The juvenile phase of maize is correlated with elevated expression of stress-related genes.
Abstract
As maize (Zea mays) plants undergo vegetative phase change from juvenile to adult, they both exhibit heteroblasty, an abrupt change in patterns of leaf morphogenesis, and gain the ability to produce flowers. Both processes are under the control of microRNA156 (miR156), whose levels decline at the end of the juvenile phase. Gain of the ability to flower is conferred by the expression of miR156 targets that encode SQUAMOSA PROMOTER-BINDING transcription factors, which, when derepressed in the adult phase, induce the expression of MADS box transcription factors that promote maturation and flowering. How gene expression, including targets of those microRNAs, differs between the two phases remains an open question. Here, we compare transcript levels in primordia that will develop into juvenile or adult leaves to identify genes that define these two developmental states and may influence vegetative phase change. In comparisons among successive leaves at the same developmental stage, plastochron 6, three-fourths of approximately 1,100 differentially expressed genes were more highly expressed in primordia of juvenile leaves. This juvenile set was enriched in photosynthetic genes, particularly those associated with cyclic electron flow at photosystem I, and in genes involved in oxidative stress and retrograde redox signaling. Pathogen- and herbivory-responsive pathways including salicylic acid and jasmonic acid also were up-regulated in juvenile primordia; indeed, exogenous application of jasmonic acid delayed both the appearance of adult traits and the decline in the expression of miR156-encoding loci in maize seedlings. We hypothesize that the stresses associated with germination promote juvenile patterns of differentiation in maize.
The juvenile phase of vegetative shoot development in angiosperms, defined by the lack of competence to flower, dedicates seedlings of annual species to successful establishment (Poethig, 2013). As development proceeds, plants gain the ability to flower, marking them as adult. Phase in many species of both annuals and woody plants also can be distinguished by heteroblastic variation between juvenile and adult leaves. Phase-associated heteroblasty in maize (Zea mays) is a developmentally rich phenomenon, spanning a variety of traits (Dudley and Poethig, 1993). For example, the shorter, rounder juvenile leaves lack trichomes but have a waxy epidermis whose cell walls are devoid of lignin and wavy in peridermal view. Adult leaves, in contrast, have a hairy epidermis lacking wax, and the walls of epidermal cells are reinforced with lignin and crenulated. They also possess the bulliform cells that curl the leaves in drought conditions. Such a variety in phase-specific traits supports the view that phase change is not simply the gain of the ability to flower but rather is a systemic maturation.
Vegetative phase change is under the control of a circuit of microRNA (miRNA)-regulated genes. The juvenile phase is defined by the duration of microRNA156 (miR156) expression, miR156 being necessary and sufficient for juvenility in Arabidopsis (Arabidopsis thaliana; Wu et al., 2009) and other plants (Wang et al., 2011). In Arabidopsis, miR156 expression is established early in embryogenesis (Nodine and Bartel, 2010). Its levels are high in germinating seedlings but then decline to end the juvenile phase (Yang et al., 2011). The Corngrass mutation of maize causes prolonged expression of two of its 12 miR156 loci, thereby extending the juvenile phase (Chuck et al., 2007). The relief of miR156 repression of target transcripts of SQUAMOSA PROMOTER-BINDING (SBP) transcription factors (Preston and Hileman, 2013) confers floral competence by activating the transcription of genes encoding several MADS transcription factors that promote flowering (Wang et al., 2009; Yamaguchi et al., 2009). SBPs also regulate flowering through the expression of miR172, which targets the transcripts of AP2 transcription factors, some of which inhibit flowering (Zhu and Helliwell, 2011). Another miR172 target in maize, GLOSSY15, is involved in heteroblasty, conferring juvenile leaf epidermis traits (Moose and Sisco, 1996). Although the regulation of miR172 and SBP as well as many of the interactions downstream are known (Huijser and Schmid, 2011), what causes the levels of miR156 to be high at germination and later decline remains an open question.
It has been demonstrated in several species that the decline in miR156 levels in the shoot is mediated by a signal from juvenile leaves and not other parts of the plant (Yang et al., 2013). Experimental manipulation of sugar levels supported the conclusion that Suc is that signal (Yang et al., 2013; Yu et al., 2013). Aside from a requirement for protein synthesis and the involvement of both transcriptional and posttranscriptional repression (Yu et al., 2013), the molecular mechanism by which sugars repress miR156 is unknown. The hormone GA3 also is capable of advancing the timing of vegetative phase change in maize (Evans and Poethig, 1995), although whether it does so by reducing miR156 levels has not yet been determined: miR156 levels are unaffected by a GA deficiency in rice (Oryza sativa) that delays phase change (Tanaka 2012). Rather, given the binding between DELLA proteins and SBPs, GA likely relieves the DELLA inhibition of SBP protein activity (Yu et al., 2012), which, in turn, activates the transcription of the SBP-responsive MADS factor genes (Mutasa-Göttgens and Hedden, 2009).
In order to identify candidate components of the networks both underlying the suite of phase-specific traits and affecting vegetative phase change in maize, we conducted a systematic comparison of gene expression in juvenile versus adult leaves. Leaves were chosen for this study because, although meristems initiate the leaf, the specification of the leaf’s phase identity occurs after the primordium is well established (Irish and Karlen, 1998; Orkwiszewski and Poethig, 2000) and because leaves have been shown to provide signals that influence miR156 levels (Yang et al., 2013). We examined leaves at plastochron 6, a stage long before any patterns of differentiation appear (Sylvester et al., 1990), taking advantage of the relatively large shoot meristem and the primordia it produces. Gene expression microarrays were employed to compare transcript levels among each of the first 12 leaves, thus spanning the set made from early postgermination at the beginning of the juvenile phase to well into the adult phase. The transcriptomes of juvenile leaf primordia were found to differ from those of adult leaf primordia largely in their higher expression of many photosynthetic and stress-responsive genes, confirming our previous results examining a smaller set of probes and leaves (Strable et al., 2008). These genes were tightly coexpressed in a small number of hierarchical clusters, and many shared sequence motifs in upstream regions, pointing to common regulation. Treatment with the stress-associated hormone jasmonic acid (JA) prolonged the juvenile phase and delayed the decline in miR156 levels, whereas reduced-JA mutants showed early phase change. Our results are consistent with the hypothesis that stress responses play a part in promoting the juvenile state in this annual species.
