Homeostasis and remodeling of the chloroplast proteome involve various “processing” and “processive” proteases.
Abstract
Chloroplasts originated from the endosymbiosis of ancestral cyanobacteria and maintain transcription and translation machineries for around 100 proteins. Most endosymbiont genes, however, have been transferred to the host nucleus, and the majority of the chloroplast proteome is composed of nucleus-encoded proteins that are biosynthesized in the cytosol and then imported into chloroplasts. How chloroplasts and the nucleus communicate to control the plastid proteome remains an important question. Protein-degrading machineries play key roles in chloroplast proteome biogenesis, remodeling, and maintenance. Research in the past few decades has revealed more than 20 chloroplast proteases, which are localized to specific suborganellar locations. In particular, two energy-dependent processive proteases of bacterial origin, Clp and FtsH, are central to protein homeostasis. Processing endopeptidases such as stromal processing peptidase and thylakoidal processing peptidase are involved in the maturation of precursor proteins imported into chloroplasts by cleaving off the amino-terminal transit peptides. Presequence peptidases and organellar oligopeptidase subsequently degrade the cleaved targeting peptides. Recent findings have indicated that not only intraplastidic but also extraplastidic processive protein-degrading systems participate in the regulation and quality control of protein translocation across the envelopes. In this review, we summarize current knowledge of the major chloroplast proteases in terms of type, suborganellar localization, and diversification. We present details of these degradation processes as case studies according to suborganellar compartment (envelope, stroma, and thylakoids). Key questions and future directions in this field are discussed.
Over 1 billion years of plastid evolution since the endosymbiosis of ancestral cyanobacteria (Douzery et al., 2004), chloroplast biogenesis has gained complexity, with large sets of the endosymbiont genes being transferred to host nuclear genomes. While only 100 endosymbiont genes remain in the plastid genome, with the corresponding proteins biosynthesized there, the nuclear genes have often gained complexity by duplication and diversification of the original endosymbiotic genes. This complexity raises numerous questions regarding (1) at what level gene expression is coordinately controlled, (2) what molecules coordinate the cross talk between chloroplasts and the nucleus, (3) how proteins get across membranes and become imported into chloroplasts, and (4) how the stoichiometries of nucleus- and chloroplast-encoded subunits within individual chloroplast protein complexes such as photosystems and Rubisco are strictly maintained. Given these questions, chloroplast biogenesis has remained a central subject in plant physiology for the last few decades (Jarvis and López-Juez, 2013). In our view, the aforementioned questions point to the importance of protein homeostasis and posttranslational modification, in which proteases play a dominant role. Therefore, we focus on the major events of proteolysis governed by chloroplast proteases, paying particular attention to what kinds of proteases are present and how they exert their functions in chloroplasts.
ADVANCES
The 20 known chloroplast proteases include two types, processive proteases and processing peptidases, that shape the chloroplast proteome.
The major processive proteases have heteromeric structures with unfolding and proteolytic domains and are involved in chloroplast proteome homeostasis by degrading unnecessary proteins.
Recent reports implicate the extraplastidic ubiquitin proteasomal system and intraplastidic prokaryotic-like proteases in regulation of the chloroplast proteome through the control of protein translocation across the envelopes.
Stromal proteases function in sequential preprotein maturation, environment-dependent metabolic regulation, and mineral homeostasis; thylakoid proteases are involved in protein maturation and quality control of photosynthetic complexes.
In this review, we describe the major protease machineries and their intraorganellar functions in proteolytic regulation of the intraplastid proteome landscape, the latter of which is addressed as three case studies covering the different chloroplast compartments (envelope, stroma, and thylakoids). More detailed descriptions of each chloroplastic protease can be found in previous reviews (Kato and Sakamoto, 2010; Clarke, 2012; Teixeira and Glaser, 2013; Kmiec et al., 2014; Adam, 2015; Nishimura and van Wijk, 2015; van Wijk, 2015). In particular, readers are encouraged to refer to the recent excellent review by van Wijk (2015), in which the regulation of proteolytic machineries is thoroughly described, not only in plastids but also in mitochondria and peroxisomes.
OVERVIEW OF THE MAJOR PROTEASES IN CHLOROPLASTS
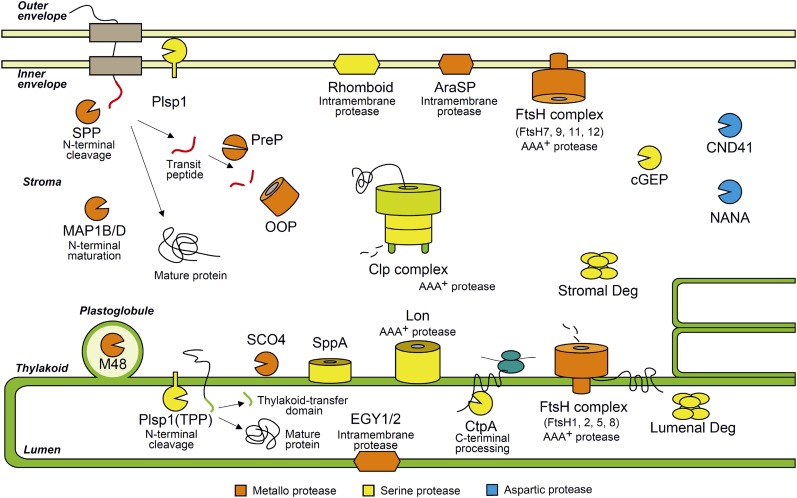
More than 20 proteolytic machineries have been identified in chloroplasts through biochemical, genetic, bioinformatic, and proteomic approaches during the last two decades (van Wijk, 2015). Figure 1 illustrates our current understanding of chloroplast proteases. Most chloroplast proteases originated from respective bacterial prototypes. During plastid evolution, they have been duplicated and diversified in terms of structure and function (Box 1). Many are metalloproteases or Ser proteases, and a few are Asp proteases. Genetic studies of model plants have demonstrated the physiological importance of these proteases (for review, see Kato and Sakamoto, 2010). Due to space constraints, we outline only the major intraplastid proteolytic machineries, with emphasis on two major processive proteases, Clp and FtsH.
Figure 1.
Intraplastid proteases in land plants. Land plant chloroplasts contain three types of proteolytic machines, namely metalloproteases, Ser proteases, and Asp proteases, which are colored in orange, yellow, and blue, respectively.
Box 1. Evolutionary Diversification of Proteases in Photosynthetic Organisms
Chloroplast proteases appear to have diverged into multiple isoforms during the evolution of photosynthetic organisms. It is noteworthy that primitive protease machineries in proteobacteria, like those in Escherichia coli, are present as homooligomeric assemblies. By contrast, the cyanobacterial proteases have gained multiple paralogous components, leading to the emergence of heterooligomeric structures. The complexity of protease organization has much increased in land plants. Representative examples include chloroplast FtsH, Deg, and Clps (see table below). The FtsH diversification has produced distinct heterohexameric protease complexes each localized at different intraplastid compartments. The chloroplast Deg proteases have acquired quite diverse domain organizations and function at specific suborganellar locations. The evolution of nonphotosynthetic organisms into photosynthetic organisms has been accompanied by the occurrence of a proteolytically inactive subunit in the stromal Clp core, although the physiological relevance of this is not known. Furthermore, plastid- or plant-specific Clp components have been generated through endosymbiosis. These structural complexities and heterogeneities represent critically important but difficult aspects of chloroplast protease studies. Structural diversification is related to specific functions and, thus, increased physiological importance. For instance, plastid Clp is essential for plant survival, growth, and development, while the proteobacterial prototype is nonessential. FtsH, Deg, and Clps all have gained crucial roles in chloroplast biogenesis and differentiation.
Box 1 Table. Multiplication of the prokaryotic-type protease subunits through evolution.
