The transcription factor AtERF11 promotes stem growth by increasing GA biosynthesis and GA response in Arabidopsis.
Abstract
The phytohormone gibberellin (GA) plays a key role in promoting stem elongation in plants. Previous studies show that GA activates its signaling pathway by inducing rapid degradation of DELLA proteins, GA signaling repressors. Using an activation-tagging screen in a reduced-GA mutant ga1-6 background, we identified AtERF11 to be a novel positive regulator of both GA biosynthesis and GA signaling for internode elongation. Overexpression of AtERF11 partially rescued the dwarf phenotype of ga1-6. AtERF11 is a member of the ERF (ETHYLENE RESPONSE FACTOR) subfamily VIII-B-1a of ERF/AP2 transcription factors in Arabidopsis (Arabidopsis thaliana). Overexpression of AtERF11 resulted in elevated bioactive GA levels by up-regulating expression of GA3ox1 and GA20ox genes. Hypocotyl elongation assays further showed that overexpression of AtERF11 conferred elevated GA response, whereas loss-of-function erf11 and erf11 erf4 mutants displayed reduced GA response. In addition, yeast two-hybrid, coimmunoprecipitation, and transient expression assays showed that AtERF11 enhances GA signaling by antagonizing the function of DELLA proteins via direct protein-protein interaction. Interestingly, AtERF11 overexpression also caused a reduction in the levels of another phytohormone ethylene in the growing stem, consistent with recent finding showing that AtERF11 represses transcription of ethylene biosynthesis ACS genes. The effect of AtERF11 on promoting GA biosynthesis gene expression is likely via its repressive function on ethylene biosynthesis. These results suggest that AtERF11 plays a dual role in promoting internode elongation by inhibiting ethylene biosynthesis and activating GA biosynthesis and signaling pathways.
Bioactive gibberellin (GA), a diterpenoid compound, is an allosteric inducer of its nuclear receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1; Ueguchi-Tanaka et al., 2005; Murase et al., 2008; Shimada et al., 2008). Acting downstream of GID1, the DELLA proteins are transcription regulators that repress GA signaling and restrict plant growth by causing transcriptional reprogramming (Ueguchi-Tanaka et al., 2007). Binding of GA to GID1 enhances the interaction between GID1 and DELLA, resulting in rapid degradation of DELLAs via the ubiquitin-proteasome pathway. In Arabidopsis (Arabidopsis thaliana), DELLAs are members of the GRAS (GA INSENSITIVE [GAI], REPRESSOR OF ga1-3 [RGA], and SCARECROW) family of regulatory proteins (Tian et al., 2004). Like all GRAS family members, DELLA contains a conserved C-terminal GRAS domain that confers the transcription regulator function. The unique DELLA domain in the N terminus of the protein is required for GA-induced degradation via GID1 binding (Dill et al., 2001; Itoh et al., 2002; Griffiths et al., 2006; Murase et al., 2008); this domain is absent in other GRAS family members. Among the five DELLAs (RGA, GAI, RGA-LIKE1 [RGL1], RGL2, and RGL3) in Arabidopsis, RGA and GAI are the major DELLAs for regulating GA-induced vegetative growth (Dill and Sun, 2001; King et al., 2001).
Recent studies also show that DELLAs integrate GA and other signaling pathways by antagonizing or enhancing functions of key regulators in other pathways via direct protein-protein interactions (Xu et al., 2014; Daviere and Achard, 2016). Most of the DELLA-interacting proteins are transcription factors or transcription regulators. Examples of DELLA-inhibited transcription factors/regulators include bHLH transcription factors, PIFs, in light signaling (de Lucas et al., 2008; Feng et al., 2008); the jasmonic acid (JA) signaling repressors, JAZs (Hou et al., 2010; Yang et al., 2012); ETHYLENE INSENSITIVE3 (EIN3), an ethylene signaling activator (An et al., 2012); and BRASSINAZOLE-RESISTANT1, a brassinosteroid signaling activator (Bai et al., 2012). DELLA-activated transcription factors include type-B ARABIDOPSIS RESPONSE REGULATORs (Marín-de la Rosa et al., 2015), ABSCISIC ACID INSENSITIVE3 (ABI3), and ABI5 (Lim et al., 2013). Other types of DELLA interactors include chromatin-remodeling complexes (Switch/Suc Nonfermenting, and a Chromodomain-Helicase-DNA-binding domain-containing protein PICKLE; Sarnowska et al., 2013; Zhang et al., 2014), RING domain proteins BOTRYTIS SUSCEPTIBLE1 INTERACTOR (BOI) and BOI-RELATED GENEs (BRG1, BRG2, and BRG3; Park et al., 2013), and subunits of the prefoldin complex for tubulin folding (Locascio et al., 2013). These findings indicate that protein-protein interaction is a central regulatory mechanism in DELLA-modulated plant development. Although a number of DELLA-interacting proteins have been reported, our current knowledge on how DELLAs regulate plant growth and development is still limited.
To uncover new regulators of the GA pathway, we performed an activation-tagging mutant screen and identified AtERF11, a member of the ERF (ETHYLENE RESPONSE FACTOR)/AP2 (APETALA2) family (Nakano et al., 2006), as a novel regulator of GA pathway. Our results show that AtERF11 promotes cell elongation by increasing bioactive GA accumulation. ERF11 also enhances GA responses by directly antagonizing DELLA function through protein-protein interaction. Interestingly, ERF11 has been reported to repress genes encoding ethylene biosynthetic enzymes (ACC synthases [ACSs]) (Li et al., 2011). Therefore, ERF11 may provide a molecular link between GA and ethylene pathways in modulating internode elongation.
RESULTS
Identification of ERF11 as a Positive Regulator of Internode Elongation through an Activation-Tagging Approach
To identify positive components of the GA signaling pathway, an activation-tagging mutant screen was employed to isolate mutants that grow taller than parental ga1-6 plants. The ga1-6 mutant is a GA-deficient semidwarf because of a missense mutation in GA1 (AtCPS) that encodes ent-copalyl diphosphate synthase (CPS) for GA biosynthesis (Sun et al., 1992; Sun and Kamiya, 1994). We reasoned that enhanced expression of positive regulators of the GA pathway would lead to a taller phenotype in this semidwarf mutant background. The ga1-6 plants (backcrossed four times to Col-0) were transformed with Agrobacterium tumefaciens that carried four copies of 35S transcriptional enhancers linked to a constitutively expressed BASTA resistance gene (Weigel et al., 2000). Approximately 12,500 T1 transformants were screened for increased final height. Among the mutants identified, mutant #279-2 was dramatically taller than the parental ga1-6 plants, an average height of 28.5 cm versus 18.3 cm (Fig. 1, A and B).
