of adenosine monophosphate–activated protein kinase (AMPK) (Fig. 1B), the major energy sensor in eukaryotic cells. In response to energy stress, AMPK is activated and functions to repress anabolic processes (such as protein/lipid/sterol synthesis) and promote catabolic processes (such as glycolysis, fatty acid oxidation, autophagy), resulting in restoration of energy balance.9 As anabolic processes are often required to support tumor growth, the AMPK pathway plays tumor suppressive roles in many cancers. Consistent with this, our data showed that NBR2 knockdown promoted anchorage-independent growth and xenograft tumor development in vivo, whereas NBR2 overexpression inhibited tumor cell proliferation. Accordingly, NBR2 is downregulated and its low expression is correlated with poor prognosis in several human cancers. Further functional studies showed that NBR2 exerted its tumor suppressive function at least in part through regulation of AMPK activity.8
Taken together, our results not only reveal the biological function of NBR2 in tumor suppression but also provide a new conceptual framework to understand the regulation of kinase signaling by lncRNAs. Several important questions remain to be addressed in future studies. First, how NBR2 regulates AMPK kinase activity remains incompletely understood. It will be particularly important to determine the crystal structure of the NBR2-AMPK complex, which undoubtedly will provide further mechanistic insight into how NBR2 regulates AMPK activation. Whether the NBR2-AMPK regulation discovered in our study can be applied to other lncRNAs that may regulate kinase function also awaits further investigation. In addition, our data show that NBR2 overexpression still partially inhibited tumor cell proliferation in AMPK-deficient cells,8 suggesting the existence of AMPK-dependent and -independent mechanisms that mediate NBR2 function in tumor suppression. Since many lncRNAs function to regulate the transcription of neighboring genes, the hypothesis that NBR2 regulates BRCA1 transcription is of particular interest. Although our current data do not support such a hypothesis,8 another study showed that NBR2 knockdown modestly downregulated BRCA1 mRNA level.10 Since these studies were conducted in different cell lines with different knockdown efficiencies, it is possible that NBR2 regulation of BRCA1 expression is context dependent, and it will be important to further clarify this question in future studies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem 1997; 272:20994-7; PMID:9261099; http://dx.doi.org/ 10.1074/jbc.272.34.20994 [DOI] [PubMed] [Google Scholar]
- 2.Xu CF, Brown MA, Nicolai H, Chambers JA, Griffiths BL, Solomon E. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet 1997; 6:1057-62; PMID:9215675; http://dx.doi.org/ 10.1093/hmg/6.7.1057 [DOI] [PubMed] [Google Scholar]
- 3.Jin H, Selfe J, Whitehouse C, Morris JR, Solomon E, Roberts RG. Structural evolution of the BRCA1 genomic region in primates. Genomics 2004; 84:1071-82; PMID:15533724; http://dx.doi.org/ 10.1016/j.ygeno.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 4.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al.. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505-16; PMID:19250911; http://dx.doi.org/ 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 5.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154:26-46; PMID:23827673; http://dx.doi.org/ 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Ouweland AM, Dinjens WN, Dorssers LC, van Veghel-Plandsoen MM, Bruggenwirth HT, Withagen-Hermans CJ, Collee JM, Joosse SA, Terlouw-Kromosoeto JN, Nederlof PM. Deletion of exons 1a-2 of BRCA1: a rather frequent pathogenic abnormality. Genetic testing and molecular biomarkers 2009; 13:399-406; PMID:19405878; http://dx.doi.org/ 10.1089/gtmb.2008.0155 [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Casado Z, Romero I, Fernandez-Serra A, Rubio L, Llopis F, Garcia A, Llombart P, Lopez-Guerrero JA. A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer. BMC medical genetics 2011; 12:134; PMID:21989022; http://dx.doi.org/ 10.1186/1471-2350-12-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, et al.. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 2016; 18:431-42; PMID:26999735; http://dx.doi.org/ 10.1038/ncb3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251-62; PMID:22436748; http://dx.doi.org/ 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, et al.. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016; 18(5):637-52; PMID:26996597; http://dx.doi.org/ 10.1016/j.stem.2016.01.024 [DOI] [PubMed] [Google Scholar]


