Expressologs identify functional orthologs and will be a powerful tool in future orthology assignment.
Abstract
The identification of functionally equivalent, orthologous genes (functional orthologs) across genomes is necessary for accurate transfer of experimental knowledge from well-characterized organisms to others. This frequently relies on automated, coding sequence-based approaches such as OrthoMCL, Inparanoid, and KOG, which usually work well for one-to-one homologous states. However, this strategy does not reliably work for plants due to the occurrence of extensive gene/genome duplication. Frequently, for one query gene, multiple orthologous genes are predicted in the other genome, and it is not clear a priori from sequence comparison and similarity which one preserves the ancestral function. We have studied 11 organ-dependent and stress-induced gene expression patterns of 286 Arabidopsis lyrata duplicated gene groups and compared them with the respective Arabidopsis (Arabidopsis thaliana) genes to predict putative expressologs and nonexpressologs based on gene expression similarity. Promoter sequence divergence as an additional tool to substantiate functional orthology only partially overlapped with expressolog classification. By cloning eight A. lyrata homologs and complementing them in the respective four Arabidopsis loss-of-function mutants, we experimentally proved that predicted expressologs are indeed functional orthologs, while nonexpressologs or nonfunctionalized orthologs are not. Our study demonstrates that even a small set of gene expression data in addition to sequence homologies are instrumental in the assignment of functional orthologs in the presence of multiple orthologs.
With the rapid advancement of next-generation sequencing technologies, sequencing a transcriptome/genome is highly feasible today within a decent time and at low cost. One important bottleneck for downstream analysis is the annotation, i.e. how accurately we can transfer gene function information from well-characterized reference genomes and model plants to these newly sequenced genomes and/or crop plants. The major reason for this uncertainty is the occurrence of multiple homologous sequences as a result of gene family expansions and polyploidization events. Orthologs are defined as genes in different species that have emerged as a result of an evolutionary speciation event. Since they are derived from a single gene in the last common ancestor, orthologs frequently share the same function in the newly evolved species. However, gene duplications after the speciation may result in a functional divergence where the ancestral function either is split between such coorthologs or the functions are otherwise transformed (see below). Thus, a multiple orthology situation has arisen in such cases, and the congruence of an evolutionary relationship and a conserved function may have been lost (Remm et al., 2001; Bandyopadhyay et al., 2006). In accordance with these previous studies, we define functional orthologs as those coorthologs that have retained highly similar functions in the two species in such a multiple orthology situation. Therefore, correct identification of functional orthologs is critical for gene annotations by extrapolating functions across species barriers.
Genes that arose following duplication events (whole-genome, segmental, or tandem duplications) are called paralogs. Paralogs that are also orthologs (i.e. which have been formed after a speciation event) are called in-paralogs (or coorthologs), in contrast to out-paralogs, which are derived from a gene duplication before an evolutionary speciation (Remm et al., 2001). The older the duplication event, the higher the chances are that the (in) paralogs will undergo functional divergence. The possible fates of such gene copies and gene groups are (1) nonfunctionalization and pseudogenization, where one ortholog retains the ancestral function while the other ortholog loses the function by acquiring deleterious mutations; and (2) neofunctionalization, where one ortholog acquires a new function by beneficial mutations, whereas the other one retains the original function. In the course of its adaption to a distinct environment, an ortholog in one species also may undergo neofunctionalization, resulting in a species-specific function for this gene. A subsequent duplication of this gene actually results in two in-paralogous copies that differ significantly in their function from the ortholog of the other species. Therefore, we define a group leading to (3) species-specific functionalization, where the whole orthologous group in one species differs from the gene in the other species and does not retain the function (i.e. either all in-paralogs acquire new roles or one ortholog has a new function), while the others lose their original role (nonfunctionalized). An extreme case of such a development is (4) species-specific nonfunctionalization, where all orthologs are pseudogenized and lose their function in one species. A fifth possible fate is (5) subfunctionalization, where the ancestral gene function is split among duplicated copies. Finally, there is (6) genetic redundancy, where all coorthologs still share the ancestral function. However, in an existing, already further evolved species, genetic redundancy and subfunctionalization or neofunctionalization will overlap and depend on the depth of phenotypic analysis. Thus, in most cases, genetic redundancy may not define an independent evolutionary category of genes per se but rather point to a lack of detailed knowledge about divergent functions of these genes.
Several automated cluster methods with varying degrees of selectivity and sensitivity have been developed to assign orthologous relationships across genomes (COG [Tatusov et al., 1997], KOG [Tatusov et al., 2003], OrthoMCL [Li et al., 2003], and Inparanoid [O’Brien et al., 2005]). These sequence-based methods are appropriate to cluster genes with high similarity and possible common ancestry, but they cannot unambiguously identify functional orthologs. One way to track the functionality of the homologous genes after species split is to dissect their expression patterns under a range of spatiotemporal and/or environmental conditions. In yeast, regulatory neofunctionalization events were identified for 43 duplicated gene pairs based on their asymmetric expression profiles, which the sequence data analysis had failed to detect (Tirosh and Barkai, 2007). In plants, most attention was paid to study how polyploidy has fueled the expression divergence of duplicated gene pairs in a single species (Blanc and Wolfe, 2004; Duarte et al., 2006; Ha et al., 2007; Throude et al., 2009; Whittle and Krochko, 2009). With the availability of multiple genome sequences, cross-species comparisons have been gaining momentum. Publicly available gene expression data were used to conduct a cross-species comparison between rice (Oryza sativa) and hybrid poplar (Populus tremula × P. tremuloides) in order to identify transcription factors associated with leaf development (Street et al., 2008). Gene coexpression network analysis was performed on 3,182 DNA microarrays from human, flies, worms, and yeast to identify core biological functions that are evolutionarily conserved across the animal kingdom and yeast (Stuart et al., 2003). A similar study conducted on six evolutionarily divergent species, Saccharomyces cerevisiae, Caenorhabditis elegans, Escherichia coli, Arabidopsis (Arabidopsis thaliana), Drosophila melanogaster, and human, concluded that functionally related genes often are coexpressed across species barriers (Bergmann et al., 2004). Taken together, all these studies indicate that combining sequence and expression data may increase the prediction ability of gene function annotation. However, such coexpression approaches are only possible if large-scale transcriptome analyses are available for both (or more) species to be compared. Thereby, less well-studied and/or newly sequenced species are not (immediately) amenable to such comparisons. Furthermore, none of these studies could experimentally prove the success rate of such prediction at the level of individual gene functions.
An alternative strategy to predict functional orthologs was established by Patel et al. (2012). These authors ranked genes from homology clusters of seven plant species based on extensive gene expression profiles obtained from comparable tissues among these species. The top ranking homolog based on expression pattern similarity was termed the expressolog, which should indicate the functional ortholog. Bandyopadhyay et al. (2006) employed protein-protein interaction data to identify functional orthologs among large S. cerevisiae and D. melanogaster paralogous gene families; in about half of the studied cases, the most conserved functions were not favored by sequence analyses.
The two well-annotated but biologically divergent Brassicaceae species Arabidopsis and Arabidopsis lyrata included in this study diverged approximately 10 million years ago (Hu et al., 2011). Both species differ substantially in several biological traits that are crucial differences in their lifestyle: life cycle (annual Arabidopsis versus perennial A. lyrata), mating system (selfing Arabidopsis versus outcrossing A. lyrata), geographical distribution (continuous distribution of Arabidopsis versus scattered distribution of A. lyrata), and genome size (125-Mb Arabidopsis versus 207-Mb A. lyrata). Furthermore, the Arabidopsis lineage has undergone three rounds of whole-genome duplication followed by differential loss of genes in the different species. Therefore, we aimed at identifying genes that exist as single-copy genes in one species but as multiple copies in the other species and thus are defined as a one-to-many situation. Due to the lack of large-scale expression data for A. lyrata, a coexpression-based approach was not possible. Instead, we studied expression pattern correlation based on a small set of 11 diverse experimental scenarios involving expression in organs (leaf, root, and flower bud) and under different stress conditions. Using such gene expression similarities, we predicted the expressolog for individual gene clusters and thereby candidates of functional orthology in the new, to be analyzed species A. lyrata. Importantly, we could prove that predicted expressologs were indeed functionally equivalent, while nonexpressologs or nonfunctionalized genes were not, using genetic complementation experiments.
RESULTS
OrthoMCL Analysis to Identify One-to-Many Situations between Arabidopsis and A. lyrata
OrthoMCL analysis between Arabidopsis and A. lyrata transcriptomes identified 2,850 gene clusters where either one-to-many or many-to-many situations were present. Of these 2,850 clusters, 613 were one Arabidopsis gene to multiple A. lyrata genes, 366 were one A. lyrata gene to multiple Arabidopsis genes, and 1,871 were multiple Arabidopsis genes to multiple A. lyrata genes. One of the major aims of this study was to experimentally check the efficiency of predicted expressologs in terms of their function. Gene-specific loss-of-function mutants are currently available for Arabidopsis but not for A. lyrata; therefore, we focused our studies on the one Arabidopsis gene to multiple A. lyrata genes group.
