Abstract
The M2 protein is a small proton channel found in the influenza A virus that is necessary for viral replication. The M2 channel is the target of a class of drugs called the adamantanes, which block the channel pore and prevent the virus from replicating. In recent decades mutations have arisen in M2 that prevent the adamantanes from binding to the channel pore, with the most prevalent of these mutations being S31N. Here we report the first crystal structure of the S31N mutant crystallized using lipidic cubic phase crystallization techniques and solved to 1.59 Å resolution. The Asn31 residues point directly into the center of the channel pore and form a hydrogen‐bonded network that disrupts the drug‐binding site. Ordered waters in the channel pore form a continuous hydrogen bonding network from Gly34 to His37.
Keywords: influenza M2, S31N, proton channel, crystal structure
Short abstract
PDB Code(s): 5C02
Introduction
The influenza A matrix 2 (M2) protein consists of a 96‐residue monomer that forms a homotetrameric channel. M2 is an amazingly small multifunctional protein, with different functions localized sequentially along the amino acid chain. The N‐terminal 22 residues, which project out of the virus, contribute to budding in influenza A virus, but are missing in influenza B virus. The transmembrane helix spanning residues 22–46 forms a tetramer that retains channel‐forming function, selectivity, and sensitivity to drug.1 A region C‐terminal to the TM helix forms a peripheral membrane‐binding helix that stabilizes envelope excision during membrane budding by stabilizing negative Gaussian curvature,2, 3 and the C‐terminal region also interacts with the M1 matrix protein.4 The M2 protein is a drug target of two FDA approved anti‐flu drugs, amantadine and rimantadine, which bind in the pore of the M2 channel.5, 6
The M2 protein mediates the equilibration of pH between the endosomal compartment and the viral interior at a critical point of the viral reproduction cycle. In some strains of influenza A virus, M2 also serves to delay acidification of the interior of the Golgi apparatus to prevent HA from prematurely undergoing an acid‐mediated conformational change.7, 8 Amantadine and rimantadine exert their antiviral effect by binding to the pore of the M2 tetramer and blocking proton conduction.5, 6 In recent years amantadine resistance from the S31N mutant has become so widespread that it has curtailed the use of this class of drugs.9
Despite the importance of the S31N mutation, almost all structural work has focused on variants of M2 with Ser at position 31. Chou and coworkers have built homology models for the S31N variant of M2 based on NMR restraints.10 Although dynamics prevented determination of the location of Asn31, they hypothesized that its side chain projects out towards the lipid tails. More recently, we used small molecule drugs that bind to the S31N mutant to lock it into a conformation that was more amenable to solution NMR structure determination, allowing the determination of the structure at moderate resolution.11 Unfortunately, although the overall structure was well defined, conformational exchange of the Asn31 side chain made it difficult to determine its orientation; we attributed this conformational exchange to the fact that the drug is intrinsically asymmetric while the protein is four‐fold symmetric. Here, we present the first crystal structure of the S31N mutant of influenza M2.
Results and Discussion
The crystal structure of the S31N mutant was obtained using lipidic cubic phase (LCP) techniques, giving crystals (Supporting Information Fig. S1) at pH 8.0 that diffract to a resolution of 1.59 Å (Table 1) in the same space group as the “wild‐type” Ser31 channel.12 A single monomer forms the asymmetric unit (Supporting Information Fig. S2), which is repeated along a crystallographic four‐fold symmetry axis to create the pore. Monoolein molecules interact with the hydrophobic membrane‐exposed face of each monomer. There are very few contacts between protein molecules in the crystal lattice; the membrane‐exposed face of each M2 monomer interacts with monoolein molecules at its N‐terminus and, to a lesser extent, with other M2 monomers near the C‐terminus [Fig. 1(B)].
Table 1.