RESULTS
Each Successive Juvenile Leaf Is Less Different; Adult Leaves Are the Same
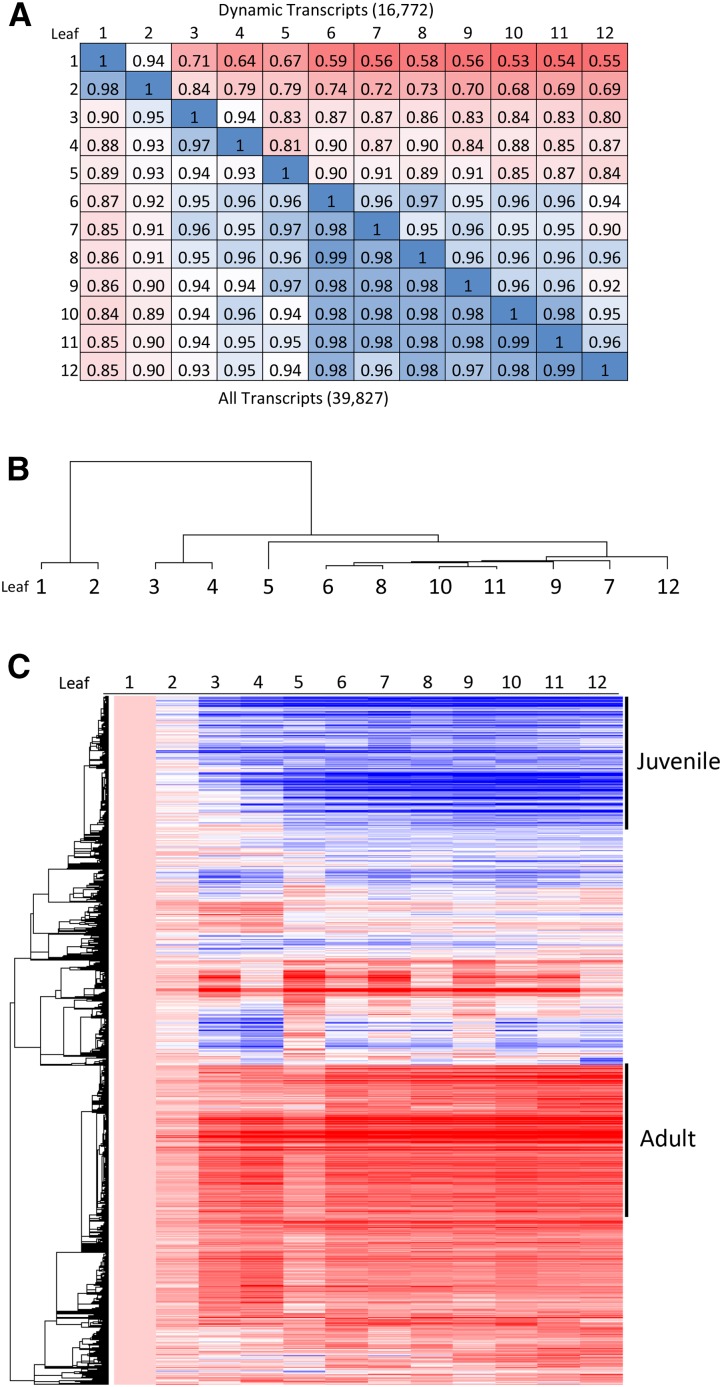
When fully expanded, juvenile and adult leaves of maize differ by a number of morphological traits, but at plastochron 6, all maize leaves are identical in appearance: tiny and pale yellow, with only the first veins apparent. Using microarray analysis, we found that, at this early stage, there were nonetheless large differences in patterns of gene expression. In pairwise comparisons of the first 12 leaf primordia, 16,772 transcripts (42.2% of the filtered gene set) showed at least a 2-fold difference in expression between any two of the 12 samples. The genome-wide similarity (Pearson r2) of expression of these so-called dynamic transcripts ranged from 0.53 (leaves 1 and 10) to 0.98 (leaves 10 and 11; Fig. 1A). The first-formed, juvenile leaves were most distinct, with steadily decreasing differences in gene expression profiles, leaves 7 to 12 being most similar. Pairwise comparisons excluding leaves 1 or 2 gave r2 values of 0.8 or greater, and comparisons among leaves 6 through 11 had values of 0.95 or greater. Likewise, hierarchical clustering separated primordia of leaves 1 and 2 as the most distinct, with leaves 3 and 4 and leaf 5 also clustering separately from the later-formed leaves (Fig. 1B). These divisions are consistent with the observed morphology in the genetic background used, in which leaf 4 is the last with wholly juvenile identity (waxy, hairless, and no lignin) and leaf 6 is the first with majority adult identity (hairs over most of leaf surface and lignin present), and with previous work suggesting that the first two maize leaves are distinct from other juvenile leaves (Bongard-Pierce et al., 1996).
Figure 1.
Genome-wide differences in gene expression among plastochron 6 leaf primordia. A, Pairwise comparisons of genome-wide 8-mm leaf primordia expression (Pearson r2), with red hue increasing with difference. B, Hierarchical clustering of leaf primordia 1 to 12 using 16,772 differentially expressed transcripts. C, Pearson hierarchical clustering in Arraystar of 16,772 variably expressed transcripts, normalized to leaf 1. Major juvenile and adult phase-specific clusters are indicated. Per row, the color scale excludes the highest and lowest 1% of values. Red color indicates highest expression, and blue color indicates lowest expression.
Examination of expression patterns among dynamically expressed genes led to our categorization as phase specific of any gene that showed either at least a 2-fold difference in expression between most juvenile and most adult samples or at least a 5-fold difference between the first two leaves and adult leaves. In comparisons of representative juvenile and adult leaf primordia, leaves 1 to 3 and 9 to 11, respectively (which also were sets of the least and most similar samples), we found that 1,107 transcripts (2.8% of the filtered gene set) showed significantly higher expression in one of the two phases (Supplemental Table S1). Some 921 transcripts were at least 2-fold more highly expressed in the juvenile leaves in all nine comparisons and/or were at least 5-fold more highly expressed in the six comparisons involving leaves 1 and 2. Some 243 transcripts were more highly expressed in the adult leaves. Gene Ontology analysis of the juvenile and adult sets of phase-specific genes identified photosynthesis as, by far, the most significantly enriched juvenile term, followed by involvement in oxidation reduction (redox) reactions and immune processes (Table I). The adult set was enriched in transcriptional regulators, also in agreement with earlier work in which a smaller set of genes was examined (Strable et al., 2008).