Figure 1.
Overexpression of ERF11 increased plant height by promoting cell elongation. A, The tall phenotype of mutant #279-2 (renamed erf11-1D ga1-6) was recapitulated by expressing 35S:HA-ERF11-GFP (ERF11-OE) in the ga1-6 background. Plant image was taken 50 d after planting. #1-1, #1-2, and #2-8 are three independent ERF11-OE ga1-6 transgenic lines (T2 generation). B, The final height of erf11-1D ga1-6 is taller than ga1-6. n = 24. C, The ERF11 transcript levels are significantly higher in erf11-1D ga1-6 than in ga1-6. Data represent RT-qPCR results using RNA isolated from 8-d-old seedlings. A GA-nonresponsive gene (At4g33380) was used to normalize different samples. Means ± se of four repeats (from two biological replicas, two technical repeats each) are shown. The level in ga1-6 was set to 1. D, HA-ERF11-GFP protein levels correlated with the plant heights in A. The immunoblot contains proteins from 8-d-old seedlings of three transgenic ERF11-OE ga1-6 lines as described in A and was probed with an anti-HA antibody. The bottom panel shows equal loading by Ponceau staining. E and F, Internodes in erf11-1D ga1-6 were longer than in ga1-6. Average internode lengths of 70-d-old ga1-6 and erf11-1D ga1-6 were calculated by dividing the length of the primary stem from the apex to the last secondary inflorescence with the total numbers of nodes. n = 24. G and H, The average epidermal cell length in the primary stem of erf11-1D ga1-6 is longer than in ga1-6. In G, n ≥ 600. H, scanning electron microscopy images of the internodes of 70-d-old plants. A representative cell in each line was outlined. I and J, erf11-1D ga1-6 and ERF11-OE ga1-6 double homozygous mutants have larger rosette leaves than ga1-6. Images in I show 33-d-old plants. In J, n = 20. K and L, The final heights (K) and internode lengths (L) of erf11 and erf4 erf11 mutants are shorter than those of the wild type. n = 24. In B, C, F, G, and J to L, data are means ± se. *P < 0.05; **P < 0.01.
The #279-2 mutant contains a single T-DNA insertion site, as the linked BASTA resistance gene segregated 3:1 in the T2 generation. Thermal asymmetric interlaced PCR (TAIL-PCR) revealed that the activation tag in #279-2 is inserted in chromosome 1 between At1g28360 and At1g28370, which encode two members of the ERF family, AtERF12 and AtERF11, respectively (Supplemental Fig. S1A). RT-qPCR further showed that the transcript levels of AtERF11 (At1g28370) were 18.3-fold higher in the mutant #279-2 than in the ga1-6 control (Fig. 1C). In contrast, the expression levels of the other two adjacent genes, At1g28360 (AtERF12) and At1g28375 (an expressed endomembrane protein), were unchanged or only 3-fold higher, respectively, in #279-2 compared to ga1-6 (Supplemental Fig. S1A). To verify whether overexpression of ERF11 causes the mutant phenotype of #279-2, we generated transgenic ga1-6 plants carrying CaMV 35S promoter:HA-ERF11-GFP (ERF11-OE lines). Indeed, the final heights of these ERF11-OE lines correlated with ERF11 protein levels in these plants (Fig. 1, A and D). The line with the highest ERF11 protein expression (#1-1) reached a similar final height as #279-2 (Fig. 1A), indicating that overexpression of ERF11 is responsible for the observed tall phenotype. Mutant #279-2 will be referred to as erf11-1D ga1-6 in the rest of this report; the homozygous erf11-1D ga1-6 double mutant in the T5 generation was used for all data presented here.
To understand better the effect of overexpression of ERF11 on stem elongation, the inflorescence stem of erf11-1D ga1-6 was characterized in more detail. erf11-1D ga1-6 produced a similar number of siliques as ga1-6 (Supplemental Fig. S1B), but with 82.3% longer internodes that contributed to the increased final height of the mutant (Fig. 1, E and F). Scanning electron microscopy analysis revealed that the longer internode in erf11-1D ga1-6 is caused by increased cell length (Fig. 1, G and H), but not greater cell numbers (Supplemental Fig. S1C). A previous study indicated that ERF11 is expressed ubiquitously in different tissues of wild-type plants; however, the highest transcript levels were detected in leaves and stems (Yang et al., 2005). Consistent with this ERF11 expression pattern, the rosette leaves of erf11-1D ga1-6 were 22.7% larger than ga1-6 (Fig. 1, I and J). In addition to the longer internode length and larger rosette size, erf11-1D ga1-6 flowered slightly earlier and displayed increased fertility compared to ga1-6 (Supplemental Table S1).
AtERF11 is a member of the ERF subfamily VIII-B-1a of ERF/AP2 transcription factors. There are eight members in this subfamily (ERF3, 4, and 7–12); each of them contains an ERF/AP2 domain and a transcription repression EAR motif (DLNxxP; McGrath et al., 2005; Nakano et al., 2006). Recently, AtERF11 was shown to inhibit ethylene biosynthesis by binding to the promoters of two ACC synthase genes ACS2 and ACS5 to repress their expression (Li et al., 2011). Furthermore, overexpression of another VIII-B-1a ERF member, AtERF4, confers reduced ethylene sensitivity in hypocotyl growth (Yang et al., 2005). Similar to erf11-1D ga1-6, overexpression of AtERF4 or AtERF8 (another close homolog of ERF11 in the same subfamily) also resulted in increased final height in the ga1-6 background (Supplemental Fig. S1D). We also generated the erf11-1D single mutant by backcrossing erf11-1D ga1-6 to the wild-type Col-0 and found that erf11-1D is taller with 26.5% longer internodes than the wild-type (Fig. 1, K and L). Moreover, the erf11 knockout mutant showed slightly shorter final height and internode length compared with wild-type Col-0; the erf11 erf4 double homozygous mutant displayed even shorter stems compared to Col-0 and the erf11 single mutant (Fig. 1, K and L). Consistent with the shorter stem phenotype, the rosette leaf length of the erf11 erf4 double mutant was slightly reduced comparing with the wild type (Supplemental Fig. S1E). These results indicated that ERF11 and its close homologs share redundant function in promoting stem elongation and rosette leaf expansion.
