Abstract
Objective
This work evaluates current 3-D image registration tools on clinically acquired abdominal computed tomography (CT) scans.
Methods
Thirteen abdominal organs were manually labeled on a set of 100 CT images, and the 100 labeled images (i.e., atlases) were pairwise registered based on intensity information with six registration tools (FSL, ANTS-CC, ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS). The Dice similarity coefficient (DSC), mean surface distance, and Hausdorff distance were calculated on the registered organs individually. Permutation tests and indifference-zone ranking were performed to examine the statistical and practical significance, respectively.
Results
The results suggest that DEEDS yielded the best registration performance. However, due to the overall low DSC values, and substantial portion of low-performing outliers, great care must be taken when image registration is used for local interpretation of abdominal CT.
Conclusion
There is substantial room for improvement in image registration for abdominal CT.
Significance
All data and source code are available so that innovations in registration can be directly compared with the current generation of tools without excessive duplication of effort.
Index Terms: Image registration, Abdomen, Computed tomography
I. Introduction
The human abdomen is an essential, yet complex body space. Bounded by the diaphragm superiorly and pelvis inferiorly, supported by spinal vertebrae, and protected by the muscular abdominal wall, the abdomen contains organs involved with blood reservation, detoxification, urination, endocrine function, and digestion, and includes many important arteries and veins. Computed tomography (CT) scans are routinely obtained for the diagnosis and prognosis of abdomen-related disease; yet no specific image registration tools for the abdomen have been developed.
General-purpose registration tools (initially designed for volumetric brain registration) are being applied to abdominal CT scans [1, 2] On abdominal CT, inter-subject variability (e.g., age, gender, stature, normal anatomical variants, and disease status) can be observed in terms of the size, shape, and appearance of each organ. Soft anatomy deformation further complicates the registration by varying the inter-organ relationships, even within individuals (e.g., pose, respiratory cycle, edema, digestive status). Hence, characterization of tools specifically on abdominal structures is necessary, as opposed to relying on brain-centric reviews [3].
This work follows the framework of Klein et al. [3], in which 14 nonlinear registration tools and one linear registration algorithm were applied to 80 MRIs of the human brain. Manual segmentations of regions are used to assess volumetric overlap and surface-based criteria separately from the intensity-based metrics that drive registration. In related work, West et al. [4] established a platform for assessing landmark-based registrations on retrospective intermodality (MR, CT, and PET) brain images, where 12 methods were evaluated based on target registration error [5]. Murphy et al. [6] compared 20 registration algorithms to 30 thoracic CT pairs in the EMPIRE10 challenge by metrics specified for pulmonary area alignment and correspondence. The VISCERAL challenge [7] provided a platform for evaluating abdominal organ segmentation on four image modalities.
This work expands on [8] by including more datasets (100 vs. 20), adjusting the label sets (the previous individual labels of the adrenal glands were separated into two labels: right and left), using a different registration framework (previously all non-rigid registrations were initialized by one affine registration tool), and presents more comprehensive statistical analyses (see the methods section) (Fig. 1). We selected 5 registration tools that have been successful in volumetric brain registrations, including FSL (FMRIB Software Library) [9], IRTK (Image Registration Toolkit) [10], NiftyReg [11], ANTs (Advanced Normalization Tools) [12], and DEEDS (DEnsE Displacement Sampling) [13] due to their academic popularity and general availability. In total, six registration methods were evaluated with two different parameter settings for ANTs. For each registration tool, we applied affine registration followed by non-rigid registration. Registration results from both stages were evaluated based on the Dice similarity coefficient (DSC [14]), mean surface distance (MSD), and Hausdorff distance (HD). We note that compared to the brain and thorax registrations, substantial registration errors can be observed in the abdomen due to the large variability and deformation; registration tools tailored for these intricacies can potentially improve the performance. We also note that the efficacy of non-rigid registrations are greatly impacted by the baseline affine registrations as a lesson learned from [8], thus we modified the registration framework to use affine and non-rigid registration from the same registration tool. The main focus of this paper is to provide a public abdomen dataset and to evaluate the common registration tools on the provided dataset.
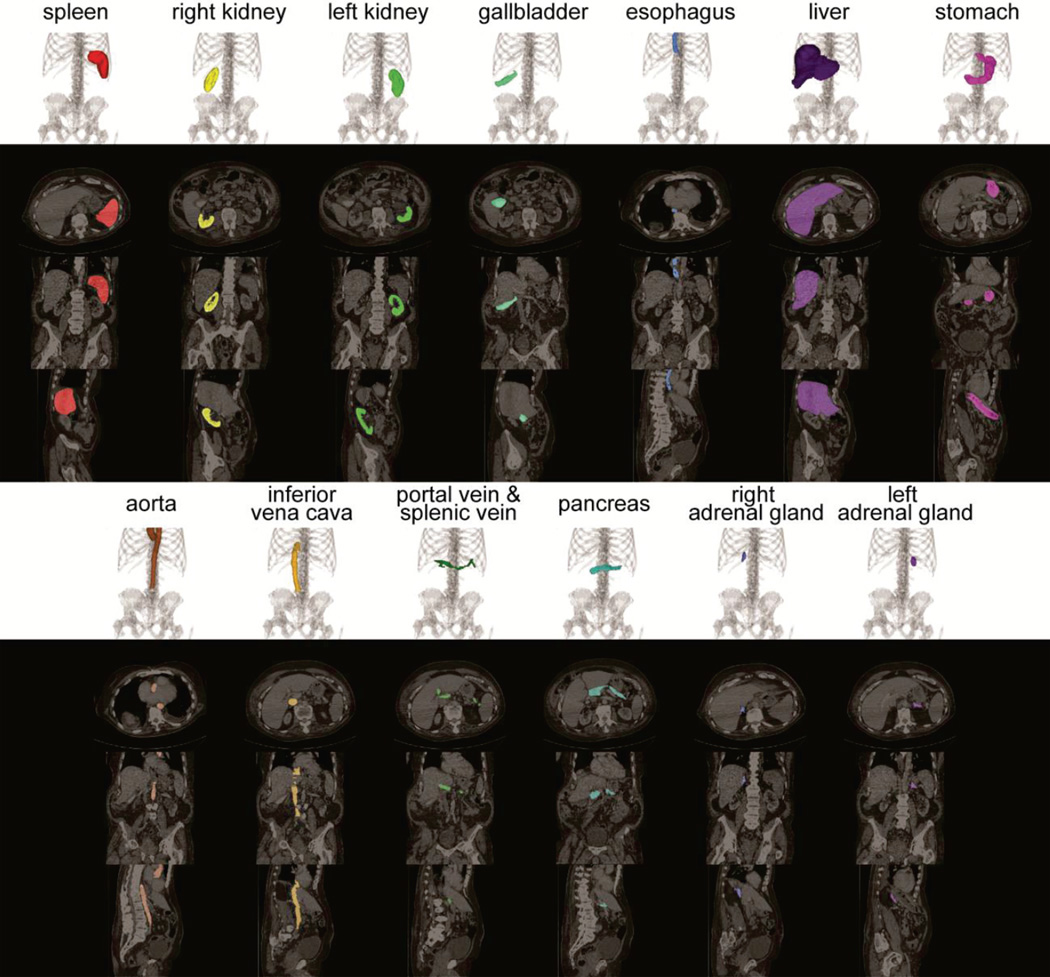
Fig. 1.
