Silencing two cytochome P450 genes reduces steroidal glycoalkaloid content and stops tuber sprouting.
Abstract
α-Solanine and α-chaconine, steroidal glycoalkaloids (SGAs) found in potato (Solanum tuberosum), are among the best-known secondary metabolites in food crops. At low concentrations in potato tubers, SGAs are distasteful; however, at high concentrations, SGAs are harmful to humans and animals. Here, we show that POTATO GLYCOALKALOID BIOSYNTHESIS1 (PGA1) and PGA2, two genes that encode cytochrome P450 monooxygenases (CYP72A208 and CYP72A188), are involved in the SGA biosynthetic pathway, respectively. The knockdown plants of either PGA1 or PGA2 contained very little SGA, yet vegetative growth and tuber production were not affected. Analyzing metabolites that accumulated in the plants and produced by in vitro enzyme assays revealed that PGA1 and PGA2 catalyzed the 26- and 22-hydroxylation steps, respectively, in the SGA biosynthetic pathway. The PGA-knockdown plants had two unique phenotypic characteristics: The plants were sterile and tubers of these knockdown plants did not sprout during storage. Functional analyses of PGA1 and PGA2 have provided clues for controlling both potato glycoalkaloid biosynthesis and tuber sprouting, two traits that can significantly impact potato breeding and the industry.
Potato (Solanum tuberosum) is the world’s fourth most important food crop after maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum). Potato steroidal glycoalkaloids (SGAs) are abundant poisons in tuber sprouts and green tubers and are described as bitter tasting, burning, scratchy, or acrid (Friedman, 2006; Ginzberg et al., 2009; Taylor et al., 2007). SGAs in the tuber are induced by exposure to light, low temperature, and mechanical injury (Valkonen et al., 1996). Producers and consumers have called for the removal of SGAs from potatoes. Only potato with SGAs of major food crops has such broad industry consensus on the need to solve this important worldwide problem. Controlling the SGA content is also important for potato breeding. Wild germplasm has been used in potato breeding programs as a source of pest and disease resistance. Since high SGA concentrations are found in most wild tuber-bearing species, introgression of wild germplasm may increase the risk for high SGA levels. Although the initial concentrations are still low in new breeding lines, dangerous levels of SGAs can accumulate due to environmental factors, pathogen infections, and postharvest treatments (Valkonen et al., 1996).
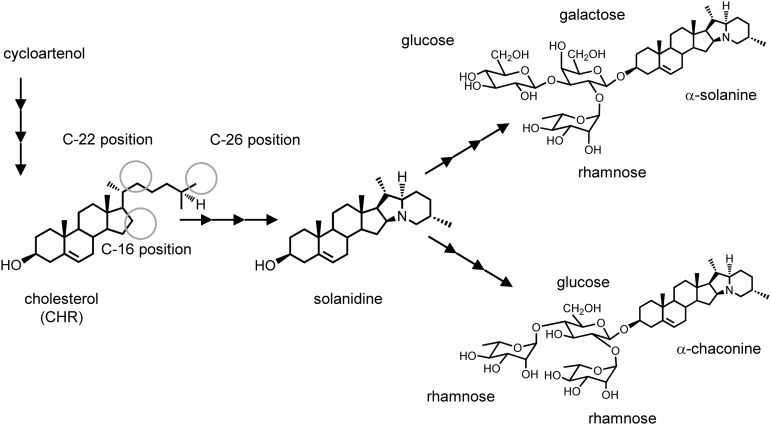
SGAs are biosynthesized from a common precursor, cholesterol (CHR; Sawai et al., 2014), but little is known about intermediates and enzymes in the SGA biosynthetic pathway. To change a biosynthetic flow to CHR and decrease SGA contents, transgenic potatoes overexpressing a heterologous soybean sterol methyltransferase gene were produced (Arnqvist et al., 2003). Three genes responsible for glycosylating potato SGA have been identified (McCue et al., 2005, 2006, 2007). However, changing the expression of the sterol methyltransferase or glycosyltransferase genes does not effectively decrease SGA levels. To control the SGA content of potato, we focused on the biosynthetic steps from CHR to the aglycone, solanidine. Few details about the biosynthetic pathway are verified; however, the pathway is hypothesized to require at least three oxidization steps at positions C16, C22, and C26 of CHR structure and the addition of one nitrogen atom at the position C26 (Fig. 1; Kaneko et al., 1977; Erich, 1983; Eich, 2008). The later step was shown to be another oxidation and an amination reaction at the position C26 (Ohyama et al., 2013). Here, we identified two cytochrome P450 monooxygenase (CYPs) genes, POTATO GLYCOALKALOID BIOSYNTHESIS1 (PGA1) and PGA2 that encode enzymes mediating two oxidation steps. Silencing PGA1 and PGA2 resulted in a significant reduction in SGA composition and the creation of novel phenotypes, including the suppression of flower development and tuber sprouting. Sprouting reduces the quality and yield of potato tubers in storage. Suppression of tuber sprouting is of significant benefit to the industry for the long-term storage of tubers. Thus, controlling tuber sprouting is another important objective in potato breeding (Sonnewald and Sonnewald, 2014).
Figure 1.
Biosynthetic pathway for SGAs in potato. The structures of CHR and solanidine, two biosynthetic intermediates of potato SGAs, are shown with the structures of the SGA products. Circles indicate putative carbon positions that are oxidized in the hypothesized pathway.