The numbers of protease subunits are indicated. For Clps, their chaperones, adaptors, and accessary proteins are not counted.
| Protease | Proteobacteria (Escherichia coli) | Cyanobacteria (Synechocystis PCC 6803) | Chloroplast (Arabidopsis) |
|---|---|---|---|
| FtsH | 1 | 4 | 9 |
| Deg | 1 | 3 | 5 |
| Clp | 1 | 4a | 9b |
A proteolytically inactive core subunit is included.
Four catalytically inactive core components are included.
Processive Proteases Clp and FtsH
Clp in the stroma and FtsH on thylakoid membranes are the major conserved ATP-dependent multimeric protease complexes catalyzing processive degradation in their respective suborganellar compartments (Box 2). Chloroplast Clp and FtsH both have two characteristic domains of bacterial origin, namely an AAA+ (for ATPase associated with various cellular activities) chaperone domain for substrate recognition and unfolding and a proteolytic domain for degradation, which are separated into individual subcomplexes for Clp or are organized together within a single protomer for FtsH. In the course of the evolution of photosynthetic organisms, these proteolytic enzymes have been converted from homomeric to heteromeric macromolecular complexes. Despite the complicated heteromultimeric compositions of these two enzyme complexes, their basic characteristics are well studied, as described below. Another ATP-dependent Ser protease is long filament phenotype (Lon), which is formed by six to seven protomers, each comprising an AAA domain and a protease domain with a catalytic SK dyad (Rotanova et al., 2004). Arabidopsis (Arabidopsis thaliana) possesses four Lon proteins, one of which, LON4, is found in the thylakoid membrane, although its precise function in chloroplasts has not been elucidated (Ostersetzer et al., 2007).
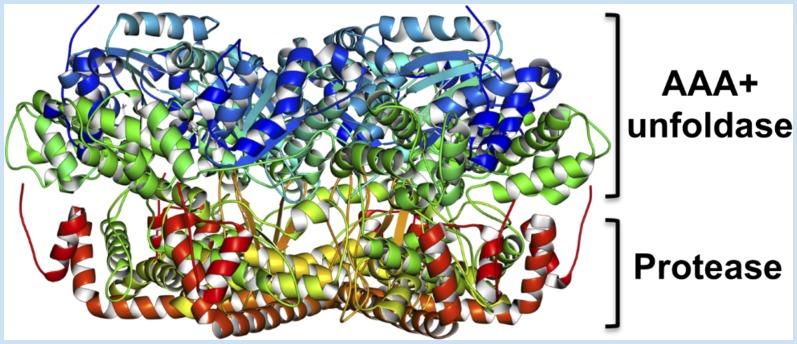
Box 2. Architecture and Action of the Processive Proteases
Three bacterial-like ATP-hydrolyzing protein-degrading enzymes, Clp, FtsH, and Lon, catalyze processive degradation. Their architecture consists of the AAA+ chaperone ring and the barrel-shaped protease complexes (see fig. below). The catalytic center is positioned inside the proteolytic chamber so as to avoid nonspecific degradation of proteins. Substrate access to the protease domain is governed by the chaperone machinery that directly recognizes the protein substrate. The protein is then unfolded and translocated continuously in an ATP-dependent manner to and inside the protease chamber, where it undergoes endoproteolytic cleavage. This chaperone-mediated energy-dependent continuous supply of the substrate proteins allows for targeted processive proteolysis. In Clp, the chaperone and the protease domains are separated into two different assemblies, and small peptide fragments can pass directly through a narrow entrance pore to the protease chamber, where they are degraded. However, folded proteins are unable to enter the pore, and substrate access is controlled at the chaperone gate. Such substrate access control and degradation mechanisms also are found for the cytosolic ubiquitin-dependent proteasomal degradation systems.
Box 2 Figure.
Stucture of the archetypal processive protease. Side view of the soluble domain structure of the hexameric FtsH protease from a thermophilic bacterium Thermus thermophiles is shown as a representative structure of the processive protease. Its characteristic AAA+ unfoldase chaperone and protease domains are indicated. The image of the FtsH structure (PDB ID: 4EIW) was obtained from the Protein Data Bank of Japan (PDBj).
Clp
Clp is an ATP-dependent Ser-type protease complex that comprises a chaperone subcomplex and a tetradecameric proteolytic core. The chaperone complex is composed of ClpC1/C2/D, each presumably constituting homooligomers, with ClpC1 as the major chaperone. The ClpC/D structure consists of an N-terminal region called the N domain as a substrate/adaptor docking site, two ATPase domains involved in substrate unfolding and translocation into the core, and an IGF motif (for Ile-Gly-Phe) and an R motif (named for its characteristic Arg residue), both required for core docking, with ClpC but not ClpD containing a uvrB/C motif of unassigned function (Nishimura and van Wijk, 2015). ClpC/D can recognize substrates directly (Rosano et al., 2011; Bruch et al., 2012; Huang et al., 2016), although recognition of a subset of proteins is mediated by a dedicated binary adaptor consisting of ClpS1 and ClpF (Nishimura et al., 2013, 2015). The basic ClpS structure has an N-terminal extension for substrate delivery to the chaperone and a C-terminal core domain for substrate recognition and chaperone binding (Dougan et al., 2002; Zeth et al., 2002; Erbse et al., 2006; Rivera-Rivera et al., 2014). ClpF is a plastid-specific ClpS1-interacting protein with a tripartite structure harboring a unique N-terminal domain for adaptor-substrate-chaperone binding, a uvrB/C motif for chaperone interaction, and a YccV-like domain of unknown function, the last two of which likely are derived from two distinct proteins of bacterial origin (Nishimura et al., 2015). The ClpS prototype functions in the N-end rule pathway, in which the half-life of a protein correlates with the identity of its N-terminal amino acid (Erbse et al., 2006; Varshavsky, 2011). The presence of another substrate recognition and delivery mechanism involving the binary adaptor in chloroplasts has been suggested (Nishimura et al., 2015).
The Clp core consists of two asymmetric rings, namely the P ring containing catalytic subunits ClpP3/P4/P5/P6 in a 1:2:3:1 ratio and the R ring containing proteolytically active ClpP1 (the only chloroplast-encoded subunit) and proteolytically inactive ClpR1/R2/R3/R4 proteins in a 3:1:1:1:1 ratio (Olinares et al., 2011). Clp core assembly and stabilization require plant-specific accessory proteins ClpT1/2 (Peltier et al., 2004; Sjögren and Clarke, 2011; Clarke, 2012; Kim et al., 2015). Loss-of-function mutants for the ClpC1 chaperone and the ClpPRT core show pale-green, seedling-lethal, or embryo-defective phenotypes, whereas knockouts for two adaptor proteins and ClpC2/D display no visible effects, underscoring their distinct contributions to plant development.