Table I. Biological process Gene Ontology of juvenile and adult subclusters, ranked by P values.
The juvenile cluster was further subdivided by those genes strongly up-regulated in leaves 1 and 2 only versus those up-regulated in all four juvenile samples. Fisher’s exact test after multiple testing correction was used to generate the P values. Enrichment represents the ratio of percentage in the set compared with percentage in the reference.
| Wilcoxon Sum Rank Tests of MapMan Categories | P | Phase |
|---|---|---|
| Major categories (bins) | ||
| Photosynthesis | 4.39E-30 | Juvenile |
| Secondary metabolism | 8.39E-12 | Juvenile |
| Cell | 2.76E-08 | Adult |
| Amino acid metabolism | 4.72E-07 | Juvenile |
| Cell wall | 2.18E-06 | Juvenile |
| Tetrapyrrole synthesis | 2.34E-06 | Juvenile |
| RNA | 4.35E-06 | Adult |
| DNA | 5.16E-05 | Adult |
| Metal handling | 3.16E-03 | Juvenile |
| Transcription factor families | ||
| SBP family | 4.81E-04 | Adult |
| B3 transcription factor family | 6.61E-04 | Adult |
| MADS box transcription factor family | 7.75E-04 | Adult |
| C2C2(Zn) GATA transcription factor family | 1.91E-03 | Juvenile |
| CCAAT box-binding factor family, HAP2 | 6.50E-03 | Adult |
| Chromatin-remodeling factors | 1.14E-02 | Adult |
| ARF, auxin response factor family | 1.43E-02 | Adult |
| C2C2(Zn) YABBY family | 1.61E-02 | Adult |
| GRAS transcription factor family | 1.84E-02 | Juvenile |
| C2C2(Zn) CONSTANS-like zinc finger family | 1.95E-02 | Juvenile |
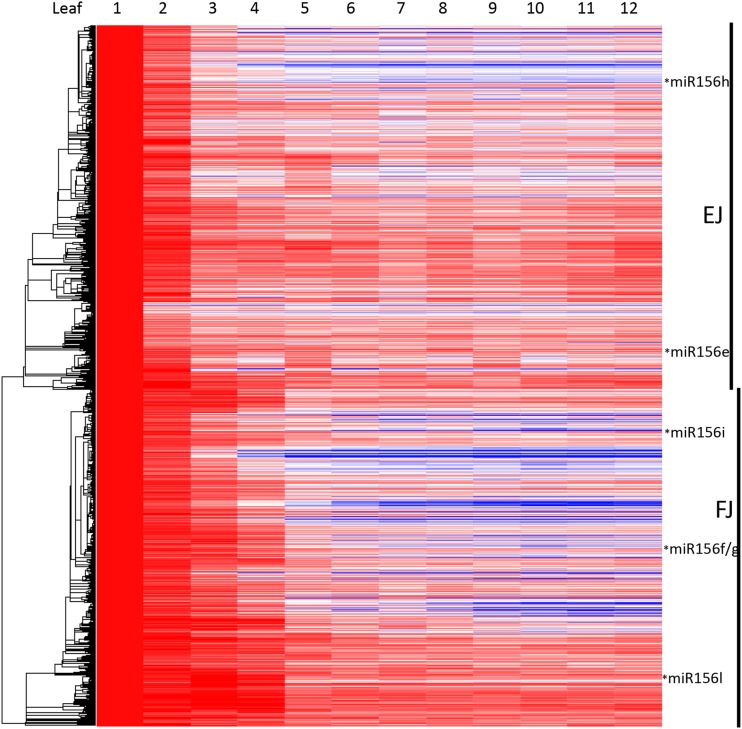
To confirm our criteria for identifying phase-specific genes, we subjected the dynamic transcripts to hierarchical clustering. Both juvenile and adult gene sets were positioned within distinct clusters (Fig. 1C). Some 86% of the adult-specific genes were located within a single 4,196-transcript cluster, and 94% of juvenile-specific genes were located within a single 3,385-transcript cluster (Fig. 1C). The latter cluster, 25.6% of which had met our criteria as juvenile-specific transcripts, cleanly subdivided into approximate halves, with an early juvenile cluster (1,760 transcripts) characterized by high expression limited to the first two leaf primordia and a so-called full juvenile cluster (1,625 transcripts) where high expression extended into the third and fourth leaf primordia (Fig. 2). Gene Ontology analysis indicated that these two juvenile subclusters were enriched in different functions: redox reactions and stress responses were especially high in the first two leaves, while photosynthetic and chlorophyll biosynthesis genes were highly expressed throughout the juvenile phase (Table I).
Figure 2.
Pearson hierarchical clustering in Arraystar of 3,385 juvenile cluster transcripts. EJ and FJ indicate constituents of early juvenile and full juvenile clusters, respectively. Transcripts of miR156 loci are located as indicated. Per row, the color scale excludes the highest and lowest 1% of values. Red color indicates highest expression, and blue color indicates lowest expression.
Transcriptional Regulators Show Phase Specificity
Maize has 12 miR156 loci, miR156a to miR156l. Whereas the expression patterns of individual loci varied substantially, their overall expression tended to decrease sharply during the juvenile phase, with highest levels in leaf 1, lower but equivalent levels in leaves 2 and 3, a step down in leaves 4 and 5, and a constant lower level in subsequent leaves. The most highly expressed loci were miR156g and miR156f, which were tightly coregulated (r2 > 0.97) and were located in the full juvenile cluster, as were miR156i and miR156l (Fig. 2). miR156h and miR156e were located in the early juvenile cluster. miR156j and the two overexpressed Corngrass1 mutants, miR156b and miR156c (Chuck et al., 2007), were minimally expressed in plastochron 6 leaf primordia (data not shown). miR172 expression was appreciable from only one locus, miR172c (Fig. 3), which increased 18-fold between leaves 1 and 5 and continued to increase in expression in successive leaves. The other four miR172 loci, including miR172e, whose loss of function is the tasselseed4 mutation, were minimally expressed (data not shown) in plastochron 6 primordia.
