C-terminally encoded peptides together with the CRA2 receptor regulate nodulation and lateral root development through ethylene-dependent and ethylene-independent pathways, respectively, in Medicago truncatula.
Abstract
C-TERMINALLY ENCODED PEPTIDEs (CEPs) control root system architecture in a non-cell-autonomous manner. In Medicago truncatula, MtCEP1 affects root development by increasing nodule formation and inhibiting lateral root emergence by unknown pathways. Here, we show that the MtCEP1 peptide-dependent increase in nodulation requires the symbiotic signaling pathway and ETHYLENE INSENSITIVE2 (EIN2)/SICKLE (SKL), but acts independently of SUPER NUMERIC NODULES. MtCEP1-dependent inhibition of lateral root development acts through an EIN2-independent mechanism. MtCEP1 increases nodulation by promoting rhizobial infections, the developmental competency of roots for nodulation, the formation of fused nodules, and an increase in frequency of nodule development that initiates at proto-phloem poles. These phenotypes are similar to those of the ein2/skl mutant and support that MtCEP1 modulates EIN2-dependent symbiotic responses. Accordingly, MtCEP1 counteracts the reduction in nodulation induced by increasing ethylene precursor concentrations, and an ethylene synthesis inhibitor treatment antagonizes MtCEP1 root phenotypes. MtCEP1 also inhibits the development of EIN2-dependent pseudonodule formation. Finally, mutants affecting the COMPACT ROOT ARCHITECTURE2 (CRA2) receptor, which is closely related to the Arabidopsis CEP Receptor1, are unresponsive to MtCEP1 effects on lateral root and nodule formation, suggesting that CRA2 is a CEP peptide receptor mediating both organogenesis programs. In addition, an ethylene inhibitor treatment counteracts the cra2 nodulation phenotype. These results indicate that MtCEP1 and its likely receptor, CRA2, mediate nodulation and lateral root development through different pathways.
Root system architecture is determined by complex interactions between intrinsic root developmental and extrinsic environmental cues. In legumes, the formation of nitrogen (N) fixing root nodules and lateral roots (LRs) predominantly determine root system architecture. Cells that become competent for root nodule and LR formation are both similarly located close to the root apical meristem (RAM; Bhuvaneswari et al., 1980; Sargent et al., 1987; Herrbach et al., 2014). The N2-fixing nodules induced by rhizobia provide considerable N to legumes and enable them to grow in N-poor environments. As a result of this symbiosis, legumes provide protein-rich food, oil, fiber, and feed to agro-ecosystems, and they contribute to sustainable agriculture (Herridge et al., 2008; Jensen et al., 2012). Similarly, the improvement of LR deployment in crops is of significant interest to breeders to maximize water, nutrient, and fertilizer acquisition and alleviate pollution issues that arise from poor fertilizer uptake (Gamuyao et al., 2012); therefore legumes provide a system to study both nodule and LR organogenesis programs and how they interact. Although both LRs and nodules are induced by common environmental cues such as low N-availability (Ruffel et al., 2008; Jeudy et al., 2010; Jin et al., 2012), the common and specific pathways that regulate these different developmental competencies are poorly understood. Therefore, understanding signaling molecules, receptors, and downstream pathways that regulate both nodule and LR development in legumes under different environmental conditions is essential to improve nutrient acquisition by the root system.
C-TERMINALLY ENCODED PEPTIDEs (CEPs) have recently been implicated in controlling major aspects of root development (Ohyama et al., 2008; Delay et al., 2013; Imin et al., 2013; Tabata et al., 2014; Djordjevic et al., 2015; Mohd-Radzman et al., 2015). CEP peptides are signaling molecules that positively regulate nodulation and negatively regulate LR emergence in several legumes (Imin et al., 2013; Mohd-Radzman et al., 2015). An archetypal gene from the Medicago truncatula CEP family, MtCEP1, is induced under low-N and expresses during nodule and LR formation (Imin et al., 2013). Biochemical and mass spectrometry analyses demonstrated that MtCEP1-overexpressing and control root cultures both secrete a mixture of hydroxylated, 15-amino acid, MtCEP1 peptides. In root cultures, MtCEP1-dependent inhibition of lateral root formation occurs independently of shoots (Mohd-Radzman et al., 2015). In Medicago, the hydroxylation patterns of MtCEP1 peptides strongly influence how CEPs modify the extent of nodule and LR formation (Imin et al., 2013; Mohd-Radzman et al., 2015). How CEP peptides simultaneously regulate both root nodule and LR formation is unknown. At early developmental stages LRs and nodules share developmental genes (Franssen et al., 2015; Larrainzar et al., 2015) and morphological features, such as reactivation of cortical, pericycle, and endodermal cell divisions (Imin et al., 2013; Herrbach et al., 2014; Xiao et al., 2014; Mohd-Radzman et al., 2015), and it is therefore possible that overlapping genetic mechanisms may be involved. In addition, these two developmental pathways are initiated in cells in close proximity to the RAM (Imin et al., 2013; Herrbach et al., 2014; Xiao et al., 2014). It is not known if MtCEP1 peptides affect LR and nodule development through the same or different pathways, or if common developmental components are involved.
Medicago nodule formation is tightly associated with Sinorhizobium meliloti Nod factor (NF) production (Lerouge et al., 1990). S. meliloti induces two, parallel, nodule-specific processes to form indeterminate nodules adjacent to root proto-xylem poles: (1) the rhizobial infection pathway, which involves infection thread formation in root hairs and cortical cells; and (2) the nodule organogenesis pathway, which involves the activation of cell divisions in root cortical, endodermal, and pericycle cell layers to form a nodule primordium and, subsequently, a nodule meristem (Timmers et al., 1999; Desbrosses and Stougaard, 2011; Oldroyd et al., 2011, 2013; Xiao et al., 2014; Djordjevic et al., 2015). Rhizobial NFs rapidly trigger nuclear calcium oscillations in root hair cells (Lévy et al., 2004; Miwa et al., 2006), which transcriptionally activate key symbiotic (SYM) genes (e.g. NODULE INCEPTION and NODULATION SIGNALING PATHWAY1 and 2 [Kaló et al., 2005; Smit et al., 2005] and MtCLV3/ESR-RELATED12 and 13 [MtCLE12 and 13; Mortier et al., 2010; Saur et al., 2011]). Nodulation is also positively and negatively controlled by complex, context-dependent interactions with various hormones and peptides (Penmetsa and Cook, 1997; Gonzalez-Rizzo et al., 2006; Penmetsa et al., 2008; Mortier et al., 2010; Plet et al., 2011; Saur et al., 2011; Mortier et al., 2012; Larrainzar et al., 2015; van Zeijl et al., 2015). Collectively, these signals, along with the NF/SYM pathway, determine how, when, and where nodules form, as well as the overall nodule number on the root system (Penmetsa and Cook, 1997; Oldroyd et al., 2011). It is also noteworthy that the vast majority of infection threads that initiate fail to make a nodule (Djordjevic et al., 1985, 1986; Penmetsa and Cook, 1997), and this highlights the influence of negative regulatory circuits mediated by ethylene-related and CLE-related pathways (Penmetsa and Cook, 1997; Mortier et al., 2010; Kassaw et al., 2015).
Ethylene has a well-established negative role in M. truncatula nodulation, which is illustrated by the ethylene-insensitive, MtEIN2-defective mutant, sickle (skl; Penmetsa et al., 2008; Larrainzar et al., 2015). Rhizobia hyperinfect skl and form numerous underdeveloped nodules that are often fused (Penmetsa and Cook, 1997). Fused nodules have been defined as having two or more nodule meristems coalescing into one nodule structure confined by a single layer of endodermis (Xiao et al., 2014). In addition, skl nodules show spatially aberrant initiation sites in roots and can develop proximal to proto-xylem and -phloem poles (Penmetsa et al., 2003), whereas in wild-type roots, a localized ethylene production adjacent to proto-phloem poles is thought to prevent nodules developing in this area (Heidstra et al., 1997). Consistently, ethylene synthesis inhibitors such as aminoethoxyvinyl-Gly (AVG) promote both proto-phloem pole- and fused-nodule formation (Heidstra et al., 1997), as well as an increase in infection thread and nodule numbers (Guinel and Larue, 1992; Xiao et al., 2014). Conversely, the ethylene precursor, 1-aminocyclopropane-1-carboxylic-acid (ACC), reduces nodule number and, at high concentration, inhibits calcium oscillations and abolishes nodulation (Penmetsa and Cook, 1997; Oldroyd et al., 2001). Recently, a comprehensive transcriptional profiling of the Mtein2/skl mutant revealed a complex MtEIN2-dependent negative control of NF-mediated responses (Larrainzar et al., 2015).