Microarray Studies on Arabidopsis and A. lyrata Plants to Dissect Organ-Dependent and Stress-Responsive Expression Patterns of Duplicated Genes
Genome-wide expression analyses were performed on Arabidopsis and A. lyrata plants to determine gene expression similarity or divergence between closely related homologs (Supplemental Table S1). Gene expression data were collected from three different tissues (shoot, root, and flower bud) and from plants subjected to salt, drought, and UV-B light stress regimes to measure gene expression patterns in different organs and for time courses of diverse stress situations. Pearson correlation analysis was performed by analyzing all organ, control, and stress-induced gene expression data obtained from Arabidopsis and A. lyrata. The mean correlation value of all-against-all comparisons between Arabidopsis and A. lyrata transcriptomes was 0.019 (Table I). On the contrary, when syntenic Arabidopsis-A. lyrata orthologous or OrthoMCL one-to-one gene groups were analyzed, much higher correlation values of 0.329 and 0.320 were obtained, which is in accordance with the expectation that the majority of orthologous gene copies still share similar functions. We excluded 263 out of 613 candidates based on probes with a cross-hybridization potential in order to avoid ambiguous measurements due to high sequence similarities of A. lyrata paralogs. An average expression threshold value of 9 (log2 scale) was introduced to exclude such gene groups, where all members are only lowly expressed close to the detection limit of our system in shoots, roots, and flower buds of both Arabidopsis and A. lyrata (see “Materials and Methods”). The final gene set comprised 272 Arabidopsis genes each having two or more A. lyrata homologs.
Table I. Pearson correlation analysis of stress-induced gene coexpression data between different groups of orthologous and nonorthologous genes of Arabidopsis and A. lyrata.
| Gene Groups | Mean | Median |
|---|---|---|
| OrthoMCL all | 0.272 | 0.313 |
| OrthoMCL one-to-one | 0.320 | 0.354 |
| OrthoMCL multiple | 0.262 | 0.300 |
| Syntenic | 0.329 | 0.369 |
| Arabidopsis versus A. lyrata | 0.019 | 0.036 |
Functional Categorization Based on Gene Expression Data and Prediction of Expressologs and Nonexpressologs
Pearson correlation coefficients for each Arabidopsis-A. lyrata pair present within an OrthoMCL gene group were calculated based on microarray data collected from all stress and control experiments. The differential expression patterns of each of the duplicated A. lyrata genes along with the related Arabidopsis copies were measured under salt, drought, and UV-B light stress conditions. The normalized expression levels of these genes were calculated in shoot, root, and flower bud tissues. Based on these analyses, we predicted functionally related expressologs and functionally diverged homologs for each of the 272 OrthoMCL gene groups (Supplemental Table S2). An A. lyrata ortholog was classified as an expressolog (1) if it was detected in the same pattern in rosette leaves, roots, and flowers like the Arabidopsis gene and (2) if its correlation regarding the stress responsiveness across all eight tested scenarios was bigger than 0.3. If the genes were not stress responsive in our conditions, the stress response correlation was not taken into account. All other cases showing detectable gene expression were denoted as nonexpressologs (Supplemental Table S2).
If the normalized organ expression value of any single member of an OrthoMCL gene group is below 9 in all organs or any of the stress scenarios studied here, we predict that the gene is nonfunctionalized under the studied conditions, since such a level is close to the detection limit. This classification cannot exclude the possibility that the gene is expressed in yet another scenario, which would indicate a neofunctionalization of the respective ortholog.
This strategy identified 34 out of 272 (12.5%) OrthoMCL gene groups where one A. lyrata ortholog retains the original function (expressolog) while the other orthologs are nonfunctional (Supplemental Table S2, group 1). One example is constituted by the three members of the chloroplast TIC complex (Arabidopsis AT1G06950, A. lyrata scaffold_100703.1, and A. lyrata fgenesh2_kg.1__669__AT1G06950.1). The normalized organ expression levels of the Arabidopsis and A. lyrata fgenesh2_kg.1__669__AT1G06950.1 gene were in a range of 12 to 15, while the A. lyrata scaffold_100703.1 gene copy had very low expression levels of 6.44, 3.63, and 4.05 in shoots, roots, and flower buds, respectively (Supplemental Table S2). The pairwise correlation analysis between the two highly expressed genes was 0.6, while it dropped to −0.48 between Arabidopsis AT1G06950 and the putatively nonfunctionalized A. lyrata copy.
In 49 (approximately 18%) gene groups, one A. lyrata homolog maintained a similar expression pattern like the Arabidopsis gene, while the other homolog showed a differential expression pattern at a significant expression level (nonexpressolog); therefore, we classified them as neofunctionalized (Supplemental Table S2, group 2). For example, two A. lyrata members (fgenesh2_kg.1__967__AT1G09240.1 and fgenesh2_kg.1__4760__AT1G56430.1) and one Arabidopsis member (AT1G09240) were detected in a gene group encoding NICOTIANAMINE SYNTHASE3. While the Arabidopsis AT1G09240 and A. lyrata fgenesh2_kg.1__967__AT1G09240.1 genes are positively correlated under drought-stressed (r = 0.84) and salt-stressed (r = 0.45) conditions, with a total stress correlation of r = 0.59, the A. lyrata fgenesh2_kg.1__4760__AT1G56430.1 gene was negatively correlated under drought-stressed (r = −0.90) and salt-stressed (r = −0.85) conditions, with a total stress correlation of r = −0.47. When the expression of these genes in different organs was studied, the loss of expression of the A. lyrata fgenesh2_kg.1__4760__AT1G56430.1 gene in flower bud further differentiated it from the Arabidopsis and the other A. lyrata genes (Supplemental Table S2). This clearly indicates that A. lyrata fgenesh2_kg.1__967__AT1G09240.1 is the predicted expressolog to Arabidopsis AT1G09240, while A. lyrata fgenesh2_kg.1__4760__AT1G56430.1 has acquired a new expression pattern and is likely neofunctionalized.
A total of 115 (approximately 42%) gene groups were categorized as species-specific functionalization, since the expression pattern of all functional A. lyrata genes in an OrthoMCL cluster was different from that of the Arabidopsis gene. Two types of divergences were recorded: (1) either all A. lyrata orthologs are neofunctionalized (nonexpressologs; 74 gene groups) or (2) one A. lyrata ortholog is a nonexpressolog while the other(s) lost the original function (nonfunctionalized; 41 gene groups; Supplemental Table S2, groups 3a and 3b). For instance, the members of the UDP-XYLOSE TRANSPORTER1 (UXT1) cluster consist of Arabidopsis AT2G28315/UXT1, A. lyrata scaffold_8500004.1, and A. lyrata fgenesh1_pm.C_scaffold_4000618. The two A. lyrata genes acquired salt and drought responsiveness and are negatively correlated to the Arabidopsis gene under salt and drought stresses (r = −0.8; Supplemental Table S2). A small group of six gene clusters showed an extreme form of species-specific functionalization, where all the A. lyrata genes present in a cluster are nonfunctionalized (Supplemental Table S2, group 4).
Subfunctionalization of genes would be indicated by a complementary expression of the coorthologs that covers the whole expression pattern of the corresponding gene in the other species (group 5). Possibly due to the limited number of 11 tested scenarios in the expression analyses, there were no clear indications for such a subfunctionalization. Instead, in 68 (25%) gene groups, both A. lyrata homologs maintained similar organ and stress expression patterns like the Arabidopsis genes and were interpreted as a group composed of genetically redundant genes based on our experimental assays. This is also reflected in the comparable correlation values between individual A. lyrata and Arabidopsis pairs residing in the same cluster. One such gene group consists of Arabidopsis AT1G06680, A. lyrata fgenesh2_kg.1__643__AT1G06680.1, and A. lyrata scaffold_401578.1. All three genes are well expressed in the three organs studied (Supplemental Table S2). They were up-regulated in the late time point of salt and drought treatment, while no response was found for UV-B light. Consistently, the overall stress correlation value between Arabidopsis AT1G06680 and A. lyrata fgenesh2_kg.1__643__AT1G06680.1 is 0.939 and that between Arabidopsis AT1G06680 and A. lyrata scaffold_401578.1 is 0.916.
