Abstract
We describe the USC Multimodal Connectivity Database (http://umcd.humanconnectomeproject.org ), an interactive web-based platform for brain connectivity matrix sharing and analysis. The site enables users to download connectivity matrices shared by other users, upload matrices from their own published studies, or select a specific matrix and perform a realtime graph theory-based analysis and visualization of network properties. The data shared on the site span a broad spectrum of functional and structural brain connectivity information from humans across the entire age range (fetal to age 89), representing an array of different neuropsychiatric and neurodegenerative disease populations (autism spectrum disorder, ADHD, and APOE-4 carriers). An analysis combining 7 different datasets shared on the site illustrates the diversity of the data and the potential for yielding deeper insight by assessing new connectivity matrices with respect to population-wide network properties represented in the UMCD.
Introduction
The endeavor to unravel the human brain connectome using neuroimaging is now well underway. The Human Connectome Project (http://www.humanconnectome.org/ and http://www.humanconnectomeproject.org/) is emblematic of this, having collected and shared high quality functional MRI, diffusion weighted MRI, structural MRI, and detailed demographic and behavioral data from 1200 subjects. Meanwhile, labs around the world continue to collect and analyze MRI-based connectivity data at a rapid pace, furthering our knowledge about neurological and neuropsychiatric disease (Crossley et al., 2014) (Deco and Kringelbach, 2014), the structural connectivity of the brain’s anatomical core (Van Den Heuvel and Sporns, 2011) (Irimia and Van Horn, 2014), the relationship between structural and functional connectivity (Goñi et al., 2014; Heuvel and Sporns, 2013), evolving landscape of systems connectivity across the lifespan (Betzel et al., 2014; Chan et al., 2014), temporal dynamics of functional brain connectivity (Allen et al., 2014; Zalesky et al., 2014), and the effects of neurostimulation (Wang et al., 2014).
All of these studies can be unified by their common underlying data format, the connectivity matrix (CM). A connectivity matrix has brain regions of interest along rows and columns and the connectivity strength between a given pair of ROIs stored in the cell where these two regions intersect (Bullmore and Sporns, 2009). The connection strength or “edge weight” is typically a structural connectivity measure such as the fiber tractography streamline density between two ROIs, or a functional connectivity measure representing the statistical similarity between two ROIs’ BOLD signal timeseries. The CM is a highly compact, distilled representation of network-wide or often brain-wide connectivity. The USC Multimodal Connectivity Database (http://umcd.humanconnectomeproject.org; formerly the UCLA Multimodal Connectivity Database; (Brown et al., 2012)) is an open repository for CMs. It is a publicly accessible website where any user can download CMs that have been shared by other researchers, upload their own CMs to share with the research community, and perform on-the-fly graph theory-based analyses of any publicly available CM. Numerous studies have been conducted using data available on the UMCD for testing reproducibility of structural network properties (de Reus and van den Heuvel, 2013), designing new community detection algorithms (Dodero et al., 2014; Richiardi et al., 2013), and developing network-based classification algorithms for autism spectrum disorder (Cheplygina et al., 2014).
Description of the tool
The site was designed as a central repository for connectivity matrices. There are a number of immediate benefits that a centralized resource provides: broad, click-of-the-mouse fast meta analyses, such as those enabled by BrainMap (http://www.brainmap.org/) or Neurosynth (http://neurosynth.org/); reproducibility of findings across study sites and varieties in data analysis methodology; and the availability of data to other researchers whose expertise may enable the re-analysis of existing data in order to yield previously undiscovered insights.
In the four years of existence, UMCD has accumulated 1887 publicly available CMs from 21 different studies. These CMs are primarily derived from individual subjects, though group average matrices are also accepted. Of these data, 1652 CMs are from functional MRI (fMRI) data, 224 from Diffusion Tensor Imaging (DTI), 5 from Structural MRI, and 6 from Diffusion Spectrum Imaging (DSI) are publicly available. These data blanket the human lifespan, from fetuses with a gestational age of 200 days to 89-year old individuals. Represented subject populations include fetuses, typically developing children and adolescents, healthy adults, and patients with ADHD (hyperactive, inattentive, and combined subtypes), Autism Spectrum Disorder (ASD), Obsessive Compulsive Disorder, and APOE-4 carrier status. The majority of functional data are dervied from task-free fMRI scans, during which subjects are awake but not receiving stimulus or explicitly performing any cognitive task.