Apart from the ethylene-mediated local inhibition of infection and nodule formation, nodule number is also negatively and systemically regulated by an autoregulation of nodulation (AON) pathway (Searle et al., 2003). In M. truncatula, AON is mediated by the interaction between nodule-specific CLE peptides, MtCLE12 and MtCLE13, and the SUPER NUMERIC NODULES (SUNN) Leu-rich repeat receptor-like kinase acting in shoots (Mortier et al., 2010; Saur et al., 2011). Similarly to skl, the sunn mutant has a supernumerary nodulation phenotype (Schnabel et al., 2005) but, unlike skl, sunn nodules form adjacent to xylem poles (Penmetsa et al., 2003). The combined negative action of ethylene and AON pathways leads to a tight restriction of the developmental competency to form symbiotic nodules (Bhuvaneswari et al., 1980; Sargent et al., 1987; Kassaw et al., 2015). Indeed, when a root is inoculated, nodule formation results from infections initiated in young, developing root hairs, and competency for further root hair infection and nodule organogenesis diminishes rapidly due to the activation of the local ethylene/EIN2-dependent (Penmetsa and Cook, 1997) and systemic CLE/AON-dependent responses (Kassaw et al., 2015). This tight control of nodulation capacity allows the determination of the duration of the competency for root nodulation and offers insights into how signaling molecules can alter this developmental competency.
Recently, a COMPACT ROOT ARCHITECTURE2 mutant (cra2) defective in a Leu-rich repeat receptor-like kinase distinctive from SUNN, was identified in M. truncatula. In contrast to SUNN, CRA2 positively controls root nodule formation systemically from shoots, and negatively regulates LR development locally (Huault et al., 2014). The cra2 phenotypes, therefore, are opposite to those observed by raising MtCEP1 levels (Imin et al., 2013). CRA2 is most closely related to the Arabidopsis (Arabidopsis thaliana) XYLEM INTERMIXED IN PHLOEM (XIP1)/CEP RECEPTOR1 (CEPR1) receptor (Bryan et al., 2012) that specifically binds CEP peptides (Tabata et al., 2014). This makes CRA2 a candidate receptor for MtCEP1 peptides.
In this study, pathways acting downstream of MtCEP1 peptides to regulate nodule and LR development were identified. A genetic analysis was used to dissect the role of the NF/SYM, EIN2, and AON pathways in the developmental responses controlled by the MtCEP1 domain1 peptide with Pro hydroxylation at the 4 and 11 positions, which is produced in roots and affects their development (Mohd-Radzman et al., 2015). We showed that MtCEP1 requires the SYM pathway and EIN2, but not SUNN, to stimulate nodulation, whereas MtCEP1 effects on LR regulation occur independently of the SYM pathway, EIN2, and SUNN. The phenotypic basis of MtCEP1-mediated increase in nodulation was then investigated in detail, by (1) measuring the developmental competency of roots and the rhizobial infection capacity and by (2) assessing nodule developmental features such as the level of fused nodule formation. The phenotypes induced by MtCEP1 were reminiscent of those of the ethylene-insensitive mutant skl, and accordingly, the ability of MtCEP1 peptide to stimulate nodule formation in the presence of a stimulator and an inhibitor of ethylene synthesis (e.g. ACC and AVG) was altered. We also tested if MtCEP1 affects overall ethylene production levels. Finally, we showed that MtCEP1 regulation of LR and nodules was dependent upon the CRA2 receptor, and that an ethylene synthesis inhibitor can counteract the cra2 nodulation phenotype.
RESULTS
The SYM Pathway Is Required for MtCEP1 Effects on Nodule But Not LR Formation
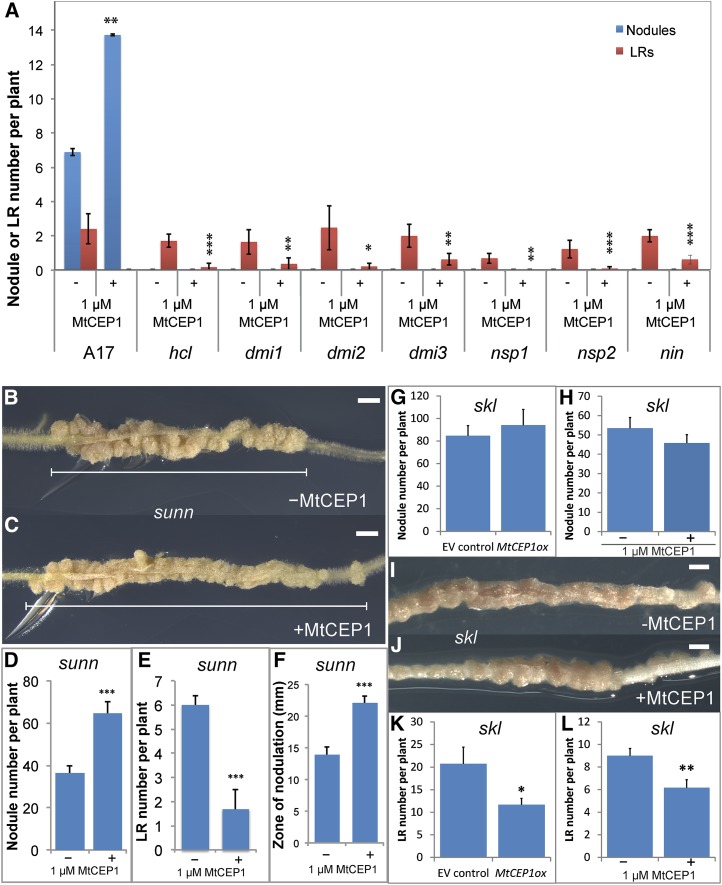
Mutants defective in the SYM pathway were treated with MtCEP1 peptides and assessed for root nodule and LR formation (Fig. 1A). In contrast to wild-type plants, which showed a significant increase in nodule number upon MtCEP1 peptide addition, no nodule was formed on roots of any of the MtCEP1 peptide-treated SYM pathway mutants tested (hcl, dmi1, dmi2, dmi3, nsp1, nsp2, and nin). The MtCEP1 peptide treatment, however, inhibited LR formation in all mutants. Therefore, MtCEP1 requires the presence of SYM pathway genes to enhance nodulation and is epistatic to the SYM pathway. In addition, the inhibitory effect of MtCEP1 on LR emergence occurs through an independent pathway.
Figure 1.
MtCEP1 requires the SYM and EIN2/SKL pathways to modulate nodule but not LR formation and acts independently of SUNN. A, Nodule number and emerged LR number were scored in the wild type and in SYM pathway mutants defective in NF perception (hcl), signal transduction (dmi1, dmi2, dmi3), and transcriptional regulation (nsp1, nsp2, nin; n ≥ 6; Student’s t test, *: P ≤ 0.05, **: P ≤ 0.01, ***: P ≤ 0.001). B to F, Effects of the MtCEP1 peptide application on sunn root phenotypes. B, C, and F, Representative images showing the effect of the MtCEP1 peptide application on the size of the root nodulation zone (scale bars = 2 mm). D and E, Effect of the MtCEP1 peptide application on nodule and LR number, respectively (n ≥ 5; Student’s t test, ***: P ≤ 0.001). G to L, The effects of raising MtCEP1 levels on skl phenotypes. G and H, Effect of MtCEP1 overexpression (n ≥ 21) or MtCEP1 peptide application (H, n ≥ 26), respectively, on the skl root nodule number. I and J, Representative images of the effect of the MtCEP1 peptide application on the width of the skl nodulation zone (scale bars = 2 mm). K and L, Effect of MtCEP1 overexpression or peptide application on the skl ELRs number. In all cases, the S. meliloti WSM1022 strain was used to nodulate plants and measurements were performed 2 weeks pi. Errors bars represent se.