FtsH
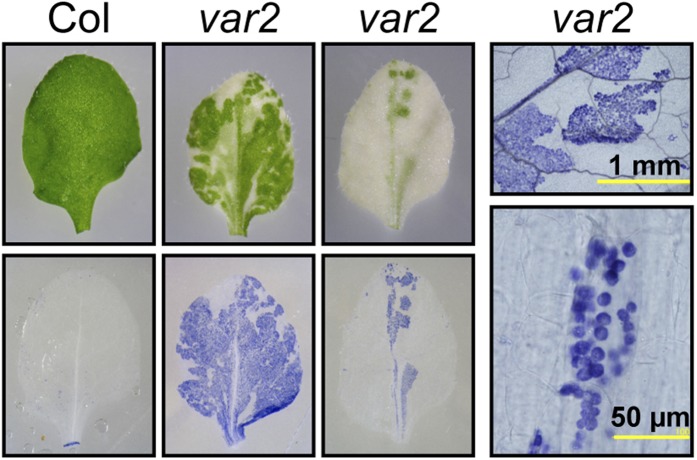
FtsH is a membrane-associated ATP-dependent zinc metalloprotease complex whose protomer consists of an AAA domain and a proteolytic domain containing a metal-binding H-E-x-x-H motif (Ito and Akiyama, 2005). Land plants possess multiple FtsH orthologs. Nine out of 12 Arabidopsis FtsH homologs are found in the chloroplast, with the four major isoforms (FtsH1/2/5/8) all thylakoid localized via their single transmembrane domains. Interchangeability and functional redundancy have been observed between FtsH5 and FtsH1 (type A) and between FtsH2 and FtsH8 (type B; Yu et al., 2004, 2005; Zaltsman et al., 2005), suggesting that FtsH exists as a hexameric heterocomplex with two types of isoforms. The stoichiometry of type A to type B subunits in an FtsH complex is estimated as 2:4 (Moldavski et al., 2012), whereas the cyanobacterial prototype comprises type A and B subunits in a 3:3 ratio with an alternating arrangement (Boehm et al., 2012). FtsH2 and FtsH5 are targeted to the thylakoid via the TAT (for twin-Arg translocation) pathway and the SEC (for secretion) pathway, respectively (Rodrigues et al., 2011). Leaf variegation phenotypes are observed in mutants for FtsH2 (also known as YELLOW VARIEGATED2 [VAR2]) and FtsH5 (VAR2; Fig. 2). FtsH2 and FtsH5 are the most abundant and the second most abundant FtsH isomers, respectively, reflecting the severity of their loss-of-function phenotypes of leaf variegation compared with the two other minor subunits, whose knockouts show wild-type-like appearance (Sakamoto et al., 2003; Zhang et al., 2010). ROS, including superoxide radicals and hydrogen peroxide (the former of which is shown in Fig. 2), are detected in the green but not the white leaf areas, specifically in the chloroplasts, of the var2 mutant grown even under normal light conditions, indicative of its persistent photooxidative stress (Kato et al., 2009). A series of genetic studies in Arabidopsis identified trans-acting factors suppressing var1/var2 leaf variegation; these included Clp subunits, translation factors, a pentatricopeptide repeat protein, a pseudouridine synthase homolog, a plastid transcriptionally active chromosome component, a prokaryotic-like peptide deformylase, ribosomal proteins, circularly permuted GTPase family proteins, and a sigma factor (Park and Rodermel, 2004; Miura et al., 2007; Yu et al., 2008, 2011; Liu et al., 2010a, 2010b, 2013; Adam et al., 2011; Wu et al., 2013; Powikrowska et al., 2014; Hu et al., 2015; Ma et al., 2015; Qi et al., 2016). Several models have been proposed to explain leaf variegation suppression, but the precise mechanism remains elusive (Miura et al., 2007; Yu et al., 2008).
Figure 2.
Leaf variegation in the Arabidopsis var2 mutant lacking FtsH2. Four-week-old wild-type Columbia (Col) and var2 leaves are shown, along with in situ detection of reactive oxygen species (ROS; superoxide radicals) visualized by NBT staining (blue color). The right two images show closeup views of variegated leaves where ROS is confined to green sectors and chloroplasts. Images are from Kato et al. (2009).
Another set of FtsH isomers, FtsH7/9/11/12, is present in the envelope (Fig. 1; Wagner et al., 2012). FtsH7/9 share high sequence similarity and, therefore, have been speculated to constitute a heteromeric protease complex. FtsH11 is involved in high-temperature tolerance, which is reminiscent of its bacterial prototype (Chen et al., 2006, 2007). High-light responses in the ftsh11 mutant are normal, unlike the variegated mutants defective in FtsH2/5. FtsH11 is a potential target for an intramembrane protein degradation pathway (Knopf et al., 2012). FtsH11 also is localized in mitochondrial inner membranes, where it seems to act in parallel with FtsH4 (Urantowka et al., 2005). FtsH11 has a unique N-terminal extension of unknown function. Although the proteolytic domain of mitochondrial FtsH11 faces the inner membrane space, similar to FtsH4 and the yeast ortholog, its membrane topology in the chloroplast is not known.
The other chloroplastic isomer, FtsH6, is dispensable for normal growth under high light and natural environmental stress conditions. However, its suborganellar location and precise function remain unclear (Wagner et al., 2012; Lu, 2016).
Endopeptidase Deg Participating in Processive Proteolysis
Deg (originally termed DegP, for degradation of periplasmic proteins; also known as high-temperature requirement A) also is well known to function together with the processive protease FtsH in chloroplasts (see below). Deg is an ATP-independent Ser-type endopeptidase that harbors an N-terminal proteolytic domain with an HDS catalytic triad and C-terminal PDZ (for PSD-95/SAP90, disc large, and ZO-1) domain(s) involved in protein-protein interaction (Schuhmann and Adamska, 2012). Deg1/5/8 and Deg2/7 are present in the lumenal and stromal sides of the thylakoid membrane, respectively (Fig. 1). The number of PDZ domains differs among Degs: there is one in Deg1/8, two in Deg2, four in Deg7, and none in Deg5 (Schuhmann and Adamska, 2012). Monomeric Deg forms a trimer, with each protomer connected via the protease domain, and trimeric units assemble through interactions between the PDZ domains into higher-ordered oligomers, including hexamers (Clausen et al., 2002). For example, Deg1 undergoes a conformational transition from its inert monomer through the trimeric intermediate to the active hexamer upon lumenal acidification (Kley et al., 2011). Deg7 has a characteristic primary structure comprising one active and one degenerated protease domain with four PDZ domains. Its trimerization is mediated by the degenerated protease domains (Schuhmann et al., 2011). Deg2 possesses two PDZ domains (PDZ1 and PDZ2) and forms a hexamer by dimerization of trimers through interactions of the PDZ2 domain with the protease domain and with PDZ1, but the hexamer is quite rigid in structure and rather inactive, whereas a higher-ordered structure is suggested to represent the active state (Sun et al., 2012). Proteolytically inactive Deg5 and proteolytically active Deg8 together constitute a stable heterohexameric complex in a 1:1 ratio (Sun et al., 2007).
Processing Proteases
Most plastid proteins are encoded in the nuclear genome and biosynthesized in the cytosol as preproteins bearing N-terminal transit peptides, followed by trans-envelope protein import TIC/TIC channels (Jarvis and López-Juez, 2013). Imported preproteins are subjected to sequential proteolytic processing for transit peptide cleavage and maturation. Many thylakoid proteins, including photosynthetic proteins of the thylakoid membrane, are subsequently sorted to their proper locations through bacterial-like SEC, TAT, and signal recognition particle (SRP) pathways. Their N-terminal bipartite transit peptides, which consist of a thylakoid transfer signal (TTS) following the transit peptide, also are cleaved by limited proteolysis (Celedon and Cline, 2013).
Stromal processing peptidase (SPP) is a metalloendopeptidase that removes transit peptides from preproteins with broader substrate specificity (Richter and Lamppa, 1998). Transit peptide degradation involves one or two isoforms of the presequence peptidase (PreP1/2), which belongs to a metalloendopeptidase family (Stahl et al., 2002; Bhushan et al., 2003, 2005; Moberg et al., 2003). Organellar oligopeptidase (OOP) is another zinc metalloprotease that participates in targeting peptide degradation. OOP degrades peptides with lengths of eight to 23 amino acid residues but fails to act on folded proteins, consistent with its catalytic cavity size, as in the case of PreP1/2 (Kmiec et al., 2013, 2014). The OOP-null mutant phenotype is wild type like, but genetic interaction with the prep1 prep2 double mutant is observed, indicating complementary functions for OOP and PreP1/2 (Kmiec et al., 2013).
Arabidopsis has three isoforms of the plastidic type I signal peptidase I family, namely Plsp1 and Plsp2A/B (Hsu et al., 2011). Thylakoidal processing peptidase (TPP) is the integral membrane protease with a lumen-facing Ser/Lys-type proteolytic domain, and it cleaves TTSs off preproteins (Chaal et al., 1998). Plsp1 functions as the TPP in maturation of a subset of the SEC/TAT substrates and is necessary for proper thylakoid formation (Endow et al., 2010; Shipman-Roston et al., 2010; Midorikawa et al., 2014). Plsp1 also is targeted to the envelope, where it participates in TOC75 precursor processing (Inoue et al., 2005; Shipman and Inoue, 2009). The biochemical properties and physiological functions of Plsp2A/B are currently unclear.