Illustration of 13 organs of interest on volumetric rendering and 2-D slices of axial, coronal and sagittal orientations.
II. Methods
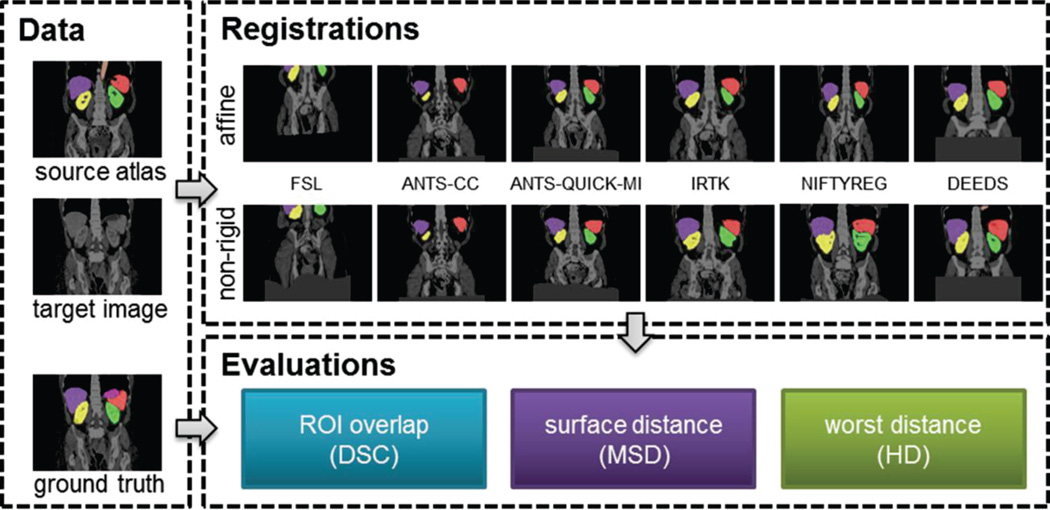
The registration evaluation process follows the flowchart in Fig. 2.
Fig. 2.
Registration pipeline. Given a pair of target image and a source atlas (image and labels), an affine registration was applied followed by a non-rigid registration for each of the six evaluated registration methods. The registered labels were validated against the ground truth (manual labels) in terms of DSC, MSD, and HD.
A. Data Acquisition
Under institutional review board supervision, 100 abdominal CT scans were collected anonymously from two clinical trials. From an ongoing colorectal cancer chemotherapy trial, the baseline sessions of the abdominal CT scans were randomly selected from 75 metastatic liver cancer patients; the remaining 25 scans were acquired from a retrospective post-operative cohort with suspected ventral hernias. All 100 scans were captured during portal venous contrast phase with variable volume sizes (512 × 512 × 53 ~ 512 × 512 × 368) and field of views (approx. 280 × 280 × 225 mm3 ~ 500 × 500 × 760 mm3). The in-plane resolution varies from 0.54 × 0.54 mm2 to 0.98 × 0.98 mm2, while the slice thickness ranged from 1.5 mm to 7.0 mm. All image scans and their associated labels were converted to NIFTI format with the DCM2NII tool of the MRIcron package [15]. The image orientations in the NIFTI header describe the relative position of patients with respect to the scanner. Due to the inconsistencies of scanning protocols, the images were re-oriented to standard orientation with the FSL package before any further processing [9].
Thirteen abdominal organs were considered regions of interest (ROI), including spleen, right kidney, left kidney, gall bladder, esophagus, liver, stomach, aorta, inferior vena cava, portal and splenic vein, pancreas, left adrenal gland, and right adrenal gland. The organ selection was essentially based on [16]. As suggested by a radiologist, we excluded the heart for lack of full appearance in the datasets, and included the adrenal glands for clinical interest. These ROIs were manually labeled by two experienced undergraduate students with 6 months of training on anatomy identification and labeling, and then verified by a radiologist on a volumetric basis using the MIPAV software [17]. A subset of 13 scans was randomly selected, and independently labeled by each of the two raters. Mean overall DSC overlap between the raters (i.e., inter-rater variability) was 0.87 ± 0.13 (0.95 ± 0.04 when considering only the spleen, kidneys, and liver).
B. Registration Pipeline
General-purpose registration software typically provides options and parameters for specific applications. Six registration methods from six registration tools were evaluated in this study, and indicated as FSL, ANTS-CC, ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS respectively. All registration commands evaluated in this study were verified by the developers of the corresponding registration software.
All tested methods follow a standard registration pipeline: For each image pair, source (moving/floating) and target (fixed/reference) images, the registration was driven by the similarity metrics between their intensity images. The registration was divided into two stages - affine registration that aligned the two images with co-linearity persevering transformation (translation/rotation/scaling/shearing), followed by a non-rigid registration that refined the local correspondence with deformation models. Based upon the transformation/deformation generated from the intensity-driven registration, the labels associated with the source image were propagated to the target space with nearest neighbor interpolation as the estimate of the target structures.
We note that before performing this large-scale study, we invited the authors of the evaluated algorithms to optimize their algorithms on a subset of our dataset (10 scans). The authors of NIFTYREG and DEEDS provided us their optimized parameters; the authors of IRTK approved our configuration with no further optimization; the authors of ANTs and FSL approved our configuration while considering their level of participation did not warrant authorship to this manuscript. The focus of parameter optimization for NIFTYREG and DEEDS lay on levels of a multi-resolution strategy, thresholds of intensity range, use of discrete optimization; default parameters, or those recommended in the example of the software documentation were used if no optimization was provided by the registration authors.
We briefly describe the registration setups for each method without detailed parameters. The full registration commands can be found in the supplementary material.
FSL used the FLIRT and FNIRT for affine and non-registration, respectively. The affine registration with 9 degrees of freedom (DOF) was initialized by a rigid registration. Both rigid and affine registrations constrained the search of rotations with “-nosearch”.