MtEIN2/SKL But Not SUNN Is Required for MtCEP1-Mediated Effects on Nodule Formation
We examined if Mtein2/sickle (skl) and sunn retain phenotypic responsiveness to MtCEP1 (Fig. 1, B–K). In sunn, MtCEP1 peptides significantly increased nodule number and reduced LR number (Fig. 1, B–F). In addition, all sunn nodules clustered in a tight region of the root (hereafter termed the “nodulation zone”) and exposure to the MtCEP1 peptide increased the width of this nodulation zone by over 50% (Fig. 1E). In contrast to sunn, MtCEP1 peptide application, or MtCEP1 overexpression, did not increase nodule number in skl (Fig. 1, G–J) or the size of the nodulation zone (Fig. 1, I and J) and was unable to rescue the nodule development phenotype of skl (Fig. 1I). The ability of the MtCEP1 peptide or MtCEP1 overexpression to inhibit LR emergence was, however, retained in skl (Fig. 1, K and L).
MtCEP1 Increases the Root Developmental Competency for Nodule Formation
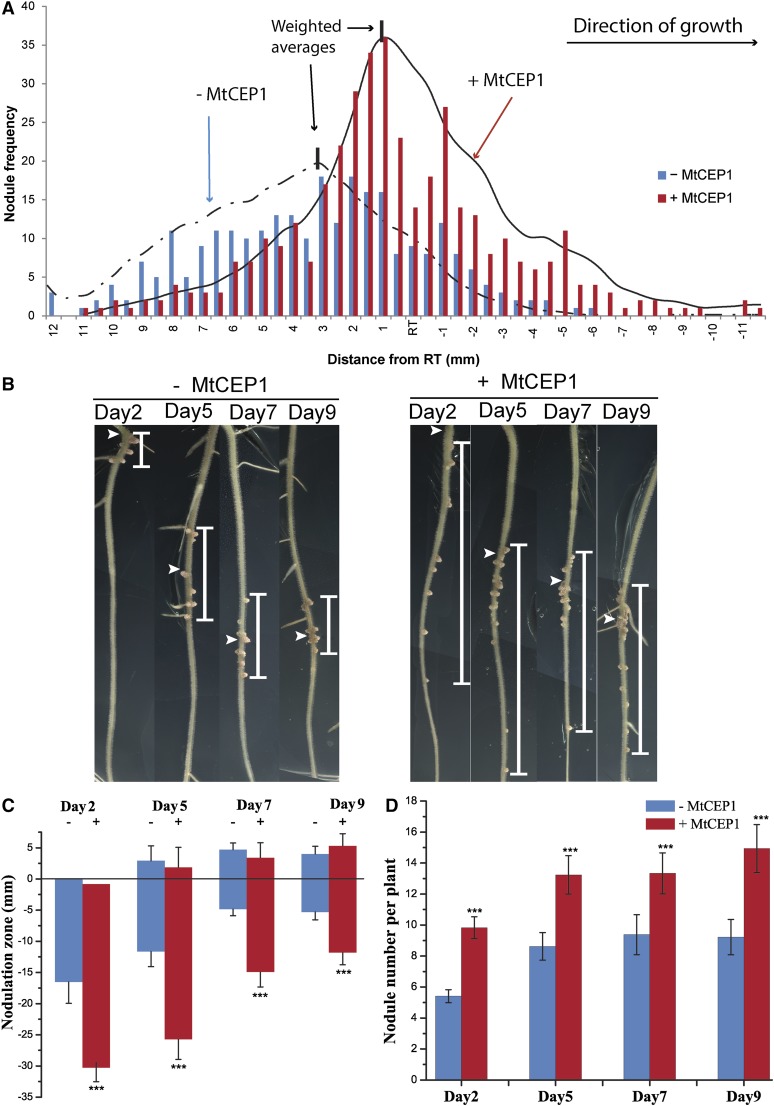
As MtCEP1 peptide treatment enabled sunn to support increased nodulation over a wider root area (Fig. 1E), this suggested that MtCEP1 increases the root developmental competency to form nodules. This feature was therefore assessed in more detail by determining the location of nodule development relative to the position of the root tip at the time of Rhizobium inoculation in a large cohort of wild-type plants in the presence or in the absence of a MtCEP1 pretreatment. In untreated plants, 81% of S. meliloti nodules occurred above the root tip position at the time of inoculation and more than 98% nodules formed within a tight 15-mm root region similar to that observed in sunn (Figs. 2A and 1, B–E). As with sunn, no nodule formed above or below this 15-mm region. The 15-mm zone corresponds to an approximately 48 h competency period, as the root growth rate is approximately 7 mm⋅d−1 in the presence or absence of MtCEP1. Similar to the response of sunn, in MtCEP1 peptide-treated roots, 98% of nodules formed within a 20-mm zone and a change in nodule distribution relative to the position of the root tip was observed compared to controls. With MtCEP1 treatment, only 64% of nodules formed above the initial position of the root tip, which represents a 21% decrease relative to controls. By contrast, 36% of nodules formed below the initial position of the root tip after MtCEP1 treatment and this represents a 1.9-fold increase compared to controls. The 20-mm zone corresponded to an approximately 72 h competency period. These results indicate that MtCEP1 increases nodule number and the duration of the root nodulation competency period, and it shifts successful nodulation toward developmentally younger root regions. Smoothed curves describing the response of treated and untreated roots (Fig. 2A) highlight the shift in the positioning of MtCEP1-treated nodules on roots. The location of the nodulation capacity peak was calculated by determining the weighted averages. The peak of nodulation formation in the untreated- and treated-roots was 3.4 mm and 0.5 mm, respectively, above the root tip position at the time of inoculation (Fig. 2A). This also indicates that MtCEP1 shifts the peak of nodulation to developmentally younger root regions.
Figure 2.
MtCEP1 increases the root developmental competency for nodulation. A, The locations of S. meliloti WSM1022 nodules were scored relative to the position of the root tip at the time of inoculation in control (blue bars) and MtCEP1 peptide-treated roots (red bars; n = 18). Most control nodules formed within a 15-mm zone (+10 mm to −5 mm relative to the root tip) and 81% of nodules formed above the position of the root tip at the time of inoculation. The MtCEP1 peptide-treatment shifted nodulation positions to developmentally younger root regions. Smoothed curves were overlayed with nodule frequency data for MtCEP1 peptide (1 μm)-treated (continuous line) or -untreated (discontinuous line) wild-type plants. The weighted average (see “Materials and Methods”) for the nodulation peak is 3.4 mm above the root tip (dashed vertical line) for untreated plants and 0.5 mm above the root tip for MtCEP1-treated plants (continuous vertical line; n = 18). B, The nodulation zone relative to the root tip at the time of inoculation is shown for seedlings at four different times pg. A wider zone of nodulation (represented by the white vertical bar) was observed on MtCEP1 peptide-treated roots inoculated with the S. meliloti WSM1022 strain on seedlings 2, 5, 7, or 9 dpg. White arrowheads indicate the root tip position at the time of inoculation. C, Quantification of the nodulation zone size with or without a MtCEP1 peptide-treatment relative to the root tip position. The root tip position is defined at 0 mm; positive numbers are above the root tip, negative numbers are below the root tip. D, Nodule number with or without a MtCEP1 peptide-treatment on seedlings at different times pg. n ≥ 16; Student’s t test, ***: P ≤ 0.001 in C and D. Error bars represent se. RT, Root tip.
The effect of MtCEP1 on the size of the root competency zone for nodulation was then assessed in seedlings at different times postgermination (pg; Fig. 2, B–D). Consistent with Figure 2A, nodules in control roots formed within a tight zone (approximately 15 mm in 2 d and 5 d postgermination [dpg] seedlings and approximately 10 mm in 7- and 9-dpg seedlings), and the MtCEP1 peptide treatment significantly widened the zone of nodule formation by 1.5- to 2-fold (Fig. 2, B and C). As expected, the MtCEP1 peptide treatment increased total nodule number in seedlings of all ages (Fig. 2D).