Procedure for sharing or accessing data
Connectivity matrices can be shared to the UMCD on the ‘Upload New Data’ page. They are typically symmetric, undirected, square CMs that represent connectivity strengths derived from processed data. The user needs to prepare the following: 1) a n × n connectivity matrix saved as a square, tab or space-delimited text file, 2) a n × 3 column text file where the (x,y,z) center of mass in MNI coordinate space is listed for each of the n nodes in the network, 3) a n × 1 column text file that lists the full name of the brain region for each node in the network, and 4) a n × 1 column text file that lists the regional abbreviation for each node.
Once each of these files have been uploaded, the user is then requested to fill in the demographic features about the CM that is being shared, along with a list of preprocessing steps. Requested demographic information include the subject age, sex, population from which that subject came (eg ‘Healthy Control’ or ‘ADHD-Inattentive’, and group size (where a ‘1’ specifies a CM from an individual and a value greater than 1 specifies an average of that number of subjects). Study-specific data parameters include the data type (fMRI, DTI, DSI, structural MRI, EEG, or MEG), the scanner manufacturer, and the MRI field strength. Numerous optional parameters describing the data preprocessing are also present (motion correction, deterministic tractography, etc.). Data can be batch uploaded by vertically stacking text files for each CM. Once data has been shared via the submit button, it is automatically added to the database and searchable on the ‘Browse All Data’ page.
The data come from scanners and centers all over the world, including both single-center studies and data from international consortia such as the INDI 1000 Functional Connectomes dataset (Biswal et al., 2010), the ADHD-200 competition (“The ADHD-200 Consortium,” 2012, p. -), and the Human Connectome Project (http://humanconnectome.org/).
The data can be accessed by visiting the “Browse All Data” page – http://umcd.humanconnectomeproject.org/umcd/default/browse - which has a rapid, easy to use search function for limiting only datasets that have the search term. A dataset can then be selected to download using the “View/Download” link. Any visitor to the site can download any publicly available dataset without an account. A registration process with a username/password combination is required for any user who wishes to share data to the site. The default sharing option is ‘public’, which makes the data viewable and downloadable by any site user. A ‘private’ option exists for users who wish to upload data so that they can access the graph theory analysis tools available on the site, without making their data accessible to the public. This is also a convenient option for users to store their data in the cloud and make it available to them wherever and whenever they may need to download their data again. Data usage statistics for each CM are kept and displayed on the ‘View’ page for that CM, including the number of times the dataset has been download and analyzed. Collectively, ~1.03 million CMs have been downloaded from the site and ~5900 analyses have been performed using the site’s built in graph theory engine. Each matrix has a unique identification number in the database which can be plugged into a URL where the matrix and related metadata can be downloaded or viewed, e.g. http://umcd.humanconnectomeproject.org/umcd/default/update/109 where “109” is the ID number for the Study Name “ADHD200_CC200” and the Network Name “KKI_2026113”.
There is no quality control process limiting the data that is added to the database. Again, the assessment of data quality is left to the user of the site. Data quality can be determined by common sense measures such as whether or not an associated publication is listed, whether the source of the data is a recognized laboratory. Quantitative measures can be assessed on the data once it is downloaded, such as looking at the distribution of edge weights, the spatial pattern of edge weights, and other graph theory measures.
Users have the option of download individual CMs via the “View/Download” link for the network of interest identified on the Browse All Data page. If the full set of data and metadata for a given study is desired, the user can download it all as one zipped file via the Browse Studies page and the Download all Data link for that study. The data is all made available under the terms of the Creative Commons Attribution Non-Commercial license. UMCD will continue to be hosted by the Laboratory of Neuroimaging at the University of Southern California. Plans are to keep the site running, available to data download, upload, and analysis for the forseeable future.