ANTS-CC and ANTS-QUICK-MI used different parameter settings with the ANTs package. The parameters were derived from the example scripts (antsRegistrationSyN and antsRegistrationSyNQuick, respectively) in the ANTs package. ANTS-CC used cross-correlation as the image similarity metric, while ANTS-QUICK-MI used mutual information. ANTS-QUICK-MI was specified to converge with fewer iterations than ANTS-CC, and thus noted with “QUICK”. Both methods applied 5 levels of multi-resolution sampling, windowed the intensity range, started with the alignment of center of mass, initialized the affine registration with rigid registration, and used symmetric normalization (SyN) transform for the non-rigid registration. Multi-thread computing was enabled to use two CPU cores for one registration process.
IRTK sequentially used rigid, affine, and non-rigid registrations. For all three procedures, the target padding value was set to −900 to reduce the impact of the background in the CT scans (air with −1024 Hounsfield units), 3 levels of multi-resolution sampling were applied. Assuming relatively homogenous orientations of patient bodies in the CT scan, the options of “translation_only” and “translation_scale” were specified for the rigid and affine registration, respectively, so that only translation (and scaling for the affine registration) adjustments were allowed, and the search over rotations was prohibited. The B-spline control spacing free-form deformation for the non-rigid registration was set to be 20, 10 and 5mm for the 3 resolution levels, respectively.
NIFTYREG used 5 levels of multi-resolution sampling for both affine and non-rigid registrations. For the non-rigid registration based on a block-matching approach and free-form deformation, an upper intensity threshold of 500 was set for both target and source image, and the maximum iteration for convergence was limited to 1000. Multi-thread computing was enabled to use two CPU cores for one registration process.
DEEDS used 5 scale levels with grid spacing ranging from 8 to 4 voxels, displacement search radii from 6 to 2 steps with quantizations between 5 and 1 voxels. The regularization weighting was set to be 0.4. Self-similarity context descriptors [18] were derived, while their Hamming distance between images were used to guide the local displacement. All scans were resampled to an isotropic resolution of 2.2 mm3, and cropped to have same dimensions. The non-rigid registration was initialized using an affine registration that was based on the same similarity metric, a similar block-matching search and trimmed least squares.
C. Running Registrations
All registrations were run on an Oracle Grid cluster of twelve 64-bit Ubuntu 14.04LTS Linux servers. Each server had 12 2.8GHz cores and 48 GB RAM. Each registration was specified with the approximated maximum memory usage based on their computational complexity; multiple registrations were allocated on the memory requirements on servers, and operated in parallel. The memory specified in GB for FSL, ANTS-CC, ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS were 20, 20, 20, 10, 10, and 5. Given 100 scans, 9900 sets of output registration can be generated for each method with a leave-one-out scheme. Specifically, for each target image among the 100 scans; the remaining 99 scans were used as source images to the target image in a pair-wise manner. However, during initial running trials, we found that FSL and ANTS-CC took an unreasonable amount of time to complete (> 6 h, see Table 1). Therefore, these two methods are only validated on a randomly selected subset of the datasets. Specifically, 20 target images and 20 source images were randomly selected without replacement from the 100 datasets, and 400 registrations were applied from all combinations of the source-target pairs. For the other four methods, i.e., ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS all 9900 registrations were applied. In total, this study used approximately 103,800 hours of CPU time for registration.
TABLE I.
Metrics on 400 registrations for all tested methods (mean ± std)
| Method | DSC | MSD (mm) | HD (mm) | Time (min) |
|---|---|---|---|---|
| FSL | 0.12 ± 0.19 | 37.92 ± 44.11 | 84.28 ± 59.96 | 951.73 ± 201.20 |
| ANTS-CC | 0.18 ± 0.21 | 27.15 ± 32.65 | 62.92 ± 44.60 | 411.60 ± 74.20 |
| ANTS-QUICK-MI | 0.27 ± 0.25 | 15.96 ± 19.22 | 49.66 ± 32.96 | 50.18 ± 21.93 |
| IRTK | 0.28 ± 0.26 | 19.07 ± 26.50 | 55.58 ± 39.26 | 220.27 ± 91.79 |
| NIFTYREG | 0.35 ± 0.29 | 15.72 ± 19.16 | 59.59 ± 42.60 | 116.91 ± 34.94 |
| DEEDS | 0.49 ± 0.26 | 8.63 ± 16.16 | 40.15 ± 32.11 | 3.73 ± 0.77 |
Note that ANTS-CC, ANTS-QUICK-MI, and NIFTYREG used two CPU cores for each registration process. The mean DSC across four large organs (liver, spleen, kidneys) is 0.19, 0.31, 0.43, 0.48, 0.55, and 0.70 for FSL, ANTS-CC, ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS, respectively.
D. Evaluation Metrics
DSC was used to evaluate the volumetric overlap between the estimated segmentation and the true segmentation. Briefly, consider A as the segmentation volume, B the ground truth volume, and |·| the L1 norm operation,
| (1) |
Surface error criteria characterize how far the surfaces of the estimated segmentation and the true segmentation are from each other. Vertices were collected from the surfaces of both the segmentation and the ground truth, based on which distances between the sets of vertices are measured in terms of their spatial coordinates. Let the vertices on the segmentation and the ground truth surface be X and Y, respectively, and d(·,·) be an indicator of distance measure. Then typically, the MSD error and HD error from the segmentation to the ground truth can be measured as below.
| (2) |
| (3) |
where sup represents the supremum, inf the infimum, avg the average. Symmetric surface differences were used in this study as they better capture errors between potentially rough surfaces, i.e.,
| (4) |
| (5) |
All metrics were evaluated in an organ-wise manner between the registered labels (estimated segmentation) and the manual labels (ground truth).
E. Statistical Analyses
For each pair of methods, permutation tests were performed to examine the statistical significance for the overall DSC and MSD across all organs. Following [3, 19], each test provided an exact p-value calculated as the percentage of N permutations that the absolute mean differences after permutation is larger than the original absolute mean differences between the metrics of two methods on a subset of independent registration pairs, where no overlap is allowed within the images (including both target and source images) associated with the selected registrations, and thus the correlation between registrations with shared scans was prevented. The tests were repeated M times with randomized selection of subsets, and an average p–value was obtained to indicate the significant difference between tested methods. Tests involving FSL or ANTS-CC (or both) selected subsets among the 20 target images and 20 source images (400 registrations) that these two methods had been applied, where 10 independent registration pairs could be obtained for each subset. Tests within the other four methods (i.e., ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS) selected 50 independent registration pairs among 100 images (9900 registrations). In both cases, we let N = 1000 for the number of permutations, and M = 10000 for the number of random selections of subsets.