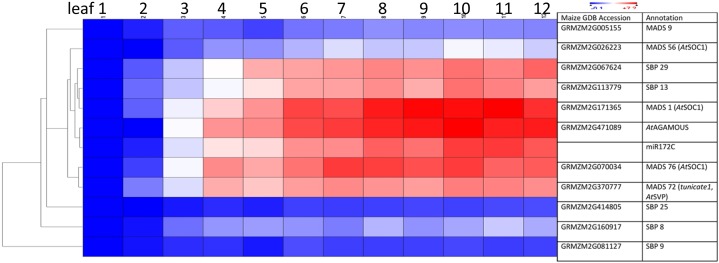
Figure 3.
Adult up-regulated transcripts include those for SPBs and are coexpressed with miR172c. Per row, the color scale excludes the highest and lowest 1% of values. Red color indicates highest expression, and blue color indicates lowest expression.
During vegetative phase change, the decline of miR156 expression results in the up-regulation of SBP transcription factors in primordia of leaves destined to become adult. Of the 32 SBPs in maize, 19 were differentially expressed at plastochron 6, including six that lack miR156 target sites. Transcript levels, of course, are only reflective of miR156 regulation by transcript cleavage; inhibition of translation also is almost assured, as has been observed in Arabidopsis miR156 regulation (Gandikota et al., 2007). Transcription factors were overrepresented among the relatively few classes of transcripts that were strongly adult up-regulated (Table I). MapMan analysis found the adult up-regulated SBP, B3, and MADS families to be the most phase biased (Table II). Genes encoding several SBP (SBP8, SBP9, SBP13, SBP25, and SBP29) and MADS (MADS1, MADS9, MADS56, MADS72, and MADS76) factors were coexpressed and within the same expression subcluster as miR172c using Pearson hierarchical clustering (Fig. 3). This is in agreement with the known SBP stimulation of MADS factors that promote maturation and flowering (Wang et al., 2009; Yamaguchi et al., 2009) and of miR172 (Zhu and Helliwell, 2011). SBP13 and SBP29 showed the greatest increases in transcript level. MADS1, MADS56, and MADS76 are putative orthologs of AtSOC1, which represses the expression of miR156 and juvenile AP2 factors by binding to their promoter sequences (Immink et al., 2012). MADS72 (tunicate1) is a putative ortholog of AtSHORT VEGETATIVE PHASE, which stimulates the transcription of miR172a by binding its promoter (Cho et al., 2012a). Transcription factors overrepresented in the juvenile phase included the CONSTANS-like zinc finger, GATA zinc finger, AP2, and GRAS families (Table II).
Table II. MapMan bins differing most greatly between leaf primordia 1 and 9.
P values indicate the false discovery rate after multiple testing correction.
| Significant Gene Ontology Process Terms | P | Enrichment |
|---|---|---|
| Juvenile | ||
| Photosynthesis | 7.8e-16 | 10.8 |
| Oxidation reduction | 4.1e-08 | 2.0 |
| Immune system process | 0.00013 | 3.2 |
| Lipid metabolic process | 0.00011 | 2.2 |
| Inorganic anion transport | 0.00073 | 4.7 |
| Early juvenile cluster | ||
| Oxidation reduction | 2.9e-15 | 2.0 |
| Response to stress | 8.9e-07 | 1.6 |
| Temperature homeostasis | 2.6e-06 | 1.7 |
| Oligopeptide transport | 2.2e-06 | 4.3 |
| Metal ion transport | 0.00034 | 1.9 |
| Full juvenile cluster | ||
| Photosynthesis | 1.9e-18 | 8.5 |
| Tetrapyrrole biosynthetic process | 7.5e-08 | 7.5 |
| Protein folding | 1.6e-05 | 2.4 |
| Translation | 0.00012 | 1.6 |
| Adult | ||
| Regulation of transcription | 1.1e-06 | 2.5 |
| Transcription, DNA dependent | 0.00096 | 2.2 |
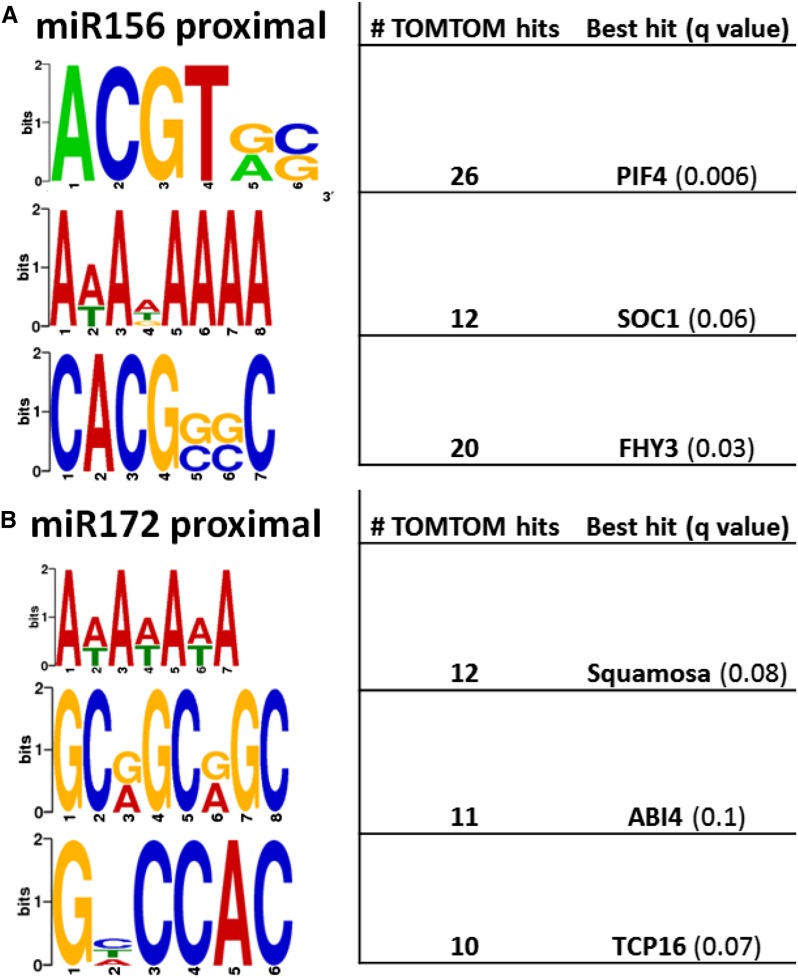
Promoter motif analysis of genes coexpressed in the hierarchical clustering with the two miRNAs showing strongest phase-biased expression, miR156f/g/i and miR172c, was conducted on the 400-bp upstream regions using SCOPE (Chakravarty et al., 2007) and DREME (Bailey and Elkan, 1994). These analyses identified motifs associated chiefly with light-responsive transcription factors in juvenile up-regulated genes (Fig. 4A) and MADS proteins in adult (Fig. 4B). The MADS binding-like motif of the proximal region of miR156-coexpressed genes (miR156 proximal), AWADAAA, is most similar to the binding recognition sequence of AtSOC1. The miR172 proximal regions were enriched in a similar motif, AWAWAWA, which also is predicted to be a recognition site for multiple MADS factors. A large proportion (43.5%) of miR156 proximal regions contained the motif ACGTRS, often as a staggered palindrome. This sequence is similar to the canonical G box motif CACGTG recognized by phytochrome-interacting factors (Martínez-García et al., 2000). Regulation of the juvenile photosynthetic cluster by light also is suggested by the motif CACGSSC, similar to the FAR-RED ELONGATED HYPOCOTYL3 (FHY3)-binding site CACGCGC (Fig. 4A); FHY3 integrates phytochrome A signaling with the circadian clock (Ouyang et al., 2011).