C-terminal processing protease, a monomeric Ser-type protease that cleaves off a C-terminal extension for maturation of the D1 precursor protein, is another type of processing peptidase in the thylakoid lumen (Anbudurai et al., 1994; Fujita et al., 1995; Oelmüller et al., 1996; Satoh and Yamamoto, 2007; Che et al., 2013). Its basic structure contains a PDZ domain for D1 C-terminal binding and a protease domain with an SK dyad for proteolysis (Liao et al., 2000).
CASE STUDIES OF PROTEOLYTIC REGULATION OF CHLOROPLAST PROTEINS
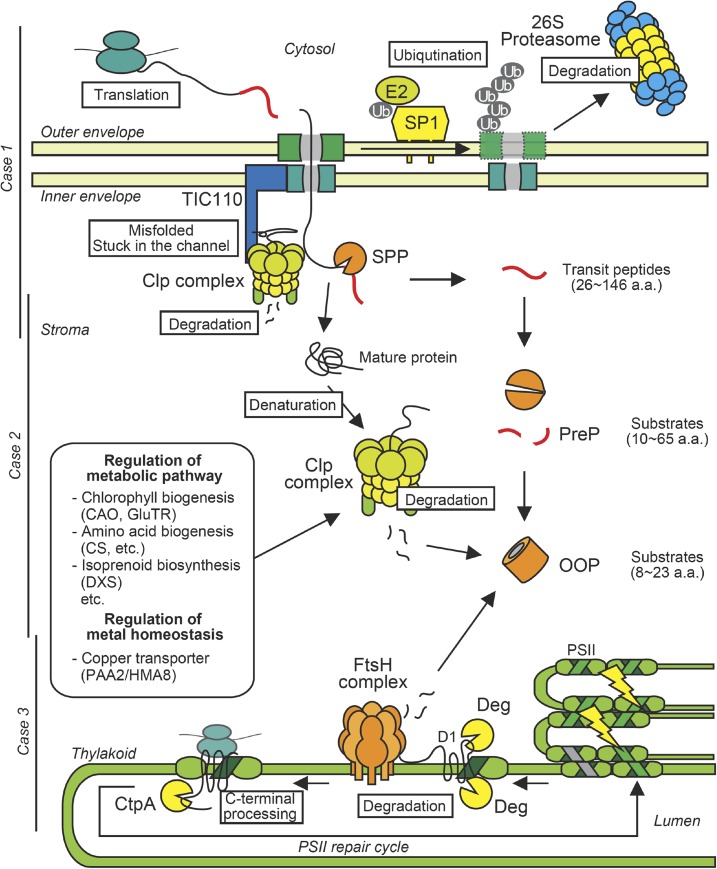
Following the above overview of the major protease machineries in the chloroplast, we here present three case studies of proteolytic regulatory processes based on their intraorganellar compartments, namely the envelopes (case 1), stroma (case 2), and thylakoid membranes (case 3). In case 1, recently discovered regulation and quality control mechanisms of protein import across the outer and inner envelopes by the actions of extraplastid as well as intraplastid protein degradation machineries are illustrated. Case 2 describes multiple proteolytic events involving the stromal processive protease or the processing peptidases. Examples of environmentally responsive proteolysis also are included. Finally, in case 3, proteolytic functions in thylakoid protein biogenesis, homeostasis, and quality control are exemplified. These proteolytic events are graphically summarized in Figure 3.
Figure 3.
Proteolytic regulation of intraplastid proteome homeostasis. The scheme shows multiple events in the regulation of chloroplast protein homeostasis, which involves not only intraplastidic but also extraplastidic protein degradation machineries. a.a., Amino acids.
Case 1: Regulation and Quality Control of Protein Import across the Envelopes (26S Proteasome and Clp)
Proteolytic Reorganization of Protein Import Machinery by the Cytosolic Ubiquitin-Proteasome System
Jarvis and coworkers demonstrated that the ubiquitin-proteasome system regulates protein import across the chloroplast outer envelope through the degradation of TOC components. Genetic screening for the suppressors of a TOC33 knockout mutant (plastid protein import1 [ppi1]) identified a RING-type ubiquitin E3 ligase, SUPPRESSOR OF PPI1 LOCUS1 (SP1; Ling et al., 2012). SP1 has a cytosolically exposed RING finger domain for ubiquitination and two transmembrane domains for integration into the outer envelope, where it recognizes TOC components, including TOC75, TOC159, and TOC33, through its intermembrane space domain. SP1 ubiquitinates these client proteins for proteasome-mediated proteolysis (Fig. 3). The TOC complexes use different receptor isoforms for the recognition of distinct sets of preproteins (Paila et al., 2015). Therefore, SP1 functions in plastid proteome remodeling through reorganization of the TOC constituents, thereby regulating organellar differentiation in response to environmental context. The sp1 mutant shows inefficient deetiolation upon illumination and delayed chloroplast-to-gerontoplast transition during dark-induced senescence, whereas the SP1 overexpressor exhibits enhanced deetiolation and senescence (Ling et al., 2012). Whether the TOC reorganization involving the ubiquitin-proteasome system accounts for proteomic changes during the other plastid differentiation stages deserves future investigation.
Interestingly, genetic studies have suggested the involvement of SP1 in abiotic stress responses through proteolytic regulation of the protein-import apparatus (Ling and Jarvis, 2015). The Arabidopsis genome encodes two SP1 homologs, SP1-LIKE1/2 (SPL1/2), both of which are outer envelope proteins. SPL1 is the closest homolog of SP1, but its overexpression failed to complement the sp1 mutation, likely due to distinct target specificity (Ling et al., 2012). Potential ligands for these E3 ligases include other outer envelope proteins such as enzymes involved in lipid metabolism, regulators for chloroplast division/movement, and an envelope-associated PHD transcription factor mediating retrograde signaling (Huang et al., 2013).
Quality Control of Protein Import by Clp
A portion of the ClpC chaperone present in chloroplasts is associated with the inner envelope membranes, where it is found in complexes with the protein translocation machinery (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). The N-terminal domain of ClpC is important for its envelope localization (Chu and Li, 2012). The ClpC protein associates with TIC110 but not with TIC40/55 (Flores-Pérez et al., 2016). Recently, it has been demonstrated that, along with ClpC chaperones, the ClpPR core also is attached to envelope membranes, whereas ClpD is localized exclusively in the stroma (Sjögren et al., 2014; Fig. 3). Through quantitative analysis of the ClpC-ClpPR stoichiometry, all envelope ClpC proteins (30% of total ClpC) are presumed to function together with the proteolytic core. Clp is unlikely to be involved in the maintenance of the TIC machinery itself (Sjögren et al., 2014). Rather, it seems to participate in a protein quality control mechanism for surveying preproteins being released from the TIC complex, in particular during transit peptide processing and subsequent refolding, to ensure the integrity of the chloroplast proteome (Sjögren et al., 2014). Indeed, the protein import efficiency is reduced in mutants for the proteolytic core as well as the chaperones. In addition, mutated Clp chaperone that is defective in protease core interaction fails to import proteins efficiently despite its normal envelope association (Flores-Pérez et al., 2016). Clp chaperones have the intrinsic ability to bind the N-terminal transit peptide as well as to renature nonnative proteins (Rosano et al., 2011; Bruch et al., 2012). ClpC interactions with the transit peptide and mature regions of preproteins being imported were detected recently (Huang et al., 2016). These observations suggest the involvement of the Clp system in clearing unprocessed or misfolded proteins generated during import. Alternatively, proteins being imported could aggregate and become stuck in the import channel. Such protein aggregates would have to be removed proteolytically for efficient protein translocation.