RESULTS
Construction and Characterization of PGA1- and PGA2-Knockdown Transgenic Potato Plants
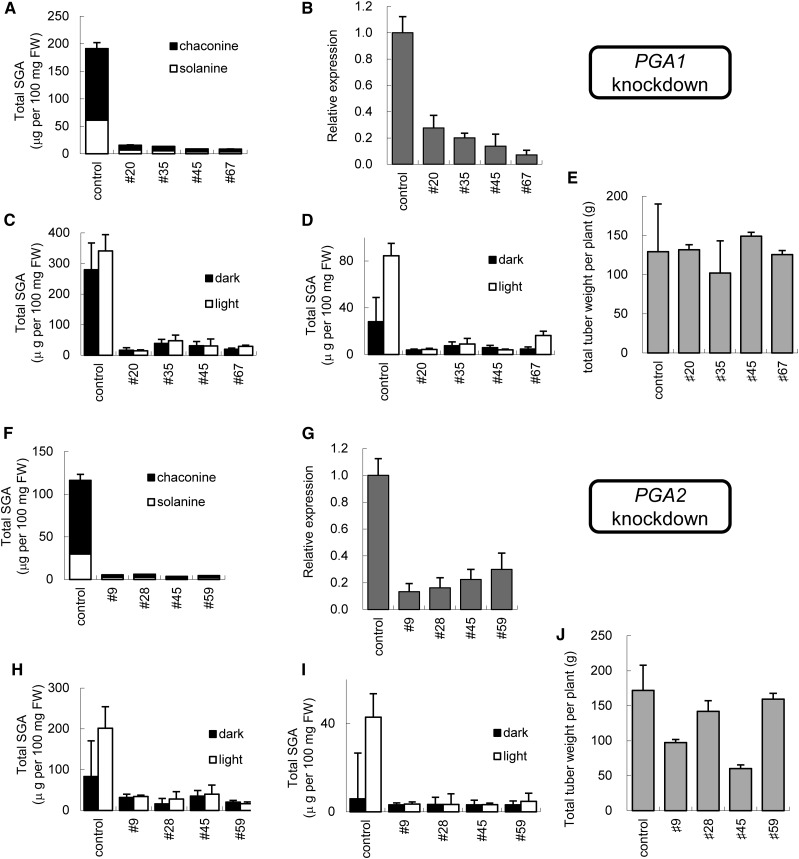
Some CYPs are associated with steroid oxidization (Ohnishi et al., 2009). SGAs can be found in most potato plant tissues and are abundant in flowers and tuber sprouts (Kozukue and Mizuno, 1985, 1989; Ginzberg et al., 2009). We chose two candidate CYP genes that were most expressed in flowers and tuber sprouts from the potato unigene databases of the Gene Index project (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/). Two CYP cDNA sequences, encoding CYP72A208 and CYP72A188 (http://drnelson.uthsc.edu/cytochromeP450.html), share 52% amino acid sequence identity and have about 45% amino acid identity to licorice CYP72A154 that is associated with the biosynthesis of glycyrrhizin, a triterpenoid saponin (Seki et al., 2011). CYP72A208 and CYP72A188 genes (designated as PGA1 and PGA2) are located on chromosomes 6 (PGSC0003DMG400026586 and PGSC0003DMG400026594, of which the deduced proteins share >99% amino acid identity) and on chromosomes 7 (PGSC0003DMG400011750), respectively (Xu et al., 2011). To analyze the contribution of PGA1 and PGA2 to SGA biosynthesis, we transformed potatoes with an RNA interference vector to create PGA1- or PGA2-knockdown transgenic potato plants (Supplemental Fig. S1, pKT226 and pKT227). The knockdown vectors were constructed from a stem-loop fragment of PGA cDNAs that did not match over 10 consecutive nucleotides. Of the 28 pKT226 transgenic lines, the in vitro shoots of four independent lines (#20, #35, #45, and #67 in Fig. 2. A–E) had a much lower SGA content than the controls (Fig. 2A), consistent with their lower PGA1 transcript levels (Fig. 2B). Similarly, the in vitro shoots of four out of 27 pKT227 transgenic lines (#9, #28, #45, and #59 in Fig. 2, F–I) had lower SGA contents (Fig. 2F) and lower levels of PGA2 mRNA expression (Fig. 2G). All PGA1- and PGA2-knockdown transgenic lines were grown in a greenhouse and produced tubers. The SGA contents of tuber peel and tuber cortex in the knockdown lines were lower than the control in both dark and light conditions (Fig. 2, C, D, H, and I), although the transgenic tuber peels also accumulated normal levels of chlorophyll and anthocyanin (Supplemental Fig. S2). Most of the PGA-knockdown plants had similar tuber yields to the control (Fig. 2, E and J), although two lines of the PGA2-knockdown plants had lower tuber yields than the control. To confirm the results, we constructed the pKT249 vector that used a different region of the PGA1 gene (Supplemental Fig. S1, pga1-2). Of the 27 pKT249 transgenic lines, three independent lines also had lower levels of SGAs and similar vegetative growth (Supplemental Fig. S3, A–C) compared to the control and the PGA1-knockdown transgenic plants (pKT226, pga1-1). These results indicate that PGA1 and PGA2 are necessary for SGA production in potato plants.
Figure 2.
SGA content and yield of the PGA1- and PGA2-knockdown plants. A and B, LC-MS analysis of SGA (α-solanine and α-chaconine) levels (A) and quantitative RT-PCR analysis of PGA1 expression levels (B) in the in vitro-grown shoots of independent PGA1-knockdown plants. C and D, LC-MS analysis of the SGA levels (total SGA = α-solanine + α-chaconine) of the PGA1-knockdown plants in the peel (C) and cortex (D) of harvested tubers with/without light exposure. E, Yields of the tubers from PGA1-knockdown plants. F and G, LC-MS analysis of SGA (α-solanine and α-chaconine) levels (F) and quantitative RT-PCR analysis of PGA2 expression levels (G) in the in vitro-grown shoots of independent PGA2-knockdown plants. H and I, LC-MS analysis of the SGAs levels of the PGA2-knockdown plants in the peel (H) and cortex (I) of harvested tubers with/without light exposure. J, Yields of the tubers from PGA2-knockdown plants. Expression levels are shown relative to levels in control plants. FW, fresh weight. Each bar of SGA content, relative expression level and yield shows the mean of three samples, with error bars indicating sd.