MtCEP1 Increases the Level of Root Hair Infection and Infection Thread Formation
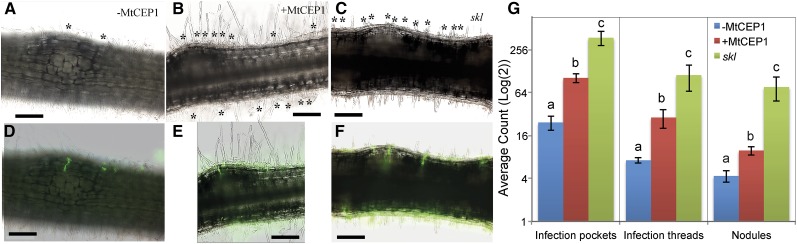
The MtCEP1-dependent increase in nodule number could be explained by elevating infection thread number. The number of infections was therefore audited on MtCEP1 peptide-treated and -untreated wild-type roots compared to skl hyperinfected roots (Fig. 3, A–F). Infection pockets (defined as “initiated infection thread foci in tightly curled root hairs”), infection threads, and nodules were counted using a S. meliloti GFP-labeled Sm1021 strain (Fig. 3G). The MtCEP1 peptide significantly increased the number of infection pockets, infection threads, and nodules compared to untreated controls. The numbers measured were found to be intermediate between the nontreated control and skl, which has an extreme hyperinfected phenotype (Fig. 3G). Similar trends were also observed using a Methylene Blue staining to assess the number of infection pockets, infection threads, and nodules in S. meliloti WSM1022-inoculated roots of wild-type and MtCEP1-overexpressing (MtCEP1ox) transgenic plants (Supplemental Figs. S1 and S2). MtCEP1ox roots have an approximately 4-fold increase in the number of infection pockets and infection threads and an approximately 3-fold increase in the total nodule number at 14 dpi compared to the empty vector control.
Figure 3.
MtCEP1 increases infection and nodulation capacities. A to C, Representative light micrographs of roots 120 h postinoculation (early nodulation stages) with a S. meliloti 1021-GFP strain in the zone of maximal nodulation susceptibility of wild-type (A and B) or ethylene-insensitive skl mutant (C) roots. Curled root hairs are indicated by asterisks. D to F, Green fluorescence images are merged with bright-field pictures shown (A–C), and infection threads penetrating into the cortex are detected in green fluorescence (bars in A–F = 200 μm). G, Quantification of infection pockets, infection threads, and nodules along root segments of equivalent length corresponding to the zone of maximal nodulation susceptibility (n = 12; ANOVA; P < 0.05). Error bars represent se.
MtCEP1 Induces the Development of Fused Nodules with an Altered Spatial Patterning
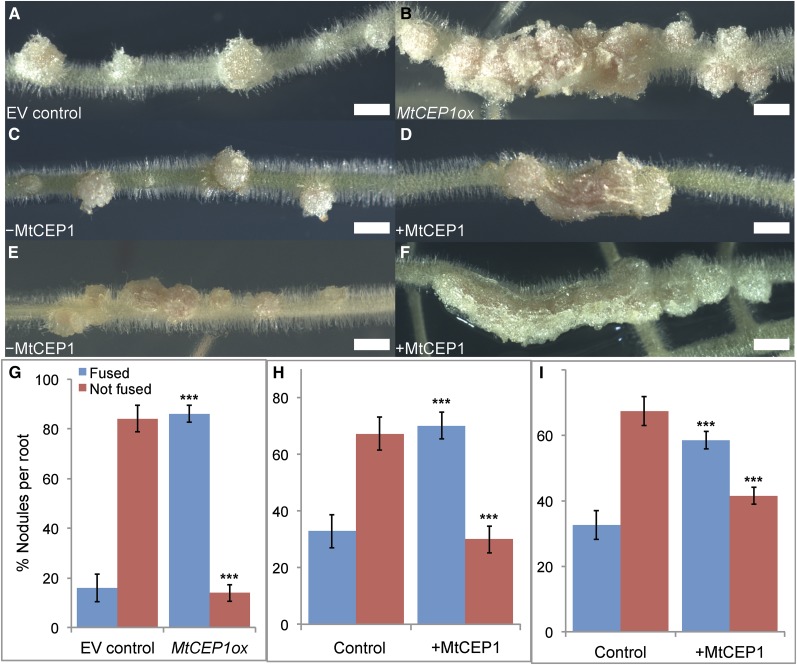
Because MtCEP1 increased infection number and the phenotypes depended on skl, we determined if it induced other phenotypic effects previously observed in the Mtein2/skl mutant (Penmetsa and Cook, 1997). We determined if the MtCEP1 peptide induced fused nodules or nodules with altered spatial development. Distinct, well-spaced nodules form on control Sm1021- or WSM1022-inoculated roots (Fig. 4, A, C, and E). MtCEP1 induced a significantly higher fused nodule number when inoculated with either strain Sm1021 or WSM1022 (Fig. 4, B, D, and F–I). Fused nodules formed along and across the root axis (Fig. 4, B, D, and F). Sm1021 formed 5.6-fold more fused nodules on roots overexpressing MtCEP1 and a significant decrease in nonfused nodules was observed (Fig. 4G). Similarly, Sm1021 or WSM1022 strains induced significantly more fused nodules and significantly fewer nonfused nodules when roots were treated with MtCEP1 peptide (Fig. 4, H and I).
Figure 4.
MtCEP1 promotes the formation of fused nodules. A and B, Representative EV and MtCEP1-overexpressing (MtCEP1ox) roots were infected with S. meliloti Sm1021. C to F, Wild-type controls (C and E) and MtCEP1 peptide-treated roots (D and F) were inoculated with the Sm1021 (C and D) or the S. meliloti WSM1022 (E and F) strain (bars in A–F = 2 mm). G, Quantification of fused nodules forming on roots transformed with MtCEP1ox compared with EV-transformed roots (n ≥ 18; Student’s t test, ***: P ≤ 0.001). H and I, Quantification of fused nodules formed on MtCEP1 peptide-treated plants compared to nontreated plants, inoculated with the Sm1021 (H) or the WSM1022 (I) strain (n ≥ 25; Student’s t test, ***: P ≤ 0.001). In G to I, error bars represent se. EV, Empty vector.
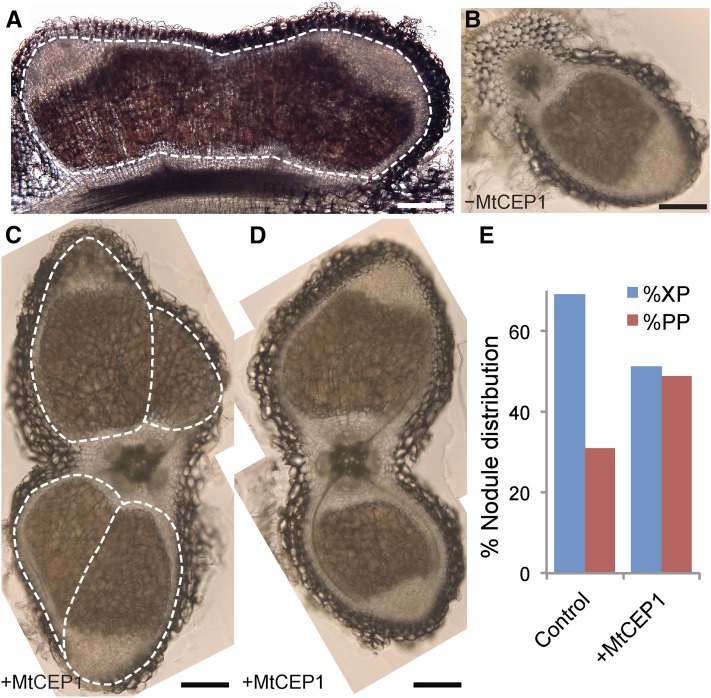
Longitudinal sections of a typical fused nodule (e.g. similar to Fig. 4D) showed two meristems, distinct vascular traces entering the fused nodule, and a single endodermal layer (Fig. 5A). In untreated roots, transverse sections confirmed that wild-type nodules preferentially formed adjacent to xylem poles (Fig. 5, B and E), whereas in MtCEP1 peptide-treated roots, nodules showed no preference for forming in the proximity of xylem or phloem poles (Fig. 5, C–E). The MtCEP1 peptide also enabled longer, wider, and multilobed nodules to form, which spanned two xylem poles and the phloem pole in between. These larger and multilobed nodules are connected by vascular bundles linked to two root xylem poles (Fig. 5, C and D). The larger nodules induced by the MtCEP1 peptide were not observed on untreated wild-type plants.
Figure 5.