Nucleotide Substitution Rate Calculation and Comparisons between Expressologs, Nonexpressologs, and Nonfunctionalized Genes in Four Different Functional Categories
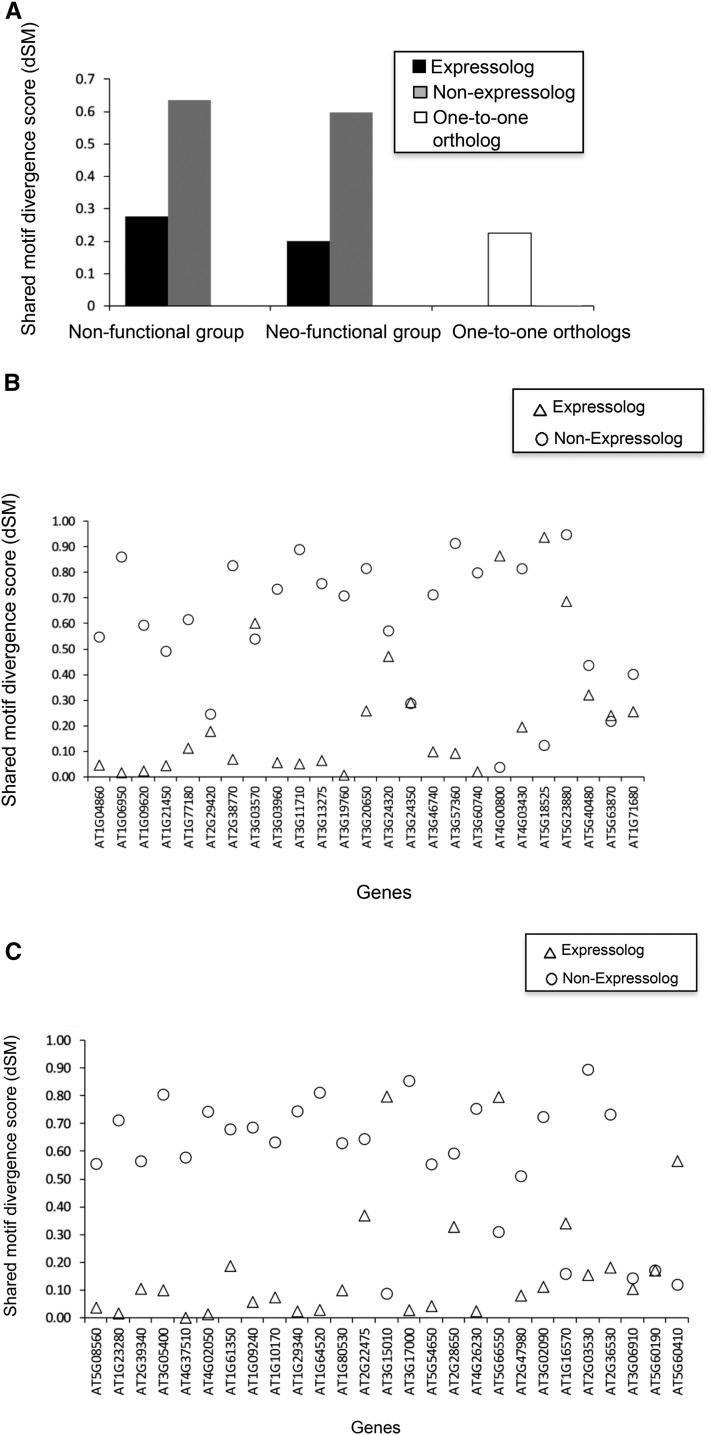
The transcription of a gene is largely controlled by its promoter. Therefore, we first tested if the promoter sequences of expressologs were more conserved than those of the predicted nonexpressologs. Such a correlation could initially support the identification of expressologs in newly sequenced species even in the absence of expression data. The shared motif divergence (dSM) method was employed to quantify the nucleotide changes in the upstream regions of A. lyrata gene groups with respect to the orthologous Arabidopsis genes. This analysis revealed that the upstream sequences of an expressologous gene group were on average less divergent compared with the divergence of nonexpressologous genes such as neofunctionalized or nonfunctionalized groups (Fig. 1A). The promoter sequence divergence score of the one-to-one gene group was comparable to that of the expressologous gene group (Fig. 1A). To avoid complication in data analyses arising due to the presence of too many A. lyrata homologs within a gene cluster or the unavailability of sufficiently long promoter sequences, a few genes were discarded from the analysis. Therefore, the number of gene groups compared in this analyses was 32 for the nonfunctionalized group, 35 for the neofunctional group, 57 for the genetically redundant group, and 81 for the species-specific group.
Figure 1.
Promoter sequences divergence analysis between expressologs and nonexpressologs in two different functional categories. A, dSM scores of expressologs, nonexpressologs, and one-to-one orthologs. The first group compares the promoter sequence divergence scores of A. lyrata expressologs and nonfunctionalized nonexpressologous genes (evolutionary group 1, Supplemental Table S2), the second group compares between the expressologs and neofunctionalized genes (evolutionary group 2, Supplemental Table S2), and the third group compares between the promoter sequence divergence scores of A. lyrata genes having single orthologous copies of Arabidopsis genes (as predicted by Ortho-MCL). B, Promoter analyses of the gene group, where at least one A. lyrata gene has been nonfunctionalized as predicted by gene expression analyses. For each Arabidopsis and A. lyrata orthologous gene pair within a gene group, we calculated the promoter dSM score of A. lyrata genes with reference to the promoter sequence of their Arabidopsis orthologous gene (on the x axis) by the dSM method. Circles represent the promoter dSM score of the A. lyrata gene copy predicted as expressologs by gene expression analyses, and triangles stand for that of nonexpressologs. C, Promoter analyses of the gene group, where at least one A. lyrata gene has been predicted to be neofunctionalized. The other parameters used were as described in B.
In addition to the overall analyses comparing promoters of expressologs versus nonexpressologs, the promoter divergence of the genes within different evolutionary gene groups was assessed. Within the nonfunctionalized gene groups, in 78% of the cases (25 out of 32 gene groups), the A. lyrata expressologs revealed less promoter divergence compared with the nonfunctionalized genes (Fig. 1B). Similarly, in 77% of the neofunctionalized gene groups (27 out of 35 gene groups), the expressologs possessed less promoter divergence than the nonexpressologs (Fig. 1C). In contrast to these two groups, where one member showed a conserved expression pattern and one member exhibited a nonconserved expression pattern, all the A. lyrata genes contained in the genetically redundant gene groups and in the species-specific functionalization groups showed either a similar (genetic redundancy group) or divergent (species-specific group) expression pattern in organs and/or stress conditions with respect to the Arabidopsis gene. Therefore, in these cases, we analyzed whether this differential behavior also was obvious among the promoters of the two members present within such a gene group in comparison with the corresponding Arabidopsis gene. To assess this question, the average promoter divergence of all A. lyrata genes compared with the respective Arabidopsis genes in the genetically redundant and species-specific functionalization groups was calculated separately. Indeed, the average dSM of the species-specific group was almost 2-fold (0.391) higher than that of the genetically redundant group (0.219). To address the promoter divergence of these two groups also at the individual gene group level, the difference of the promoter dSM between Arabidopsis-A. lyrata 1 (dSM1) and Arabidopsis-A. lyrata 2 (dSM2; ΔdSM = dSM1 − dSM2] present within the same gene group was calculated. If there was an overlap with the expression-based classification, lower ΔdSM values would be expected for the genetically redundant gene groups than for the species-specific gene groups. If we consider a conservative ΔdSM cutoff of less than 0.2, meaning high promoter similarity, then in 53% (43 out of 81) of the species-specific groups, the two A. lyrata promoter sequences are not comparable with respect to their sequence divergence from the Arabidopsis promoter. Thus, in about half of the cases, the promoters of the species-specific groups have undergone a strong change, in agreement with their changing expression pattern, whereas in the other half, the promoter divergences were not indicative of the expression patterns (Supplemental Fig. S1). In the case of the genetically redundant gene pairs, 40% (23 out of 57) of the gene groups also showed a high differential divergence of promoters of the coorthologs compared with the Arabidopsis gene, in contrast to the similar and conserved expression patterns observed.
One such example from the genetic redundancy group consists of Arabidopsis AT1G06680, A. lyrata fgenesh2_kg.1__643__AT1G06680.1, and A. lyrata scaffold_401578.1 genes. While the two A. lyrata genes are highly correlated to the Arabidopsis gene with respect to their organ expression and their stress-responsive gene expression pattern (r = 0.98), the promoters of the two A. lyrata genes reveal a differential sequence divergence from the Arabidopsis gene, with a ΔdSM = 0.38 (AT1G06680-A. lyrata fgenesh2_kg.1__643__AT1G06680.1 dSM = 0.003, and AT1G06680-A. lyrata scaffold_401578.1 dSM = 0.382).
The gene group Arabidopsis AT2G31160, A. lyrata fgenesh1_pg.C_scaffold_4001226, and A. lyrata fgenesh2_kg.163__1__AT2G31160.1 provides an example from the species-specific category, which shows a high promoter conservation of the A. lyrata gene promoters in comparison with the Arabidopsis gene despite the changed expression pattern. Both A. lyrata coorthologs were induced by salt stress, in contrast to the Arabidopsis copy contributing to the low correlation of the total stress responses (r = −0.0581 and r = −0.1191). Furthermore, the two A. lyrata copies were different among themselves, with one copy being expressed at a very low level in all organs (Supplemental Table S2). Nevertheless, ΔdSM was 0 and the dSM levels for both A. lyrata-Arabidopsis comparisons were very low (dSM = 0.007).
It is evident from our analyses that, while promoter divergence analysis can be used as an additional tool for annotation purposes, experimental classification as expressologs/nonexpressologs provides more accurate functional information and mode of functional divergence, such as nonfunctionalization, neofunctionalization, or species-specific functionalization and genetic redundancy, which the promoter analyses cannot fully offer.