Indifference-zone ranking considers two metrics as equal when they are within a delta of one another, where the delta characterizes the practical difference [20]. We performed two groups of indifference-zone ranking to examine the practice significances for DSC and MD in an organ-wise manner among the non-rigid registrations of the tested methods. The first group included all methods with 400 registrations, while the second group had ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS evaluated with 9900 registrations. For each organ, let i and j be the row and column index of an L × L matrix (L is 6 and 4 for the first and second group, respectively), Lij was assigned with the values of −1, 0, or 1, for the cases when the evaluation measure for the ith method was at least delta less than, within delta of, or at least delta greater than that of the jth method. The outputs were then averaged across all registrations. The delta value was specified for each organ on each subject based on the surface area of organs. The surface area of an organ label was calculated by summing up the face areas in contact with the background across the foreground voxels; it was adjusted by a constant coefficient to yield a delta value that represents the practical difference of the evaluation metric. For DSC, we used a mean delta value of 0.05 for DSC across all organs and 7 mm for MSD. A higher indifference-zone score represents a better DSC performance, while a lower score was favorable for the MSD performance.
III. Results
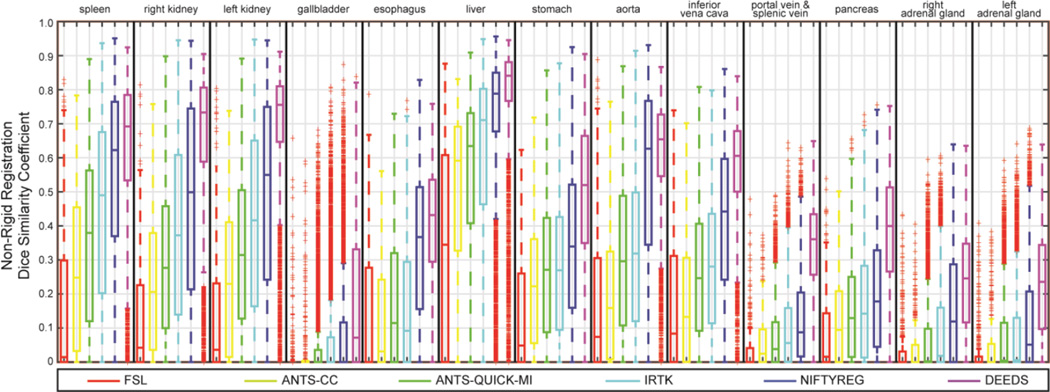
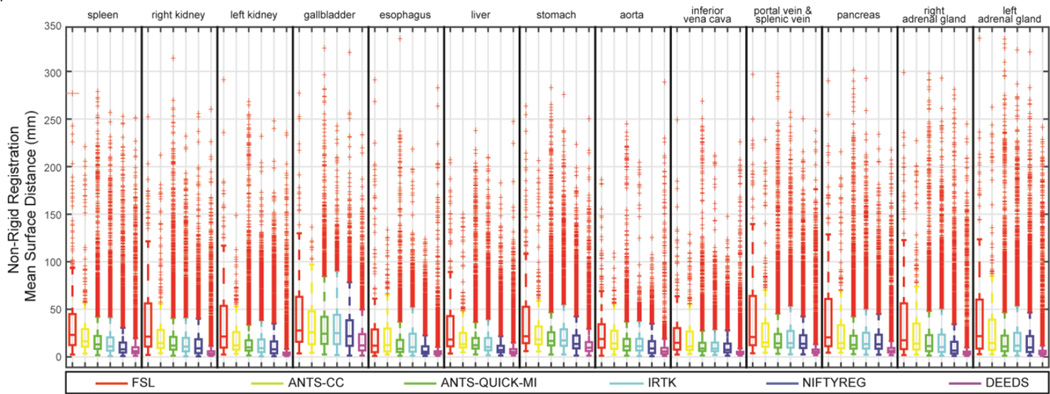
Registrations were successful in terms of software error codes except for 6 out of 9900 ANTS-QUICK-MI failed without producing output. The evaluated metrics of the non-rigid outputs on each organ were illustrated in Fig.s 3, 4, and 5 in terms of DSC, MSD, and HD, respectively. Note that the affine outputs were presented in the supplementary material.
Fig. 3.
Boxplot of DSC values on 13 organs for the non-rigid outputs of six registration methods.
Fig. 4.
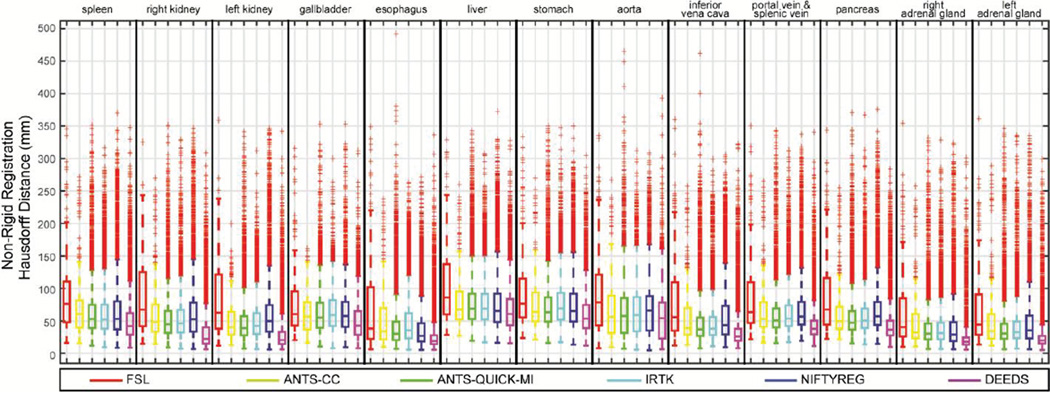
Boxplot of MSD values on 13 organs for the non-rigid outputs of six registration methods.
Fig. 5.
Boxplot of HD values on 13 organs for the non-rigid outputs of six registration methods.
Regarding the overall performance across all registration methods, over half of the registrations have the DSC values lower than 0.7 for the majority of the organs. The MSD and HD boxplots clearly illustrate the overbearing amount of outliers with up to 500 mm.