Data Collection and Refinement Statistics
| Crystal | |
|---|---|
| Space group | I 4 |
| Cell dimensions | |
| a=b, c (Å) | 28.65, 68.38 |
| α=β=γ (°) | 90 |
| Data collection | |
| Wavelength (Å) | 0.979338 |
| Resolution (Å) | 1.59 (1.64–1.59) |
| No. reflections | 11589 (613) |
| Redundancy | 3.2 (2.2) |
| I/σI | 17.2 (3.3) |
| Completeness (%) | 98.9 (94.3) |
| R merge | 0.041 (0.243) |
| Refinement | |
| Resolution (Å) | 1.59 |
| R work | 0.1571 |
| R free | 0.1949 |
| Number of nonhydrogen atoms | 276 |
| Protein | 194 |
| Ligands | 58 |
| Water | 21 |
| B factors | |
| Protein | 21.90 |
| Ligands | 32.60 |
| Solvent | 36.30 |
| RMS deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.71 |
| Ramachandran favored | 100% |
| Ramachandran outliers | 0% |
Figure 1.
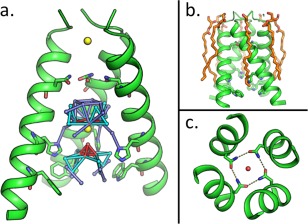
(a) The M2 pore; the M2 tetramer is shown with the front monomer removed. Full occupancy waters are shown as red spheres, partial occupancy waters are shown in cyan and dark blue, and chloride ions are shown in yellow. Hydrogen bonds between pore waters are shown as colored sticks (red, cyan, and dark blue). Asn31, His37 and Trp41 are shown as sticks. (b) The M2 tetramer (green) is surrounded by monoolein molecules (orange) within the crystal lattice. (c) Top‐down view of the channel pore; Asn31 residues form a network of hydrogen bonds (dashed lines). A single water (red sphere) is located in the middle of these residues.
Solvent network and partially occupied waters
As in other NMR and crystal structures of the M2 channel,6, 12 a continuous pore runs through the membrane‐spanning region of the structure, interrupted only at Val27. In the S31N mutant, the polar Asn side chain points towards the center of the channel; each carboxamide nitrogen of the Asn residue forms a hydrogen bond with the carbonyl of a neighboring Asn31 side chain. A water molecule was observed at the center of the Asn31 tetrad, bridging the cyclic hydrogen‐bonding arrangement [Fig. 1(C)]. This water might serve a special role in transmitting protons into the channel, although dynamic motions would be required to diffuse past Val27, as described previously.13, 14, 15 Four Å below this water is a cluster of waters that form a continuous water wire leading to the proton‐shuttling residue His37 [Fig. 1(A)]. Most of these waters exist at half occupancy. The presence of two partial‐occupancy water networks in the channel could result from averaging of the water densities across the four‐fold symmetry axis at the center of the channel. A strong spherical density was observed along the four‐fold axis among the water clusters. Of the ions in the buffer, only chloride fit well into this density. A single chloride at a similar position along the symmetry axis has also been proposed to play a role in stabilizing the M2 tetramer.16
Resistance to adamantane drugs and inhibitor binding
The mechanism by which the S31N mutation confers resistance to amantadine can be seen upon overlaying the newly solved LCP structure with the previously solved crystal structure of wild‐type M2 in the presence of amantadine [Fig. 2(A)]. Hydrogen bonding between symmetrically repeated Asn31 residues within the homotetrameric channel narrows the portion of the channel to which the adamantine drugs bind, providing steric hindrance that reduces the drugs' affinity to that site. Substitution of Ser31 with Asn also adds an extra hydrophilic atom to a site that interacts with the hydrophobic adamantine cage.
Figure 2.
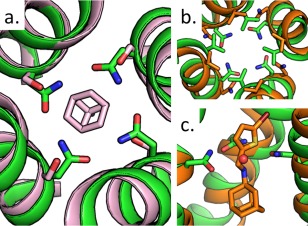
(a) Overlay of S31N mutant structure 5C02 (green) onto the drug‐bound structure 3C9J (pink) with Asn31, Ser31, and amantadine shown as sticks. (b) Top‐down view of 5C02 aligned to inhibitor‐bound solution NMR S31N structure 2MUV (orange), Val27 and Asn31 shown as sticks. (c) Side view of N‐[(5‐bromothiophen‐2‐yl)methyl]adamantan‐1‐amine inhibitor from 2MUV in channel pore, with water from 5C02 shown as a red sphere.