Identification of Genetic Mutants for Experimental Validation of Predicted Expressologs
To confirm the functionality of our predicted expressologs, we applied genetic complementation assays using the A. lyrata gene variants transformed to Arabidopsis loss-of-function mutants for the group of one Arabidopsis-multiple A. lyrata candidate genes. We scanned the insertion mutant repositories to identify mutant lines corresponding to our list of 272 genes. Additionally, we checked the available literature for appropriate mutants. Out of 272 queried one-to-many Arabidopsis genes, homozygous mutant SALK lines were obtained for 147 genes. All these 147 insertion lines were grown under greenhouse conditions, but no obvious morphological phenotypes could be observed for any of these lines studied.
However, four published Arabidopsis mutants, cls8-1, tso2-1, stabilized1-1 (sta1-1), and manganese transporter11 (mtp11), could be used for our analyses (Supplemental Table S3). Their mutant phenotypes could be clearly reproduced, and the corresponding A. lyrata homologous gene copies along with their native promoters were amplified for genetic complementation assay. Based on our expressolog classification, the corresponding Arabidopsis-A. lyrata gene groups represented one case of a possible neofunctionalization (CLS8/RNR1) and one case of pseudogenization (STA1). Two cases (TSO2 and MTP11) were indicative of genetic redundancy.
Example 1: Potential Neofunctionalization by Acquiring Changes in the Regulatory Region of the Gene and in the Coding Region
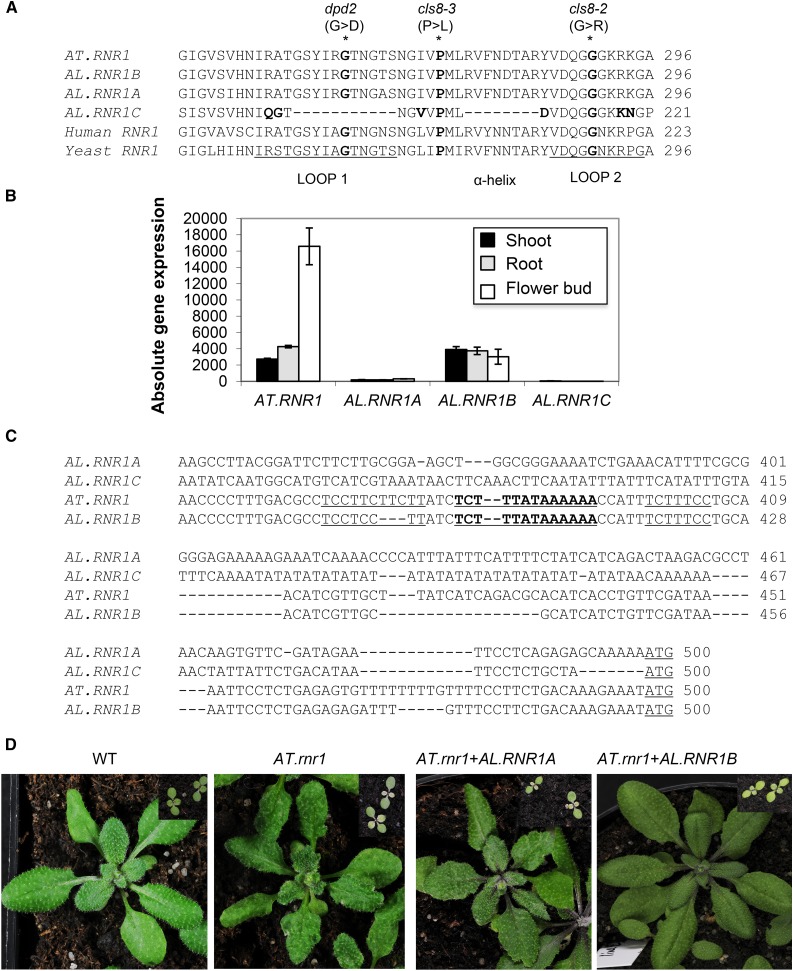
Neofunctionalization is predicted for the A. lyrata genes encoding the large subunit of ribonucleotide reductase, which catalyzes the reduction of ribonucleoside diphosphates to deoxyribonucleotides, the rate-limiting step in the de novo synthesis of deoxyribonucleotide triphosphates (Sauge-Merle et al., 1999). In Arabidopsis, the large subunit is encoded by a single-copy gene, AT.RNR1 (AT2G21790; CLS8), while in A. lyrata, three homologous copies exist, AL.RNR1A (Al_scaffold_0007_128/AL7G01310), AL.RNR1B (fgenesh2_kg.4__104__AT2G21790.1/AL4G01010), and AL.RNR1C (scaffold_200715.1/AL2G07030). Nonsense point mutations in Arabidopsis caused visible early and late developmental phenotypes such as bleached first true leaves and crinkled rosette leaves with white pits on the surface (Garton et al., 2007; Supplemental Table S3). All Arabidopsis RNR1 sequences were aligned to the yeast and human RNR proteins to analyze whether any sequence alteration could be observed in the two catalytically important 10- to 15-amino acid stretches called LOOP1 and LOOP2 (Xu et al., 2006). Single-amino-acid, nonsynonymous mutations located at the LOOP1/LOOP2 region cause the phenotypic defects in Arabidopsis (dpd2, cls8-2, and cls8-3). Therefore, we focused our analysis mostly on this region. A stretch of 11 amino acids was missing in the LOOP1 region of AL.RNR1C, although no such change was noticed in AT.RNR1, AL.RNR1A, AL.RNR1B, human, and yeast copies (Fig. 2A). In addition, AL.RNR1C was not detected in any of the expression analyses; therefore, it was also denoted as a nonfunctional copy based on the expression data (Supplemental Table S2). Correlation analysis indicated that AL.RNR1B is most closely related to AT.RNR1 (r = 0.83) based on its stress-responsive gene expression pattern. Since it was also expressed in all organs, like the Arabidopsis gene, AL.RNR1B was predicted as the expressolog (Table II; Supplemental Table S2). AL.RNR1A also reported a good, albeit lower stress-related correlation (r = 0.64). However, its organ expression level was close to or below the detection level of the microarray analysis, and a detailed examination of all three types of stress experiments indicated that only salt responsiveness was partially retained by AL.RNR1A, leading to an expression above the detection threshold (Fig. 2B; Supplemental Table S2). Thus, AL.RNR1A could be a neofunctionalized coortholog that is active only in certain stress scenarios.
Figure 2.
Sequence, gene expression, and genetic complementation analyses of ribonucleotide reductase large subunit (RNR1) gene copies in Arabidopsis and A. lyrata. A, Alignment of the Arabidopsis RNR1 amino acid sequences along with those of human and yeast sequences. Biologically important LOOP1 and LOOP2 regions are depicted. Part of the highly conserved LOOP1 region is missing, and two nonsynonymous amino acid changes were detected in the LOOP2 region of AL.RNR1C. However, the AL.RNR1B coding sequence (CDS) is identical to that of AT.RNR1. These regions play important roles in enzymatic function by controlling the specificity of the incoming deoxyribonucleotide triphosphate. The biological importance of this region is emphasized by the identification of three mutations that caused severe developmental defects (indicated by asterisks at top). Another allele, cls8-1, affects a distant region leading to amino acid change G718E but showing the same mutant phenotype (Supplemental Table S3). B, Expression patterns of the four Arabidopsis RNR1 genes in root, shoot, and flower bud. Background-corrected and multiplicatively detrended signal intensities were imported to GeneSpring (G3784AA; version 2011) to calculate normalized gene expression values (for details, see “Materials and Methods”). C, Comparison of core promoter regions (250 bp upstream from ATG) indicates the loss of the AT.RNR1-like TATA box (boldface and underlined) and the Y patch (underlined) in the case of AL.RNR1A and AL.RNR1C homologs. This analysis was done in the plant promoter database (http://133.66.216.33/ppdb/cgi-bin/index.cgi#Homo). D, Genetic complementation of Arabidopsis rnr1/cls8-1 with AL.RNR1B and AL.RNR1A gene copies. The phenotype of the AL.RNR1B (predicted expressolog) complemented plants resembles that of the wild type (WT). However, the plants complemented by AL.RNR1A (predicted pseudogene) show the mutant phenotype, such as the yellowish first true leaves (in the insets) and crinkled mature leaves.
Table II. Gene expression similarity (r) and promoter sequence divergence (dSM score) for selected genes analyzed by genetic complementation assay.
| Gene Name | Gene Identifier | Promoter Divergence Score | Correlation Based on Stress Expression Data | Syntenic Gene | Predicted Expressolog | Functional Ortholog Based on Genetic Complementation |
|---|---|---|---|---|---|---|
| AT.RNR1 | AT2G21790 | – | – | – | – | – |
| AL.RNR1A | Al_scaffold_0007_128 | 0.685 | 0.64 | No | No | No |
| AL.RNR1B | fgenesh2_kg.4__104__AT2G21790.1 | 0.333 | 0.83 | Yes | Yes | Yes |
| AL.RNR1C | scaffold_200715.1 | 0.676 | 0.65 | No | No | Not tested |
| AT.STA1 | AT4G03430 | – | – | – | – | |
| AL.STA1A | fgenesh2_kg.6__3353__AT4G03430.1 | 0.197 | 0.75 | Yes | Yes | Yes |
| AL.STA1B | scaffold_700051.1 | 0.815 | −0.50 | No | No | No |
| AT.TSO2 | AT3G27060 | – | – | – | – | – |
| AL.TSO2A | fgenesh2_kg.5__483__AT3G27060.1 | 0.155 | 0.56 | Yes | No | Yes |
| AL.TSO2B | scaffold_703867.1 | 0.828 | 0.86 | No | Yes | Yes |
| AT.MTP11 | AT2G39450 | – | – | – | – | – |
| AL.MTP11A | fgenesh2_kg.4__2026__AT2G39450.1 | 0.307 | – | Yes | Yesa | Yes |
| AL.MTP11B | fgenesh2_kg.463__5__AT2G39450.1 | 0.297 | – | No | Yesa | Yes |
Because of very high sequence similarity, microarray probes were not gene specific; hence, expression similarity was assessed by RT-qPCR analyses.