Figure 4.
Promoter motifs found in the upstream 400 bp of transcripts most closely coexpressed with miR156f/g/i (A) or miR172c (B). The 10 most enriched motifs identified by the SCOPE and DREME programs, queried against known binding sequences in TOMTOM and with hits at q ≤ 0.1 (indicating the minimum false discovery rate that would include the hit), are presented. All motifs except ACGTRS originate from DREME.
Phloem-Mobile Stress Signals Delay Vegetative Phase Change
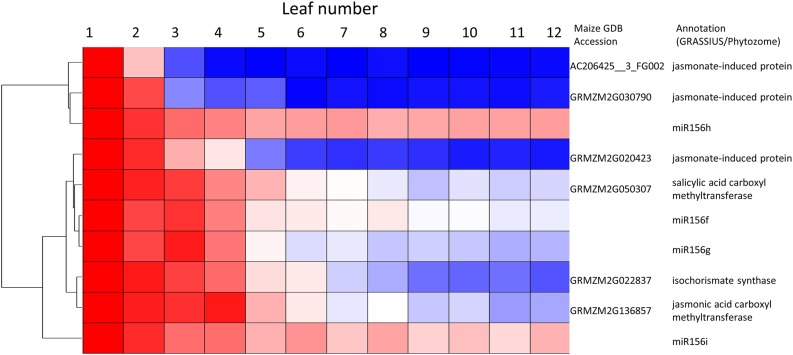
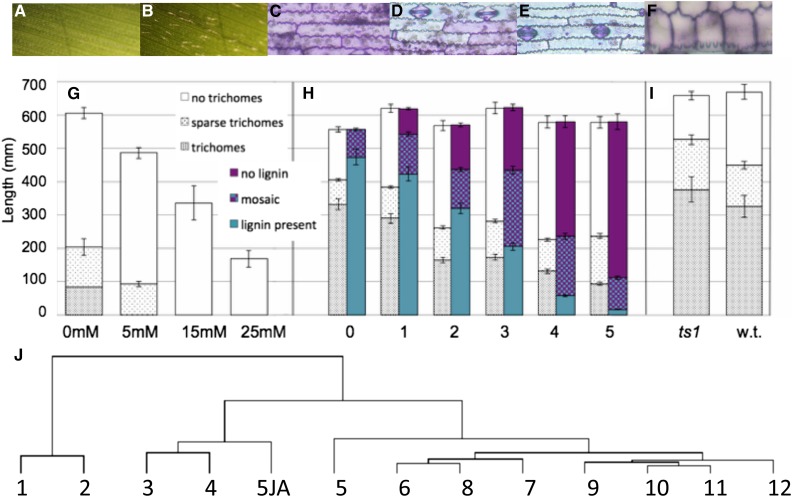
The enrichment of the early juvenile phase in stress signaling transcripts (Table I; Fig. 5) prompted us to hypothesize that the potential stresses that accompany seedling establishment in light might be among the factors that promote juvenility. The abundance of transcripts encoding enzymes that catalyze the synthesis or modification of SA, which is produced in response to biotrophic pathogens, and JA, which is produced in response to necrotrophic pathogens and insects and to abiotic stresses such as drought, suggested high levels of these phytohormones in juvenile primordia. Genes encoding methyltransferases that synthesize methyl salicylate and methyl jasmonate were strongly expressed; methyl salicylate has been demonstrated to be a phloem-mobile form of SA (Park et al., 2007). Three JA-induced proteins (JIPs) were among the most highly expressed juvenile transcripts (Supplemental Table S1). The expression of a gene encoding a 23-kD JIP, which has been shown to localize to phloem (Hause et al., 1996), strongly correlated with that of several miR156 loci (r2 = 0.93 with miR156f). Because of the up-regulation of JA-associated genes in our data and reduced miR156 levels in JA-deficient tasselseed1 mutants (Hultquist and Dorweiler, 2008; Acosta et al., 2009), JA emerged as a likely signaling candidate. Application of JA to the apical whorl of seedlings was indeed sufficient to delay the appearance of adult traits in transition leaves, which, in accordance with the basipetal pattern of differentiation in maize leaves, display juvenile traits at the tip and adult traits at the base (Fig. 6). Epidermal trichomes, staining patterns signifying the presence of lignin in cell walls (O’Brien et al., 1964), and bulliform cells all were displaced basipetally in a JA dosage-dependent manner. In contrast, those adult traits appeared precociously in tasselseed1 mutants. Treatment with 30% hydrogen peroxide similarly delayed vegetative phase change, while 100 mm SA had a modest but nonsignificant juvenilizing effect (Supplemental Fig. S1).
Figure 5.
Juvenile-specific coexpression of miR156 with selected JA- and salicylic acid (SA)-associated transcripts, normalized to leaf 1 expression. Per row, the color scale excludes the highest and lowest 1% of values. Red color indicates highest expression, and blue color indicates lowest expression.
Figure 6.