Comparative leaf proteomic studies of the Clp core mutants have shown a drastic loss of photosynthetic machineries that consist of numerous nucleus-encoded, imported proteins (for review, see Nishimura and van Wijk, 2015). Systematic down-regulation of plastid-encoded proteins for photosynthesis but not for other functions such as translation, transcription, and fatty acid biosynthesis have been observed in the Clp core mutant. This fact is explainable by the accelerated degradation of plastid-encoded partner proteins that cannot assemble with nucleus-encoded photosynthetic proteins because of their inefficient import. Because a lack of functional photosynthetic complexes engenders severe phenotypes from chlorosis to albinism that resemble the Clp chaperone or core mutant phenotypes, the major function of plastid Clp, in terms of effects on growth and development, might be to eliminate aggregated, misfolded, or unprocessed preproteins of such photosynthesis machineries during protein import into the chloroplast (for a more extensive discussion, see Nishimura and van Wijk, 2015).
Case 2: Protein Degradation in Stroma-Facing Compartments (SPP, PrePs, OOP, and Clp)
Sequential Proteolytic Events for Preprotein Processing and Transit Peptide Removal
The aberrant accumulation of peptides generated by protein degradation or preprotein processing can affect plastid function and physiology and, therefore, must be removed. Upon arrival in the stroma, SPP binds with a broad spectrum of specificity to the transit peptides and removes them by a single endoproteolytic reaction, with subsequent release of the mature proteins but not the transit peptides (Richter and Lamppa, 1999, 2002). Recent proteome analysis showed that a substantial number of stromal proteins have multiple distinct N termini, implying the presence of multistep N-terminal processing mechanisms or simply reflecting SPP’s imprecise site specificities (Rowland et al., 2015). The cleaved transit peptides are subjected to additional trimming into smaller fragments by SPP and are degraded further by PrePs and OOPs in the stroma (Teixeira and Glaser, 2013; Fig. 3). Transit peptides are 26 to 146 amino acids long (Zybailov et al., 2008). Structural and biochemical studies have reported that PreP1 degrades various peptide substrates of 10 to 65 amino acids (Moberg et al., 2003; Ståhl et al., 2005), whereas OOP cleaves peptide fragments ranging from eight to 23 amino acids (Kmiec et al., 2013). Consequently, both proteases are able to function in transit peptide degradation, with OOP acting in parallel or downstream to PreP. Furthermore, given the sizes of the degradation products generated by the proteases such as Clp and FtsH, PrePs and OOP can function downstream of these proteolytic machineries. However, whether and how the transit peptides are degraded in vivo by PrePs and OOP (together with the other proteases) await further investigation.
Regulation of Metabolic Pathways
Chlorophyllide a oxygenase (CAO) is a key enzyme converting chlorophyll a to chlorophyll b during chlorophyll biogenesis. CAO protein stability is subject to negative feedback regulation in response to chlorophyll b metabolite levels (Yamasato et al., 2005). CAO degradation involves the Clp system (Nakagawara et al., 2007). CAO is localized in thylakoid and inner envelope membranes (Eggink et al., 2004), where it is accessible to Clp. A short degradation signal, the CAO degron, is located in the N-terminal domain of CAO (Sakuraba et al., 2007, 2009). A plausible model for metabolite-dependent CAO degradation has been proposed; in the model, the CAO degron would be located within the interior region in the absence of chlorophyll b but would be exposed to the exterior through a structural change in the presence of chlorophyll b such that the degron could be recognized by the protease (Sakuraba et al., 2009).
Glutamyl-tRNA reductase (GluTR) catalyzes the initial step generating Glu-1-semialdehyde from glutamyl-tRNA (Glu-tRNA) during tetrapyrrole biogenesis. GluTR is recognized directly through its N-terminal region by the ClpF-ClpS1 binary adaptor system as well as by ClpC chaperones for proteolysis (Nishimura et al., 2015; Apitz et al., 2016). GluTR is localized in the stroma and thylakoids. Its thylakoid localization is mediated in part by GluTR-binding protein (GBP; Czarnecki et al., 2011). GluTR abundance in the thylakoid is reduced in the absence of GBP, suggesting an accelerated destabilization of GluTR. A possible mechanism has been proposed in which Clp-dependent proteolysis and GBP-assisted stabilization together fine-tune GluTR accumulation to optimize chlorophyll and heme biosynthesis (Apitz et al., 2016). Glu-tRNA is a substrate not only for GluTR in the tetrapyrrole pathway but also for chloroplast ribosomes in translation; these two enzymes consume it competitively. The observation that the ClpS1-null mutant is sensitive to a translational inhibitor has led to the inference that ClpS1 can modulate chloroplast protein biosynthesis by regulating Glu-tRNA flux through GluTR degradation (Nishimura et al., 2013; Nishimura and van Wijk, 2015).
Degradation of a key metabolic enzyme, deoxyxylulose 5-phosphate synthase (DXS), in the methylerythritol 4-phosphate pathway for isoprenoid biosynthesis is regulated by metabolite levels (Guevara-García et al., 2005; Han et al., 2013). Furthermore, inactive DXS is likely recognized by a J protein, J20, which functions as an adaptor delivering misfolded or damaged client proteins to Hsp70 chaperone for either refolding or proteolysis depending on intracellular contexts (Pulido et al., 2013). The decision of dysfunctional DXS refolding versus degradation is proposed to involve the actions of stromal ClpB3 and ClpC1 chaperones; ClpB3 may function together with Hsp70 in DXS reactivation, while ClpC1 likely recognizes the enzyme for proteolysis (Pulido et al., 2016). Regulatory mechanisms of the nonfunctional protein fate decision involving multiple molecular chaperones deserve further investigation.
Several metabolic enzymes in the shikimate pathway of aromatic amino acid biosynthesis, including chorismate synthase, are potential targets for ClpS1-dependent proteolysis, perhaps suggesting the involvement of plastid Clp in the multistep regulation of various metabolic pathways (Nishimura et al., 2013; Nishimura and van Wijk, 2015). During plastid differentiation from etioplasts to chloroplasts in leaves, multiple enzymes in amino acid metabolism are decreased considerably in abundance, whereas Calvin cycle enzymes and thylakoid proteins accumulate to higher levels (Kleffmann et al., 2007). Possible involvement of the Clp system in this selective protein reduction awaits experimental verification.
Regulation of Metal Homeostasis
The copper transporter PAA2/HMA8 is an integral thylakoid membrane protein that delivers copper ions to plastocyanin (PC) in the lumen (Abdel-Ghany et al., 2005). PAA2/HMA8 abundance is down-regulated in response to high copper concentrations (Tapken et al., 2012). This copper-induced down-regulation involves proteolysis by the stromal Clp system rather than thylakoid-located FtsH protease (Tapken et al., 2015).
How the Clp system recognizes and degrades PAA2/HMA8 remains an important question. Genetic studies showing that its proteolytic regulation requires the ClpC chaperone and the ClpPR core but not the adaptor protein ClpS1 suggest that the chaperone directly recognizes the substrate depending on the copper level. PAA2/HMA8 has eight transmembrane domains, with the N-terminal domain, three internal loop structures, and the C-terminal tail exposed to the stromal side (Bernal et al., 2007). In particular, the N-terminal domain contains a metal-binding motif, which potentially induces a conformational change in response to elevated copper levels. This change might well trigger PAA2/HMA8 recognition by the Clp chaperone, similar to its CAO recognition. Another copper transporter, PAA1/HMA6, is present in the inner envelope membrane (Shikanai et al., 2003). Its accumulation is unaffected by high-copper conditions (Tapken et al., 2012). It is noteworthy that some of the ClpS1 targets isolated through affinity purification are metal-binding or metal-related proteins (Nishimura et al., 2013).