Since the low expression level of AL.RNR1A in unstressed conditions is an important signature for possible promoter mutations, we checked the presence/absence of important transcriptional regulators in the promoter regions of the RNR1 genes. While overlapping, intact AT.RNR1-like TATA element and Y patches were predicted for AL.RNR1B, these were disrupted in both the AL.RNR1A and AL.RNR1C copies (Fig. 2C). Finally, to check the reliability of expression-based prediction about gene functionality, we cloned the expressologous (AL.RNR1B) and nonexpressologous (AL.RNR1A) gene copies and tested for complementation of the AT.rnr1/AT.cls8-1 mutant (Supplemental Table S3). Recovery of the wild-type phenotype was observed in the case of AL.RNR1B complemented plants. However, AL.RNR1A complemented plants did not revert to the mutant phenotype, which indicates that the AL.RNR1A homolog does not retain the RNR1 function (Fig. 2D). Although three independent transgenic lines each clearly differentiated the complementing from the noncomplementing orthologs, we confirmed the presence of the transgene insertion of AL.RNR1A by PCR (Supplemental Fig. S2); expression of the AL.RNR1A transgene was not detected by reverse transcription (RT)-PCR, probably due to its low expression level, as observed in A. lyrata.
Example 2: Event 1 of Genetic Redundancy
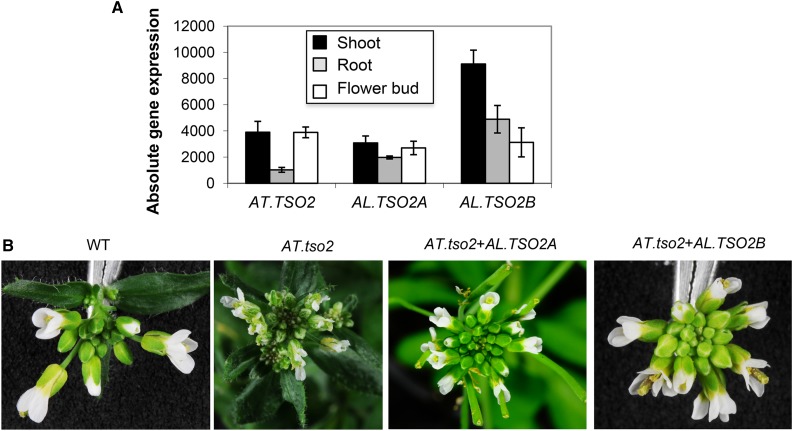
Interestingly, the gene(s) encoding the small subunit of ribonucleotide reductase (RNR2) also were among the genes of the one Arabidopsis-multiple A. lyrata in addition to the genes encoding its large subunit (see above). The small subunit-related genes are AT.TSO2 (AT3G27060), AT.RNR2A (AT3G23580), and AT.RNR2B (AT5G40942). However, among these three subunits, TSO2 is biologically the most active copy. In A. lyrata, TSO2 is found to be duplicated, resulting in AL.TSO2A and AL.TSO2B. The phenotype of AT.tso2-1 revealed similar developmental defects to AT.rnr1, such as irregular leaves and homeotic transformations (Wang and Liu, 2006). Multiple sequence alignment of AT.TSO2, AL.TSO2A, and AL.TSO2B reveals only one nonsynonymous change between AT.TSO2 and AL.TSO2A, while 28 nonsynonymous changes were noticed between AT.TSO2 and AL.TSO2B outside the region of important enzymatic function (Supplemental Fig. S3). The two A. lyrata copies are well expressed in different organs, like the Arabidopsis gene (Fig. 3A). Correlation analysis based on its stress response pattern indicated that AL.TSO2B is closest to AT.TSO2 (r = 0.85; Table II) and, therefore, is predicted as the expressolog. However, AL.TSO2A also showed a reasonably good correlation (r = 0.55; Supplemental Table S2). This indicates that AL.TSO2A and AL.TSO2B are possibly redundant to each other within the resolution provided by our expression study. The promoter comparisons revealed that the TATA box and the Y patch were preserved in both AL.TSO2A and AL.TSO2B. Both AL.TSO2A and AL.TSO2B copies were cloned along with their native promoter and transformed into the AT.tso2-1 plants. The transformed plants restored the wild-type phenotype in both cases and thus proved that AL.TSO2A and AL.TSO2B are functionally redundant in the analyzed context and orthologous to AT.TSO2 (Fig. 3B).
Figure 3.
Gene expression and genetic complementation analyses of ribonucleotide reductase small subunit (TSO2) gene copies in Arabidopsis and A. lyrata. A, Expression of the three Arabidopsis TSO2 genes in root, shoot, and flower bud. All the copies are expressed well above background. B, Genetic complementation of Arabidopsis tso2-1 with the AL.TSO2A or AL.TSO2B gene copy. Both AL.TSO2A and AL.TSO2B were predicted as expressologs by our analysis, although the sequence analysis indicated several nonsynonymous changes in the AL.TSO2B copy compared with the Arabidopsis gene (Supplemental Fig. S3A). The result of the complementation assay supported this prediction. WT, Wild type.
Example 3: Pseudogenization by Acquiring Changes in the Coding Region of the Gene
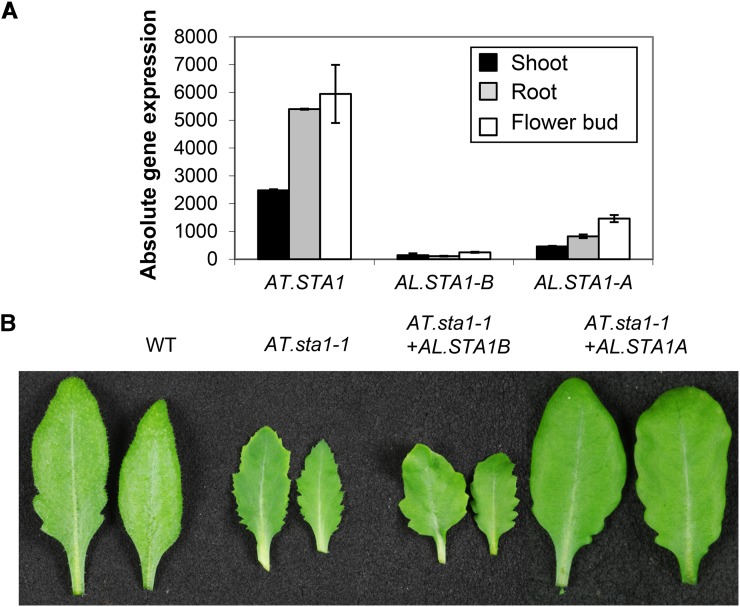
STA1 is a pre-mRNA splicing factor. The gene function is similar to that of the human U5 small ribonucleoprotein and to the yeast pre-mRNA splicing factors Prp1p and Prp6p (Lee et al., 2006). Arabidopsis harbors a single gene (AT4G03430), while in A. lyrata, two copies, AL.STA1A and AL.STA1B, have been identified by our OrthoMCL analysis. The Arabidopsis loss-of-function mutant shows many developmental and stress-related phenotypes, such as smaller plant height, smaller leaf size, and higher sensitivity to abscisic acid, compared with the wild type (Lee et al., 2006). The expression level of AL.STA1B was below the detection limit of our microarray analysis in all three organs and in all stress scenarios (Supplemental Table S2). On the contrary, AL.STA1A was expressed above the detection limit and was similarly regulated under diverse stress conditions, like AT.STA1 (r = 0.75; Fig. 4A; Supplemental Table S2). Therefore, we predicted that, while AL.STA1A was the expressolog, AL.STA1B had presumably been pseudogenized (Table II). We checked the coding regions of AL.STA1B for additional indications of its pseudogenization. Although AT.STA1 does not contain any intron, a 43-nucleotide-long intron was predicted for the AL.STA1A gene model, while three introns of 50, 44, and 324 nucleotides were predicted for the AL.STA1B gene model. Therefore, we sequenced the AL.STA1B cDNA to verify such splicing events in this A. lyrata gene. However, the AL.STA1B cDNA sequence indicated that it was also an intronless gene like AT.STA1. Additionally, we detected the insertion of one A nucleotide at position 1,352 of the AL.STA1B CDS, which causes a premature stop codon and possible pseudogenization of this gene copy (Supplemental Fig. S4). To check the accuracy of this prediction, we cloned both AL.STA1A and AL.STA1B copies and transformed them in At.sta1-1 plants (Supplemental Table S3). The wild-type phenotype could be recovered for AL.STA1A-transformed plants, while plants harboring the AL.STA1B construct still exhibited the mutant phenotype (Fig. 4B). Three independent transgenic lines each clearly differentiated the complementing from the noncomplementing ortholog. Furthermore, the presence of the transgene insertion of the noncomplementing AL.STA1B was confirmed by PCR (Supplemental Fig. S5); expression of the AL.STA1B transgene was not detected by RT-PCR, probably due to its low expression level, as observed in A. lyrata.