MtCEP1 promotes nodule formation adjacent to phloem poles. A, Longitudinal section of a fused (multimeristematic) nodule comprising two nodule meristems and a single endodermal layer. At least two independent vascular bundles connect the fused nodule to the root. B to D, Transverse sections of a wild-type nodule forming near a xylem pole (B) or of nodules forming in the presence of MtCEP1 peptide (C and D). MtCEP1 peptide enables larger phloem pole-oriented nodules to form, often with multilobes as in (C). Bars in A to D = 500 μm. E, Quantification of nodule spatial distribution in MtCEP1 peptide-treated and untreated plants, in percentage, relative to XP poles or PP poles. (n ≥ 43; χ2 test, P ≤ 0.05.) WSM1022 was used as the inoculum. XP, Proto-xylem; PP, proto-phloem.
MtCEP1 Can Alleviate the Effects of Increased ACC Concentrations
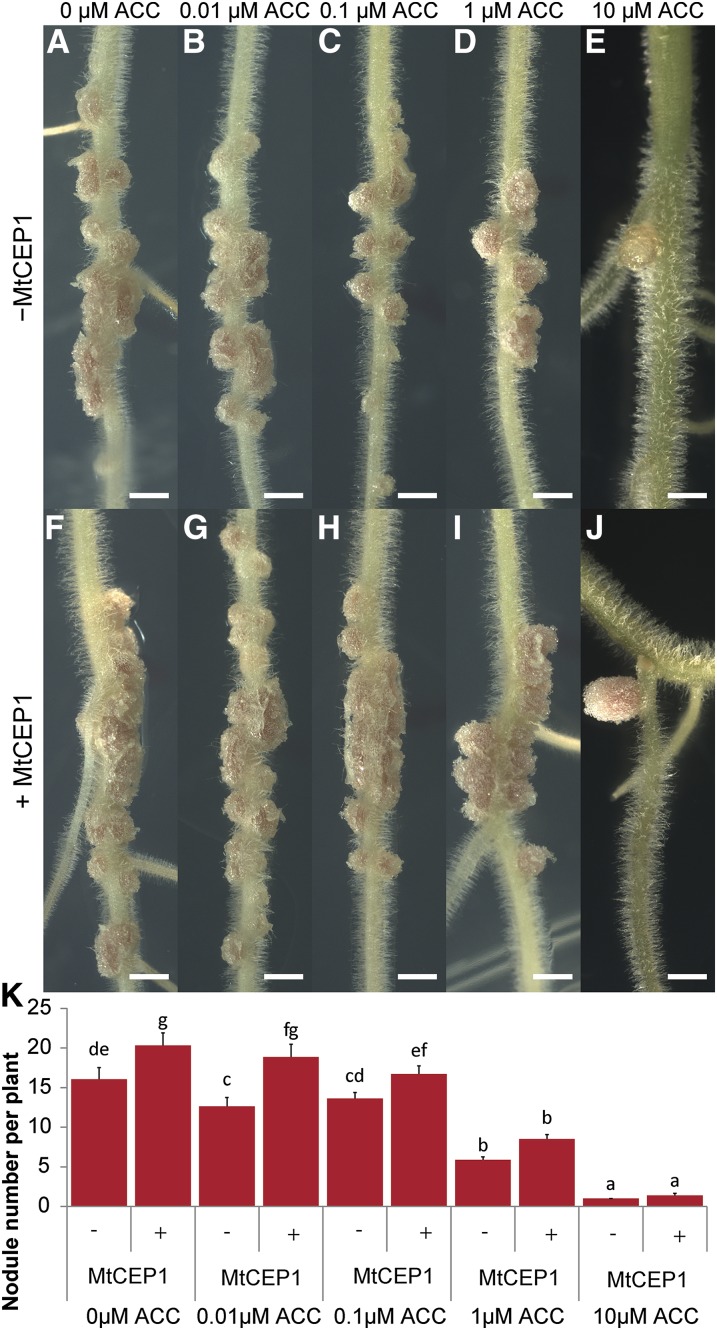
The observed increase in nodule and infection thread numbers and the formation of fused nodules in response to MtCEP1 are typical features observed in skl/ein2 (Penmetsa and Cook, 1997). In addition, MtEIN2 is required for MtCEP1-mediated effects on root nodulation (Fig. 1, G–J). This suggests that MtCEP1 stimulation of nodulation may involve a suppression of the MtEIN2-mediated ethylene signaling. To test this, we determined if MtCEP1 suppressed the effects of increasing concentrations of the ethylene precursor ACC. As expected, increasing ACC concentrations reduced nodulation levels progressively (Fig. 6, A–E). The MtCEP1 peptide cotreatment, however, significantly increased nodule numbers at 0.01 and 0.1 μm ACC (Fig. 6, A–C, versus Fig. 6, F–H and K) and enabled the formation of fused nodules up to 1 μm ACC concentrations (Fig. 6, F–I). ACC at 10 μm severely suppressed nodule number and only white nodules formed. By contrast, the MtCEP1 peptide-treatment allowed the development of larger pink nodules, suggesting an active N-fixation metabolic activity (Fig. 6, E and J).
Figure 6.
MtCEP1 alleviates ACC suppression of nodule formation. A to J, Plants were grown in N-free Fåhraeus medium without (−MtCEP1; A–E) or with MtCEP1 peptide (+MtCEP1; F–J) with increasing concentrations of the ethylene precursor ACC (0–10 μm). Nodules were counted 2 weeks postinoculation with the WSM1022 strain (bars = 2 mm). K, Quantification of the nodule number in response to increasing ACC concentrations, with or without MtCEP1 (n ≥ 22; two-way ANOVA, P ≤ 0.001). Error bars represent se.
MtCEP1 Enhancement of Nodulation Does Not Occur in the Presence of AVG
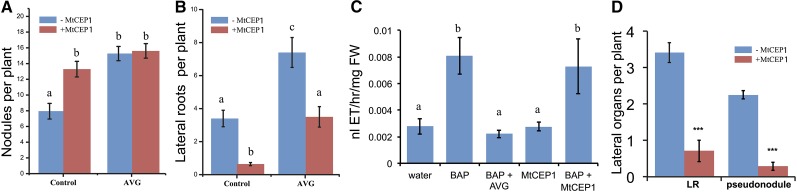
We tested the ability of MtCEP1 to affect development in the presence of an ethylene synthesis inhibitor as well as the effect of MtCEP1 on processes regulated by ethylene signaling. We first tested if the ethylene synthesis inhibitor, AVG, affected MtCEP1-stimulation of nodulation or inhibition of LR emergence. No MtCEP1 peptide-dependent enhancement of nodulation was observed in the presence of AVG, which significantly increased nodule numbers in both presence and absence of MtCEP1 (Fig. 7A). By contrast, AVG did not affect the ability of MtCEP1 to suppress LR emergence (Fig. 7B), in agreement with results obtained in the skl mutant (Fig. 1, J and K).
Figure 7.
The MtCEP1 effect on nodule number but not LR or pseudonodules numbers is impaired by AVG. A, Quantification of nodule number in S. meliloti WSM1022-inoculated plants grown on N-free Fåhraeus medium with or without the MtCEP1 peptide and/or AVG (1 μm). Nodules were counted 2 weeks postinoculation. Error bars represent se (n ≥ 6; one-way ANOVA, P ≤ 0.05). B, Quantification of emerged LRs in plants grown on a 5 mm KNO3 Fåhraeus medium with or without the MtCEP1 peptide and/or AVG (1 μm). LR number was quantified 14 d after germination. Error bars represent se (n ≥ 20; one-way ANOVA, P ≤ 0.05). C, Quantification of the ethylene production in single seedlings treated with or without the MtCEP1 peptide or the cytokinin BAP (0.01 μm), and/or the ethylene synthesis inhibitor AVG (1 μm). BAP and AVG were used as positive and negative controls, respectively, for the detection of increased or decreased ethylene levels. Ethylene was measured using laser-based trace gas detection 24 h after treatment. Error bars represent confidence interval (α < 0.05; n ≥ 6). D, Quantification of TIBA-induced pseudonodule and LR numbers with or without the MtCEP1 peptide. The numbers of LRs (left side) and pseudonodules (right side) were counted 2 weeks post-TIBA addition. Error bars represent se (n ≥ 52; Student’s t test, ***: P ≤ 0.001). FW, Fresh weight.