Overexpression of ERF11 Causes Elevated Bioactive GA4 and Reduced Ethylene Levels in the Growing Internodes
To determine whether the longer internodes of erf11-1D ga1-6 are caused by increased levels of bioactive GAs, we first analyzed the transcript levels of GA biosynthesis and catabolism genes that are important for vegetative growth (Mitchum et al., 2006; Rieu et al., 2008a, 2008b) by RT-qPCR. We found that expression of several GA biosynthesis genes, including GA20ox1, GA20ox2, and GA3ox1, was up-regulated in the internodes of erf11-1D ga1-6 compared with ga1-6, while a GA catabolism gene, GA2ox6, was down-regulated in erf11-1D ga1-6 (Fig. 2A). In contrast, GA3ox2 mRNA levels were not altered (Fig. 2A). GA analysis further showed that the GA4 level in the rosette leaves of erf11-1D ga1-6 was about 2-fold higher than that in ga1-6 (Fig. 2B). Our previous study showed that a 2-fold reduction in GA4 levels in the Arabidopsis rosette leaves could lead to a 2-fold reduction in the final plant height (Mitchum et al., 2006).
Figure 2.
Elevated bioactive GA levels and reduced ethylene levels in erf11-1D ga1-6. A, Relative mRNA levels of GA biosynthesis genes (left panel) and GA catabolism gene (right panel) in the internodes of ga1-6 and erf11-1D ga1-6. Data represent means ± se of four repeats. **P < 0.01. The levels in ga1-6 were set to 1. B, GA4 levels (ng/g dry weight) in erf11-1D ga1-6 were elevated comparing to ga1-6. GA4 contents in rosette leaves of 33-d-old plants were analyzed using two biological repeats. Due to very low levels of bioactive GA4 content in the ga1-6 background, we were unable to measure GA4 accurately in one of the ga1-6 repeats (test 2), which had trace amounts of GA4 estimated to be <0.27 ng/g dry weight. C, RGA protein levels were reduced in the internodes of erf11-1D ga1-6 compared to ga1-6. Proteins were extracted from internodes of 50-d-old plants, and immunoblotting was performed with affinity-purified anti-RGA antibodies. The bottom panel shows equal loading by Ponceau staining. D, The RGA transcript levels were similar in the internodes of ga1-6 and erf11-1D ga1-6. Data represent means ± se of four repeats. E, The ACS2 transcript levels in the internodes of erf11-1D ga1-6 were reduced in comparison to ga1-6. Data represent means ± se of four repeats. **P < 0.01. F, Ethylene production (pL/mg dry weight/h) from the internodes of 50-d-old plants. Data represent means ± se of four repeats. Similar results were obtained using another set of samples. **P < 0.01.
Consistent with the elevated GA levels in erf11-1D ga1-6, the amounts of RGA protein (an Arabidopsis DELLA) in the internodes of erf11-1D ga1-6 were much lower than in ga1-6 (Fig. 2C), even though RGA mRNA levels were not altered by erf11-1D (Fig. 2D). Taken together, overexpression of ERF11 in erf11-1D ga1-6 caused elevated bioactive GA4 levels through up-regulation of GA biosynthesis genes and down-regulation of a GA catabolism gene, which subsequently led to DELLA degradation and internode growth.
Being an EAR-containing transcription repressor, ERF11 is unlikely to up-regulate expression of GA biosynthesis genes directly. Instead, the elevated GA levels and longer internodes of erf11-1D ga1-6 may be due to reduced ethylene levels because ERF11 is known to inhibit ethylene biosynthesis by down-regulating ACS2 and ACS5 transcription (Li et al., 2011) and enhanced ethylene signaling decreases bioactive GA levels in Arabidopsis rosette plants (Achard et al., 2007). To test this possibility, the transcript levels of ACS2 in the internodes of erf11-1D ga1-6 and ga1-6 were analyzed by RT-qPCR. As predicted, expression of ACS2 was approximately 5-fold lower in the internodes of erf11-1D ga1-6 than in ga1-6 (Fig. 2E). Moreover, the ethylene production in internodes of erf11-1D ga1-6 was 60% lower than in ga1-6 (Fig. 2F). Consistently, erf11-1D displayed reduced ethylene response (Supplemental Fig. S1F), whereas erf11 and erf4 erf11 showed increased ethylene response (Supplemental Fig. S1G). These results indicated that the taller phenotype caused by overexpression of ERF11 is due to elevated bioactive GA4 levels and decreased ethylene levels.
ERF11 Positively Regulates GA Responses
The above data indicated that ERF11 promotes bioactive GA accumulation. Interestingly, our hypocotyl elongation assays showed that ERF11 also enhances GA response (Fig. 3; Supplemental Fig. S2A). The erf11 single mutant displayed slightly reduced GA response compared to the wild type, and the erf11 erf4 double mutant showed a further reduction in GA response (Fig. 3, A and C). Consistent with these results, overexpression of ERF11 (due to erf11-1D) caused an elevated GA response; both the double mutant erf11-1D ga1-6 and the single erf11-1D mutant displayed increased GA response compared to ga1-6 and Col-0, respectively (Fig. 3, B and D; Supplemental Fig. S2A). We also examined RGA protein levels in seedlings of ga1-6 and erf11-1D ga1-6 in response to GA treatments. Supplemental Figure S2B shows that RGA levels decreased in both lines in response to GA treatments, although RGA accumulated to lower levels in erf11-1D ga1-6 than in ga1-6 when untreated or with 0.01 µM GA4. This is consistent with the elevated GA content in erf11-1D ga1-6. These results indicate that ERF11 functions as a positive regulator of both GA production and GA signaling.
Figure 3.