“On-the-fly” network analysis and visualization
Any CM on the UMCD can be analyzed “on the fly” via the ‘Analyze Network’ page. Here, the user can select a CM of interest. They can then specify the graph theory analysis parameters for weighted or binary edges, and the percent of edges in the matrix to include. It is important for the user to keep in mind that certain parameters may be less appropriate for certain types of matrices. For example, including 100% of edges in a functional connectivity matrix may include negative connection strengths, which the will be handled by shifting the entire distribution to be positive. Once an analysis is submitted, a report is generated that displays the network’s global network metrics, including characteristic path length, clustering coefficient, small world attributes, and nodal network metrics, including degree, betweenness centrality, clustering coefficient, regional efficiency, participation coefficient, and modularity membership. All these measures are calculated using functions in the NetworkX Python library, with links to the NetworkX documentation alongside each that network property in the “Analyzed Network” report. For each nodal network measure, the network is shown in both an interactive 3D display (rendered using WebGL) and static 2D network figures that show nodes and edges as balls and sticks, with node diameter proportional to a given nodal network measure. Should the user wish to qualitatively compare each of these measures for two CMs side by side, they can so on the ‘Compare Networks’ page. A more detailed description of the network analysis is provided in (Brown et al., 2012).
An illustrative meta-analysis
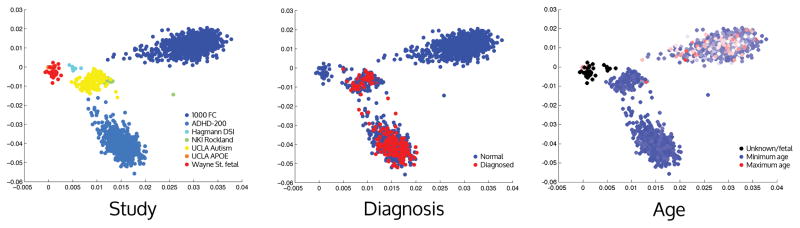
As neuroimaging data continue to accumulate at an accelerating pace in the near future, it is imperative to maintain unified standards for dataset compilation and sharing. These efforts will maximize our potential to make broad comparisons of brain connectivity patterns across different stages of development and aging, disease conditions, cognitive states, and imaging modalities. One eventual goal of this rich compilation of data will be to help assess where individuals lie within the spectrum of the population (Dosenbach et al., 2010). As a demonstration of a spectrum, the CMs from 7 different studies in the UMCD comprising 1863 individuals were compared for similarity using a principal component analysis. From the plot, it can be seen that CMs from the same dataset tend to cluster near each other. The predominant source of variation appears to be driven by the method used to process the data, as clouds from the 1000 Functional Connectomes (Figure 1, left) and ADHD-200 (dataset 2, cornflower blue) samples, both of which are task-free fMRI functional connectivity, show substantial separation. The components intersect on the UCLA autism spectrum disorder data (ASD; dataset 5), which contains a mixture of tf-fMRI and DTI-based matrices.
Figure 1.
A representation of the similarity of 1819 connectivity matrices (CMs) from 7 different studies shared on the UMCD, including both fMRI-based functional CMs and diffusion-weighted MRI-based structural CMs. Each CM is plotted in two dimensions representing the first two components of a Principal Component Analysis of all 1819 CMs. In the left panel, each CM color coded according to the study it came from. In the middle panel, CMs are color coded in blue if they came from healthy control subjects and in red if they came from diagnoses of autism spectrum disorder (middle cloud) or ADHD (lower cloud). In the right panel, CMs are color coded continuously according to age, with the youngest subjects in dark blue, intermediate age subjects in white, and the oldest subjects in red.
The potential of this approach for data discovery and as a diagnosis aid is considerable. While the datasets combined in this analysis come from multiple different imaging modalities and have heterogeneous processing methods, future meta-analyses will have to consider the impact of this variability and employ data aggregation methods that expose meaningful biological commonalities. As an example in this analysis, it is noteworthy that the tf-fMRI networks from the Wayne State fetal tf-fMRI study (Thomason et al., 2014) appear more similar to structural connectivity networks that other functional connectivity networks. It produces the testable hypothesis that fetal brain connectivity may be locally distributed and more closely resemble anatomical connectivity in adults.
Future directions and outstanding issues in connectomics research
When considering psychiatric disease, it would be informative to see where the connectivity pattern of an individual falls along a spectrum among children and adolescents that are typically developing or have received diagnoses of ADHD or ASD. One potential future application is a brain connectivity search engine where an individual is scanned, their CM is obtained, and a search is run to find the CMs in the database that most closely match the query. It would be imprudent to use this tool exclusively to diagnose a subject but could add to evidence about the similarity of this individual to a broad database of subjects with similar demographics, symptoms, behavior, and genetics.