Metabolite Analyses of PGA1- and PGA2-Knockdown Transgenic Potato Plants
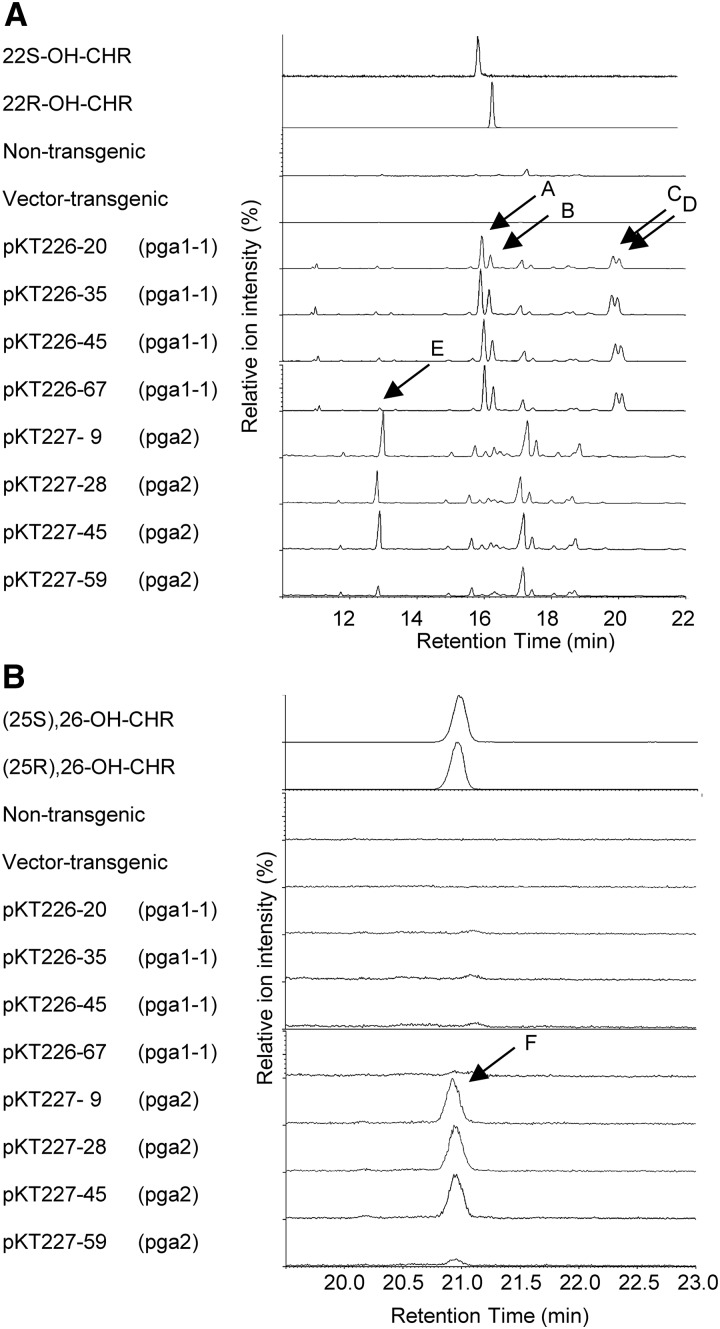
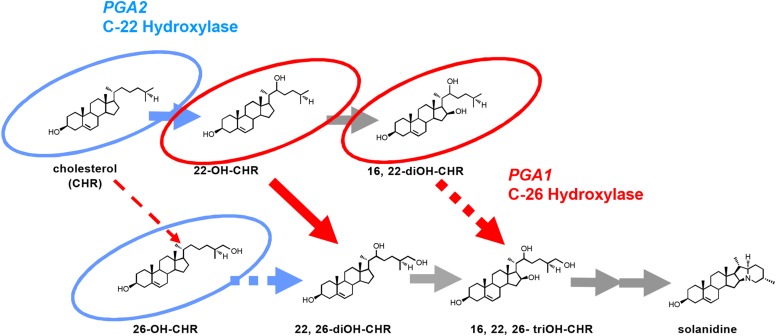
To characterize the enzymes encoded by the PGA1 and PGA2 genes, we analyzed metabolites that accumulated in the PGA1- and PGA2-knockdown plants by gas chromatography-mass spectrometry (GC-MS). The extracted ion chromatogram (EIC) of 173 m/z sterols that accumulated in the PGA1-knockdown plants (pga1-1) had four new peaks (Fig. 3A, peaks A–D), compared to samples from nontransgenic plants and vector control plants. The retention time (Rt) and mass spectrum of peaks A and B matched with those of authentic 22S-hydroxycholesterol (22S-OH-CHR) and 22R-hydroxycholesterol (22R-OH-CHR), respectively (Fig. 3A, EIC = 173 m/z; Supplemental Figs. S4A and S5A). From the mass spectra of peaks C and D, we inferred that the two peaks corresponded to 22-hyroxycholesterol (22-OH-CHR) derivatives that are presumed to be enantiomers of 16,22-dihydroxycholesterols (Supplemental Figs. S4A and S5A). In contrast, the PGA2-knockdown plants accumulated several to 10 times more CHR than control plants (peak E as shown in Fig. 3A and Supplemental Fig. S4B) and one minor product whose Rt and mass spectrum matched those of authentic 26-hydroxycholesterol (26-OH-CHR; peak F as shown in Fig. 3B, EIC = 468; Supplemental Figs. S4C and S5B). CHR is the precursor for SGA biosynthesis (Sawai et al., 2014). As the PGA2-knockdown transgenic plants accumulated larger amounts of CHR compared to the control plants, we inferred that PGA2 must use CHR as its substrate. The PGA1-knockdown plants did not accumulate significant amounts of CHR. Therefore, we hypothesized that PGA2, a C-22 hydroxylase, oxidizes CHR to 22-OH-CHR and that PGA1, a C-26 hydroxylase, must metabolize 22-OH-CHR (Fig. 4).
Figure 3.
Accumulated compounds in PGA1- and PGA2-knockdown plants. A and B, GC-MS EIC (m/z = 173; A) and EIC (m/z = 456; B) chromatograms of accumulated compounds in the nontransgenic plant, the vector-transgenic plant, and the PGA1- and PGA2-knockdown plants. Peaks A to F were not present in control plants. The retention times and mass spectra (Supplemental Fig. S4A) of peaks A and B compare well with those of authentic 22S-OH-CHR and 22R-OH-CHR, respectively. The mass spectra of peaks C and D also match those of authentic 22S-OH-CHR and 22R-OH-CHR (Supplemental Fig. S4A). Peaks C and D were expected to be 22-OH-CHR derivatives, as shown in Supplemental Figure S5A. Peak E is coincident with that of CHR (Supplemental Fig. S4B). Peak F is coincident with that of 26-OH-CHR (Supplemental Fig. S4C). The enantiomer form of peak F was not determined.
Figure 4.
Proposed pathways for SGA biosynthesis catalyzed by PGA1 and PGA2. Compounds that accumulated in the PGA1-knockdown plants (red circles) and the PGA2-knockdown plants (light blue circles) are shown. Solid arrows are indicated as metabolite flows of PGA1 (red) and PGA2 (light blue). Dashed and gray arrows are putative. OH-CHR, hydroxycholesterol.
In Vitro PGA1 and PGA2 Enzymatic Activity Assay
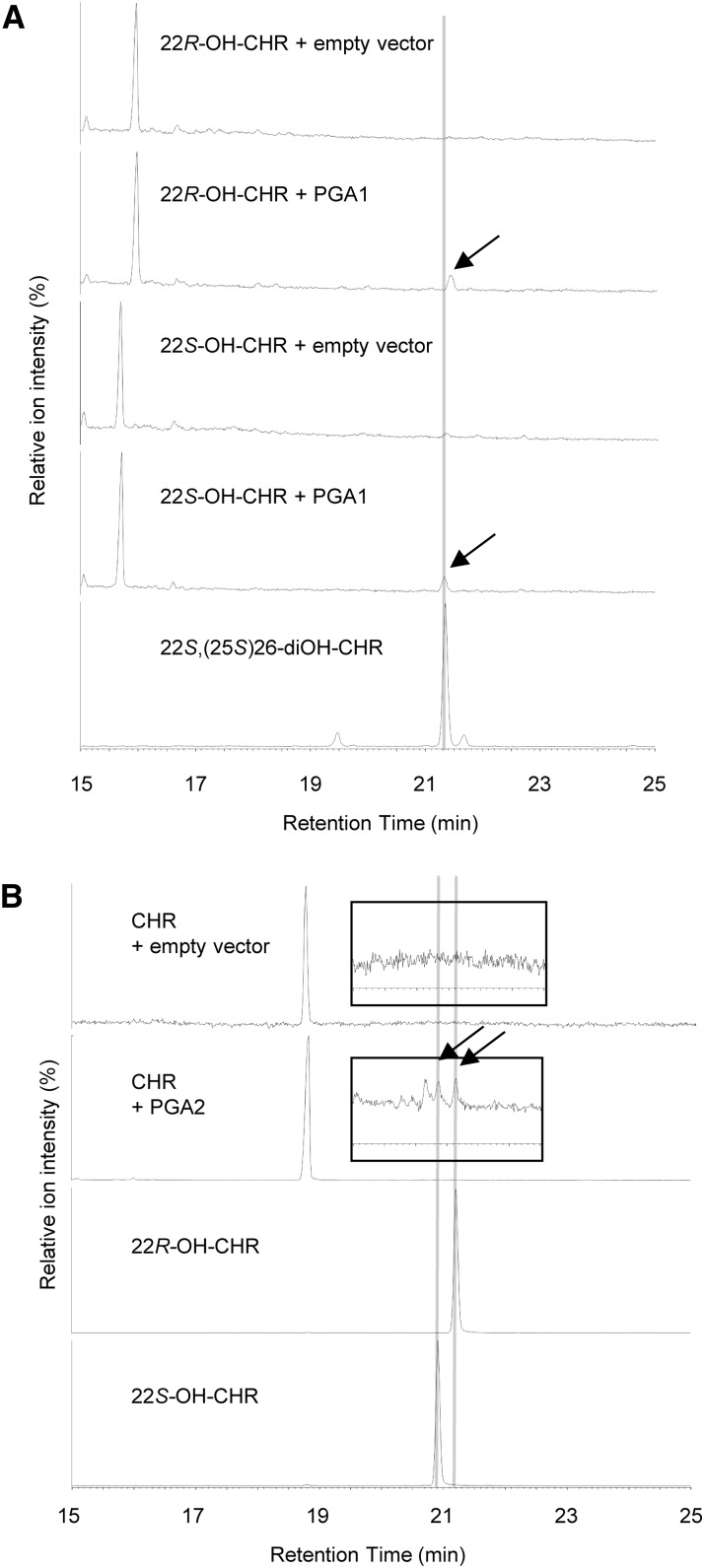