Figure 2(B,C) compare the recently solved structure to the previously solved inhibitor‐bound solution NMR structure of S31N.17 Though the conformations of the asparagine residues in the NMR structure could not be resolved, the position of the inhibitor (compound 11, N‐[(5‐bromothiophen‐2‐yl)methyl]adamantan‐1‐amine) within the channel is well defined by NOEs. The Asn31 residues of the crystal structure form a hydrogen bonded ring that is too tight to accommodate inhibitors, so it is likely that a conformational shift of the protein backbone is necessary for inhibitor binding. The location of the single water at the center of the S31N crystal structure approximately maps to the position of the nitrogen in the bound inhibitor molecule. This supports the suggestion that the ammonium group of M2 inhibitors occupy the same positions in the channel as ordered waters in the apo structure. The availability of a high‐resolution structure of the S31N should also be quite helpful for design of drugs that target drug‐resistant mutants bearing this mutation.
Materials and Methods
The peptide corresponding to the M2(22–46) S31N mutant of A/Udorn/307/1972 was prepared and crystallized and the structure solved as described in the Supporting Information.
Accession numbers
The Protein Data Bank accession number for the 1.59 Å structure of AM2(22–46) S31N is 5C02.
Supporting information
Supporting Information
Acknowledgments
We thank Magdalena Korczynska and Nathan Joh for helpful discussions concerning data collection and structure determination. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines. Use of the Advanced Photon Source, operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported under Contract No. DE‐AC02‐06CH11357.
References
- 1. Ma CL, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, Magavern E, Jing XH, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH (2009) Identification of the functional core of the influenza A virus A/M2 proton‐selective ion channel. Proc Natl Acad Sci USA 106:12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA (2010) Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J Virol 84:5078–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GCL (2013) Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J Am Chem Soc 135:13710–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen BJ, Leser GP, Jackson D, Lamb RA (2008) The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol 82:10059–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cady SD, Schmidt‐Rohr K, Wang J, Soto CS, DeGrado WF, Hong M (2010) Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF (2008) Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451:596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciampor F, Bayley PM, Nermut MV, Hirst EMA, Sugrue RJ, Hay AJ (1992) Evidence that the amantadine‐induced, M2‐mediated conversion of influenza‐A virus hemagglutinin to the low pH conformation occurs in an acidic transgolgi compartment. Virology 188:14–24. [DOI] [PubMed] [Google Scholar]
- 8. Grambas S, Hay AJ (1992) Maturation of influenza‐A virus hemagglutinin—Estimates of the pH encountered during transport and its regulation by the M2 protein. Virology 190:11–18. [DOI] [PubMed] [Google Scholar]
- 9. Dong G, Peng C, Luo J, Wang C, Han L, Wu B, Ji G, He H (2015) Adamantane‐resistant influenza A viruses in the world (1902–2013): Frequency and distribution of M2 gene mutations. PLoS One 10:e0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pielak RM, Schnell JR, Chou JJ (2009) Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci USA 106:7379–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Wu Y, Ma C, Fiorin G, Wang J, Pinto LH, Lamb RA, Klein ML, Degrado WF (2013) Structure and inhibition of the drug‐resistant S31N mutant of the M2 ion channel of influenza A virus. Proc Natl Acad Sci USA 110:1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomaston JL, Alfonso‐Prieto M, Woldeyes RA, Fraser JS, Klein ML, Fiorin G, DeGrado WF (2015) High‐resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc Natl Acad Sci USA 112:14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khurana E, Peraro MD, DeVane R, Vemparala S, DeGrado WF, Klein ML (2009) Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc Natl Acad Sci 106:1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yi M, Cross TA, Zhou HX (2008) A secondary gate as a mechanism for inhibition of the M2 proton channel by amantadine. J Phys Chem B 112:7977–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang R, Li H, Swanson JM, Voth GA (2014) Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc Natl Acad Sci USA 111:9396–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei C, Pohorille A (2013) Activation and proton transport mechanism in influenza A M2 channel. Biophys J 105:2036–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Y, Canturk B, Jo H, Ma C, Gianti E, Klein ML, Pinto LH, Lamb RA, Fiorin G, Wang J, DeGrado WF (2014) Flipping in the pore: Discovery of dual inhibitors that bind in different orientations to the wild‐type versus the amantadine‐resistant S31N mutant of the influenza A virus M2 proton channel. J Am Chem Soc 136:17987–17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