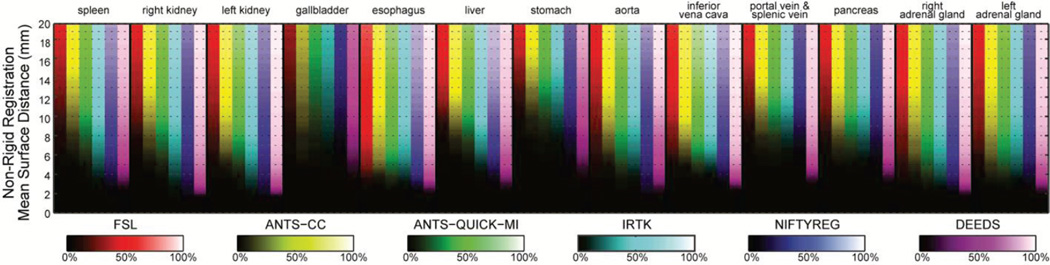
When comparing registration methods with each other, DEEDS presented the best overall DSC of non-rigid registration across all organs (Fig. 3). For non-rigid registration, NIFTYREG presented slightly higher median DSC over ANTS-QUICK-MI and IRTK, while FSL and ANTS-CC demonstrated overall inferiority compared to the other three methods. On the MSD and HD boxplots, the dominance of any registration tool is not visually apparent given the substantial outliers for all methods. To evaluate the results that were not catastrophic failures (i.e., those that could meaningfully contribute to a multi-atlas approach [1, 2]), Fig. 6 presents MSD results in the form of cumulative percentage, where a higher portion of samples below a certain MSD upper bound was more favorable, where DEEDS yields the highest percentage of registrations with lower MSD. Table I presents the overall performance of DSC, MSD, and HD averaged across all organs for all tested methods on the subset of 400 registrations, while Table II shows the metrics for ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS on all 9900 registrations; DEEDS demonstrates the best overall performance in both cases. The computation time was also collected in Table I, where ANTS-QUICK-MI and NIFTYREG could complete in approximately 1h and 2h, respectively using 2 CPU cores, and DEEDS had the lowest computational time (< 4 min).
Fig. 6.
Brightness-coded cumulative percentages based on MSD values on 13 organs for the non-rigid outputs of six registration methods. Six methods were represented in 6 difference colors. Each column indicates a cumulative curve for the associated organ with the underlying registration method; it demonstrated the percentage of included registration outputs along the increase of the MSD upper bound with its brightness transition from bottom to top. A column with quicker transition from dark to bright indicates more registration outputs with small MSD.
TABLE II.
Metrics on 9900 registrations for four registration methods (mean ± std)
| Method | DSC | MSD (mm) | HD (mm) |
|---|---|---|---|
| ANTS-QUICK-MI | 0.23 ± 0.23 | 20.68 ± 26.14 | 57.44 ± 39.85 |
| IRTK | 0.26 ± 0.26 | 20.36 ± 24.01 | 58.71 ± 37.33 |
| NIFTYREG | 0.35 ± 0.29 | 16.98 ± 21.58 | 62.52 ± 44.29 |
| DEEDS | 0.47 ± 0.26 | 9.79 ± 17.44 | 43.18 ± 35.08 |
Note that the mean DSC across 4 large organs (liver, spleen, kidneys) is 0.38, 0.46, 0.55, and 0.68 for ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS, respectively.
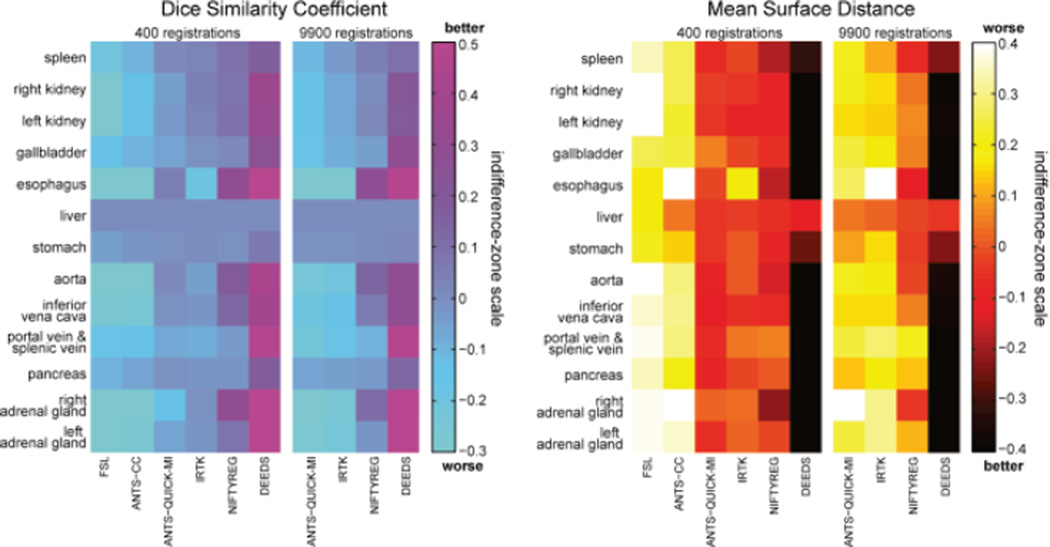
The permutation tests found that the superiority of DEEDS in non-rigid registration was significantly better (p < 0.05) than all other methods in DSC, and the majority of the others in MSD (Tables III and IV). The indifference-zone ranking also indicated that DEEDS yielded the best registration performance in an organ-wise manner. NIFTYREG presented the second best results, closely followed by ANTS-QUICK-MI and IRTK, while FSL and ANTS-CC were last (Fig. 7).
TABLE III.
Averaged p-values of permutation tests between 6 methods performed on 400 registrations
| Method | FSL | ANTS-CC | ANTS-QUICK_MI | IRTK | NIFTYREG | DEEDS |
|---|---|---|---|---|---|---|
| FSL | 0.340 | 0.026 | 0.057 | 0.014 | 0.002 | |
| ANTS-CC | 0.371 | 0.098 | 0.052 | 0.016 | 0.001 | |
| ANTS-QUICK-MI | 0.077 | 0.266 | 0.515 | 0.236 | 0.010 | |
| IRTK | 0.216 | 0.183 | 0.524 | 0.249 | 0.003 | |
| NIFTYREG | 0.144 | 0.169 | 0.517 | 0.465 | 0.019 | |
| DEEDS | 0.032 | 0.030 | 0.230 | 0.044 | 0.106 |
Note the entries in the upper triangular part represent p-values tested on DSC, while those in the lower triangular part were tested on MSD. The shaded entry indicates significant difference (p < 0.05) between the correspondent methods of the row and column.
TABLE IV.
Averaged p-values of permutation tests between 4 methods performed on 9900 registrations
| Method | ANTS-QUICK-MI | IRTK | NIFTYREG | DEEDS |
|---|---|---|---|---|
| ANTS-QUICK-MI | 0.174 | 0.000 | 0.000 | |
| IRTK | 0.501 | 0.002 | 0.000 | |
| NIFTYREG | 0.255 | 0.272 | 0.000 | |
| DEEDS | 0.024 | 0.019 | 0.071 |
Note the entries in the upper triangular part represent p-values tested on DSC, while those in the lower triangular part were tested on MSD. The shaded entry indicates significant difference (p < 0.05) between the correspondent methods of the row and column.
Fig. 7.