To confirm the catalytic functions of PGA1 and PGA2, we performed in vitro conversion assays using recombinant proteins produced in insect cells by baculovirus-mediated expression. In the presence of NADPH, PGA1 metabolized 22S-OH-CHR and 22R-OH-CHR but not CHR into products with Rts of 21.3 and 21.4 min, respectively (Fig. 5A). From their mass spectra, the former product is identical to 22S,(25S)26-diOH-CHR, and the latter product is presumed to be 22R,(25S)26-diOH-CHR. In contrast, PGA2 metabolized CHR into two products with Rts of 20.9 and 21.2 min that are identical to 22S-OH-CHR and 22R-OH-CHR, respectively (Fig. 5B; Supplemental Fig. S6). These results clearly indicate that PGA1 and PGA2 encode a C26-hydroxylase and a C22-hydroxylase, respectively, and are consistent with the observations of the accumulated compounds in the knockdown transgenic plants (Fig. 4).
Figure 5.
In vitro oxidation of 22-OH-CHR and CHR by PGA1 and PGA2 expressed in insect cells, respectively. A, GC-MS analysis (TIC) of the reaction products from an in vitro assay containing 22-OH-CHRs as substrates and microsomal fractions isolated from PGA1-expressing Sf9 cells, respectively. Arrows indicate products (Rt = 21.4 and 21.3 min) using 22R-OH-CHR and 22S-OH-CHR, respectively. The later peak is coincident with that of authentic 22S,(25S)26-diOH-CHR. B, GC-MS analysis (TIC) of the reaction products from an in vitro assay containing CHR as substrates and microsomal fractions isolated from PGA2-expressing Sf9 cells, respectively. Enlargements of the chromatograms corresponding to the same retention times as the original are indicated in the inserts. Arrows indicate products using CHR as the substrate. The peaks are coincident with those of authentic 22S-OH-CHR and 22R-OH-CHR. Mass spectra of these reaction products are shown in Supplemental Figure S6.
Phenotypes of PGA1- and PGA2-Knockdown Transgenic Potato Plants
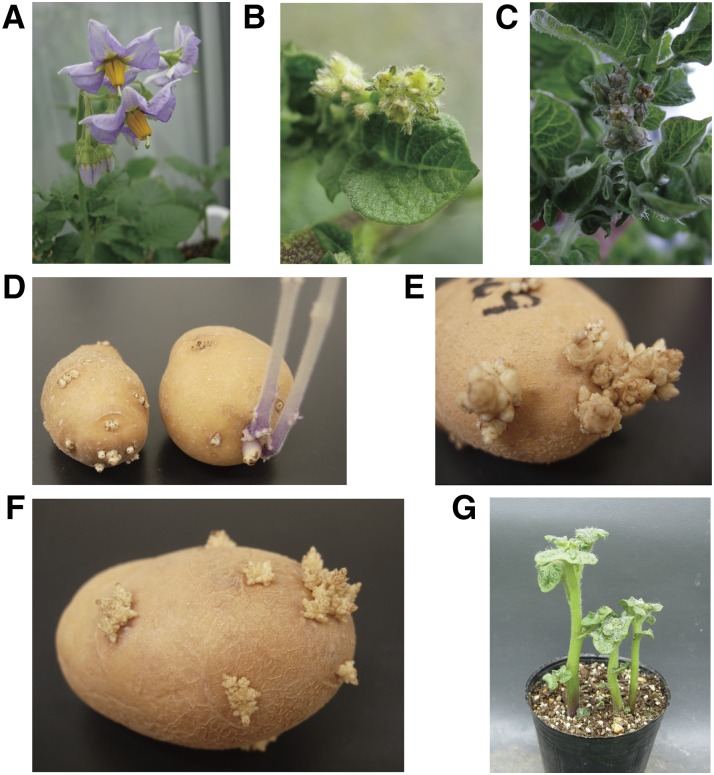
The PGA1- and PGA2-knockdown transgenic plants demonstrated two distinct phenotypes. First, the PGA1-knockdown plants (pKT226-20, -35, -45, and -67) and the PGA2-knockdown plants (pKT227-9, -28, -45, and -59) produced abnormal flowers that were sterile (Fig. 6, A–C). Flower initiation in these plants seemed normal, but development stopped and the flowers withered. The knockdown transgenic plants produced no pollen and were thus infertile. The alternative series of PGA1-knockdown lines constructed with another silencing region from the PGA1 gene (pga1-2; pKT249-6, -11, and -20; Supplemental Figs. S1 and S3D) showed the same phenotype. The second distinctive phenotype was that the tubers of the PGA1- and PGA2-knockdown transgenic plants could not sprout without treatment. Sprouting initiates normally, but the sprout clusters did not grow even after more than 3 months at 20°C or for 1 year at 4°C after the cessation of usual dormancy in the control plants (Fig. 6, D–H). Interestingly, the sprout clusters could grow after the tubers were planted in soil (Fig. 6G) but not in water. Two independent series of PGA1-knockdown lines constructed with two different regions in the PGA1 gene (pKT226 lines and pKT249 lines; Supplemental Fig. S1) showed the same phenotype. We confirmed the reproducibility of the nonsprouting tuber phenotype three times for the PGA1-knockdown plants, pKT226 lines, and twice for the PGA2-knockdown plants, pKT227 lines, during different growing seasons. After placing excised sprout tips on tissue culture media, the sprouts could also grow (Supplemental Fig. S7), suggesting that the sprout clusters were still alive and had only stopped growing.
Figure 6.