Loss-of-function erf mutants displayed reduced GA response, whereas overexpression of ERF11 increased GA response. A and B, Hypocotyl elongation assays of 4-d-old seedlings grown in different GA concentrations under continuous light (50 µmol m−2 s−1). *P < 0.05; **P < 0.01. C, erf11 and erf11 erf4 displayed shorter hypocotyl length in the presence of 0.05 µM GA4, but not in the untreated control (–GA). Image shows 4-d-old seedlings. D, erf11-1D ga1-6 showed longer hypocotyl length with 0.05 and 1 µM GA4 treatment, but not in the untreated control. Image shows 4-d-old seedlings.
ERF11 Antagonizes DELLA Function via Protein-Protein Interaction
To place ERF11 in the GA signaling pathway, genetic interactions between ERF11 and RGA were examined by double mutant analysis. rga-Δ17 is a transgenic line that expresses a dominant active form of RGA, which lacks a 17-amino acid motif within the DELLA domain that is required for GA-induced degradation (Dill et al., 2001). rga-Δ17 displays a severe dwarf phenotype, while erf11-1D has longer internodes than the wild type. The use of rga-Δ17 in the double mutant analysis allowed us to uncouple the GA response activity from GA biosynthesis so that we could examine the direct role of ERF11 in the GA response. Because homozygous rga-Δ17 plants are sterile and grow extremely slowly, we compared phenotypes of the semidwarf hemizygous rga-Δ17 in the homozygous erf11-1D background (referred to as erf11-1D rga-Δ17) to the hemizygous rga-Δ17. We found that the final height of erf11-1D rga-Δ17 was 30% taller than rga-Δ17 (Fig. 4A; Supplemental Fig. S3A), and the internode length of erf11-1D rga-Δ17 was 46% longer than that of rga-Δ17 (Supplemental Fig. S3B), whereas the average number of siliques of erf11-1D rga-Δ17 was similar to that of erf11-1D (Supplemental Fig. S3C). These results indicated that erf11-1D partially rescued the dwarf phenotype of rga-Δ17, suggesting that erf11-1D either inhibits rga-Δ17 protein accumulation or activity, or acts downstream of RGA. However, rga-Δ17 protein levels in the internodes of rga-Δ17 and erf11-1D rga-Δ17 were similar (Fig. 4B), indicating that the longer internodes in erf11-1D rga-Δ17 were not caused by reduced rga-Δ17 accumulation.
Figure 4.
ERF11 interacts with RGA and inhibits its function. A, erf11-1D partially rescued the dwarf phenotype of rga-∆17. Image shows 60-d-old plants. B, Similar rga-∆17 protein levels were present in the upper growing internodes of 70-d-old rga-∆17 and erf11-1D rga-∆17. The immunoblot was probed with affinity-purified anti-RGA antibodies. The bottom panel shows equal loading by Ponceau staining. C, Interactions between ERFs and RGA in yeast two-hybrid assays. A truncated RGA protein (amino acids 187 to 587) that contains the C-terminal GRAS domain was used as the bait. ERFs were used as the prey. FL, Full-length; –EAR, EAR motif deleted. For each strain, 2 μL yeast cells with OD600 values of 0.25 and 0.025 were spotted on control media (+His) and –His media +1 mM 3-AT (a competitive inhibitor of His-3 enzyme). Empty prey and bait vectors were included as negative controls. D, co-IP of ERFs and RGA in planta. HA-RGA was transiently expressed alone or coexpressed with cMyc-tagged GUS, ERF11 (–EAR), or ERF8 (–EAR) in N. benthamiana. The total protein extracts were immunoprecipitated with anti-cMyc antibody-conjugated agarose beads; the input and IP samples were analyzed by immunoblotting using antibodies for HA and cMyc, separately. E, erf11-1D reduced the transcript levels of DELLA target genes bHLH137, bHLH154, and Exp-PT1 in the internodes of 70-d-old rga-∆17 plants. RT-qPCR data represent means ± se of four repeats. **P < 0.01. In A, B, and E, rga-∆17 is hemizygous in both rga-∆17 and erf11-1D rga-∆17.
We then tested whether erf11-1D inhibits RGA activity by direct protein-protein interaction, a known regulatory mechanism for DELLA and its interactors (Xu et al., 2014; Daviere and Achard, 2016). Our yeast two-hybrid assays showed that the GRAS domain of RGA directly interacts with ERF11 minus the EAR motif (Fig. 4C). No interaction was detected between RGA and the full-length ERF11, presumably because the transcription repression mediated by the EAR motif interfered with reporter gene expression. We also observed interactions between RGA and three ERF11 close homologs ERF4, ERF8, and ERF10 (in subgroup VIII-B-1a). However, RGA did not interact with ERF88, which belongs to a different subgroup VIII-B-1b (Fig. 4C), suggesting that RGA specifically interacts with ERFs in the VIII-B-1a subfamily. RGA and GAI are the major DELLA proteins that control stem elongation (Dill and Sun, 2001; King et al., 2001). We found that GAI also interacted with ERF11, ERF4, ERF8, and ERF10 in yeast two-hybrid assays (Supplemental Fig. S4).
To confirm ERF11-RGA interaction in planta, we performed coimmunoprecipitation (co-IP) assays by transiently coexpressing 35S:cMyc-ERF11 (or ERF8) and 35S:HA-RGA constructs in leaves of Nicotiana benthamiana through Agrobacterium-mediated transformation. Tissues infiltrated with 35S:HA-RGA alone or coinfiltrated with 35S:cMyc-GUS-NLS served as negative controls. Immunoprecipitation was performed using anti-cMyc antibody-conjugated agarose beads. Figure 4D shows that HA-RGA was coimmunoprecipitated when it was coexpressed with cMyc-ERF11 (–EAR) or cMyc-ERF8 (–EAR), but not when it was expressed alone or coexpressed with cMyc-GUS-NLS. These co-IP assays further support the idea that ERF11 and its close homologs directly interact with RGA. To test whether overexpression of ERF11 inhibits DELLA function, we then examined transcript levels of DELLA target genes (Zentella et al., 2007) in rga-Δ17 and erf11-1D rga-Δ17 by RT-qPCR analysis (Fig. 4E). We found that expression of three of the DELLA target genes (bHLH137, bHLH154, and Exp-PT1) was down-regulated in the internodes of erf11-1D rga-Δ17, suggesting that ERF11 and DELLA interfere with each other's function by direct protein-protein interaction.