We then tested if MtCEP1 affected ethylene production over a 24-h period (Fig. 7C) using a very sensitive laser-based detection system enabling ethylene to be measured from single seedlings. Negative and positive controls were defined by measuring ethylene levels in response to a BAP treatment with or without AVG, respectively. As expected, BAP increased ethylene production and AVG counteracted this increase (Cary et al., 1995). In response to a MtCEP1 treatment, however, no significant change in ethylene content was detected and the effect of BAP on ethylene production was also not affected (Fig. 7C), suggesting that the MtCEP1 peptide does not affect the overall root pool of ethylene. Finally, the MtCEP1 peptide effect on pseudonodules induced by the auxin transport inhibitor TIBA was examined (Rightmyer and Long, 2011). Interestingly, pseudonodules form independently of the SYM pathway but are MtEIN2-dependent (Rightmyer and Long, 2011). We reasoned that if MtCEP1 reduced signaling through MtEIN2, then pseudonodule formation but not LR formation would be inhibited by the MtCEP1 peptide. A MtCEP1 and TIBA cotreatment indeed significantly reduced pseudonodule number, but not LR number (Fig. 7D).
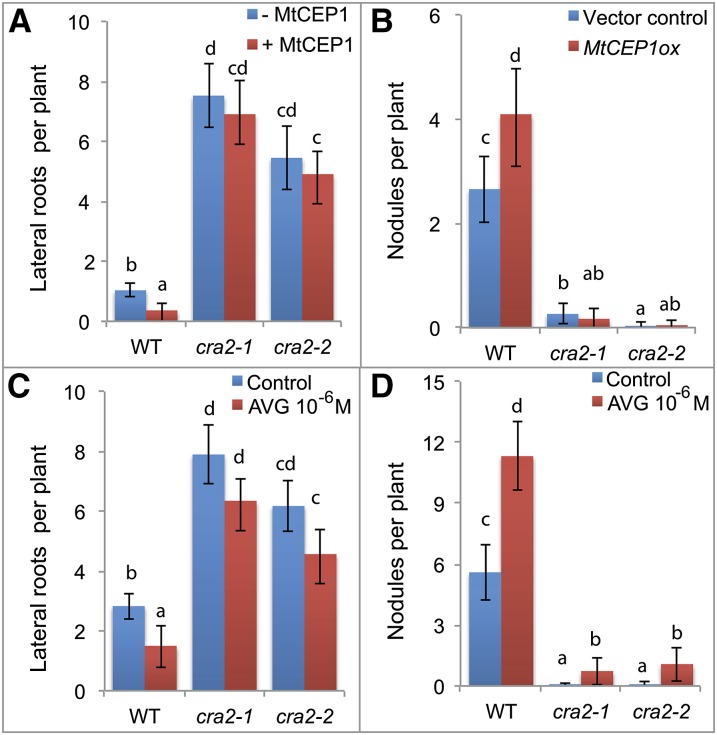
LR and Nodule Development in cra2 Is Unaffected by MtCEP1 and the cra2 Nodulation Phenotype Can Be Counteracted by AVG
The CRA2 receptor mutant has opposite LR and nodulation phenotypes to those imposed by MtCEP1 peptide application (Imin et al., 2013; Huault et al., 2014; Mohd-Radzman et al., 2015), and CRA2 is most closely related to the AtCEP1 peptide receptor XIP1/CEPR1 in Arabidopsis (Huault et al., 2014). Therefore, we tested if MtCEP1 affected LR and nodule formation in cra2 (Fig. 8, A and B). In contrast to wild-type plants, MtCEP1 peptides failed to decrease LR development or to increase nodule number in two independent cra2 mutant alleles. To determine if ethylene levels affect cra2 LR and nodulation phenotypes, an AVG treatment was performed, which enabled a significant increase in cra2 nodule number but did not significantly affect LR number (Fig. 8, C and D). The ability of AVG to counteract the low cra2 nodulation phenotype is in agreement with a model where MtCEP1 peptides would inhibit the ethylene/MtEIN2 pathway, depending on CRA2, to increase nodulation.
Figure 8.
CEP1 effect on nodulation and LRs depends on CRA2 and the cra2 nodulation phenotype is counteracted by AVG. A, LR and B, nodule number formed on wild-type and cra2 plants with or without MtCEP1. Due to the variability of A. rhizogenes composite plant root system architecture, the LR phenotype was assessed on MtCEP1 peptide-treated 7-d-old seedlings, whereas the nodulation phenotype was determined in MtCEP1 overexpressing roots compared to the empty vector control, 15 d after S. meliloti inoculation (n > 20). C, LR and D, nodule number of wild-type and cra2 plants with or without AVG (1 μm). The LR phenotype was assessed 7 d posttreatment, and nodulation 15 d after S. meliloti inoculation (n > 20). In all graphs, error bars represent confidence intervals (α = 0.05), and a Kruskal-Wallis test (α < 0.05) was performed to identify significant differences, indicated by letters.
DISCUSSION
Legumes have evolved mechanisms to simultaneously regulate the initiation, emergence, and growth of root nodules and LRs. The modulation of root lateral organ formation depends on intrinsic developmental and extrinsic environmental cues such as N availability. Previous work showed independently that MtCEP1 peptides and the MtCRA2 receptor play an unexpected major role in controlling both LR and nodule formation (Imin et al., 2013; Huault et al., 2014; Djordjevic et al., 2015; Mohd-Radzman et al., 2015). A common feature of LR and nodule development in M. truncatula is that these lateral organs are initiated from root cells located close to the RAM. Because MtCEP1 affects both LR and nodule developmental processes but not main root growth, this root region near the M. truncatula RAM is particularly receptive to MtCEP1 peptides (Imin et al., 2013). Prior work also showed that MtCEP1 acted rapidly to affect LR organogenesis in a shoot-independent manner (Imin et al., 2013; Mohd-Radzman et al., 2015). In addition, MtCEP1 and MtCRA2 both show gene expression in the vicinity of the root tip, which could enable a direct interaction between them (Imin et al., 2013; Huault et al., 2014), and MtCRA2 is also expressed in shoots, which is a prerequisite to be involved in systemic regulations from roots (Huault et al., 2014). The potential involvement of MtCRA2 and the underlying downstream pathways that enable MtCEP1 peptides to control these developmental processes and, therefore, to shape root system architecture depending on environmental conditions were, however, not yet described.
Here, we show that the SYM pathway is required for, and epistatic to, the MtCEP1 regulation of nodule number and that the MtCEP1-dependent inhibition of LR emergence occurs through an independent pathway. This clearly separates the MtCEP1-dependent downstream pathway stimulating nodulation from the MtCEP1 negative effects on LR emergence. Furthermore, MtCEP1 was unable to promote nodule formation in the ethylene-insensitive ein2/skl mutant but could still regulate LR number, whereas both MtCEP1 effects on LR and nodule formation were maintained in sunn. This shows that ethylene signaling through MtEIN2/SKL is an additional requirement for MtCEP1 action on nodulation, but not on LRs. Consistent with this model, the ethylene biosynthesis inhibitor AVG abolished the MtCEP1-dependent stimulation of nodule number but did not affect MtCEP1 inhibition of LR number. Hence, collectively, the genetic and pharmacological evidence presented here indicate that MtCEP1 affects LR and nodule number by independent pathways.
A model to explain the positive effect of the MtCEP1 peptide on nodulation (Fig. 9) is that the peptide dampens MtEIN2/SKL-dependent ethylene signaling, therefore attenuating its negative effect on rhizobial infections. Consistent with such MtEIN2/SKL-related action, MtCEP1 peptide also increases the number of infection events, as well as fused nodule formation and nodule number, which are phenotypes that resemble those of skl (Penmetsa and Cook, 1997; Penmetsa et al., 2003, 2008; Xiao et al., 2014). Additionally, similarly to skl phenotypes, MtCEP1 peptides increased the root competency zone for nodulation, the frequency of nodules forming in the proximity of phloem poles, and the number of nodules that spanned two xylem poles and the phloem pole in between. Furthermore, MtCEP1 provokes an attenuation of the ACC inhibition of nodulation and AVG inhibits the MtCEP1-dependent increase of nodule number. Finally, TIBA-induced and EIN2-dependent pseudonodule formation is inhibited by MtCEP1. This raises the possibility that MtCEP1 may affect ethylene metabolism and/or signaling, which is supported by our finding that MtCEP1 has no detectable effect on the root ethylene production.