Phenotypes of the PGA1- and PGA2-knockdown plants. A, Flowers of the control plant. B, Flowers of the PGA1-knockdown plant (pKT226-35). C, Flowers of the PGA2-knockdown plant (pKT227-45). D, Sprouted potatoes of the PGA1-knockdown plant (pKT226-67, left) and control (right) 2 weeks after the cessation of plant dormancy in the control. E and F, Sprouted potatoes of the PGA1-knockdown plant (pKT226-67; E) and the PGA2-knockdown plant (pKT227-45; F) 3 months and 2 months after the cessation of plant dormancy in the control, respectively. G, Sprouted potatoes of the PGA1-knockdown plant (pKT226-67) 3 weeks after being planted into soil. The sprouted plants grew normally.
DISCUSSION
PGA1 and PGA2 Catalyze Hydroxylation Steps from CHR
SGAs were initially proposed to be biosynthesized from CHR (Heftmann 1983), and only recently the CHR synthase gene, SSR2, has been identified and found to be necessary for SGA biosynthesis (Sawai et al., 2014). Our in vitro enzymatic activity assay results demonstrated that PGA2 and PGA1 catalyze C-22 hydroxylation and C-26 hydroxylation, respectively. The results were clearly supported by analysis of the compounds that accumulated in the PGA-knockdown plants. PGA1 and PGA2 catalyze early hydroxylation steps in the SGA biosynthetic pathway (Fig. 4). In the biosynthetic and catabolic pathways for brassinosteroid plant hormones, the C-22 hydroxylase and the C-26 hydroxylase are encoded by different genes; for example, in Arabidopsis (Arabidopsis thaliana), DWF4 and BAS1 belong to the CYP90B and CYP734A subfamilies, respectively (Ohnishi et al., 2009). In tomato (Solanum lycopersicum), CYP90B2 was previously reported to be a C-22 hydroxylase (Ohnishi et al., 2006b). The likely orthologs were found in the potato genome (CYP90B, PGSC0003DMG400014902; CYP734A, PGSC0003DMG400001060; Xu et al., 2011). PGA1 and PGA2 belong to the CYP72A subfamily, indicating that the SGA biosynthetic pathway evolved independently from the brassinosteroid biosynthetic pathway. The CYP72A subfamily contains many CYPs that have not been characterized yet, although some CYP72As appear to function in secondary metabolism (Seki et al., 2011; Fukushima et al., 2013). Of CYP genes, the expressed sequence tags of PGA1 and PGA2 were most observed both in flowers and tuber sprouts. We found 13 other candidate CYP genes relatively expressed in the tissues using the expression database (https://solgenomics.net/) and made the knockdown transgenic plants (Supplemental Table S1). Except for the transgenic plants decreasing expression of CYP88B1, the other transformants did not demonstrate decrease of SGA contents (see below; Umemoto and Sasaki, 2013). However, the recombinant CYP88B1 produced in insect cells did not exhibit the C-16 hydroxylation activity. These results suggested that CYP88B1 should mediate the remaining oxidation step in the SGA biosynthetic pathway.
Recently, 10 tomato genes involved in the SGA pathway were reported using comparative coexpression analysis coupled with chemical profiling and gene cluster analysis (Itkin et al., 2013). GAME4 encoding CYP88B1 was reported to be involved in the SGA pathway after construction and analysis of the corresponding knockdown potato (Itkin et al., 2013), but the function and enzyme activity of the gene have not been identified yet. Two other tomato candidate genes (GAME8 and GAME6), the functions of which have not been analyzed in detail, are orthologous to potato PGA1 and PGA2, respectively. GAME8 was also reported to be a wound-inducible gene (Bartoszewski et al., 2000) coincident with inducing SGA accumulation by mechanical injury (Valkonen et al., 1996). The PGA2 gene is in a gene cluster on chromosome 7 (Itkin et al., 2013; Cárdenas et al., 2015), but the PGA1 genes are tandemly located on chromosome 6 of S. tuberosum Group Phureja DM1-3 (Xu et al., 2011), not in a specialized gene cluster for metabolism as described (Nützmann and Osbourn, 2014). As the SSR2 gene is also located on chromosome 2 (Sawai et al., 2014), specialized gene clusters for the SGA biosynthesis are though not to be complete.
The Phenotype of PGA1- and PGA2-Knockdown Transgenic Potato Plants
Transgenic potatoes in which either the PGA1 or PGA2 genes were silenced contained significantly less SGA; however, the knockdown plants grew normally in the vegetative stages and produced yields of tubers in the greenhouse similar to control plants. These results suggest that SGAs may be dispensable to the growth of potato, at least under unstressed conditions. The results are coincident with that of GAME4-knockdown transgenic potato (Itkin et al., 2013; Umemoto and Sasaki, 2013) and those of SSR2-knockdown and SSR2-disrupted potatoes (Sawai et al., 2014). Characterization of the inheritance of specific SGAs and SGA content has been limited, despite the importance of these phytochemicals (Valkonen et al., 1996; Ginzberg et al., 2009; Manrique-Carpintero et al., 2014). The SGA content of most commercial varieties of potato tubers is well below the human safety limit (200 mg/kg fresh weight), and a high SGA content is frequently associated with the introduction of wild germplasm to the breeding program (Sinden et al., 1984). Our results open the way for breeding of SGA-free potatoes. Using recombinant gene techniques, the PGA1- and PGA2-knockdown plants contained significantly less SGA. As an alternative to transgenic technology, functionless mutants can also be obtained from mutation breeding, using such techniques as TILLING (targeting induced local lesions in genome; Elias et al., 2009) and genome editing techniques (Sawai et al., 2014; Nicolia et al., 2015).
The above-described reports about GAME4- and SSR2-suppressed or disrupted potatoes indicated that these plants were phenotypically identical to control plants, although the plants had significantly lower SGA contents. These findings imply that SGAs are dispensable for flowering and sprouting morphogenesis; however, the PGA1- and PGA2-knockdown plants were sterile and the sprout clusters did not grow. For the PGA1-knockdown plants, two independent series of silenced lines (pga1-1 and pga1-2) were constructed using different silencing regions (Supplemental Fig. S2) and both lines had the same phenotype, further strengthening the hypothesis that SGA biosynthesis and sprouting are somehow associated. We obtained tomato transgenic plants in which the PGA1-orthologous gene and PGA2-orthologous gene were silenced and found that the transgenic plants had lower levels of the predominant tomato SGA, α-tomatine (Supplemental Fig. S8). Both of the knockdown tomato lines exhibited retarded growth, dwarfing, abnormal flowers, and sterility (Supplemental Fig. S8, A–C). The transgenic tomato plants did not produce any fruit. These more severe growth phenotypes are similar to those of transgenic tomatoes that suppressed GAME1 (Itkin et al., 2011). The tomato SGA biosynthetic gene GAME1 encodes tomatidine galactosyltransferase and is orthologous to potato SGT1 and the GAME1-suppressed plants formed small flower buds, suggesting a potential influence on glycoalkaloid metabolism. The PGA1- and PGA2-knockdown plants might accumulate morphogenesis inhibitors that can be removed by planting tubers in soil. This hypothesis is supported by the observation that floral and sprout tissues contain the greatest levels of SGAs (Kozukue and Mizuno, 1985, 1989; Ginzberg et al., 2009). Unpalatable secondary metabolites found in wild species such as capsaicin from bell pepper and cucurbitacins from cucumber (Cucumis sativus) were often lost during domestication as demonstrated by characterizing the responsible genes (Stewart et al., 2005; Shang et al., 2014). The transgenic potato sprouting and sterility phenotypes may interfere with the domestication of wild potato without SGAs.