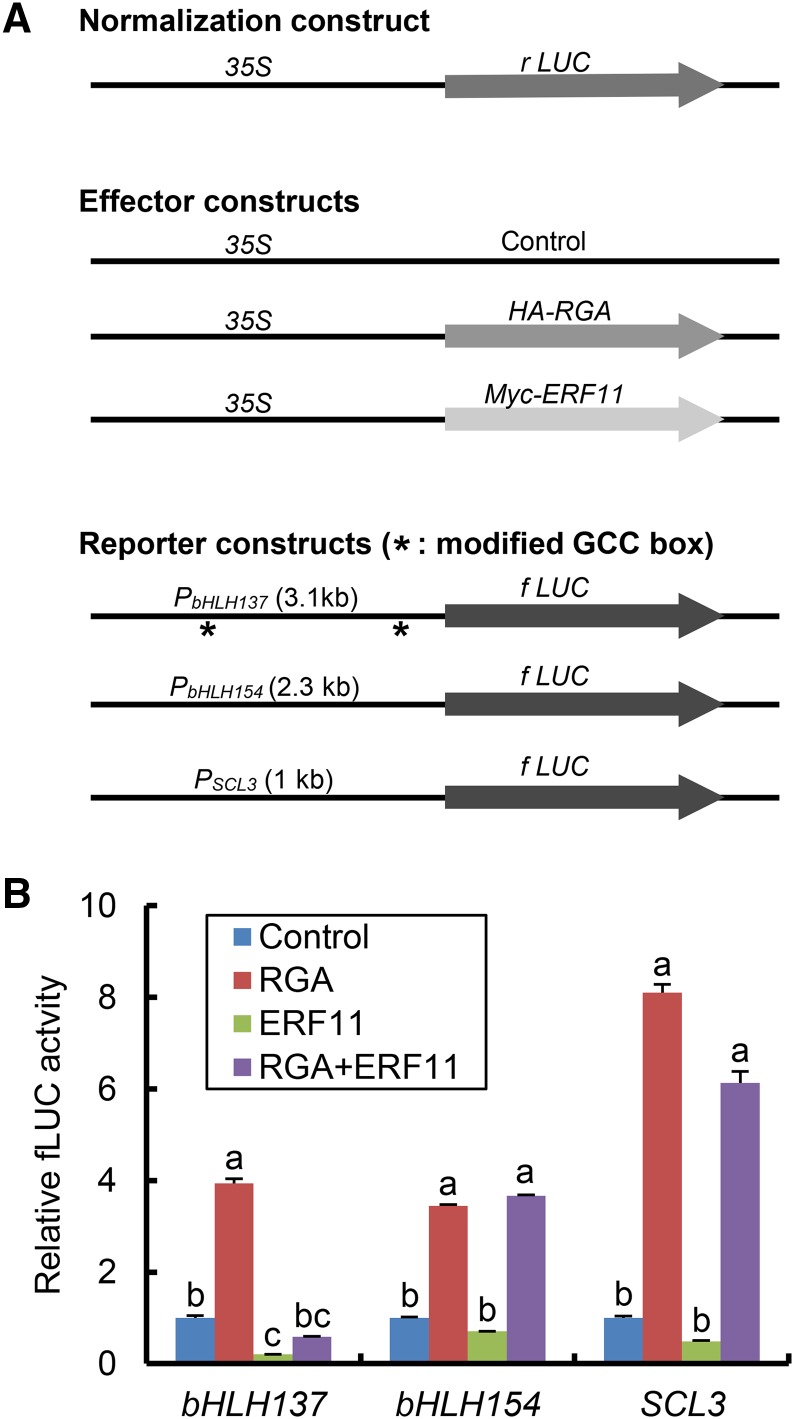
To examine further the direct antagonistic interaction between ERF11 and DELLA, these proteins were expressed alone or coexpressed in tobacco leaves by agroinfiltration to test whether they antagonistically modulate transcription of bHLLH137 and bHLH154 promoters using the dual luciferase (LUC) reporter assay (Fig. 5). The reporter constructs contain promoter sequences of bHLH137, bHLH154, and SCL3 genes, respectively, which were fused to the firefly LUC gene (fLUC; Fig. 5A). The SCL3 promoter was included as a control; SCL3 is an RGA-induced target gene (Zentella et al., 2007), but its expression was not altered by erf11-1D (Fig. 4E). The 35S:Renilla LUC (rLUC) was used as an internal standard in the assay. Two effectors are 35S:HA-RGA and 35S:myc-ERF11. As expected, expression of RGA alone induced all three promoters of RGA target genes (4-fold for bHLH137, 3-fold for bHLH154, and 8-fold for SCL3) compared to the empty effector control (Fig. 5B). ERF11 alone repressed expression of bHLH137 by 5-fold, and coexpression of RGA and ERF11 displayed antagonistic effects on the expression of this promoter (Fig. 5B). In contrast, ERF11 did not significantly repress expression of SCL3, indicating that the inhibitory effect of ERF11 on the bHLH137 promoter is specific. We also found that bHLH154 expression was not affected by ERF11 in the dual luciferase assay (Fig. 5B), suggesting that this gene may not be a direct target of ERF11. The optimal cis-element for ERFs has been identified to be the GCC box (AGCCGCC), although AtERF3 and AtERF4 can also bind similar sequences with single nucleotide substitutions (Ohme-Takagi and Shinshi, 1995; Fujimoto et al., 2000). Interestingly, using pDRAW32 DNA analysis software (http://www.acaclone.com), we found that the bHLH137 promoter contains two modified GCC elements (Fig. 5A), whereas the promoters of bHLH154 and SCL3 genes lack any putative ERF binding sequences. Taken together, our results show that ERF11 directly represses RGA-induced bHLH137 expression.
Figure 5.