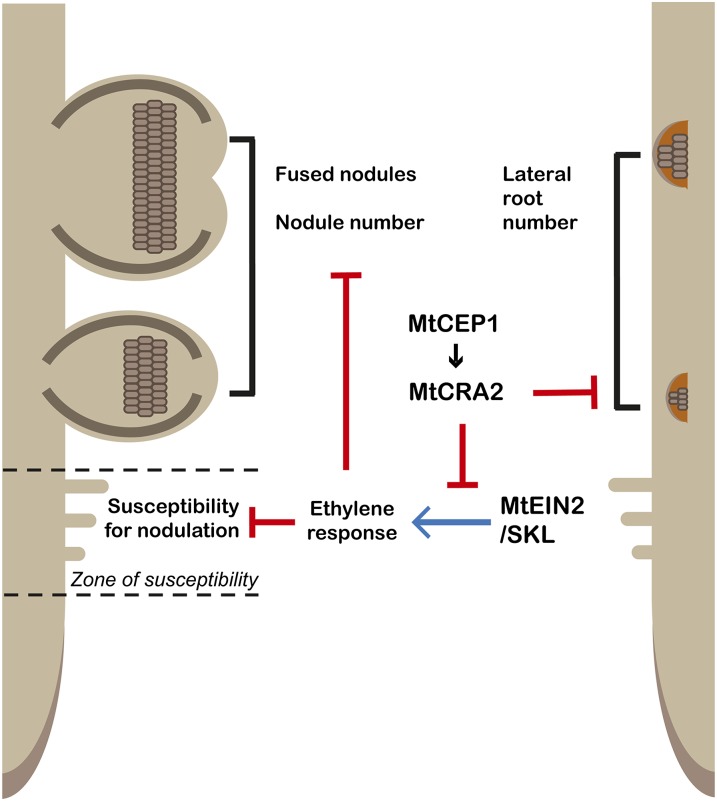
Figure 9.
A model for MtCEP1/MtCRA2 interaction during nodule and lateral root development. The MtCEP1 peptide requires the MtCRA2 receptor to regulate two different pathways determining M. truncatula root system architecture: LR development is inhibited independently of the EIN2/SKL pathway, whereas root nodule number is increased depending on the modulation of the EIN2/SKL signaling pathway. The MtCEP1 interaction with MtCRA2 is hypothesized to dampen the EIN2/SKL-mediated ethylene response during nodulation. This consequently leads to an increased number of rhizobial infection events, an extended developmental susceptibility of roots for nodulation, the formation of fused nodules, and the formation of nodules close to proto-xylem and -phloem poles.
Finally, the cra2 mutant, which is characterized by an increased LR number and a decrease in nodule number, is unresponsive to MtCEP1 peptide-dependent effects in these two developmental contexts, suggesting that CRA2 is a receptor for MtCEP1. In addition, treatment with an ethylene inhibitor can counteract the low cra2 nodulation phenotype. This supports a model where MtCEP1-CRA2 interactions modulate MtEIN2/SKL ethylene signaling to regulate nodule number (Fig. 9). This also implies that the CRA2 receptor is pivotal for MtCEP1 regulation of root lateral organ formation. It remains to be determined how the same peptide-receptor module can activate different downstream pathways depending on the root lateral organ considered. Because CEP gene expression is primarily regulated by environmental cues, and notably by N-availability (Delay et al., 2013; Imin et al., 2013), the MtCEP-CRA2 regulatory pathway is likely an important route for plants to control root system architecture in response to environmental influences such as N-availability. The widespread distribution of CEP genes in seed plants and their absence in early plant lineages (Delay et al., 2013; Ogilvie et al., 2014) suggests that CEP-dependent regulatory pathways are likely to operate widely. It is also expected that CEP signaling pathways contribute to the root system plasticity of various plant species. For instance, cepr1 phenotypes in Arabidopsis are somewhat different compared to cra2 phenotypes in M. truncatula (Tabata et al., 2014, Huault et al., 2014). In addition, the phenotypic effects of CEP peptides on lateral root and main root growth are different in Arabidopsis and Medicago (Delay et al., 2013; Imin et al., 2013; Tabata et al., 2014). Therefore, modulating CEP pathways may be a way to generate different root system architectures in various plants depending on their environments. In summary, the results support a model where CRA2 is a receptor for MtCEP1 peptides, and ethylene-MtEIN2/SKL is a downstream pathway required for MtCEP1 action on nodule formation.
MATERIALS AND METHODS
Medicago truncatula Lines and Growth Conditions
Seeds of M. truncatula wild-type genotypes Jemalong A17 or R108 and mutant lines were stratified and germinated on Fåhraeus medium (Kusumawati et al., 2008; Imin et al., 2013) and transferred to petri plates containing Fåhraeus medium without AVG (unless specified). Six seedlings per plate were grown in a growth chamber at 22°C with a 16 h photoperiod and a photon flux density of 100 μmol m–2 s–1 (Imin et al., 2013). To minimize light exposure to roots, and to optimize the phenotypic effects of CEP peptides on roots, the plates were 3/4 covered in black paper pockets. The following mutants were used in the A17 genotype: sickle1-1 (Penmetsa and Cook, 1997), sunn-4 (Schnabel et al., 2005), hcl-1 (B56), dmi1-4 (FN1), dmi2-1 (TR25), dmi3-1 (TRV25), nsp1-2 (C54), nsp2-2, and nin-1 (Catoira et al., 2000; Ané et al., 2002; Smit et al., 2005; Arrighi et al., 2006); and in the R108 genotype: cra2-1 and cra2-2 (Huault et al., 2014).
Chemicals
The MtCEP1 Domain1 Hyp 4, 11 peptide (Mohd-Radzman et al., 2015) was synthesized at the Biomolecular Resource Facility (Australian National University) or by GL BioChem with a greater than 95% purity, and validated by HPLC and mass spectrometry (Djordjevic et al., 2011). The peptide was resuspended in sterile water and added to media at 1 m. AVG (Olchemin), ACC, BAP (Sigma-Aldrich), or TIBA was added to media at the concentrations indicated.
Nodulation Assays
Standardly, 4-d-old germinated seedlings were grown on an N-free Fåhraeus medium before being inoculated with Sinorhizobium meliloti to induce nodule formation. The MtCEP1 peptide was added to the growth medium to provide continuous exposure. The S. meliloti strain WSM1022 (OD600 = 0.1), previously grown overnight at 28°C in a Bergersen’s modified medium, was centrifuged, resuspended in sterile water, and flood-inoculated onto roots (Imin et al., 2013). WSM1022 was used to assess the size of the root competency zone for nodulation, which is defined as the distance between the position of the first and the last emerged nodules on the primary root. The position and frequency of nodule formation relative to the root tip position at the time of inoculation was determined as described previously in Sargent et al. (1987). To determine the peak of nodulation, the distance of the nodule from the initial position of the root tip (A) and the nodulation frequency at each individual value (B) was used to calculate the weighted nodulation average using the formula = SUMPRODUCT(arrayA,arrayB)/SUM(arrayB) in Excel. The S. meliloti Sm1021 strain was used for nodulation on R108 and cra2 plants (Huault et al., 2014). In a separate experiment, MtCEP1 peptide-treated and control seedlings 2, 5, 7, and 9 dpg were used to assess the size of the nodulation zone using the WSM1022 strain.
Composite Plants
The MtCEP1 overexpression or the empty vector constructs were transformed into Agrobacterium rhizogenes strain ARqua1 and grown with spectinomycin (100 mg⋅L−1) and streptomycin (100 mg⋅L−1; Imin et al., 2013). Root transformation was performed as previously described in Saur et al. (2011) using kanamycin 25 mg⋅L−1 for selection. Given the highly branched phenotype of cra2 mutants and the heterogeneity of composite plant production, especially in R108, it was not possible to obtain meaningful results when comparing cra2 plants that were transformed with MtCEP1ox or vector control. Thus, for the observation of lateral root phenotypes of cra2 and R108, MtCEP1 synthetic peptide was used instead.