Case 3: Thylakoid Protein Biogenesis, Homeostasis, and Quality Control (Plsp1, Deg, and FtsH)
Regulation and Quality Control of Lumenal Targeting Peptide Removal
After import, most if not all lumenal proteins are sorted from stroma through thylakoid-localized SEC/TAT/SRP machineries to the thylakoid lumen. Protein sorting requires TTS, which must be removed by TPP for protein assembly, posttranslocational release from the membrane, and, therefore, proper thylakoid formation (Inoue et al., 2005; Shipman-Roston et al., 2010; Midorikawa and Inoue, 2013).
Plsp1 is the TPP that functions in cleaving off TTS in the lumen (Shipman and Inoue, 2009). Knockout of Plsp1 causes the accumulation of processing intermediates of the SEC substrates, PC and OE33/PsbO, and the TAT substrate, OE23/PsbP (Shipman-Roston et al., 2010; Albiniak et al., 2012). The absence of Plsp1 and the elimination of TPP cleavage sites seem not to affect the SEC pathway itself but impede substrate release from the membrane; indeed, PC and PsbO intermediates are stuck in the thylakoid membrane despite their proper membrane targeting (Shackleton and Robinson, 1991; Frielingsdorf and Klösgen, 2007; Midorikawa and Inoue, 2013). It is noteworthy that precursor PsbO is stably present in a 440-kD complex that is not fully characterized but is redox sensitive, protease tolerant, and distinct from the PSII or SEC machinery, whereas unprocessed PC is prone to light-driven proteolysis by unknown protease(s). The PC degradation activity must be metal independent but is dependent on stromal proteins, the proton motive force across the thylakoid, and ATP hydrolysis. Thylakoid-retained PC cleavage intermediates are exposed in part to the stroma (Midorikawa and Inoue, 2013), potentially allowing the access of stromal proteases, as in the case of PAA2/HMA8 for Clp (Tapken et al., 2015). The PsbP intermediate is detected as a monomer in the stroma when Plsp1 is missing (Shipman-Roston et al., 2010; Midorikawa and Inoue, 2013) but exists within the membrane, presumably through an association with the TAT machinery, when its TPP processing site is abolished (Frielingsdorf and Klösgen, 2007; Midorikawa and Inoue, 2013). An earlier report has shown that TAT-dependent substrates can be returned to the stroma and that disruption of the TPP sites of the TAT substrates causes their unprocessed intermediates to accumulate in the thylakoids, where they are likely to be trapped in the sorting machinery (Di Cola and Robinson, 2005). A possible function of thylakoid Plsp1 in preventing reverse translocation has been proposed (Midorikawa and Inoue, 2013). Interestingly, a lumenal protein lacking the lumenal target peptide is prone to mislocalization in stroma, where it is readily degraded (Halperin and Adam, 1996). This observation represents an example of a lumenal protein quality control mechanism.
PSII Repair Cycle on the Thylakoid Membrane
Excess light exposure generates ROS, which can cause irreversible inactivation of photosynthetic machineries. The main target of photodamage is the D1 subunit of the PSII reaction center. D1 protein turns over very rapidly through a cyclical mechanism called the PSII repair cycle. Photodamaged PSII core complex migrates from grana to stromal thylakoids, where it is partially disassembled, enabling protease to access D1 for degradation. Following the insertion of de novo-synthesized D1, PSII is reassembled and migrates back into the grana (Chi et al., 2012; Nath et al., 2013; Yoshioka-Nishimura and Yamamoto, 2014).
FtsH and Deg proteases act in a cooperative manner in D1 degradation, in which lumenal and stromal Deg isomers function prior to FtsH (Kato et al., 2012; Fig. 3). D1 has five transmembrane-spanning helices (A–E). Deg5/8 and possibly Deg1 endoproteolytically cleave damaged D1 at a lumen-facing loop connecting the transmembrane helix domains C and D (CD loop), whereas Deg7 and Deg2 is likely to be responsible for the respective cleavage events occurring at stroma-exposed loops between transmembrane helices B and C and transmembrane helices D and E (Haussühl et al., 2001; Kapri-Pardes et al., 2007; Sun et al., 2007, 2010; Kato et al., 2012). D1 fragments resulting from cleavage at the lumenal CD loop are detected predominantly during high-light exposure (Kato et al., 2012), consistent with Deg activation through the oligomerization triggered by light-dependent lumenal acidification (Kley et al., 2011). D1 fragmentation by lumenal Deg can be initiated through disruption of the manganese cluster of the oxygen-evolving complex by blue light, fitting with the two-step model for photoinhibition (Kato et al., 2015), in which the manganese cluster in the oxygen-evolving complex is primarily damaged and the PSII reaction center is then inactivated by chlorophyll-absorbed light energy (Hakala et al., 2005; Ohnishi et al., 2005). Whether Deg functions before, during, or after PSII migration remains unknown. Deg-generated D1 fragments are degraded further through FtsH-dependent processive proteolysis. The four major isoforms FtsH1/2/5/8 together function in this process, but whether FtsH6 is involved is not known. FtsH can initiate D1 degradation even in the absence of the Deg proteases, possibly by pulling the stroma-oriented N-terminal tail of the D1 subunit, but the degradation efficiency is enhanced in the presence of Deg proteases (Kato et al., 2012). In cyanobacteria, the D1 N-terminal tail is exposed outside the PSII complex. The absence of this N-terminal tail inhibits D1 degradation, likely because of failure in N-terminal recognition by FtsH (Komenda et al., 2007).
The repair cycle of photodamaged PSII is modulated through its reversible phosphorylation. The PSII core proteins D1, D2, CP43, and PsbH can be phosphorylated; the phosphorylation sites are all located in their stroma-facing N termini (Puthiyaveetil and Kirchhoff, 2013). The phosphorylation and dephosphorylation of these proteins require the thylakoid-associated Ser/Thr kinase STATE TRANSITION8 (STN8; Bonardi et al., 2005) and the stroma/thylakoid-localized PROTEIN PHOSPHATASE 2C-TYPE PSII CORE PHOSPHATASE (PBCP; Samol et al., 2012). Lack of STN8 results in enhanced Deg-dependent D1 fragmentation under high-light conditions (Kato and Sakamoto, 2014), whereas PBCP defects cause delayed D1 degradation in high light (Samol et al., 2012). Levels of smaller, intermediary D1 fragments are correlated with ROS accumulation, suggestive of their cytotoxicity. These observations have led to the idea that PSII phosphorylation prevents excessive D1 degradation to avoid the accumulation of cytotoxic cleavage intermediates. Furthermore, given that phosphorylation of PSII core proteins can promote PSII migration and disassembly through structural remodeling of the thylakoid membrane to facilitate D1 access to the proteases (Kirchhoff, 2013), it is plausible that PSII phosphorylation fine-tunes the repair cycle by balancing D1 presentation and degradation (Kato and Sakamoto, 2014). How PSII phosphorylation modulates D1 proteolysis requires further investigation.
Proteolytic Remodeling of the Photosynthetic Machineries in Response to Environmental Stimuli
In the unicellular alga Chlamydomonas reinhardtii, FtsH participates together with Clp and unidentified lumenal proteases in the selective degradation of the cytochrome b6/f complex and its biogenesis factors upon nitrogen starvation (Wei et al., 2014). Their mechanisms of action in cytochrome b6/f degradation, however, are apparently different, because destabilization of the cytochrome b6/f complex triggered by biogenesis and assembly problems is blocked in the absence of FtsH but not in the absence of Clp (Majeran et al., 2000; Malnoë et al., 2014). The nitrogen-depletion-stimulated cytochrome b6/f degradation is presumably controlled by a signaling pathway involving nitric oxide (Wei et al., 2014). FtsH-dependent cytochrome b6/f degradation is found in sulfur-starving algal cells as well (Malnoë et al., 2014). In addition, algal FtsH participates in PSII destruction during phosphorus-starvation and sulfur-starvation conditions, presumably through a proteolytic mechanism similar to the repair cycle (Malnoë et al., 2014).