Indifference-zone map for DSC and MSD. For both metrics, the indifference-zone ranking was applied on 400 registrations for all six methods, and 9900 registrations for ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS. A higher value for the DSC indifference-zone map indicates better performance, while a lower value is more favorable for MSD.
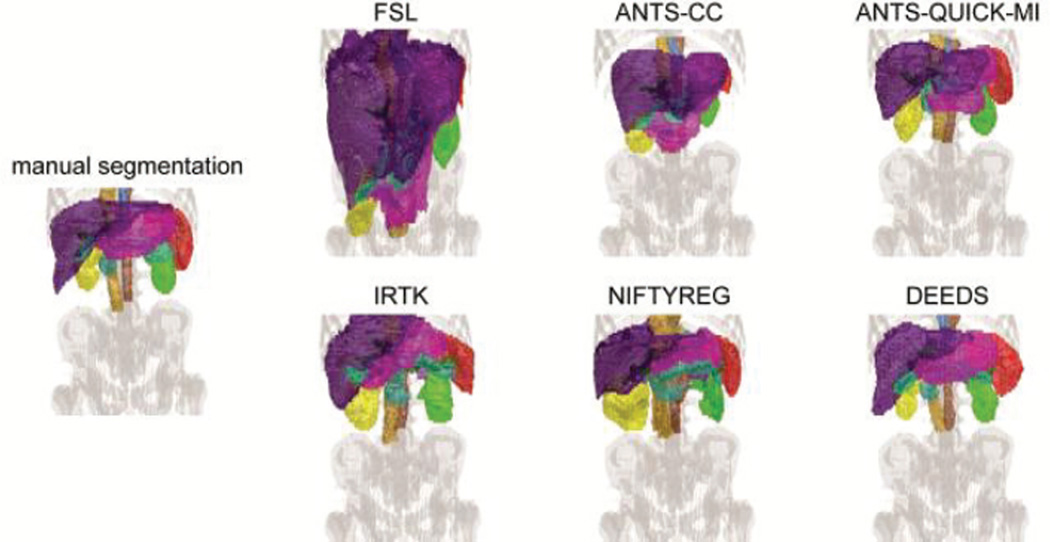
One registration sample with median overall DSC performance is shown in Fig. 8. The volumetric rendering of the registered labels from 6 methods was demonstrated and compared with the manual labels of the target scan to provide a qualitative sense of the registration quality. While large misalignment from all methods can be identified without much effort, ANTS-QUICK-MI, IRTK, NIFTYREG, and DEEDS have the majority of the registered organs located at the close positions, and scaled in similar sizes with respect to those in the target image. Visually, the organ shapes of the target are best captured by DEEDS.
Fig. 8.
Volumetric rendering on a single subject with median overall DSC performance. The organ color scheme follows that in Fig. 1.
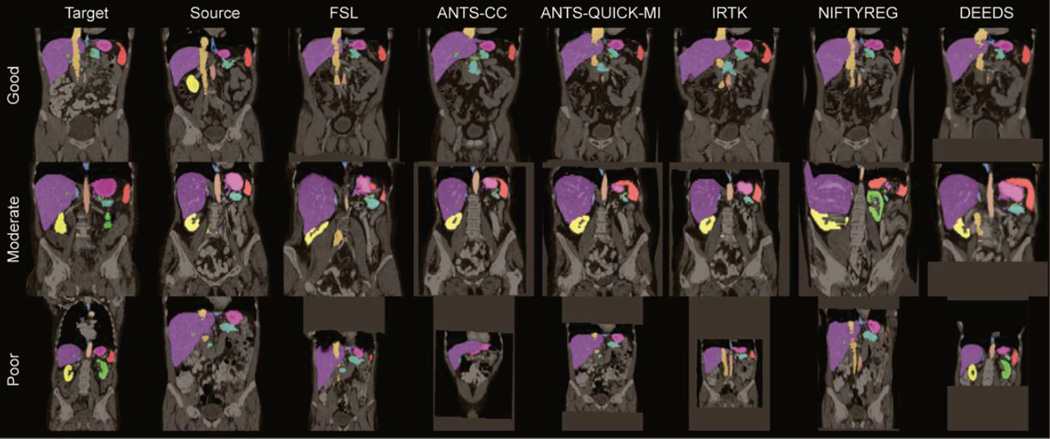
Three pairs of registrations were selected with the top 5% (good), ± 5% around median (moderate), and bottom 5 % (poor) overall DSC performance, respectively. Registration results on these cases are illustrated in Fig. 9, where a coronal slice for each case is selected for the target, source, and all registered images. Based on the overlaid organ labels and the underlying images, DEEDS presents the overall best registrations. Meanwhile, the registration performance is substantially affected by the similairities between the target and source images including the image FOVs, patient body sizes, organ shapes, and secondary organ complexities (intestines and vessels). On the other hand, we found there are still many catastrophic failures remaining after removing the subsets with large mismatches of those variables (results not shown here for brevity). Many other underlying features can have great impact on the registrations, and require further investigation.
Fig. 9.
Illustrations of six registration methods on three registration pairs with good, moderate, and poor performances.
IV. Discussion
In this study, we analyzed 6 registration methods from 5 different general-purpose image registration toolkits and applied them to abdominal CT scans. Evaluating the volumetric overlap and surface errors on the registered labels on 13 organs of interests showed that the current registration tools were generally far from ideal, where (1) median accuracy was below 0.7 for the majority of organs, and (2) massive outliers indicating catastrophic registration failures were observed. Registration performance is found to be negatively affected by the dissimilarities between the target and source images including the image FOVs, patient body sizes, and organ shape, where fundamental body misalignments were observed (Fig. 9). Additional challenges come from the implicit discontinuity within the abdomen given the secondary structures (e.g., fat, muscles, bones, intestines in this study). Their variations caused large deformations between different organs of interest so that an affine registration can hardly align all organs at the same time. In addition, their extensive presence and large coverage across the abdomen could mislead the registration algorithms and generate undesirable deformation; for the same reason, small organs could be registered to the secondary structures or other large organs.
We note that the registration results in this study could be biased towards the tested datasets. First, all scans were contrast enhanced, where organs could be more distinguishable from muscle and fat tissue. Registrations between non-contrasted scans may demonstrate additional challenges not shown with our datasets. Second, the population of patients had a greater chance of sharing specific abnormalities, e.g., enlarged spleen and liver, defected abdominal wall. In fact, these patients could also have multiple other diseases, have been treated with different surgical procedures, and demonstrate various other abnormalities (atrophied kidney, missing gallbladder). We consider the registration evaluation on our datasets to be biased towards challenging cases. Datasets among healthy subjects may yield better registration outcomes. On the other hand, contrasted CT scans on patients with all sorts of abdominal diseases are the most common image format acquired in traditional clinical trials. We consider the registration evaluation performed in this study valuable for translational research.