Expectation of Improvement of Potato Storage
Unlike cereal seeds, sprouting of potato tubers must be initiated following a period of dormancy. Producers can treat stored potatoes with chemicals to prevent the growth of potato eyes or buds in some nations, but long-term storage is another economically important aspect of the potato industry, especially for the processing of chips and frozen foods (Korpan et al., 2004; Sonnewald, 2001). No triggers for tuber sprouting have been identified so far although tuber sprouting was hypothesized to be controlled by physiological, hormonal, and environmental factors (Sonnewald and Sonnewald, 2014). The cessation of tuber sprouting that the PGA1- and PGA2-knockdown plants demonstrated seems not to prolong the length of dormancy. Transgenes related to carbohydrate metabolism (Hajirezaei and Sonnewald, 1999; Hajirezaei et al., 2003; Debast et al., 2011) or plant hormone cytokinin catabolism (Hartmann et al., 2011) delayed or inhibited tuber sprouting, but controllable tuber sprouting during at least the first year has not been observed in the transgenic plants. Harvested tubers of the PGA1- and PGA2-knockdown never sprouted, but it is noteworthy that the sprout clusters of the knockdown plants began to grow when placed in soil (Fig. 6G). There are no reported relationships between SGA biosynthesis and tuber sprouting. The new phenotype reported here is not understood completely but offers the possibility that potato storage could be controlled without the use of postharvest chemicals by introducing defects in gene function.
MATERIALS AND METHODS
Chemicals
Authentic samples of α-solanine, α-chaconine, α-tomatine, and the two 22-hydroxycholesterols were purchased from Sigma-Aldrich. CHR was purchased from Tama Biochemical Co. The two 26-hydroxycholesterols and 22S,(25S)26-dihydroxycholesterol were kindly provided by A. Yoshimoto and B. Watanabe, respectively. A list of the primer sets used to generate these constructs is shown in Supplemental Table S1.
Cloning of PGA1 and PGA2 cDNAs
The potato cDNA template was prepared from mRNA isolated from sprouts of Solanum tuberosum cv Sassy using an RNeasy plant kit (Qiagen) and SuperScript first-strand synthesis system for RT-PCR (Life Technologies). Transcripts corresponding to the PGA1 and PGA2 genes were amplified with primer sets U841/U842 and U875/U876 designed from the potato unigene sequences (TC135549 and TC141445 in the Potato Gene Index database, respectively). The PCR products of PGA1 and PGA2 were cloned into the pCR4-TOPO plasmid (Life Technologies).
Gene-Silenced Potatoes
In vitro grown plants were cultured at 20°C under a 16-h-light/8-h-dark condition. Plants were grown in a greenhouse under long-day conditions to analyze their flowering and tuberization. Gene-knockdown potatoes were generated by RNAi with Agrobacterium tumefaciens-mediated transformation of tuber discs obtained from cv Sassy (Momma 1990). The control vector, pKT19, for vector-transgenic plants in Figure 2 was generated by inserting the GUS gene into the pBin19 (Frisch et al., 1995)-based binary vector (Umemoto et al., 2001) between the Cauliflower mosaic virus 35S promoter and the nopaline synthase terminator. To silence the transcripts, potato plants were transformed with RNAi constructs designed from the protein-coding regions of the genes. The amplification products were cloned into the pCR4-TOPO plasmid. The RNAi targeting PGA1 was constructed as follows. A 372-bp fragment of PGA1 cDNA that did not match over consecutive 10 nucleotides with PGA2 was PCR amplified using primer set U724/U725. The fragments were inserted in opposite orientations with the intron of the Arabidopsis (Arabidopsis thaliana) PDS gene (Masclaux et al., 2004) by XbaI/SacI digestion into the pKT19 vector, substituting for the GUS gene, under the control of the 35S promoter in the T-DNA region to produce the RNAi binary vector pKT226 (Supplemental Fig. S1). Two RNAi binary vectors were constructed similarly; pKT249 targeted another region of the PGA1 gene, and pKT227 targeted PGA2. A 402-bp fragment of PGA1 cDNA was PCR amplified using primer set U869/U870. A 325-bp fragment of PGA2 cDNA was PCR amplified using primer set U726/U727. The stem regions of the PGA1- or PGA2-knockdown constructs (Supplemental Fig. S1) do not contain more than 10 oligonucleotides coincident with the other construct. Transgenic potato plants carrying the pKT19 (vector-transgenic), pKT226 (pga1-1 RNAi), pKT249 (pga1-2 RNAi), and pKT226 (pga2 RNAi) constructs were generated using A. tumefaciens GV3101 pMP90 cells. Transformants were individually selected by genomic PCR of the potato shoots with primer set NP2/NP3 targeting the kanamycin resistance gene in the T-DNA region integrated into the potato genome. Tomatoes (Solanum lycopersicum cv Micro-Tom) were transformed using A. tumefaciens GV3101 pMP90 cells with pKT226 and pKT227 as previously reported (Sun et al., 2006). Integration of the T-DNA region into tomato genomic DNA was investigated by genomic PCR of the tomato leaves targeting the kanamycin resistance gene on the T-DNA region integrated into the tomato genome. Sprout tips cut from tubers of the PGA1-knockdown plants were placed on tissue culture media (Murashige and Skoog, 1962) without plant hormones. The vectors targeting other candidate genes were constructed using primer sets as shown in Supplemental Table S2.
Quantitative RT-PCR
Quantitative RT-PCR analysis of PGA1, PGA2, and the EF1α control (Nicot et al., 2005) was carried out using primer sets as listed in Supplemental Table S2. Total RNA prepared from shoots of five independent lines of in vitro-cultured plants. The cDNA templates were prepared as described above and amplified using LightCycler Nano (Roche) with GeneAce SYBR qPCR Mix alpha No ROX (Nippon Gene). Each assay was repeated three times.
In Vitro Enzyme Activity Assay
The in vitro enzyme activity assays with homogenates of insect cells expressing PGAs were conducted as previously reported (Ohnishi et al., 2006a). PGA1 and PGA2 cDNAs were cloned into the pDEST8 vector (Life Technologies) and were then used to generate the corresponding recombinant Bacmid DNAs by transforming Escherichia coli DH10Bac (Life Technologies). Preparation of the recombinant baculovirus DNAs that contained PGA1 and PGA2 cDNAs and transfection of Spodoptera frugiperda 9 insect cells were carried out according to the manufacturer’s instructions (Life Technologies). Heterologous expression of PGA1 and PGA2 proteins in the insect cells and spectrometric analyses were performed as described (Saito et al., 2004).