The aforementioned macronutrient-responsive proteolytic regulation mechanisms involving algal FtsH and Clps might well be conserved in land plant chloroplasts. Furthermore, the observations that a lack of stromal Clp induces the up-regulation of thylakoid-localized FtsH and SppA proteases (Rudella et al., 2006) and that the loss of FtsH engenders the up-regulation of other chloroplast proteases, including Clp and SppA, and the recruitment of stromal Clp to the thylakoid (Kato et al., 2012) suggest that they play mutual compensatory roles, thereby implying conservation of their coordinated actions in the remodeling of the photosynthetic apparatus upon stress.
FUTURE PERSPECTIVES
Complete Sets of Substrates, Degron, and Substrate Recognition Mechanisms?
Specific degradation signals, namely degrons, are necessary for chaperones and/or adaptor proteins of the proteolytic systems to recognize protein substrates for destruction (e.g. ClpS), but such degron sequences are poorly understood in plastids (see Outstanding Questions). A recent study has identified a novel N-terminal degron for FtsH-driven proteolysis in bacteria (Bittner et al., 2015), raising the question of whether such a target recognition mechanism is conserved in chloroplasts. Meanwhile there have been several well-known examples of proteolytic regulation of specific chloroplast proteins. Specifically, light-harvesting antenna size has long been known to be strictly regulated in response to elevated light intensities, and apoproteins of LHCII are thought to be proteolytically degraded (Jansson, 1994). However, the responsible protease is unknown. Comprehensive substrate identification is necessary for determination of a common sequence signature or regulatory motif. Using substrate-trapping approaches, multiple substrates for Clp and FtsH proteases have been isolated in bacteria and in mammalian mitochondria (Flynn et al., 2003; Neher et al., 2003; Westphal et al., 2012; Bhat et al., 2013; Feng et al., 2013; Graham et al., 2013; Bittner et al., 2015). Trapping strategies use mutated, inactive forms of the protease or chaperone domains. By substituting catalytically active with catalytically inactive degradation machineries fused to a specific tag, the in vivo substrates are confined within and purified in complexes with protease assemblies from the cell. Similar methodologies are worth pursuing for identification of substrates for chloroplast proteases.
Regulatory Circuits Involving Multiple Proteases to Optimize Interorganellar Proteostasis
Chloroplast proteases can function in a compensatory manner (see “Outstanding Questions”). FtsH protease has long been known to interact genetically with the Clp system (Park and Rodermel, 2004; Yu et al., 2008; Wu et al., 2013). Knowledge of this interaction is based on the observation that the loss of Clp chaperone or protease core suppresses the leaf variegation phenotype resulting from the lack of FtsH2, a major component of the FtsH machinery. It is particularly interesting that ClpC2 is not the major ClpC chaperone but its reduction is somehow sufficient for the suppression of a variegated leaf phenotype resulting from FtsH2 loss of function, despite the fact that ClpC1 is abundantly present in the suppressor mutant (Park and Rodermel, 2004). More importantly, the Clp chaperone and proteolytic core are both up-regulated and recruited to the thylakoid membrane in the mutant lacking FtsH2, suggesting their complementary roles for FtsH deficiency (Kato et al., 2012). FtsH isomers are up-regulated in the Clp core mutant, consistent with their functional compensation (Rudella et al., 2006). Similar responses to the Clp core defect have been observed for PrePs (Zybailov et al., 2008). These findings suggest that the proteases share a common substrate recognition mechanism or that they can recognize identical substrates in a distinct manner. Further experimentation is needed to examine these proposed compensatory mechanisms.
OUTSTANDING QUESTIONS
Despite recent elucidation of substrate proteins for the major chloroplast proteases, we still need to develop a systematic approach for identifying in vivo targets to fully understand substrate recognition mechanisms.
Mechanistic insights into functional interactions among multiple plastid proteases require further investigation.
A possible regulatory mechanism of interorganellar proteome homeostasis involving dual-targeted proteases deserves particular attention and future evaluation.
Furthermore, dual targeting proteases could potentially regulate proteome homeostasis in both organelles (see “Outstanding Questions”). FtsH11, Lon4, PrePs, and OOP are targeted to both mitochondria and chloroplasts (Moberg et al., 2003; Kmiec et al., 2013). Given that the distribution of a dual-targeted protein is altered by organellar stress and dysfunction (Nargund et al., 2012), allocation of these protease and peptidase machineries between the chloroplast and mitochondrion could be regulated at the protein import step, depending on the status of organellar protein homeostasis. For the coordinated biogenesis and the maximum function of two organelles, reprogramming of the nuclear transcriptome by proteases in both organelles might contribute to the optimization of interorganellar proteomes.
Acknowledgments
We apologize to researchers whose relevant studies were not cited in this review because of page limitations and thank the members of the Sakamoto laboratory for helpful discussion related to chloroplast proteases.
Glossary
- TAT
twin-Arg translocation
- SEC
for secretion
- ROS
reactive oxygen species
- TTS
thylakoid transfer signal
- PC
plastocyanin
Footnotes
This work was supported by the Japan Science and Technology Agency (Core Research for Environmental Science and Technology grant to W.S.), the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research no. 15J03092 and postdoctoral fellowship to K.N.), and the Ohara Foundation (to W.S. and Y.K.).
Articles can be viewed without a subscription.
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z. (2015) Plastid intramembrane proteolysis. Biochim Biophys Acta 1847: 910–914 [DOI] [PubMed] [Google Scholar]
- Adam Z, Frottin F, Espagne C, Meinnel T, Giglione C (2011) Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita M, Nielsen E, Keegstra K (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiniak AM, Baglieri J, Robinson C (2012) Targeting of lumenal proteins across the thylakoid membrane. J Exp Bot 63: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB (1994) The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc Natl Acad Sci USA 91: 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz J, Nishimura K, Schmied J, Wolf A, Hedtke B, van Wijk KJ, Grimm B (2016) Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol 170: 2040–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M, Testillano PS, Alfonso M, del Carmen Risueño M, Picorel R, Yruela I (2007) Identification and subcellular localization of the soybean copper P1B-ATPase GmHMA8 transporter. J Struct Biol 158: 46–58 [DOI] [PubMed] [Google Scholar]
- Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P (2013) Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol 88: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Lefebvre B, Ståhl A, Wright SJ, Bruce BD, Boutry M, Glaser E (2003) Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep 4: 1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Ståhl A, Nilsson S, Lefebvre B, Seki M, Roth C, McWilliam D, Wright SJ, Liberles DA, Shinozaki K, et al. (2005) Catalysis, subcellular localization, expression and evolution of the targeting peptides degrading protease, AtPreP2. Plant Cell Physiol 46: 985–996 [DOI] [PubMed] [Google Scholar]
- Bittner LM, Westphal K, Narberhaus F (2015) Conditional proteolysis of the membrane protein YfgM by the FtsH protease depends on a novel N-terminal degron. J Biol Chem 290: 19367–19378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Yu J, Krynicka V, Barker M, Tichy M, Komenda J, Nixon PJ, Nield J (2012) Subunit organization of a Synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 24: 3669–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Bruch EM, Rosano GL, Ceccarelli EA (2012) Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N-terminus of a protein. BMC Plant Biol 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JM, Cline K (2013) Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim Biophys Acta 1833: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaal BK, Mould RM, Barbrook AC, Gray JC, Howe CJ (1998) Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana: implications for the origin and catalytic mechanism of the enzyme. J Biol Chem 273: 689–692 [DOI] [PubMed] [Google Scholar]
- Che Y, Fu A, Hou X, McDonald K, Buchanan BB, Huang W, Luan S (2013) C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc Natl Acad Sci USA 110: 16247–16252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Burke JJ, Velten J, Xin Z (2006) FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J 48: 73–84 [DOI] [PubMed] [Google Scholar]
- Chen J, Xin Z, Burke J (2007) The conserved role of FtsH11 protease in protection of photosynthetic system from high temperature stress in higher plants. Photosynth Res 91: 308 [Google Scholar]
- Chi W, Sun X, Zhang L (2012) The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim Biophys Acta 1817: 239–246 [DOI] [PubMed] [Google Scholar]