JA effects on phase-specific leaf traits. A to F. Phase-specific traits. A, No trichomes on juvenile leaves. B, Trichomes on adult leaves. C to F, Toluidine Blue O-stained epidermal peals. C, Juvenile. D, Transition. E, Adult. F, Bulliform cells (adult). G, A single treatment of increasing concentrations of JA at seedling emergence increasingly reduced the area with trichomes and the final length of leaf 5, the first transition leaf. n = 10. H, Increasing doses of 5 mm JA, 2 d apart, delayed the appearance of adult traits in leaf 6. n = 12. I, Leaf 6 of JA-deficient tasselseed1 (ts1) mutants develop adult-specific trichomes earlier than wild-type (w.t.) sibs. n = 8. Mutant leaf 6 stained entirely blue with Toluidine Blue O with adult-type crenulation (data not shown). J, Hierarchical clustering using combined juvenile- and adult-specific sets of genes. JA-treated leaf 5 (5JA) clusters with leaves 3 and 4; untreated leaf 5 clusters with adult leaves.
To determine whether JA-extended juvenility was associated with a prolonged period of high expression at miR156 loci, plastochron 6 primordia of leaf 5 from seedlings that had previously been treated with a single application of 15 mm JA were subjected to microarray analysis. Overall, JA-treated leaf 5 was much more similar to untreated leaf 3 (r2 of 0.81) or leaf 4 (r2 of 0.79) than to untreated leaf 5 (r2 of 0.69), mirroring the effect on leaf size (JA-treated leaf 5 was only as long as untreated leaf 3), and hierarchical clustering using phase-specific genes placed treated leaf 5 with leaves 3 and 4 (Fig. 6G). Significantly, the full juvenile miR156f, miR156g, and miR156i were each 2-fold higher in expression, while miR172c was more than 12-fold lower, in JA-treated plants (Supplemental Table S2), compared with untreated leaf 5. Transcripts of SBPs and other transcription factors also were broadly lower. Unsurprisingly, a JIP-encoding gene was among the most highly expressed in treated leaves, with a greater than 170-fold increase compared with untreated leaf 5, exceeded only by the expression of a lipoxygenase and an O-methyltransferase gene.
Juvenile Expression of Diverse Stress-Associated Genes
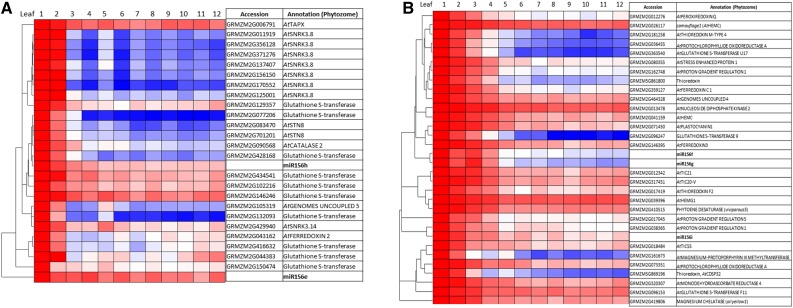
In addition to genes involved in stress hormone production, other stress-related genes showed highly elevated expression, in two distinct patterns. Genes encoding six of the maize SNF1-related kinases (SnRKs), which are central effectors of energetic stress responses, were tightly coexpressed in leaves 1 and 2 (Fig. 7A). Several glutathione S-transferase genes (GSTs), which conjugate glutathione to other proteins to maintain redox homeostasis, were similarly up-regulated in the early juvenile leaf primordia (Fig. 7). In contrast, several m- and f-type thioredoxin genes were highly expressed in all juvenile leaf primordia and coexpressed with miR156 (Fig. 7B).
Figure 7.
Leaf primordium expression patterns for selected redox and stress transcripts from the early juvenile cluster (A) and selected redox and photosynthetic transcripts coexpressed with miR156f/g/i loci (B). Per row, the color scale excludes the highest and lowest 1% of values. Red color indicates highest expression, and blue color indicates lowest expression.
DISCUSSION
The data presented here support the hypothesis that stress is among the factors that promote and maintain the juvenile phase in maize seedlings and that, once stress is relieved, the plant converts to adult patterns of differentiation. Although the first four or five leaves of maize, which are those with juvenile traits, are initiated during embryogenesis, even the first leaf shows minimal differentiation in the dry seed (Liu et al., 2013); thus, juvenility might be expected to be imposed during the process of germination. The most obvious stressor specific to juvenile leaves is photooxidative stress: upon emergence from the soil, seedling leaves must immediately commence the management of absorbed light energy. Whereas throughout the growing season each new leaf expanding into light is likely also to experience such stress, in the case of the seedling, the entire shoot undergoes deetiolation. Chloroplasts are a major source of reactive oxygen species (ROS) in plant cells, and chloroplast-generated ROS are a main pathway of retrograde regulation (Estavillo et al., 2013; Trotta et al., 2014). ROS peroxidation of membrane lipids forms phytoprostanes, which induce some of the same stress-responsive networks as JA (Stotz et al., 2013). Stress responses acting to prolong the juvenile phase are not difficult to reconcile with demonstrations that miR156 expression is sugar repressible (Yang et al., 2013; Yu et al., 2013). Many abiotic stresses converge on metabolic signaling pathways (Radomiljac et al., 2013; Tomé et al., 2014), and metabolism is linked generally to oxidative stress through redox regulation of Calvin cycle enzymes (Michelet et al., 2013). Several conserved gene families involved in the production of possible cell-autonomous juvenilizing factors that may regulate development downstream of abiotic stress pathways emerged from this study.
SnRKs, the plant homologs of yeast SNF1 and mammalian AMPK, are central effectors of energetic stress responses. Plant SnRKs have widely diversified beyond the ancestral SnRK1 subfamily, with distinct plant-specific SnRK2 and SnRK3 subfamilies that likely integrate metabolism with other stress responses (Halford and Hey, 2009). Our data identify SnRKs as present in both the adult and juvenile clusters, with a tightly coexpressed early juvenile group of six SnRK3s as especially prominent. No phase-change phenotype has yet been described for SnRK3 mutants, although SnRK1 regulation clearly affects developmental timing; this has been demonstrated by multiple studies in which overexpression delayed flowering and vegetative phase change (Baena-González et al., 2007; Tsai and Gazzarrini, 2012; Williams et al., 2014), and SnRK3s have been demonstrated to interact with SnRK1 in the regulation of sugar responses (Yan et al., 2014). Furthermore, interaction has been shown between SnRKs and the B3 transcription factor FUSCA3, which binds to the promoters of miR156 loci (Gazzarrini et al., 2004).