Visualization and Quantification of Infection Thread Formation
Infection thread and pocket formation was visualized using a GFP-labeled S. meliloti Sm1021 strain (Fournier et al., 2015) or a Methylene Blue staining for the S. meliloti WSM1022 strain (Vasse and Truchet, 1984). Methylene Blue staining was used both as an independent method for assessing infection thread and pocket formation and as the preferred method when transgenic roots expressing GFP were analyzed. Germinated seedlings were grown on Fåhraeus medium supplemented with or without the MtCEP1 peptide. After 3 d of growth, plants were flood-inoculated with either Sm1021 constitutively expressing GFP (OD600 = 0.01) or WSM1022 (OD600 = 0.001). The starting OD was carefully calibrated to minimize overcolonization of the root by rhizobia without compromising nodule number. Infection thread quantification for the GFP-labeled Sm1021 strain was microscopically accessed using approximately 15-mm root-nodulation-zone root segments at 96- and 120-h postinoculation (model no. DM550 B; Leica Microsystems), scoring infection pockets and infection threads separately. Nodule numbers were scored at 14 d postinoculation. For Methylene Blue staining, root segments corresponding to the approximately 15-mm root nodulation zone were observed 72 and 96 h postinoculation and stained with 0.01% (w/v) Methylene Blue for 30 min, then transferred to distilled water for 2 h for partial destaining. Root segments were briefly rinsed with 1 m NaCl, then replaced by distilled water for imaging.
Vibratome Sectioning and Microscopy
Root segments were embedded in 3% agarose and 100 μm sections were obtained using a vibratome (1000 Plus; Vibratome). The sections were then mounted on glass slides with water and observed with a DMBL microscope (Leica Microsystems). The root samples were observed using an SZX16 stereomicroscope (model no. SZX2-FGFPA; Olympus), and for higher magnifications with an SMZ1500 microscope (Nikon).
Induction of Pseudonodules
Induction of pseudonodules was done according to the protocol described in Rightmyer and Long (2011). Germinated M. truncatula seedlings were first grown on an N-free Fåhraeus medium with or without the MtCEP1 peptide. After 5 d, seedlings were flooded with 200 μm TIBA (Rightmyer and Long, 2011). The pseudonodules were then counted after 3 weeks.
Ethylene Measurement
Ethylene accumulation over 24 h was measured using an ETD-300 laser-based detection system (SensorSense) on 4-d-old seedlings grown in 5 mL glass vials and exposed to BAP (10 nm) in combination with AVG (Cayman Chemical; 1 m), or MtCEP1 (1 m). Measurements were conducted in sampling mode with a flow rate of 2.5 L/min and a 6-min sample period. The greater sensitivity of the laser-based system (Cristescu et al., 2013) enabled measurements to be made from individual seedlings, in contrast to other equipment types, which require pooled samples to generate detectable signals.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: for MtCEP1, EMBL: AL378645.1, CR495947.1, and AL378646.1; Affymetrix: Mtr.7265.1.S1_at; JCVI: contig_59554_1.1 (Mt3.5v5).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Assessment of the MtCEP1 peptide effect on infection thread and infection pocket formation.
Supplemental Figure S2. MtCEP1ox effect on infection thread and infection pocket formation.
Supplementary Material
Acknowledgments
Douglas Cook, Julia Frugoli, and Jean-Michel Ané provided sickle, sunn, and SYM pathway mutants, respectively. Ulrike Mathesius (Australian National University, Canberra) provided the GFP-labeled S. meliloti Sm1021 strain. The authors declare no conflict of interest.
Footnotes
Articles can be viewed without a subscription.
References
- Ané J-M, Lévy J, Thoquet P, Kulikova O, de Billy F, Penmetsa V, Kim D-J, Debellé F, Rosenberg C, Cook DR, et al. (2002) Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol Plant Microbe Interact 15: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Arrighi J-F, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet E-P, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvaneswari TV, Turgeon BG, Bauer WD (1980) Early events in the infection of soybean (Glycine max L. Merr) by Rhizobium japonicum: I. Localization of infectible root cells. Plant Physiol 66: 1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AC, Obaidi A, Wierzba M, Tax FE (2012) XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235: 111–122 [DOI] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH (1995) Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol 107: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet E-P, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu SM, Mandon J, Arslanov D, De Pessemier J, Hermans C, Harren FJM (2013) Current methods for detecting ethylene in plants. Ann Bot 111: 347–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA (2013) CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J Exp Bot 64: 5383–5394 [DOI] [PubMed] [Google Scholar]
- Desbrosses GJ, Stougaard J (2011) Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Innes RW, Wijffelman CA, Schofield PR, Rolfe BG (1986) Nodulation of specific legumes is controlled by several distinct loci in Rhizobium trifolii. Plant Mol Biol 6: 389–401 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Mohd-Radzman NA, Imin N (2015) Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot 66: 5171–5181 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Wong A, Singh M, Bhalla P, Kusumawati L, Imin N (2011) Border sequences of Medicago truncatula CLE36 are specifically cleaved by a subtilisin present in the extracellular fluids of Medicago and soybean. J Exp Bot 62: 4649–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic MA, Schofield PR, Rolfe BG (1985) Tn5 mutagenesis of Rhizobium trifolii host-specific nodulation genes result in mutants with altered host-range ability. Mol Gen Genet 200: 463–471 [Google Scholar]
- Fournier J, Teillet A, Chabaud M, Ivanov S, Genre A, Limpens E, de Carvalho-Niebel F, Barker DG (2015) Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol 167: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen HJ, Xiao TT, Kulikova O, Wan X, Bisseling T, Scheres B, Heidstra R (2015) Root developmental programs shape the Medicago truncatula nodule meristem. Development 142: 2941–2950 [DOI] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinel FC, Larue TA (1992) Ethylene inhibitors partly restore nodulation to pea mutant E 107 (brz). Plant Physiol 99: 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Yang WC, Yalcin Y, Peck S, Emons AM, van Kammen A, Bisseling T (1997) Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124: 1781–1787 [DOI] [PubMed] [Google Scholar]
- Herrbach V, Remblière C, Gough C, Bensmihen S (2014) Lateral root formation and patterning in Medicago truncatula. J Plant Physiol 171: 301–310 [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18 [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F (2014) Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet 10: e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–5409 [DOI] [PubMed] [Google Scholar]
- Jensen E, Peoples M, Boddey R, Gresshoff P, Hauggaard-Nielsen H, Alves B, Morrison M (2012) Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agron Sustain Dev 32: 329–364 [Google Scholar]
- Jeudy C, Ruffel S, Freixes S, Tillard P, Santoni AL, Morel S, Journet E-P, Duc G, Gojon A, Lepetit M, Salon C (2010) Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytol 185: 817–828 [DOI] [PubMed] [Google Scholar]
- Jin J, Watt M, Mathesius U (2012) The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol 159: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kassaw T Jr., Jr WB, Frugoli J (2015) Multiple autoregulation of nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 mutants. Plants (Basel) 4: 209–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumawati L, Imin N, Djordjevic MA (2008) Characterization of the secretome of suspension cultures of Medicago species reveals proteins important for defense and development. J Proteome Res 7: 4508–4520 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu HJ, Hwang HJ, Oh M, Kim GB, Surendrarao AK, Chasman D, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between Nod factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Downie JA (2006) Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J 48: 883–894 [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Binos S, Truong TT, Imin N, Mariani M, Djordjevic MA (2015) Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J Exp Bot 66: 5289–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S (2012) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J 70: 367–376 [DOI] [PubMed] [Google Scholar]
- Ogilvie HA, Imin N, Djordjevic MA (2014) Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, Lichtenzveig J, Gish J-C, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, et al. (2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J 55: 580–595 [DOI] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Rightmyer AP, Long SR (2011) Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol Plant Microbe Interact 24: 1372–1384 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, Tillard P, Jeudy C, Martin-Magniette ML, van der Merwe MJ, Kakar K, Gouzy J, Fernie AR, et al. (2008) Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol 146: 2020–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent L, Huang SZ, Rolfe BG, Djordjevic MA (1987) Split-root assays using Trifolium subterraneum show that Rhizobium infection induces a systemic response that can inhibit nodulation of another invasive Rhizobium strain. Appl Environ Microbiol 53: 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IM, Oakes M, Djordjevic MA, Imin N (2011) Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol 190: 865–874 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- van Zeijl A, Op den Camp RH, Deinum EE, Charnikhova T, Franssen H, Op den Camp HJ, Bouwmeester H, Kohlen W, Bisseling T, Geurts R (2015) Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol Plant 8: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Vasse JM, Truchet GL (1984) The Rhizobium-legume symbiosis: observation of root infection by bright-field microscopy after staining with methylene blue. Planta 161: 487–489 [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141: 3517–3528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.