Figure 4.
Gene expression and genetic complementation analyses of STA1 gene copies in Arabidopsis and A. lyrata. A, Expression patterns of the three Arabidopsis STA1 genes in root, shoot, and flower bud. B, Comparison of the leaf morphology of Arabidopsis sta1-1 plants with AT.sta1-1+AL.STA1B and AT.sta1-1+AL.STA1A complemented lines. While the leaf size and margins of AL.STA1A complemented plants look like the wild type (WT), the AL.STA1B transformed lines resemble the mutant.
Example 4: Event 2 of Genetic Redundancy
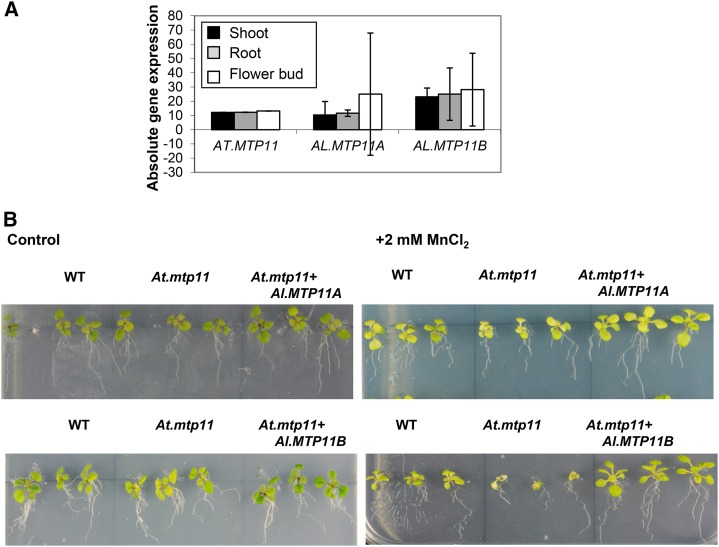
MTP11 is a member of the large cation diffusion family and is involved in Mn2+ transport and tolerance (Gustin et al., 2011). It exists as a single-copy gene in Arabidopsis (AT2G39450), while duplicated copies of AL.MTP11A and AL.MTP11B were identified in A. lyrata. The loss-of-function AT.mtp11 plants are more sensitive to Mn2+ ions. Under Mn2+-stressed conditions, they grow less vigorously compared with wild-type plants (Delhaize et al., 2007). The study of phylogenetic relationships and high sequence homology indicate that AL.MTP11A and AL.MTP11B share a recent origin. Since the two A. lyrata homologs are highly similar (98% identity at the CDS level), it was not possible to design gene-specific microarray probes. Therefore, we assessed their gene expression pattern by RT-qPCR analysis. Both homologs were well expressed in different organs, and the expression levels were comparable to that of the AT.MTP11 gene; thus, they are predicted to be genetically redundant with respect to this data set (Fig. 5A; Table II). To confirm the functional equivalence of AL.MTP11A and AL.MTP11B, we transformed the full-length genes along with their native promoters into the Arabidopsis mutant plants. Phenotypic assay of the knockout and transformed lines revealed that, while the growth of the mutant line was compromised on plates containing 2 mm MnCl2, the growth of both transgenic lines was similar to that of Arabidopsis wild-type plants (Fig. 5B; Supplemental Fig. S6), indicating genetic redundancy in this context and the functional equivalence of both A. lyrata gene copies.
Figure 5.
Gene expression and genetic complementation analyses of MTP11 gene copies in Arabidopsis and A. lyrata. A, Organ expression patterns of the two A. lyrata MTP11 homologous genes assessed by RT-qPCR analysis. B, Genetic complementation of the Arabidopsis mtp11 loss-of-function mutant with AL.MTP11A and AL.MTP11B gene copies. Seedlings were grown on Murashige and Skoog medium (1% Suc, 1× Murashige and Skoog medium, and 1.2% phytoagar) for 8 d and then transferred to agarose medium supplemented with 2 mm Mn2+. As predicted by the expression data, both homologs could complement the mutant phenotype. WT, Wild type.
DISCUSSION
Plants are sessile and are subject to varying environmental stresses. Gene duplication is the event by which a plant can gain novel adaptive genes that enable them to meet their specific ecological needs (Conant and Wolfe, 2008; Ha et al., 2009; Van de Peer et al., 2009). All plant genomes sequenced to date have undergone at least one round of whole-genome duplication (Fischer et al., 2014). While gene duplication is evolutionarily advantageous for the polyploidized plants, it imposes a challenge to transfer gene function annotation across species barriers by simple sequence comparisons. Since prediction of a correct annotation is key to any genome sequencing project and translational approaches, a number of sequence homology-based methods have been developed (Gabaldón, 2008; Kuzniar et al., 2008). While these tools are effective for single-copy genes and for genome-wide comparisons, additional support is required for large multigene families. OrthoMCL is one such tool, which is commonly used in genome-wide comparisons. Therefore, this method was employed to analyze Arabidopsis and A. lyrata CDS, identifying 2,850 genes (6.5% of Arabidopsis transcripts) that exist as one-to-many or many-to-many copies between these two species. Such uncertainty in predicting functional orthologs may be even worse in crop species of the Brassica lineage, which have undergone one round of whole-genome triplication in addition to whole-genome duplication events shared with the Arabidopsis lineage. Therefore, in such a situation, where CDS-based analyses are limited with respect to assigning functional orthology, additional support is required.
Two major approaches have been proposed to address this issue. The most popular is gene coexpression analysis, which has been used successfully for well-characterized genomes for which large-scale expression data are already available (Stuart et al., 2003; Bergmann et al., 2004; Movahedi et al., 2011; Mutwil et al., 2011). The second method also relies on extensive gene expression profiles obtained from comparable tissues among the species compared and subsequent implementation of a ranking system of genes based on expression similarity, with top-ranked genes called expressologs (Patel et al., 2012). However, both methods are dependent on the availability of large sets of highly comparable expression data obtained from diverse tissues and conditions for all of the species of interest, which is not feasible for newly sequenced genomes. Similarly, protein-protein interaction network data, which can assist the identification of functional orthologs among large paralogous gene families, would not be available in these cases (Bandyopadhyay et al., 2006). Therefore, in this study, we have tested the utility of a relatively small set of gene expression data for the prediction of functionally related orthologs. We compared the expression pattern in three organs and conducted Pearson correlation analysis based on data obtained from stress gene expression experiments in Arabidopsis and A. lyrata. In contrast to a very low basic mean correlation value of 0.019 obtained from all-against-all comparisons between the Arabidopsis and A. lyrata transcriptomes, a much higher correlation value of 0.329 was obtained when syntenic Arabidopsis-A. lyrata orthologous gene groups were analyzed. This finding proves that, although our gene expression data set is small, it is appropriate to identify functionally related genes across species. The correlation analyses also revealed that the OrthoMCL one-to-one gene group holds a higher correlation value (rmean = 0.354) than the OrthoMCL all (rmean = 0.313) and OrthoMCL many-to-many (rmean = 0.300) gene groups (Table I). This illustrates some limitations of OrthoMCL analyses in predicting functional orthologs in one-to-many or many-to-many situations.
The analyses of transcriptional expression patterns and correlations in this study show that functional categorization and prediction of expressologs based on gene expression patterns are possible for one-to-many orthologous relationships. In all tested gene groups, we could identify the functional fate of duplicated A lyrata genes. In 12.5% of the gene groups, at least one A. lyrata gene copy had been putatively nonfunctionalized. It should be noted, however, that the putative nonfunctionalization is based on very low expression levels for a limited set of expression data and that these genes might well be expressed under other, untested conditions. These limitations may well apply to the other categories as well. Approximately 18% of the gene groups suggest a functional divergence on the basis of at least one conserved coortholog, since at least one A. lyrata gene has undergone neofunctionalization. The biggest group (42%) is composed of genes that show species-specific gene expression patterns, and three forms of such expression patterns were recorded. The two Arabidopsis species are phylogenetically very close and diverged only 10 million years ago (Hu et al., 2011). However, they have adapted to distinct environments and, therefore, show many differences in terms of their life cycles as well as in their reproductive and ecological habitats (Mitchell-Olds, 2001; Clauss and Koch, 2006). Therefore, such species-specific gene expression patterns may have evolved in relation to these different lifestyles.
In 25% of the cases, our analyses consist of duplicated clusters for which both copies exhibit similar expression patterns and, therefore, were assumed to be genetically redundant within the tested conditions. Although no clear evidence of subfunctionalization was noticed in this study, it is possible that more extended gene expression analyses may identify such candidates among the currently classified genetically redundant group (MacCarthy and Bergman, 2007).