ERF11 repressed, whereas RGA induced, transcription of the bHLH137 promoter in tobacco transient expression assay by agroinfiltration. A, Schematics of the normalization control, reporter, and effector constructs. 35S:Renilla LUC (rLUC) served to normalize transformation efficiency. In the reporter constructs, the firefly LUC gene (fLUC) was placed under the control of different promoters of RGA target genes (bHLH137, bHLH154, and SCL3). 35S:RGA and 35S:ERF11 served as two effector constructs, respectively. The positions of two modified GCC box sequences in bHLLH137 promoter are labeled by asterisks: AGCCGCT at –2 kb and ACCCGCC at –0.2 kb. The empty effector vector was used as a negative control. B, RGA induced expression of all three target gene promoters, whereas ERF11 only repressed bHLH137 expression. Each reporter construct and the 35S:rLUC construct were introduced into tobacco leaves in the presence of the empty effector constructs (Control) or 35S:RGA and/or 35S:ERF11 (with the same molar ratios) by agroinfiltration. The relative fLUC activity (normalized by rLUC activity) in the empty effector control was set to 1. Data represent the average value ± se of eight biological replicas. Different letters above the bars indicate significant difference (P < 0.01).
DISCUSSION
The data in this report revealed that ERF11 enhances GA responses by two mechanisms: (1) increasing bioactive GA levels by inducing expression of the GA biosynthesis genes GA3ox and GA20ox and (2) promoting GA responses by antagonizing the activity of the GA signaling repressor DELLA via direct protein-protein interaction (Fig. 6). Our transient expression assay showed that ERF11 represses whereas DELLA induces transcription of the target gene bHLH137 (Fig. 5), indicating that ERF11 and DELLA interfere with each other's function. The second mechanism should reduce DELLA function immediately, whereas the first mechanism acts slower in regulating DELLA activity as its effect on DELLA is via alteration in GA biosynthesis. ERF11 is unique in that it promotes both GA biosynthesis and GA signaling. All of the previously reported elevated GA-signaling mutants down-regulate GA biosynthesis via the negative feedback mechanism (Sun and Gubler, 2004). Our data further suggest that induction of GA biosynthesis gene expression by ERF11 is likely an indirect effect mediated by decreasing ethylene levels (Fig. 6). erf11-1D conferred a reduction in the amounts of ethylene in the inflorescence stems, consistent with a recent report showing ERF11 represses ACS transcription (Li et al., 2011). The exact molecular mechanism of how ethylene inhibits GA biosynthesis gene expression requires further investigation.
Figure 6.
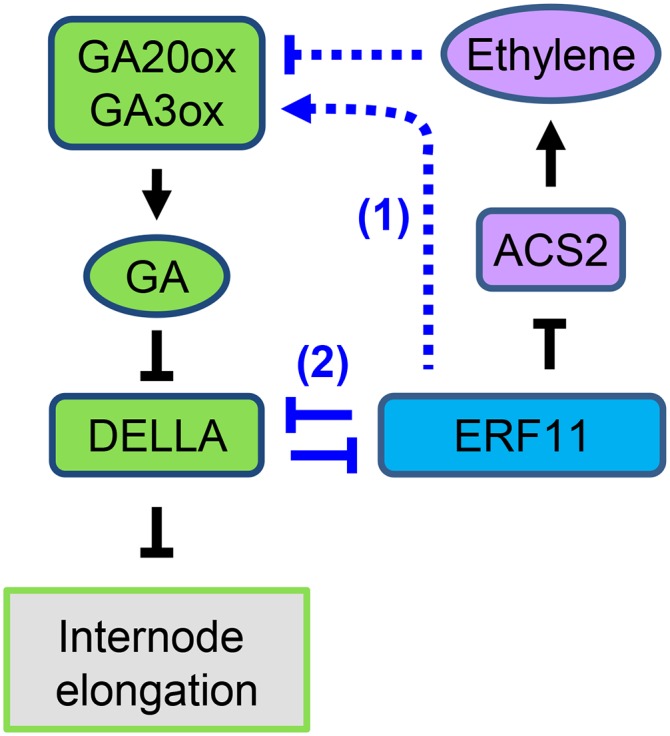
Model for antagonistic interaction between ERF11 and DELLA in regulating GA and ethylene pathways and internode elongation. The arrows and T-bars highlighted in blue represent new links that are revealed in this study. ERF11 promotes internode elongation by enhancing GA responses through two mechanisms: (1) increasing GA accumulation (and therefore DELLA degradation) indirectly via its inhibitory effect on ethylene biosynthesis; and (2) inhibiting DELLA function by direct protein-protein interaction. DELLA and ERF11 antagonize each other's function in regulating downstream gene expression, as shown in the transient expression assay. In (1), ERF11-induced GA accumulation is likely an indirect effect of reduction of ethylene production via inhibition of ACS2 expression by ERF11. The reduced ethylene levels then result in repression of GA3ox and GA20ox expression, likely through the ethylene signaling pathway.
In Arabidopsis, there are 122 ERF/AP2 family members. AtERF11 belongs to the subfamily VIII-B-1a (McGrath et al., 2005; Nakano et al., 2006). All eight members in this subfamily (ERF3, 4, and 7–12) contain a transcription repressor EAR motif near their C terminus. Interestingly, our study and previous reports reveal that three of the EAR-containing ERFs (ERF4, ERF7, and ERF11) regulate multiple hormone pathways. Overexpression of AtERF4 confers reduced sensitivity to ethylene, abscisic acid (ABA), and JA in hypocotyl or root growth, whereas the loss-of-function erf4-1 mutant displays increased JA response (McGrath et al., 2005; Yang et al., 2005). Overexpression or silencing of AtERF7 resulted in reduced or increased ABA response in stomatal closure, respectively, indicating that AtERF7 negatively regulates ABA responses (Song et al., 2005). Our data showed that overexpression of ERF11, ERF4, or ERF8 partially rescued the dwarf phenotype of the GA-deficient ga1-6 mutant. Phenotype analyses of the loss-of-function erf11 and erf4 single and double mutants further confirmed that ERF4 and ERF11 positively regulate GA response in hypocotyl elongation. Similar to ERF11, ERF4, 8, and 10 interact with DELLA in co-IP and/or yeast two-hybrid assays. Future studies will determine whether all eight VIII-B-1a subfamily ERFs regulate GA and/or other hormone pathways by a similar mechanism as illustrated for ERF11. Our yeast two-hybrid results indicate that RGA does not interact with ERF88 in the VIII-B-1b subfamily. Interestingly, a group-VII ERF/AP2 (RELATED TO APETALA2.3) was shown recently to be a DELLA-interacting protein, playing a role in GA and ethylene-regulated apical hook development (Marín-de la Rosa et al., 2014). Future studies will address whether any of the other ERF/AP2 subfamily members are DELLA-interacting proteins.