Several GSTs, which conjugate glutathione to other proteins to maintain redox homeostasis, also were highly juvenile up-regulated. Two are putative homologs of AtGSTU17, which substantially increases leaf number when mutated and decreases it when overexpressed (Chen et al., 2012). Interestingly, several GSTs were identified as down-regulated in a miR156-overexpressing line of switchgrass (Panicum virgatum; Fu et al., 2012). The juvenile up-regulation in maize, taken together with the GST overexpression phenotypes in Arabidopsis and switchgrass, suggest that miR156 shares stress-responsive regulatory pathways with a subset of GSTs and also that some GST activity likely promotes vegetative phase change. GSTs could conceivably encourage maturation by ameliorating developmentally inhibitory stress downstream of oxidative products like phytoprostanes (Stotz et al., 2013) or through broader interactions between antioxidants and immunity (Han et al., 2013). The extensively diversified plant thioredoxins link redox homeostasis and development through the activity of chloroplast proteins. Several m- and f-type thioredoxins were highly coexpressed with miR156 in juvenile primordia. The m-type thioredoxins have been shown to inhibit CEF (Courteille et al., 2013) and to be required for plasmodesmatal flow and SAM maintenance (Benitez-Alfonso et al., 2009). The tetrapyrrole pathway ultimately produces chlorophyll, and synthetic intermediates of heme are major regulators of retrograde signaling (Estavillo et al., 2013; Terry and Smith, 2013). Thioredoxin control of retrograde signaling through the tetrapyrrole pathway is well established (for review, see Serrato et al., 2013) and also is suggested in our data by coexpression or thioredoxins and tetrapyrrole synthesis enzymes (Fig. 6).
It has been shown that JA induces DELLA proteins, which function to target the JA ZIM domain proteins that repress JA-inducible gene expression (Wild et al., 2012); thus, JA may promote DELLA repression of SBP activity, thereby preventing flowering. JA and methyl jasmonate treatment has been shown to decrease the expression of SBP genes both with and without a miR156-binding site in grape (Vitis vinifera; Hou et al., 2013). JA and GA signaling through DELLAs is known to be antagonistic (Yang et al., 2012), with JA deficiency increasing GA sensitivity and vice versa. DELLAs also closely regulate oxidative stress responses by promoting the expression of detoxifying elements, including superoxide dismutases and GSTs (Achard et al., 2008).
miR156 and miR172 regulate phase change in woody species as well as Arabidopsis and maize (Wang et al., 2011). If stress is a common inducer of juvenility, acting by promoting high levels of miR156 in trees, other mechanisms would be expected to be involved in sustaining the juvenile phase, which can persist for decades. Stress as an inducer of juvenility in maize provides an appealing explanation for the phenomenon of rejuvenation by shoot apex culture in maize (Irish and Karlen, 1998); indeed, stress-associated genes were among those identified as most highly up-regulated in culture-reset leaves (Strable et al., 2008).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays) B73 and Mo17 (seeds kindly provided by P. Schnable) were crossed to generate hybrid seeds, which were planted and grown in a temperature-controlled greenhouse and illuminated for 14 h daily under 1-kW metal halide and sodium lights (www.osram.com). Seeds segregating for tasselseed1 seeds were a gift from Josh Strable. Plants were grown in Compost Plus growing mix (www.beautifullandproducts.com). Leaf primordia at the developmental stage of plastochron 6, measured as a length of 8 ± 2 mm, were selected for analysis, based on unpublished RT-PCR comparison of the expression of genes identified in a previous study of gene expression during vegetative phase change in maize (Strable et al., 2008). In this genetic background, leaves 1 to 4 displayed only juvenile traits, leaves 5 to 7 were transition leaves, with successive leaves displaying increasing amounts of adult tissue, and leaf 8 and above were entirely adult in morphology.
RNA Isolation and cDNA Synthesis
Leaf primordia were isolated by dissection of shoot tips and flash frozen in liquid nitrogen. Total RNA was extracted by Trizol (www.lifetechnologies.com) and purified on RNeasy columns (www.qiagen.com) using the manufacturers’ protocols. Double-stranded cDNA was synthesized using SuperScript III reverse transcriptase with an oligo(dT) primer according to the NimbleGen gene expression protocol booklet.
Microarray Design and Data Normalization
A custom microarray designed by us in consultation with Roche NimbleGen (NimbleGen), representing the primary transcripts of genes in the maizeGDB.org RefGen_v2 filtered gene set (39,656 genes) and 171 known maize miRNA precursor transcripts from mirbase.org (Supplemental Table S1), was used in these experiments. Arrays contained 120,000 60-mer probes, with three probes corresponding to each gene or miRNA transcript, which, in turn, were represented in triplicate. Array data were scaled in Arraystar 4 (www.dnastar.com) using robust multiarray average analysis with quantile normalization. Twelve arrays (three biological replicates with four technical replicates each) were initially hybridized. The r2 values among biological replicates were considered sufficiently high (0.98 or greater) that all subsequent samples were assayed by three technical replicates.
To identify differentially expressed genes, pairwise Student’s t tests with Benjamini-Hochberg multiple testing correction were performed. Transcripts less than 2-fold differentially expressed at 99% confidence in all pairwise comparisons (22,888 transcripts) were removed from the analysis. Additionally, 167 transcripts that were consistently more highly expressed in odd-numbered leaf primordia (1.5-fold higher than immediately preceding and succeeding even-numbered primordia) were removed from the analysis. Leaves to be used as standards in juvenile (leaves 1–3) and adult (leaves 9–11) comparisons were selected based on morphological indicators and genome-wide comparisons. Genes that were at least 2-fold differentially expressed at 99% confidence in nine comparisons (leaves 1, 2, and 3 compared with leaves 9, 10, and 11) or at least 5-fold differentially expressed in six comparisons (leaves 1 and 2 compared with leaves 9, 10, and 11) were considered to be juvenile or adult specific. Pearson hierarchical clustering was performed using Arraystar.
Gene Ontology Analysis
Singular enrichment analysis was performed using agriGO (Du et al., 2010). Gene sets were queried against the maize V5a reference set using standard settings (Fisher’s exact test with significance threshold of 0.05, Yekutieli multitest adjustment).