As the field of neuroimaging connectomics matures, the UMCD will need to mature along with it. The connectivity matrices that are currently stored are a very simple format for data representation that does not have the flexibility necessary to represent details about different properties of the connections between regions, such as the fiber connection length, fiber integrity, or various connectivity branching patterns that can give rise to the same connectivity “strength” (Mitra, 2014). Previous efforts have been made to create file formats with various levels of extensibility, such as the subject specific “netmats” that are derived from data in the Human Connectome Project, the more extensive CIFTI file format designed as part of the HCP (https://www.nitrc.org/projects/cifti/), or the connectome file format (Gerhard et al., 2011). The UMCD will continue to serve as a repository for “distilled” formats of these data, containing relatively sparse and lightweight descriptions of connectivity such as the regional coordinates, connectivity weights, and data processing details, along with pointers to locations on the web where more detailed information about a dataset can be obtained, such as the raw data or the regional parcellation file. The UMCD will also continue to add new graph theory measures to the analysis report as they become ubiquitous in the field. One example of this the rich club characteristics (Van Den Heuvel and Sporns, 2011) of a network, which can be expected in a forthcoming release.
Comparison of networks is another important area of active development in neuroimaging connectomics. While the basics of statistical comparison of global and nodal network properties are largely agreed upon, many open questions remain. How can dynamic functional networks derived with a sliding window approach be compared to one another (Allen et al., 2014) (Zalesky et al., 2014)? How do we cope with degeneracy, the ability to networks with different topology to perform the same cognitive operation (Fornito et al., 2015)? How can networks derived from subjects performing a task be compared with task-free networks (Cole et al., 2014)? The UMCD will continue to serve as a repository for sharing CMs, rapidly locating them, and performing a basic visualization of network properties. Software designed for in-depth statistical comparisons such as the CONN toolbox (https://www.nitrc.org/projects/conn) will tackle new challenges in brain network analysis. Other standalone programs provide a more in-depth visualization of brain networks than what the UMCD is intended for. Powerful features offered by programs like the BrainNet Viewer (Xia et al., 2013), the Connectome Visualization Utility (LaPlante et al., 2014), and the Connectome Workbench (http://www.humanconnectome.org/software/connectome-workbench.html) include visualization of groupwise statistical differences in networks, detailed neuroanatomical connectivity maps, and alternative connectivity summaries such as circular “connectograms”. An example workflow for a connectivity researcher might go as follows: 0) derive CMs from their neuroimaging data using standard neuroimaging processing software, 1) analyze networks with the Brain Connectivity Toolbox, 2) visualize and interpret their results with the BrainNet Viewer, and 3) publish their study and share their CMs on the UMCD with a link to the paper describing the results, where other researchers can locate the shared data, download it if desired to visualize and/or analyze it, or perform a subsequent meta-analysis, and reference the original manuscript. The continuity and diversity of this software ecosystem will help ensure that connectomics data is broadly utilized.
Highlights.