In rice (Oryza sativa), group VII ERFs (OsSUB1A, SNORKEL1, and SNORKEL2) that lack the EAR motif have been shown to regulate internode elongation that is modulated by ethylene and GA (Xu et al., 2006; Hattori et al., 2009). Interestingly, these OsERFs (transcription activators) function differently from AtERF11. In submergence-tolerant rice, ethylene inhibits internode growth under submergence conditions. In this case, ethylene induces OsSUB1A expression (Xu et al., 2006), which in turn inhibits GA response by promoting SLR1 (rice DELLA) and SLRL1 transcript and protein accumulation (Fukao and Bailey-Serres, 2008). Ectopic expression of OsSUB1A in Arabidopsis leads to reduced GA response and increased ABA response (Fukao et al., 2011). In contrast to the submergence-tolerant rice, deepwater rice responds to submergence by rapid internode growth. In this case, ethylene promotes internode elongation by increasing GA levels via induction of SNORKEL1 and SNORKEL2 expression, although it is unclear how SNORKEL1 and SNORKEL2 promote GA accumulation (Hattori et al., 2009). Therefore, different members of the group VII ERFs play distinct roles in mediating ethylene and GA responses in controlling internode elongation.
In summary, increasing numbers of ERFs have been shown to regulate plant development in response to hormonal signals or abiotic stresses. Our work reveals that AtERF11 promotes internode elongation by promoting both GA biosynthesis and signaling pathways.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The ga1-6 semidwarf mutant plant used for activation tagging was generated by crossing the original ga1-6 in the Landsberg erecta (Ler) background (Koornneef and van der Veen, 1980; Sun and Kamiya, 1994) four times into Columbia-0 (Col-0). The activation-tagging mutant pools were generated by transforming ga1-6 (4x Col-0) with pSKI015 (Weigel et al., 2000), and the mutant #279-2 was identified in the T1 generation as a BASTA-resistant plant that was taller than the parental plant. #279-2 displayed a 3:1 segregation ratio of the BASTA resistance in the T2 generation; homozygous lines were identified by screening in the T3 generation (renamed erf11-1D ga1-6). The erf11 (SALK_116053) T-DNA insertion mutant was requested from the Arabidopsis Stock Center, and the erf4-1 (Salk_073394) mutant was provided by Dr. Kemal Kazan (McGrath et al., 2005). The homozygous double mutant erf4 erf11 was generated by crossing erf11 to erf4. The erf11-1D rga-∆17 double mutant was generated by crossing a rga-∆17 transgenic line (#18-2-1) in the Col-0 background (Dill et al., 2001) to erf11-1D. Genotyping primers are listed in Supplemental Table S2.
For growth on media, seeds were plated on 1× or 0.5× Murashige and Skoog (MS) medium containing 2% or 1% Suc and 0.7% agar, and incubated at 22°C under constant light (50–70 µmol m−2 s−1). For growth on soil, seeds were sown on MetroMix 200 (Sun Gro Horticulture) and incubated at 22°C under 16 h light. The procedure for the hypocotyl elongation assay was described previously (Zhang et al., 2011). For ethylene-mediated seedling triple-response assay, the detailed procedure was described before (Zhou et al., 2007). Seeds were stratified at 4°C for 72 h and then germinated on half-strength MS at 22°C for 80 h in the dark with or without 20 μL/L ethylene gas. Dark-grown seedlings were photographed and hypocotyl length was measured using software Image J. Transgenic Arabidopsis lines were generated by the floral dip method (Clough and Bent, 1998). For selection, 10 μg/mL of BASTA or 50 μg/mL kanamycin was included in the MS medium.
All statistical analyses were performed using Student’s t tests with the statistical package JMP Pro 10.0.2 (SAS Institute).
Plasmid Constructs
All the primers used in this study are listed in Supplemental Table S2. The PCR-amplified fragments in all constructs were sequenced to ensure that no mutations were introduced. Detailed information on plasmid construction is described in Supplemental Methods.
TAIL-PCR
Genomic DNA was extracted by using the CTAB method (Weigel and Glazebrook, 2002). TAIL-PCR was carried out as described previously (Liu et al., 1995). The degenerate primers used for TAIL-PCR were AD1, AD2, and AD3 (Liu and Whittier, 1995) and AD4 and AD6 (Liu et al., 1995). The T-DNA specific primers LB8, LB7, and JL-202 (Alonso et al., 2003) were used in the primary, secondary, and tertiary TAIL-PCR reactions, respectively. Specific TAIL-PCR products were gel-purified and sequenced and BLAST searched against the National Center for Biotechnology Information database to identify the T-DNA insertion site.
Quantitative Real-Time RT-PCR Analysis
The real-time RT-qPCR analyses were performed with an Eppendorf realplex2 Mastercycler ep gradient S. Total RNA isolation, cDNA synthesis, and qPCR analyses were performed as described previously (Zentella et al., 2007). At4g33380, whose expression remains constant (Rieu et al., 2008b), was used as the control to normalize the qPCR data. Unless specified otherwise, the qPCR data are the means of four repeats (two biological repeats and two technical replicates of each set of samples).
GA Measurements
Rosette leaves of 33-d-old soil-grown ga1-6 and erf11-1D ga1-6 plants were harvested and immediately frozen in liquid nitrogen and lyophilized. GAs were purified and quantified according to Plackett et al. (2012) using a 6410 Triple Quad LC-MS (Agilent Technologies) with an Agilent 1200 series rapid resolution liquid chromatography system fitted with a Zorbax SB-Phenyl column (1.8 µm, 2.1 × 50 mm).
Immunoblot Analyses
Total proteins from seedlings or internodes were extracted as described before (Silverstone et al., 2001). The extracted proteins were fractionated by 8% SDS-PAGE and analyzed by immunoblot analysis using anti-HA antibodies (Covance) or crude anti-RGA antibodies (Silverstone et al., 2001).
Yeast Two-Hybrid Assays
The yeast two-hybrid assay was performed using the ProQuest system (Invitrogen) in the yeast strain pJ69-4A (James et al., 1996). Varying concentrations of 3-AT (0, 1, 2.5, 5, 10, and 25 mM) were included in medium lacking Trp, Leu, and His. For each combination, 2 μL yeast cells with OD600 values of 0.25 and 0.025, respectively, were spotted on media plates.