Taken together, our findings indicate that expressologs strongly reduce the existing uncertainty associated with the CDS homology-based methods to assign functional orthologs in the presence of multiple orthologs. However, there are still limitations. One major challenge of this approach is to identify comparable biological tissues or experimental conditions (severity of the applied stresses, time points, etc.) to measure comparable expression patterns of the targeted genes across species. For phylogenetically and/or ecologically distant species, it might be challenging to find comparable conditions, and further studies are required to test the efficiency of predicted expressologs in these situations.
We also examined whether a comparative analysis of promoter sequences could be used as a satisfying alternative to predict functional orthology, when comparative gene expression data are not or not yet available for a species (e.g. in the case of newly sequenced genomes). Therefore, promoter divergence analyses were performed to determine in how many instances the promoters of predicted expressologs were less divergent than those of the nonexpressologs or nonfunctionalized genes. If the gene predicted as an expressolog based on our gene expression analyses harbors less promoter dSM than that of the nonexpressolog, then we would assume that the functional orthology prediction based on gene expression patterns and promoter divergence analyses overlapped with each other. We investigated individual OrthoMCL gene clusters and found that, in 78% of the studied cases for the nonfunctional group and 77% of the neofunctionalized gene group, the A. lyrata expressologs indeed had less promoter divergence than the nonexpressologs or nonfunctionalized genes. The overlap between predictions made by expressolog and promoter divergence is even much lower for the genetic redundancy (60%) and species-specific (47%) categories. Thus, in the absence of gene expression data, the determination of promoter divergence can be complementary to the limitations of CDS-based methods such as OrthoMCL. However, a considerable number of genes could still not be correctly annotated. Furthermore, there are some other serious limitations that have to be considered in the case of promoter sequence analysis. (1) Determination of the boundaries of the promoter regions is a critical issue, since cis-elements have been reported in Arabidopsis to be located several kilobases upstream of the transcription start (Rombauts et al., 2003). Moreover, small 5′ exons or divergent untranslated region sizes can result in the comparison of completely unrelated sequences. (2) With the increase in the phylogenetic distance of the species compared, an altered sequence composition of the cis-elements may jeopardize their classification and the deduction of promoter divergence scores. (3) Unlike gene expression analysis, promoter analyses cannot study the mode of gene function diversification. For example, the promoter divergence analyses in the case of genetic redundancy or species-specific categories could not reveal any distinction between these two divergent categories. However, detailed gene expression analyses revealed that, in the case of genetic redundancy, the two or more A. lyrata genes were regulated in the same direction as the Arabidopsis gene, whereas in the event of species-specific expression, the two or more A. lyrata genes were regulated in a diverse manner from the Arabidopsis gene. Therefore, expressologs even based on a small set of expression analyses, as in our study, are a reliable and superior tool that can supplement the genome annotation pipeline for a more accurate transfer of gene functions.
Although previous studies and our data support the hypothesis that large-scale and even small-scale gene expression data could provide clues about gene functionality, no studies have been conducted so far to check the reliability of such prediction in planta. Here, we show that this concept is valid by testing it experimentally for the evolutionary categories nonfunctionalization and neofunctionalization and for two genes of the genetic redundancy class. We provide in planta evidence that the two genes of the two Arabidopsis species having closest expression patterns (expressologs) are functionally comparable (functional orthologs).
Two of the case studies implied genetic redundancy based on the expressolog classification. CDS-based prediction, promoter analysis, and expressolog prediction for AL.MTP11A and AL.MTP1B had pointed toward their possible functional similarity. On the contrary, the identification of 28 nonsynonymous nucleotide changes and a high promoter divergence between AT.TSO2 and AL.TSO2B indicate their possible functional divergence, although our gene expression analysis predicted AL.TSO2A and AL.TSO2B as expressologs. The functional equivalence as predicted by the expressolog classification was eventually corroborated by the genetic complementation analysis. Two further cases referred to pseudogenization and to neofunctionalization events as deduced from the gene expression pattern. While the A. lyrata genome annotation project did not identify these pseudogenes and nonexpressologs, our promoter sequence and expression analyses indicate possible mechanisms and directions by which these genes have evolved. Again, we could experimentally verify the authenticity of this prediction by transforming the Arabidopsis loss-of-function mutants with the corresponding A. lyrata pseudogenized and neofunctionalized genes, which did not lead to complementation.
In conclusion, we could experimentally verify functional orthologs among the one Arabidopsis-to-two (many) A. lyrata gene groups. These annotations could not be deduced using sequence-based algorithms only; instead, they were predicted based on comparative expression analyses. This success emphasizes the strength and added value of an expressolog/nonexpressolog classification based on an even limited set of expression data in order to predict the functional orthologs in such one-many gene groups.
MATERIALS AND METHODS
OrthoMCL Analysis
OrthoMCL version 1.4 with an initial cutoff E value of 1e−05 was used for the BLASTP comparisons between the transcriptomes of Arabidopsis (Arabidopsis thaliana) and Arabidopsis lyrata. The inflation parameter was set to 1.5, and all other parameters were set to default values as recommended by the developers. When exactly one Arabidopsis and exactly one A. lyrata identifiers were identified in an OrthoMCL cluster, this was defined as a one-to-one situation. In the case of one-to-many situations, one Arabidopsis and multiple A. lyrata identifiers or vice versa were present in a cluster. For all further analyses, we focused on one Arabidopsis to multiple A. lyrata groups to tap the possibility of experimental verification by utilizing Arabidopsis loss-of-function mutants.
Collection of Tissues from Stress-Induced Plants and from Different Organs of Arabidopsis and A. lyrata
Four-week-old Arabidopsis Columbia-0 and 6-week-old A. lyrata ssp. lyrata soil-grown plants were treated with either 250 or 500 mm NaCl solution by flush flooding (to soak the soil for a short period), while the control group was watered. Leaf tissues were harvested at 3 and 27 h posttreatment. To assess the effective salt exposure to the plants, the raw soil electrical conductivity (EC) was measured before tissue harvesting by the use of a 5TE sensor attached to Procheck hand-held datalogger (Decagon). Since direct EC measurement in soil was not reproducible because of the presence of particles and air pockets, the soil EC in solution was measured by mixing a ratio of 1:5 soil:water (Supplemental Fig. S7). The effective salt concentrations that the plants were subjected to were within the moderate range of soil salinity (www.saltlandgenie.org.au). For drought treatment, leaf samples were collected 8 and 11 d after withdrawal of regular watering to the soil. Plants were exposed to UV-B radiation plus photosynthetically active radiation (400–700 nm) of 140 µmol m−2 s−1. The biological effective UV-B radiation, weighted after generalized plant action spectrum (Caldwell, 1971) and normalized at 300 nm, was 1.31 and 2.62 kJ m−2 for 4- and 8-h time points, respectively. To collect root tissues, Arabidopsis and A. lyrata plants were hydroponically grown for 6 weeks on a raft following standard procedures (Conn et al., 2013). Unopened flower buds were cut at the pedicels and collected from soil-grown plants.
Microarray Design
The Arabidopsis array was customized by printing biological (43,603) and replicated (50 × 5) probe groups available commercially from Agilent Technologies (identifier 029132). The design of A. lyrata probes was done by uploading the total transcriptome (32,670) to the Agilent e-array facility (https://earray.chem.agilent.com/earray/). One probe per target sequence was generated for 32,386 transcripts, while no probes were reported for 284 sequences. These sequences were either repeat masked out or did not pass the required quality check. The specificity of the designed probes was further confirmed by blasting against the A. lyrata transcriptome. In addition to the main probe group, a replicated probe group of 477 selected genes was printed on the array for multiplicative detrending (identifier 030951). The mean probe length was 60. Both Arabidopsis and A. lyrata arrays were printed in 8 × 60 K format (Supplemental Table S1).
RNA Extraction, Array Hybridization, and Scanning
RNA was extracted using a combination of Trizol (Invitrogen) and the RNAeasy kit (Qiagen; Das et al., 2010). Quality was checked by Bioanalyzer analysis (Agilent Technologies). Approximately 100 ng of total RNA was used for complementary RNA synthesis and subsequent Cy3 labeling using the one-color low-amp quick amplification labeling kit (Agilent Technologies). Array hybridization, washing, and scanning were done according to the recommended procedures by Agilent Technologies.
Array Data Analysis
Data were extracted using an Agilent scanner and an Agilent Feature Extraction program. Background-corrected and multiplicatively detrended hybridization signals were imported to GeneSpring (G3784AA; version 2011) for log2 transformation and data normalization. The normalization conditions used were as follows: threshold raw signals to 1.0; normalization algorithm, scale; percentile target, 75. For stress data, the normalized signal intensity values were baseline corrected to the median of all samples. However, to get normalized expression data in different organs, baseline transformation was turned off. To get information about the differential expression of genes in diverse stressed conditions, a Z score (i.e. the number of sd changes between a control and a respective treatment) was calculated. To know how tightly orthologous Arabidopsis and A. lyrata genes were related in terms of gene expression, we calculated the total Pearson correlation values for OrthoMCL all, OrthoMCL one-to-one, OrthoMCL multiple, and syntenic gene groups (Table I). Also, Pearson correlations for individual Arabidopsis-A. lyrata OrthoMCL pairs were calculated to predict possible expressologs and nonexpressologs based on expression similarity or divergence.