Among the tested registration methods in the presented parameter settings, DEEDS provided the best overall performance, with median DSC, MSD, and HD as 0.49, 4.93 mm and 31.72 mm, respectively for all organs. The DSC metric is in favor of large structures; small disagreement in small structures can result in large decrease in DSC in the context of [1, 2, 21]. We can consider the reasonable DSC values for large (liver, spleen, kidneys), medium (pancreas, stomach, aorta, inferior vena cava), and small (gallbladder, esophagus, portal and splenic vein, adrenal glands) organs to be 0.95, 0.85, and 0.6 respectively. Based on these criteria, even the best registration in this study did not provide sufficient accuracy to extract the organs of interest. The massive registration failures further discouraged the direct individual use of the registration tools in clinical applications. However, if combined with pre-processing and post-processing procedures, registrations with this level of overall accuracy are encouraging and could achieve robust results. Essentially, multi-atlas techniques [22] can be used to augment local interpretation of abdominal CT scans (e.g., segmentation) by using multiple atlas-to-target registrations. Great care must be taken to account for the registration outliers, where atlas selection [23–26] and statistical fusion [27–29] are the keys for robust multi-atlas segmentation (MAS). From the perspective of MAS, registration is the bottleneck, especially in the abdomen; a better registration tool can yield better segmentation performance.
Based on the results shown in this study, many opportunities are open for future investigation and development for a registration tool tailored for abdomen.
First, although the presented registration configurations were approved by all the developers of the tested registration methods, further optimization could be possible, e.g., in terms of levels of the multi-resolution strategy, thresholds of intensity range, use of block matching strategy in affine initialization, regularization on deformation, and etc. Across the tested registrations, a good combination of the similarity metrics (mutual information, cross-correlation, sum of squared distance, and Hamming distances of the self-similarity context) and transformation models (B-splines and diffeomorphism) has been covered for deformation, while registrations using other transformation models (e.g., demons [30], optical flow [31]) could be evaluated by experts with these approaches in continuing analysis via the newly released public dataset.
Second, contributions in abdominal segmentation also provide some hints toward the potential development of abdominal registration algorithms. While using existing registration tools for segmentation, many efforts have been focused on standardizing the abdomen space. Wolz et al. [1] constrained a FOV with 25 cm along the cranial-caudal axis before registration. Linguraru et al. [21] initialized the registration by aligning a single landmark (xiphoid process). Okada et al. [32] and Zhou et al. [33] normalized the abdominal space using pre-segmented diaphragm and rib cage. Recent efforts on organ localizing [34] and organ hierarchical modeling [35] provide the options to minimize the impact of the substantial registration errors. Piece-wise registrations/segmentations have been demonstrated with better performance than their body-wise counterparts [36, 37]. These pre-processing techniques provide extra features other than intensity-based similarity metrics, and can potentially benefit registrations for capturing the most desirable organ deformation.
Third, we see a new direction in fundamental design for the registration method towards the challenging problems in the abdomen. DEEDS yields the best performance in this study, and it is different from other methods mainly by using discrete optimization. Instead of relying on differentiable similarity metric in traditional continuous optimization, DEEDS subdivides the image domain into non-overlapping cubic blocks, and calculates the displacement for each block followed by displacement regularization between blocks. This type of discrete design can capture a large range of potential deformations, and thus coped well with the discontinuous pattern between structures of interest in the abdomen. Further exploration in the discrete optimization can be expected to benefit the abdominal registrations.
Last, we consider that a structured challenge regarding registration in the abdomen using the presented datasets will further boost the development of abdomen-specific and/or general registration algorithms. We have already set up the infrastructure on Sage Synapse as a publically available challenge for researchers to evaluate their registration and segmentation algorithms (https://www.synapse.org/#!Synapse:syn3193805/wiki/89480). Note the challenge page was originally established for a MICCAI 2015 challenge, while all functionalities remain active. More comprehensive benchmarks to evaluate the efficacy of capturing the abdominal organs will be required to solidify the impact of this potential challenge.
V. Conclusion
This manuscript presents the current state of the art for registration performance at 13 abdominal organs on CT scans by evaluating six academically popular registration methods without extensive optimizations. In this study, we (1) recommend a best registration method to the registration users for their abdomen-related applications, and (2) suggest future directions for registration developers towards more robust and accurate registration algorithms in the abdomen. Specifically, DEEDS is currently the best choice for registration users to perform abdominal organ segmentation. Registration developers can focus on the perspectives of discrete optimization, non-intensity-based feature derivation, and parameter configurations.
Supplementary Material
Acknowledgments
We thank Brian Avants and Jesper Andersson for their discussion and suggestions to this work. This research was supported by NIH 1R03EB012461, NIH 2R01EB006136, NIH R01EB006193, ViSE/VICTR VR3029, NIH UL1 RR024975-01, NIH UL1 TR000445-06, NIH P30 CA068485, and AUR GE Radiology Research Academic Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN.
Contributor Information
Zhoubing Xu, Email: zhoubing.xu@vanderbilt.edu, Department of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN 37235 USA.
Christopher P. Lee, Email: christopher.p.lee@vanderbilt.edu, Department of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN 37235 USA.
Mattias P. Heinrich, Email: heinrich@imi.uni-luebeck.de, Institute of Medical Informatics, Universität zu Lübeck, Lübeck 23562, Germany.
Marc Modat, Email: m.modat@ucl.ac.uk, Translational Imaging Group, Centre for Medical Image Computing, University College London, UK.
Daniel Rueckert, Email: d.rueckert@imperial.ac.uk, Biomedical Image Analysis Group, Imperial College London, UK.
Sebastien Ourselin, Email: s.ourselin@ucl.ac.uk, Translational Imaging Group, Centre for Medical Image Computing, University College London, UK.
Richard G. Abramson, Email: richard.abramson@vanderbilt.edu, Department of Radiology and Radiological Science, Vanderbilt University, Nashville, TN 37235 USA.
Bennett A. Landman, Email: bennett.landman@vanderbilt.edu, Department of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN 37235 USA.