Coimmunoprecipitation of ERFs and RGA, and Dual Luciferase Assay
Transient expression and co-IP assays were performed as described previously (Zhang et al., 2011) with the following modifications: After cells were resuspended in infiltration media, Agrobacterium tumefaciens GV3101 strains carrying individual expression constructs were mixed to make a final OD600 of about 0.8 for each strain and infiltrated into 28-d-old Nicotiana benthamiana (tobacco) leaves by needle-less syringe; 40 h transiently transformed tobacco leaves were harvested for co-IP assay without cross-linking. Two grams of leaves was ground into a fine powder in liquid nitrogen, followed by resuspension in 5 mL extraction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 5 mm β-mercaptoethanol, and 1× Protease Inhibitors [Sigma P-9599]). The powder was ground in extraction buffer for 10 min on ice until there was no visible debris. The homogenates were centrifuged at 20,000g at 4°C for 20 min. The co-IP was performed using 10 µL anti-cMyc agarose-conjugated beads (A7470; Sigma-Aldrich) by incubating with supernatant for 2 h at 4°C.
The dual luciferase assays were also performed using the transient expression system in tobacco. The control constructs and the reporter and effector constructs were individually transformed into Agrobacterium strain GV3101. Then, each reporter- and rLUC-containing strain was coinfiltrated into tobacco leaves with various combinations of effector strains. Forty hours after infiltration, tobacco leaves were harvested for protein extraction, and luciferase activities were measured using the dual-luciferase reporter assay system (Promega). Relative promoter activity was calculated as the ratio of fLUC to rLUC activities for each sample. Six biological repeats were conducted for each effector combination.
Ethylene Measurement
The top 10 internodes of the main stems were detached, and the flower clusters and siliques were removed. For each set of measurements, 15 stems (per genotype) were placed in a 22-mL gas chromatography vial containing 0.5 mL of 0.5× liquid MS media. The vials were capped and then placed in a 16-h/8-h light/dark cycle incubator at 22°C for 5 h. The accumulated ethylene was measured and calculated based on comparison to a 1 mL/L ethylene standard (Woeste et al., 1999). Data were from three replicates, and each experiment was repeated at least once with comparable results.
Cell Length Measurement by Scanning Electron Microscopy
The 10th to 20th internodes from the bottom on the main stem of 70-d-old ga1-6 and erf11-1D ga1-6 plants were fixed overnight with FAA solution (5% acetic acid, 45% ethanol, and 5% formaldehyde) and dehydrated with a graded ethanol series (30%, 50%, 70%, 90%, and 100%) (Tsukaya et al., 1993). After dehydration, the samples were chemically dried with HMDS (hexamethyldisilazane) and then were imaged on an FEI XL30 ESEM.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: ERF11 (At1g28370), ERF4 (At3g15210), ERF8 (At1g53170), ERF10 (At1g03800), ERF12 (At1g28360), ERF88 (At1g12890), ACS2 (At1g01480), GA1 (At4g02780), RGA (At2g01570), GAI (At1g14920), GA3ox1 (At1g15550), GA3ox2 (At1g80340), GA20ox1 (At4g25420), GA20ox2 (At5g51810), GA2ox6 (At1g02400), AtGID1A (At3g05120), MYB (At3g11280), bHLH137 (At5g50915), bHLH154 (At2g31730), WRKY27 (At5g52830), SCL3 (At1g50420), EXP-PT1 (At2g45900), XERICO (At2g04240), RING (At4g19700), EXP-PT (At4g33380), and unknown protein (At1g28375).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of ERF Overexpression Lines and erf Loss-of-Function Mutants.
Supplemental Figure S2. erf11-1D Caused an Elevated GA Response and Reduced Levels of RGA Protein.
Supplemental Figure S3. erf11-1D Partially Rescued rga-∆17 Phenotypes.
Supplemental Figure S4. Interactions between ERFs and GAI in Yeast Two-Hybrid Assays.
Supplemental Table S1. Silique and Flowering Time Phenotypes of ga1-6 and erf11-1D ga1-6.
Supplemental Table S2. List of Primers and Their Uses.
Supplementary Material
Acknowledgments
We are grateful to Joe Kieber and Gyeong Mee Yoon for their advice with ethylene analyses and for providing the facility for ethylene measurements. We thank Benke Kuai for his generous support of some of the ethylene-related studies, and Tomoe Nose and Yumiko Takebayashi for technical support in LC-MS/MS hormone analysis. We also thank Kemal Kazan for erf4-1, Jian-Min Zhou for ein3 and EIN3-OE lines, Chi-Kuang Wen for helpful discussions, and Michelle Gignac at the SEM facility at Duke for help with scanning electron microscopy analysis.
Glossary
- GA
gibberellin
- JA
jasmonic acid
- TAIL-PCR
thermal asymmetric interlaced PCR
- co-IP
coimmunoprecipitation
- ABA
abscisic acid
Footnotes
Articles can be viewed without a subscription.
This work was supported by the National Science Foundation (IOS-0641548 and MCB-0923723), the U.S. Department of Agriculture (2014-67013-21548), the National Institutes of Health (R01 GM100051), and the Canada Research Chair-National Sciences and Engineering Research Council (grant no. 00003714).
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun T, Wang Z-Y (2012) Brassinosteroid, gibberellin, and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P (2016) A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant 9: 10–20 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, Choi G (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D (2013) Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Curr Biol 23: 804–809 [DOI] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet 11: e1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D, Blázquez MA (2014) Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol 166: 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Nguyen KT, Park E, Jeon JS, Choi G (2013) DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L, Benlloch R, Nilsson O, Thomas SG, Hedden P, Phillips AL (2008a) Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, Phillips AL, Hedden P (2008b) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, et al. (2013) DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol 163: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Naito S, Redei GP, Komeda Y (1993) A new class of mutations in Arabidopsis thaliana, acaulis1, affecting the development of both inflorescences and leaves. Development 118: 751–764 [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Woeste KE, Vogel JP, Kieber JJ (1999) Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol Plant 105: 478–484 [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X (2014) Shedding light on integrative GA signaling. Curr Opin Plant Biol 21: 89–95 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun TP (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R (2014) The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo J-O, Lim J, Kamiya Y, Yamaguchi S, Sun T (2011) SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen CK (2007) RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol 145: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.