Microsomal fractions of the insect cells expressing PGA1 and PGA2 were obtained from the infected cells (100 mL of suspension cultured cells). Infected cells were washed with PBS buffer and suspended in buffer A consisting of 20 mm potassium phosphate (pH 7.25), 20% (v/v) glycerol, 1 mm EDTA, and 1 mm DTT. The cells were sonicated and cell debris was removed by centrifugation at 10,000g for 15 min. The pellet was homogenized with buffer A to provide the microsomal fractions. The microsomal fractions were stored at –80°C before the enzyme assays. PGA1 and PGA2 activities were reconstituted by mixing the PGA1 and PGA2-containing microsomes with purified Arabidopsis NADPH-P450 reductase (Mizutani and Ohta, 1998). The reaction mixture consisted of 100 mm potassium phosphate, pH 7.25, 50 pmol/mL recombinant P450 protein, 0.1 unit/mL NADPH-P450 reductase, 1 mm NADPH, and 25 μm of substrate sterols. Reactions were initiated by addition of NADPH and were performed at 30°C for 3 h. The reaction products were extracted three times with an equal volume of ethyl acetate. The organic phase was collected and evaporated. The residue was trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide at 80°C for 30 min. GC-MS was conducted using a GC-MS-QP2010 Ultra (Shimadzu) with a DB-1ms capillary column (30 m × 0.25 mm, 0.25 µm film thickness; J&W Scientific) to analyze the PGA1 reaction products and with a DB-5ms capillary column (30 m × 0.25 mm, 0.25 µm film thickness; J&W Scientific) to analyze the PGA2 reaction products. The injection temperature was 250°C. The column temperature program for analysis of the PGA1 reaction products was as follows: 80°C for 1 min, followed by a rise to 300°C at a rate of 20°C min−1, and a hold at 300°C for 20 min. The column temperature program for analysis of the PGA2 reaction products was as follows: 180°C for 1 min, followed by a rise to 280°C at a rate of 20°C min−1, followed by a rise to 300°C at a rate of 2°C min−1, and a hold at 300°C for 20 min. The carrier gas was He, and the flow rate was 1.0 mL min−1; the interface temperature was 280°C with a splitless injection.
LC-MS Analysis of α-Chaconine, α-Solanine, and α-Tomatine
Extractions and LC-MS (liquid chromatography-mass spectrometry) analyses of the plant materials were performed as previously described with minor modifications (Sasaki, 2011). Fresh plant materials (100 mg) were homogenized with a mixer mill at 4°C in a 1 mL solution containing 80% (v/v) methanol and 0.1% (v/v) formic acid containing 10 μg brassinolide as an internal standard. After centrifugation, 25 μL of supernatant was diluted with 475 μL 0.1% (v/v) formic acid solution and filtered with a MultiScreen Solvinert (Millipore). An aliquot (10 μL) was analyzed by LC-MS using 10 mm ammonium hydrogen carbonate in water (pH 10):acetonitrile (2:3, v/v) as eluent at a flow rate of 0.2 mL min−1 at 40°C. LC-MS was performed with a Shimadzu LCMS-2010EV apparatus operating in ESI mode attached to an XBridge Shield RP18-5 column (150 mm × 2.1 mm i.d.; Waters). Quantifications of α-solanine and α-chaconine were calculated from the ratio of peak area at m/z 868 and 852, and 1034 of positive ion scans using a calibration curve of authentic samples (coefficients of determination: r2 > 0.999), respectively. The total SGA content of potato is the sum of the α-solanine and α-chaconine contents.
Extraction and GC-MS analysis for steroids
In vitro shoots (starting volume: 5 mL) were extracted, and GC-MS analyses were conducted with the same conditions as described previously (Choi et al., 2014). Peaks were identified by comparing the Rts and mass spectra with those of the authentic standards.
Accession Numbers
The GenBank/EMBL/DDBJ accession numbers for PGA1 and PGA2 are AB839752 and AB839753, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PGA1- and PGA2-Knockdown Vectors.
Supplemental Figure S2. Tubers of PGA1-Knockdown Plants with Light Exposure.
Supplemental Figure S3. SGA Content, Yield, and Phenotype of the PGA1-Knockdown Plants (pKT249).
Supplemental Figure S4. Mass Spectra of Accumulated Compounds in PGA1- and PGA2-Knockdown Plants.
Supplemental Figure S5. Proposed Assignment of Fragment Ions of Hydroxycholesterol Derivatives.
Supplemental Figure S6. Mass Spectra of PGA2 Metabolites and Authentic Standards of Sterols.
Supplemental Figure S7. In Vitro Growth of Sprout Tips on Tissue Culture Media.
Supplemental Figure S8. Phenotype and SGA Content of the PGA Ortholog-Knockdown Tomato Plants.
Supplemental Table S1. Candidates of CYP Genes and Knockdown Vectors in the Current Study.
Supplemental Table S2. Oligonucleotides Used in the Current Study.
Supplementary Material
Acknowledgments
We thank Eric Bonnel for suggestions, Masako Ohtsuka, Maki Watanabe, and Chie Yoshida for technical assistance, Katsunori Sasaki for glycoalkaloid analysis, Yasushi Yoshioka for providing the A. tumefaciens strain, and Yoshinori Fujimoto for providing 26-OH-CHRs.