References
- 1.Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. Medical Imaging, IEEE Transactions on. 2013;32:1723–1730. doi: 10.1109/TMI.2013.2265805. [DOI] [PubMed] [Google Scholar]
- 2.Xu Z, Burke RP, Lee CP, Baucom RB, Poulose BK, Abramson RG, Landman BA. Efficient multi-atlas abdominal segmentation on clinically acquired CT with SIMPLE context learning. Medical image analysis. 2015 doi: 10.1016/j.media.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-C, Christensen GE, Collins DL, Gee J, Hellier P. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West J, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CR, Jr, Kessler RM, Maciunas RJ, Barillot C, Lemoine D, Collignon A. Comparison and evaluation of retrospective intermodality brain image registration techniques. Journal of computer assisted tomography. 1997;21:554–568. doi: 10.1097/00004728-199707000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Fitzpatrick JM, West JB. The distribution of target registration error in rigid-body point-based registration. Medical Imaging, IEEE Transactions on. 2001;20:917–927. doi: 10.1109/42.952729. [DOI] [PubMed] [Google Scholar]
- 6.Murphy K, Van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Han X, Heinrich MP, Schnabel JA, Jenkinson M, Lorenz C, Modat M, McClelland JR, Ourselin S, Muenzing SEA, Viergever MA, de Nigris D, Collins DL, Arbel T, Peroni M, Li R, Sharp GC, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Song G, Tustison N, Avants B, Gee JC, Staring M, Klein S, Stoel BC, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim JPW. Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. Medical Imaging, IEEE Transactions on. 2011;30:1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- 7.del Toro OAJ, Goksel O, Menze B, Müller H, Langs G, Weber M-A, Eggel I, Gruenberg K, Holzer M, Kotsios-Kontokotsios G. VISCERAL–VISual Concept Extraction challenge in RAdioLogy: ISBI 2014 challenge organization. Proceedings of the VISCERAL Challenge at ISBI. 2014:6–15. [Google Scholar]
- 8.Lee CP, Xu Z, Burke RP, Baucom R, Poulose BK, Abramson RG, Landman BA. Evaluation of five image registration tools for abdominal CT. SPIE Medical Imaging. 2015:94131N–94131N-7. doi: 10.1117/12.2081045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. Medical Imaging, IEEE Transactions on. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 11.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine. 2010;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinrich MP, Jenkinson M, Brady M, Schnabel J. MRF-based deformable registration and ventilation estimation of lung CT. Medical Imaging, IEEE Transactions on. 2013;32:1239–1248. doi: 10.1109/TMI.2013.2246577. [DOI] [PubMed] [Google Scholar]
- 14.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 15.Rorden C, Brett M. Stereotaxic display of brain lesions. Behavioural neurology. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu A, Ohno R, Ikegami T, Kobatake H, Nawano S, Smutek D. Segmentation of multiple organs in non-contrast 3D abdominal CT images. International Journal of Computer Assisted Radiology and Surgery. 2007;2:135–142. [Google Scholar]
- 17.McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, Trus BL. Medical image processing, analysis and visualization in clinical research. Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. 2001:381–386. [Google Scholar]
- 18.Heinrich MP, Jenkinson M, Papież BW, Brady M, Schnabel JA. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer; 2013. Towards realtime multimodal fusion for image-guided interventions using self-similarities; pp. 187–194. [DOI] [PubMed] [Google Scholar]
- 19.Menke J, Martinez TR. Using permutations instead of student's t distribution for p-values in paired-difference algorithm comparisons. Neural Networks, 2004. Proceedings. 2004 IEEE International Joint Conference on. 2004:1331–1335. [Google Scholar]
- 20.Bechhofer RE. A single-sample multiple decision procedure for ranking means of normal populations with known variances. The Annals of Mathematical Statistics. 1954:16–39. [Google Scholar]
- 21.Linguraru MG, Sandberg JK, Li Z, Shah F, Summers RM. Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Medical physics. 2010;37:771–783. doi: 10.1118/1.3284530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iglesias JE, Sabuncu MR. Multi-atlas segmentation of biomedical images: A survey. Medical image analysis. 2015;24:205–219. doi: 10.1016/j.media.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z, Asman AJ, Shanahan PL, Abramson RG, Landman BA. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014. Springer; 2014. SIMPLE IS a Good Idea (and Better with Context Learning) pp. 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langerak TR, Van Der Heide U, Kotte AN, Viergever M, Van Vulpen M, Pluim JP. Label fusion in atlas-based segmentation using a selective and iterative method for performance level estimation (SIMPLE) Medical Imaging, IEEE Transactions on. 2010;29:2000–2008. doi: 10.1109/TMI.2010.2057442. [DOI] [PubMed] [Google Scholar]
- 25.Cardoso MJ, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, Fox NC, Ourselin S A. s. D. N. Initiative. STEPS: Similarity and Truth Estimation for Propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Medical image analysis. 2013;17:671–684. doi: 10.1016/j.media.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46:726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Rohlfing T, Brandt R, Menzel R, Maurer CR. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Sabuncu MR, Yeo BT, Van Leemput K, Fischl B, Golland P. A generative model for image segmentation based on label fusion. Medical Imaging, IEEE Transactions on. 2010;29:1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. Medical Imaging, IEEE Transactions on. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. Neuroimage. 2009;45:S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 31.Urschler M, Werlberger M, Scheurer E, Bischof H. Robust optical flow based deformable registration of thoracic CT images. Medical Image Analysis for the Clinic: A Grand Challenge. 2010:195–204. [Google Scholar]
- 32.Okada T, Linguraru MG, Yoshida Y, Hori M, Summers RM, Chen Y-W, Tomiyama N, Sato Y. Abdominal Imaging. Computational and Clinical Applications. Springer; 2012. Abdominal multi-organ segmentation of CT images based on hierarchical spatial modeling of organ interrelations; pp. 173–180. [Google Scholar]
- 33.Zhou X, Kitagawa T, Hara T, Fujita H, Zhang X, Yokoyama R, Kondo H, Kanematsu M, Hoshi H. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2006. Springer; 2006. Constructing a probabilistic model for automated liver region segmentation using non-contrast X-ray torso CT images; pp. 856–863. [DOI] [PubMed] [Google Scholar]
- 34.Criminisi A, Robertson D, Konukoglu E, Shotton J, Pathak S, White S, Siddiqui K. Regression forests for efficient anatomy detection and localization in computed tomography scans. Medical image analysis. 2013;17:1293–1303. doi: 10.1016/j.media.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer; 2013. Abdominal multi-organ ct segmentation using organ correlation graph and prediction-based shape and location priors; pp. 275–282. [DOI] [PubMed] [Google Scholar]
- 36.Xu Z, Gertz AL, Baucom RB, Poulose BK, Abramson RG, Landman BA. Improving Multi-Atlas Abdominal Organ Segmentation via Organ-Wise Normalization. MICCAI 2015 Workshop and Challenge: Multi-Atlas Labeling Beyond Cranial Vault. 2015 (in press) [Google Scholar]
- 37.del Toro OAJ, Müller H. Medical Computer Vision. Large Data in Medical Imaging. Springer; 2014. Multi-structure atlas-based segmentation using anatomical regions of interest; pp. 217–221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.