Pathway Visualization
Differentially expressed genes were visualized in MapMan (Thimm et al., 2004) using the 2012 B73 genome mapping. The difference in expression between leaf primordia 1 and 9 was used for visualization and for Benjamini-Hochberg corrected Wilcoxon sum rank tests.
Promoter Motif Analysis
Enriched promoter sequences (motifs) were identified using SCOPE (Chakravarty et al., 2007) and MEME (Bailey and Elkan, 1994). Motifs were compared with transcription factor binding site sequences in the 2014 JASPAR plant database (Mathelier et al., 2014) using TOMTOM (Gupta et al., 2007).
Seedling Treatment and Scoring
Emerging seedlings (first leaf partially expanded) received a single application of 100 µL of varying concentrations of JA (Cayman Chemical; aqueous solution), SA (Sigma Life Science; 40% ethanol solution), or hydrogen peroxide (Sigma Life Science; aqueous solution) to the apical whorl, which serves as a natural funnel. Mock treatments consisted of application of the same volume of solute. A 25 mm JA treatment caused approximately 20% mortality, while 50 mm JA killed all treated seedlings. Additional treatments with JA occurred at 2-d intervals. Leaves were examined for the presence and density of macrohairs once fully expanded. Epidermal cell wall characteristics (presence or absence of lignin, degree of cell wall crenulation, and presence of bulliform cells) were scored from peels stained with Toluidine Blue O.
Accession Numbers
Expression data can be found in the Gene Expression Omnibus under accession number GSE7495.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Treatment of seedlings with SA or hydrogen peroxide.
Supplemental Table S1. Adult-specific transcripts.
Supplemental Table S2. Selected genes showing change in expression in leaf 5 primordia in response to JA treatment compared to untreated leaf 5.
Supplementary Material
Acknowledgments
We thank Abby Long for help with the microarray experiments and the reviewers for their thoughtful comments.
Glossary
- miRNA
microRNA
- JA
jasmonic acid
- SA
salicylic acid
- JIP
jasmonic acid-induced protein
- ROS
reactive oxygen species
Footnotes
This work was supported by the National Science Foundation (grant no. 0820562) and by the Avis Cone Summer Fellowships (to B.B.).
References
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 23: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey T, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongard-Pierce DK, Evans MM, Poethig RS (1996) Heteroblastic feature of leaf anatomy in maize and their genetic regulation. Int J Plant Sci 157: 331–340 [Google Scholar]
- Chakravarty A, Carlson JM, Khetani RS, Gross RH (2007) A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics 8: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Kim JJ, Lee JH, Kim W, Jung JH, Park CM, Ahn JH (2012A) SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett 586: 2332–2337 [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Courteille A, Vesa S, Sanz-Barrio R, Cazalé AC, Becuwe-Linka N, Farran I, Havaux M, Rey P, Rumeau D (2013) Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol 161: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley M, Poethig RS (1993) The heterochronic Teopod1 and Teopod2 mutations of maize are expressed non-cell-autonomously. Genetics 133: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Chan KX, Phua SY, Pogson BJ (2013) Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front Plant Sci 3: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MM, Poethig RS (1995) Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol 108: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DG, Stewart CN Jr, Tang Y, et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 2: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Han Y, Mhamdi A, Chaouch S, Noctor G (2013) Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ 6: 1135–1146 [DOI] [PubMed] [Google Scholar]
- Hause B, Demus U, Teichmann C, Parthier B, Wasternack C (1996) Developmental and tissue-specific expression of JIP-23, a jasmonate-inducible protein of barley. Plant Cell Physiol 37: 641–649 [DOI] [PubMed] [Google Scholar]
- Hou H, Li J, Gao M, Singer SD, Wang H, Mao L, Fei Z, Wang X (2013) Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 8: e59358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hultquist JF, Dorweiler JE (2008) Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta 229: 99–113 [DOI] [PubMed] [Google Scholar]
- Immink RG, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish EE, Karlen S (1998) Restoration of juvenility in maize shoots by meristem culture. Intl J Plant Sci 159: 695–701 [Google Scholar]
- Liu WY, Chang YM, Chen SC, Lu CH, Wu YH, Lu MY, Chen DR, Shih AC, Sheue CR, Huang HC, et al. (2013) Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci USA 110: 3979–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez-Pérez ME, Francia F, Danon A, Marchand CH, Fermani S, Trost P, et al. (2013) Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 4: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH (1996) Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10: 3018–3027 [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Nodine M, Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24: 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TP, Feder N, McCully ME (1964) Polychrome staining of plant cell walls by Toluidine Blue O. Protoplasma LIX: 367–373 [Google Scholar]
- Orkwiszewski JA, Poethig RS (2000) Phase identity of the maize leaf is determined after leaf initiation. Proc Natl Acad Sci USA 97: 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2013) Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol 105: 125–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Hileman LC (2013) Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front Plant Sci 4: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomiljac JD, Whelan J, van der Merwe M (2013) Coordinating metabolite changes with our perception of plant abiotic stress responses: emerging views revealed by integrative-omic analyses. Metabolites 3: 761–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Fernández-Trijueque J, Barajas-López JD, Chueca A, Sahrawy M (2013) Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front Plant Sci 4: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Mueller S, Zoeller M, Mueller MJ, Berger S (2013) TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J Exp Bot 64: 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable J, Borsuk L, Nettleton D, Schnable PS, Irish EE (2008) Microarray analysis of vegetative phase change in maize. Plant J 56: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tanaka N. (2012) Gibberellin is not a regulator of miR156 in rice juvenile-adult phase change. Rice (N Y) 5: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt RS (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco-Llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, et al. (2014) The low energy signaling network. Front Plant Sci 5: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Rahikainen M, Konert G, Finazzi G, Kangasjärvi S (2014) Signalling crosstalk in light stress and immune reactions in plants. Philos Trans R Soc Lond B Biol Sci 369: 20130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS (2011) miRNA control of vegetative phase change in trees. PLoS Genet 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Niu F, Liu WZ, Zhang H, Wang B, Lan W, Che Y, Yang B, Luan S, Jiang YQ (2014) Arabidopsis CIPK14 positively regulates glucose response. Biochem Biophys Res Commun 450: 1679–1683 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Conway SR, Poethig RS (2011) Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62: 487–495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.