The USC Multimodal Connectivity Database is home to 1887 publicly available connectivity matrices
Available datasets include the 1000 Functional Connectomes, the ADHD-200 data, and fMRI/DTI studies of autism spectrum disorder, APOE-4 carriers and non-carriers, fetal fMRI, and diffusion spectrum imaging structural connectivity
Over one million connectivity matrices have been downloaded to date
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking Whole-Brain Connectivity Dynamics in the Resting State. Cereb Cortex. 2014;24:663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Byrge L, He Y, Goñi J, Zuo XN, Sporns O. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage. 2014;102(Pt 2):345–357. doi: 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski A-M, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kötter R, Li S-J, Lin C-P, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SARB, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng G-J, Veijola J, Villringer A, Walter M, Wang L, Weng X-C, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang Y-F, Zhang H-Y, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Rudie JD, Bandrowski A, Horn JDV, Bookheimer SY. The UCLA multimodal connectivity database: a web-based platform for brain connectivity matrix sharing and analysis. Front Neuroinform. 2012;6:28. doi: 10.3389/fninf.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–98. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS. Decreased segregation of brain systems across the healthy adult lifespan. PNAS. 2014;111:E4997–E5006. doi: 10.1073/pnas.1415122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheplygina V, Tax DMJ, Loog M, Feragen A. Network-Guided Group Feature Selection for Classification of Autism Spectrum Disorder. In: Wu G, Zhang D, Zhou L, editors. Machine Learning in Medical Imaging, Lecture Notes in Computer Science. Springer International Publishing; 2014. pp. 190–197. [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and Task-Evoked Network Architectures of the Human Brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014:awu132. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Kringelbach ML. Great Expectations: Using Whole-Brain Computational Connectomics for Understanding Neuropsychiatric Disorders. Neuron. 2014;84:892–905. doi: 10.1016/j.neuron.2014.08.034. [DOI] [PubMed] [Google Scholar]
- De Reus MA, van den Heuvel MP. Estimating false positives and negatives in brain networks. NeuroImage. 2013;70:402–409. doi: 10.1016/j.neuroimage.2012.12.066. [DOI] [PubMed] [Google Scholar]
- Dodero L, Murino V, Sona D. Joint laplacian diagonalization for multimodal brain community detection, in: 2014 International Workshop on Pattern Recognition in Neuroimaging. Presented at the 2014 International Workshop on Pattern Recognition in Neuroimaging; 2014. pp. 1–4. [DOI] [Google Scholar]
- Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Barch DM, Petersen SE, Schlaggar BL. Prediction of Individual Brain Maturity Using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- Gerhard S, Daducci A, Lemkaddem A, Meuli R, Thiran J-P, Hagmann P. The Connectome Viewer Toolkit: An open source framework to manage, analyze, and visualize connectomes. Front Neuroinform. 2011;5:3. doi: 10.3389/fninf.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi J, van den Heuvel MP, Avena-Koenigsberger A, de Mendizabal NV, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran JP, Sporns O. Resting-brain functional connectivity predicted by analytic measures of network communication. PNAS. 2014;111:833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvel MP, van den Sporns O. An Anatomical Substrate for Integration among Functional Networks in Human Cortex. J Neurosci. 2013;33:14489–14500. doi: 10.1523/JNEUROSCI.2128-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Van Horn JD. Systematic network lesioning reveals the core white matter scaffold of the human brain. Front Hum Neurosci. 2014;8:51. doi: 10.3389/fnhum.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante RA, Douw L, Tang W, Stufflebeam SM. The Connectome Visualization Utility: Software for Visualization of Human Brain Networks. PLoS ONE. 2014;9:e113838. doi: 10.1371/journal.pone.0113838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra PP. The Circuit Architecture of Whole Brains at the Mesoscopic Scale. Neuron. 2014;83:1273–1283. doi: 10.1016/j.neuron.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Greicius M. Two Test Statistics for Cross-Modal Graph Community Significance, in: 2013 International Workshop on Pattern Recognition in Neuroimaging (PRNI). Presented at the 2013 International Workshop on Pattern Recognition in Neuroimaging (PRNI); 2013. pp. 70–73. [DOI] [Google Scholar]
- The ADHD-200 Consortium. A Model to Advance the Translational Potential of Neuroimaging in Clinical Neuroscience. Front Syst Neurosci. 2012;6 doi: 10.3389/fnsys.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason ME, Brown JA, Dassanayake MT, Shastri R, Marusak HA, Hernandez-Andrade E, Yeo L, Mody S, Berman S, Hassan SS, Romero R. Intrinsic Functional Brain Architecture Derived from Graph Theoretical Analysis in the Human Fetus. PLoS ONE. 2014;9:e94423. doi: 10.1371/journal.pone.0094423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Sporns O. Rich-Club Organization of the Human Connectome. J Neurosci. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Rogers LM, Gross EZ, Ryals AJ, Dokucu ME, Brandstatt KL, Hermiller MS, Voss JL. Targeted enhancement of cortical-hippocampal brain networks and associative memory. Science. 2014;345:1054–1057. doi: 10.1126/science.1252900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y. BrainNet Viewer: A Network Visualization Tool for Human Brain Connectomics. PLoS ONE. 2013;8:e68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. PNAS. 2014;111:10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]