Real-Time RT-qPCR Analysis
Since AL.MTP11A and AL.MTP11B homologs are highly identical, the designed array probes were cross-hybridizing to each other. Therefore, the expression levels of these two homologs were measured in shoots, roots, and flower buds of A. lyrata by quantitative RT-PCR analyses. First-strand cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen), and SYBR Green fluorescence was used to measure the expression level of the targeted genes in A. lyrata. The transcript abundance of AL.MTP11A and AL.MTP11B homologs was calculated in geNORM using AL.UBQ5 and AL.S16 as reference genes (Vandesompele et al., 2002; Supplemental Table S4).
To design gene-specific primers, we targeted two single-nucleotide polymorphisms (there are only 20 single-nucleotide polymorphisms over the entire CDS region among AL.MTP11A and AL.MTP11B) and designed gene-specific real-time RT-qPCR primers based on only one nucleotide sequence divergence at the 3′ end. Indeed, by restriction enzyme digestion and subsequent sequencing of the amplified PCR product, we could confirm the identity of the amplified gene products (Supplemental Fig. S4, A and B). Gene-specific primers for AL.STA1A were designed from the CDS region located after insertion of the premature stop codon to avoid the possibility of getting amplification from truncated mRNA.
Promoter Divergence Analysis
The DNA distance matrices for upstream sequences of A. lyrata genes (neofunctionalized/nonfunctionalized versus expressologs) with respect to the upstream sequence of their Arabidopsis orthologs were calculated based on 1,000-bp upstream sequences from the start codon. We obtained the dSM scores for the upstream sequences of A. lyrata genes based on the motif divergence method SSM (Castillo-Davis et al., 2004). For a pair of sequences, the SSM calculates functional regulatory changes within the sequences and provides the dSM score that quantifies the fraction of unaligned regions between the sequences.
Gene Amplification, Gateway Cloning, Plant Transformation, and Selection
Gene-specific loss-of-function mutants were obtained either from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/) or from individual laboratories (Scholl et al., 2000; Supplemental Table S3). High-fidelity Phusion polymerase (New England Biolabs) was used to amplify genes plus native promoters of the approximately 2- to 2.5-kb upstream 5′ region and 0.5-kb 3′ downstream region of A. lyrata. Gateway recombination sequences were always tagged to the 5′ end of the gene-specific primers (Supplemental Table S4). Amplified PCR products were eluted from gels, cloned in pDONR221 vector, and subsequently recombined to a modified, promotorless pAlligator2 vector (35S promoter deleted by restriction with HindIII and EcoRV, blunting with T4 DNA polymerase, and religation; Bensmihen et al., 2004). Since the promoter and 3′ untranslated regions of AL.MTP11A and AL.MTP11B were highly identical, the full-length gene sequence for genetic complementation of both genes was amplified by identical primer pairs; the cloned fragments were analyzed by restriction digestion and sequencing to distinguish AL.MTP11A and AL.MTP11B isolates.
Cloning of the correct sequences was always confirmed by sequencing the entire insert. Finally, the expression clones were mobilized to competent Agrobacterium tumefaciens pGV3101/pMP90 strains, and Arabidopsis plants were transformed by the floral dipping method (Clough and Bent, 1998). Transformed T1 seeds were selected by observing the green fluorescence of the GFP reporter gene; at least three independent T1 plants were subsequently phenotyped.
Expression data from Agilent microarray hybridization are deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the following expression numbers: GSE80099, Arabidopsis transcriptomic responses against drought stress; GSE80100, Arabidopsis root and flower bud transcriptomes; GSE80108, A. lyrata ssp. lyrata root and flower bud transcriptomes; GSE80110, A. lyrata ssp. lyrata transcriptomic responses against drought stress; GSE80111, Arabidopsis transcriptomic responses against UV-B light stress; GSE80112, A. lyrata transcriptomic responses against UV-B light stress; GSE80114, Arabidopsis transcriptomic responses against salt stress; and GSE80115, A. lyrata transcriptomic responses against salt stress.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Promoter sequence divergence analysis between expressologs and nonexpressologs in the genetic redundant and species-specific functional categories.
Supplemental Figure S2. Confirmation of the presence of the transgene insertion of AL.RNR1A in three independent transgenic plants by PCR analysis.
Supplemental Figure S3. Sequence alignments of AT.TSO2 and AL.TSO2 homologous genes.
Supplemental Figure S4. Pseudogenization due to the insertion of one A nucleotide at position 1,352.
Supplemental Figure S5. Confirmation of the presence of the transgene insertion of AL.STA1B in three independent transgenic plants each by PCR analysis.
Supplemental Figure S6. Distinction of AL.MTP11A and AL.MTP11B homologs.
Supplemental Figure S7. Measurements of soil salinity for the stress assays used in the microarray-based gene expression analyses.
Supplemental Table S1. Summary of Arabidopsis and A. lyrata array design features.
Supplemental Table S2. Classification of gene expression patterns and categorization of Arabidopsis-A. lyrata gene groups.
Supplemental Table S3. Arabidopsis mutants used in this study for genetic complementation with A. lyrata homologs.
Supplemental Table S4. Oligonucleotides used in this study for different purposes.
Supplementary Material
Acknowledgments
We thank Glenn Thorlby, Zhongchi Liu, and Byeong-ha Lee for providing mutant seeds; Birgit Geist, Wei Zhang, and Elisabeth Becker for technical assistance; Andreas Albert, Werner Heller, Soumita Poddar, and Debarun Acharya for helpful discussions; and Jörg Durner for continuous encouragement during the course of this study.
Glossary
- CDS
coding sequence
- EC
electrical conductivity
Footnotes
This work was supported by the Alexander von Humboldt Foundation (postdoctoral fellowship to M.D.), the University Grant Commission (grant no. MRP–MAJOR–BIOT–2013–18380), the Council of Scientific and Industrial Research (grant no. 38[1386]/14/EMR–II), an FRPDF grant (Presidency University), and the Department of Biotechnology, India (grant no. BT/PR10778/PBD/16/1070/2014).
Articles can be viewed without a subscription.
References
- Bandyopadhyay S, Sharan R, Ideker T (2006) Systematic identification of functional orthologs based on protein network comparison. Genome Res 16: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bergmann S, Ihmels J, Barkai N (2004) Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM. (1971) Solar UV irradiation and the growth and development of higher plants. In Giese AC. ed, Photophysiology, Vol 6 Academic Press, New York, pp 131–177 [Google Scholar]
- Castillo-Davis CI, Hartl DL, Achaz G (2004) cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res 14: 1530–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA (2006) Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci 11: 449–459 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, et al. (2013) Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Reichman JR, Haberer G, Welzl G, Aceituno FF, Mader MT, Watrud LS, Pfleeger TG, Gutiérrez RA, Schäffner AR, et al. (2010) A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus. Plant Mol Biol 72: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51: 198–210 [DOI] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Fischer I, Dainat J, Ranwez V, Glémin S, Dufayard JF, Chantret N (2014) Impact of recurrent gene duplication on adaptation of plant genomes. BMC Plant Biol 14: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T. (2008) Large-scale assignment of orthology: back to phylogenetics? Genome Biol 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ (2007) crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase—leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50: 118–127 [DOI] [PubMed] [Google Scholar]
- Gustin JL, Zanis MJ, Salt DE (2011) Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim ED, Chen ZJ (2009) Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci USA 106: 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ (2007) External factors accelerate expression divergence between duplicate genes. Trends Genet 23: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA (2008) The quest for orthologs: finding the corresponding gene across genomes. Trends Genet 24: 539–551 [DOI] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu J, Zhu JK (2006) STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCarthy T, Bergman A (2007) The limits of subfunctionalization. BMC Evol Biol 7: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. (2001) Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol Evol 16: 693–700 [Google Scholar]
- Movahedi S, Van de Peer Y, Vandepoele K (2011) Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant Physiol 156: 1316–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer EL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RV, Nahal HK, Breit R, Provart NJ (2012) BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J 71: 1038–1050 [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Florquin K, Lescot M, Marchal K, Rouzé P, van de Peer Y (2003) Computational approaches to identify promoters and cis-regulatory elements in plant genomes. Plant Physiol 132: 1162–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauge-Merle S, Falconet D, Fontecave M (1999) An active ribonucleotide reductase from Arabidopsis thaliana: cloning, expression and characterization of the large subunit. Eur J Biochem 266: 62–69 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street NR, Sjödin A, Bylesjö M, Gustafsson P, Trygg J, Jansson S (2008) A cross-species transcriptomics approach to identify genes involved in leaf development. BMC Genomics 9: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Throude M, Bolot S, Bosio M, Pont C, Sarda X, Quraishi UM, Bourgis F, Lessard P, Rogowsky P, Ghesquiere A, et al. (2009) Structure and expression analysis of rice paleo duplications. Nucleic Acids Res 37: 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2007) Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol 8: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K (2009) The flowering world: a tale of duplications. Trends Plant Sci 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy ND, Paepe AD, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 34.1–34.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z (2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Krochko JE (2009) Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell 21: 2203–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Faber C, Uchiki T, Fairman JW, Racca J, Dealwis C (2006) Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc Natl Acad Sci USA 103: 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.