Glossary
- SGA
steroidal glycoalkaloid
- CHR
cholesterol
- GC-MS
gas chromatography-mass spectrometry
- EIC
extracted ion chromatogram
- 22S-OH-CHR
22S-hydroxycholesterol
- 22R-OH-CHR
22R-hydroxycholesterol
- 22-OH-CHR
22-hyroxycholesterol
- 26-OH-CHR
26-hydroxycholesterol
- LC-MS
liquid chromatography-mass spectrometry
References
- Arnqvist L, Dutta PC, Jonsson L, Sitbon F (2003) Reduction of cholesterol and glycoalkaloid levels in transgenic potato plants by overexpression of a type 1 sterol methyltransferase cDNA. Plant Physiol 131: 1792–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewski G, Mujer C, Smigocki A, Niemirowicz-Szczytt K (2000) A wound inducible cytochrome P450 from tomato. Acta Physiol Plant 22: 269–271 [Google Scholar]
- Cárdenas PD, Sonawane PD, Heinig U, Bocobza SE, Burdman S, Aharoni A (2015) The bitter side of the nightshades: Genomics drives discovery in Solanaceae steroidal alkaloid metabolism. Phytochemistry 113: 24–32 [DOI] [PubMed] [Google Scholar]
- Choi H, Ohyama K, Kim YY, Jin JY, Lee SB, Yamaoka Y, Muranaka T, Suh MC, Fujioka S, Lee Y (2014) The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 26: 310–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debast S, Nunes-Nesi A, Hajirezaei MR, Hofmann J, Sonnewald U, Fernie AR, Börnke F (2011) Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol 156: 1754–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eich E. (2008) Terpenoids (Isoprenoids) Solanaceae and Convolvulaceae: Secondary Metabolites. Springer, Berlin, Heidelberg [Google Scholar]
- Elias R, Till BJ, Mba C, Al-Safadi B (2009) Optimizing TILLING and Ecotilling techniques for potato (Solanum tuberosum L). BMC Res Notes 2: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erich H. (1983) Biogenesis of steroids in solanaceae. Phytochemistry 22: 1843–1860 [Google Scholar]
- Friedman M. (2006) Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J Agric Food Chem 54: 8655–8681 [DOI] [PubMed] [Google Scholar]
- Frisch DA, Harris-Haller LW, Yokubaitis NT, Thomas TL, Hardin SH, Hall TC (1995) Complete sequence of the binary vector Bin 19. Plant Mol Biol 27: 405–409 [DOI] [PubMed] [Google Scholar]
- Fukushima EO, Seki H, Sawai S, Suzuki M, Ohyama K, Saito K, Muranaka T (2013) Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol 54: 740–749 [DOI] [PubMed] [Google Scholar]
- Ginzberg I, Tokuhisa J, Veilleux R (2009) Potato steroidal glycoalkaloids: biosynthesis and genetic manipulation. Potato Res 52: 1–15 [Google Scholar]
- Hajirezaei M, Sonnewald U (1999) Inhibition of potato tuber sprouting: Low levels of cytosolic pyrophosphate lead to non-sprouting tubers harvested from transgenic potato plants. Potato Res 42: 353–372 [Google Scholar]
- Hajirezaei MR, Börnke F, Peisker M, Takahata Y, Lerchl J, Kirakosyan A, Sonnewald U (2003) Decreased sucrose content triggers starch breakdown and respiration in stored potato tubers (Solanum tuberosum). J Exp Bot 54: 477–488 [DOI] [PubMed] [Google Scholar]
- Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S (2011) Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol 155: 776–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heftmann E. (1983) Biogenesis of steroids in Solanaceae. Phytochemistry 22: 1843–1860 [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. (2011) GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Terada S, Yoshida N, Mitsuhashi H (1977) Structure of barogenin from Solanum tuberosum. Phytochemistry 16: 791–793 [Google Scholar]
- Korpan YI, Nazarenko EA, Skryshevskaya IV, Martelet C, Jaffrezic-Renault N, El’skaya AV (2004) Potato glycoalkaloids: true safety or false sense of security? Trends Biotechnol 22: 147–151 [DOI] [PubMed] [Google Scholar]
- Kozukue N, Mizuno S (1985) Studies on glycoalkaloids of potatoes (part II). Analysis of glycoalkaloid content in potato tissues and tubers. J Jpn Soc Hortic Sci (Suppl) 54: 496–497 [Google Scholar]
- Kozukue N, Mizuno S (1989) Studies on glycoalkaloids of potatoes (Part IV). Changes of glycoalkaloid content in four parts of a sprouted potato tuber and in potato tubers during storage. J Jpn Soc Hortic Sci 58: 231–235 [Google Scholar]
- Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I, Veilleux RE (2014) Allelic variation in genes contributing to glycoalkaloid biosynthesis in a diploid interspecific population of potato. Theor Appl Genet 127: 391–405 [DOI] [PubMed] [Google Scholar]
- Masclaux F, Charpenteau M, Takahashi T, Pont-Lezica R, Galaud JP (2004) Gene silencing using a heat-inducible RNAi system in Arabidopsis. Biochem Biophys Res Commun 321: 364–369 [DOI] [PubMed] [Google Scholar]
- McCue KF, Allen PV, Shepherd LV, Blake A, Maccree MM, Rockhold DR, Novy RG, Stewart D, Davies HV, Belknap WR (2007) Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry 68: 327–334 [DOI] [PubMed] [Google Scholar]
- McCue KF, Allen PV, Shepherd LV, Blake A, Whitworth J, Maccree MM, Rockhold DR, Stewart D, Davies HV, Belknap WR (2006) The primary in vivo steroidal alkaloid glucosyltransferase from potato. Phytochemistry 67: 1590–1597 [DOI] [PubMed] [Google Scholar]
- McCue KF, Shepherd LVT, Allen PV, Maccree MM, Rockhold DR, Corsini DL, Davies HV, Belknap WR (2005) Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase. Plant Sci 168: 267–273 [Google Scholar]
- Mizutani M, Ohta D (1998) Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol 116: 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momma T. (1990) Recent study for genetic engineering of soybean glycinin gene. Plant Tissue Culture Lett. 7: 57–63 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nicolia A, Proux-Wéra E, Åhman I, Onkokesung N, Andersson M, Andreasson E, Zhu LH (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204: 17–24 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Osbourn A (2014) Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26: 91–99 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, Szekeres M, Mizutani M (2006a) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Watanabe B, Sakata K, Mizutani M (2006b) CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci Biotechnol Biochem 70: 2071–2080 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yokota T, Mizutani M (2009) Insights into the function and evolution of P450s in plant steroid metabolism. Phytochemistry 70: 1918–1929 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Okawa A, Moriuchi Y, Fujimoto Y (2013) Biosynthesis of steroidal alkaloids in Solanaceae plants: involvement of an aldehyde intermediate during C-26 amination. Phytochemistry 89: 26–31 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K. (2011). Purification and analytic method of glycoalkaloids kinds using a liquid chromatography. JP Patent Publication (Kokai) 2011-027429 A [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K, Muranaka T (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23: 4112–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, et al. (2014) Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346: 1084–1088 [DOI] [PubMed] [Google Scholar]
- Sinden S, Sanford L, Webb R (1984) Genetic and environmental control of potato glycoalkaloids. Am J Potato Res 61: 141–156 [Google Scholar]
- Sonnewald S, Sonnewald U (2014) Regulation of potato tuber sprouting. Planta 239: 27–38 [DOI] [PubMed] [Google Scholar]
- Sonnewald U. (2001) Control of potato tuber sprouting. Trends Plant Sci 6: 333–335 [DOI] [PubMed] [Google Scholar]
- Stewart C Jr, Kang BC, Liu K, Mazourek M, Moore SL, Yoo EY, Kim BD, Paran I, Jahn MM (2005) The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J 42: 675–688 [DOI] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Taylor MA, McDougall GJ, Stewart D (2007) Potato flavour and texture. In Vreugdenhil D, Bradshaw J, Gebhardt C eds, Potato Biology and Biotechnology. Elsevier, Amsterdam, The Netherlands, pp 525–540 [Google Scholar]
- Umemoto N, Sasaki K (2013). Protein having glycoalkaloid biosynthetic enzyme activity and gene encoding the same. US Patent Application No. 20130167271 A1 [Google Scholar]
- Umemoto N., Tsukahara M., Yoshioka M. (2001). New promoter sequence and use thereof. JP Patent Publication (Kokai) 2001-161373 A [Google Scholar]
- Valkonen JPT, Keskitalo M, Vasara T, Pietilä L, Raman KV (1996) Potato Glycoalkaloids: A Burden or a Blessing? Crit Rev Plant Sci 15: 1–20 [Google Scholar]
- Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, et al. ; Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.