- Chu CC, Li HM (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol 158: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK. (2012) The chloroplast ATP-dependent Clp protease in vascular plants: new dimensions and future challenges. Physiol Plant 145: 235–244 [DOI] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M (2002) The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10: 443–455 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Hedtke B, Melzer M, Rothbart M, Richter A, Schröter Y, Pfannschmidt T, Grimm B (2011) An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23: 4476–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola A, Robinson C (2005) Large-scale translocation reversal within the thylakoid Tat system in vivo. J Cell Biol 171: 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B (2002) ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683 [DOI] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H (2004) The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA 101: 15386–15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink LL, LoBrutto R, Brune DC, Brusslan J, Yamasato A, Tanaka A, Hoober JK (2004) Synthesis of chlorophyll b: localization of chlorophyllide a oxygenase and discovery of a stable radical in the catalytic subunit. BMC Plant Biol 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow JK, Ruppel NJ, Inoue K (2010) Keep the balloon deflated: the significance of protein maturation for thylakoid flattening. Plant Signal Behav 5: 721–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, et al. (2013) Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res 12: 547–558 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Bédard J, Tanabe N, Lymperopoulos P, Clarke AK, Jarvis P (2016) Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol 170: 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11: 671–683 [DOI] [PubMed] [Google Scholar]
- Frielingsdorf S, Klösgen RB (2007) Prerequisites for terminal processing of thylakoidal Tat substrates. J Biol Chem 282: 24455–24462 [DOI] [PubMed] [Google Scholar]
- Fujita S, Inagaki N, Yamamoto Y, Taguchi F, Matsumoto A, Satoh K (1995) Identification of the carboxyl-terminal processing protease for the D1 precursor protein of the photosystem II reaction center of spinach. Plant Cell Physiol 36: 1169–1177 [Google Scholar]
- Graham JW, Lei MG, Lee CY (2013) Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195: 4506–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-García A, San Román C, Arroyo A, Cortés ME, de la Luz Gutiérrez-Nava M, León P (2005) Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-d-erythritol 4-phosphate pathway. Plant Cell 17: 628–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakala M, Tuominen I, Keränen M, Tyystjärvi T, Tyystjärvi E (2005) Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochim Biophys Acta 1706: 68–80 [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z (1996) Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol 30: 925–933 [DOI] [PubMed] [Google Scholar]
- Han M, Heppel SC, Su T, Bogs J, Zu Y, An Z, Rausch T (2013) Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS ONE 8: e62467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussühl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J 20: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Endow JK, Ruppel NJ, Roston RL, Baldwin AJ, Inoue K (2011) Functional diversification of thylakoidal processing peptidases in Arabidopsis thaliana. PLoS ONE 6: e27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Zhu Y, Wu W, Xie Y, Huang J (2015) Leaf variegation of Thylakoid Formation1 is suppressed by mutations of specific σ-factors in Arabidopsis. Plant Physiol 168: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PK, Chan PT, Su PH, Chen LJ, Li HM (2016) Chloroplast Hsp93 directly binds to transit peptides at an early stage of the preprotein import process. Plant Physiol 170: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ling Q, Jarvis P (2013) The ubiquitin-proteasome system regulates chloroplast biogenesis. Commun Integr Biol 6: e23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Baldwin AJ, Shipman RL, Matsui K, Theg SM, Ohme-Takagi M (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J Cell Biol 171: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Jansson S. (1994) The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta 1184: 1–19 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z (2007) The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W (2009) The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Ozawa S, Takahashi Y, Sakamoto W (2015) D1 fragmentation in photosystem II repair caused by photo-damage of a two-step model. Photosynth Res 126: 409–416 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2014) Phosphorylation of photosystem II core proteins prevents undesirable cleavage of D1 and contributes to the fine-tuned repair of photosystem II. Plant J 79: 312–321 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sun X, Zhang L, Sakamoto W (2012) Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol 159: 1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kimber MS, Nishimura K, Friso G, Schultz L, Ponnala L, van Wijk KJ (2015) Structures, functions, and interactions of ClpT1 and ClpT2 in the Clp protease system of Arabidopsis chloroplasts. Plant Cell 27: 1477–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H. (2013) Structural constraints for protein repair in plant photosynthetic membranes. Plant Signal Behav 8: e23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol 143: 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kley J, Schmidt B, Boyanov B, Stolt-Bergner PC, Kirk R, Ehrmann M, Knopf RR, Naveh L, Adam Z, Clausen T (2011) Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat Struct Mol Biol 18: 728–731 [DOI] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Berntsson RPA, Murcha MW, Branca RMM, Radomiljac JD, Regberg J, Svensson LM, Bakali A, Langel U, et al. (2013) Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc Natl Acad Sci USA 110: E3761–E3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Glaser E (2014) Shredding the signal: targeting peptide degradation in mitochondria and chloroplasts. Trends Plant Sci 19: 771–778 [DOI] [PubMed] [Google Scholar]
- Knopf RR, Feder A, Mayer K, Lin A, Rozenberg M, Schaller A, Adam Z (2012) Rhomboid proteins in the chloroplast envelope affect the level of allene oxide synthase in Arabidopsis thaliana. Plant J 72: 559–571 [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichy M, Prásil O, Knoppová J, Kuviková S, de Vries R, Nixon PJ (2007) The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. Plant Cell 19: 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DI, Qian J, Chisholm DA, Jordan DB, Diner BA (2000) Crystal structures of the photosystem II D1 C-terminal processing protease. Nat Struct Biol 7: 749–753 [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Ling Q, Jarvis P (2015) Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr Biol 25: 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Rodermel SR, Yu F (2010a) A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S (2010b) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng M, Wang R, Wang R, An L, Rodermel SR, Yu F (2013) Genetic interactions reveal that specific defects of chloroplast translation are associated with the suppression of var2-mediated leaf variegation. J Integr Plant Biol 55: 979–993 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2016) Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Wu W, Huang W, Huang J (2015) Down-regulation of specific plastid ribosomal proteins suppresses thf1 leaf variegation, implying a role of THF1 in plastid gene expression. Photosynth Res 126: 301–310 [DOI] [PubMed] [Google Scholar]
- Majeran W, Wollman FA, Vallon O (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë A, Wang F, Girard-Bascou J, Wollman FA, de Vitry C (2014) Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell 26: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa T, Endow JK, Dufour J, Zhu J, Inoue K (2014) Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J 80: 592–603 [DOI] [PubMed] [Google Scholar]
- Midorikawa T, Inoue K (2013) Multiple fates of non-mature lumenal proteins in thylakoids. Plant J 76: 73–86 [DOI] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg P, Ståhl A, Bhushan S, Wright SJ, Eriksson A, Bruce BD, Glaser E (2003) Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J 36: 616–628 [DOI] [PubMed] [Google Scholar]
- Moldavski O, Levin-Kravets O, Ziv T, Adam Z, Prag G (2012) The hetero-hexameric nature of a chloroplast AAA+ FtsH protease contributes to its thermodynamic stability. PLoS ONE 7: e36008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A (2007) Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J 49: 800–809 [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM (2012) Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath K, Jajoo A, Poudyal RS, Timilsina R, Park YS, Aro EM, Nam HG, Lee CH (2013) Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Lett 587: 3372–3381 [DOI] [PubMed] [Google Scholar]
- Neher SB, Flynn JM, Sauer RT, Baker TA (2003) Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev 17: 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Apitz J, Friso G, Kim J, Ponnala L, Grimm B, van Wijk KJ (2015) Discovery of a unique Clp component, ClpF, in chloroplasts: a proposed binary ClpF-ClpS1 adaptor complex functions in substrate recognition and delivery. Plant Cell 27: 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Asakura Y, Friso G, Kim J, Oh SH, Rutschow H, Ponnala L, van Wijk KJ (2013) ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ (2015) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 1847: 915–930 [DOI] [PubMed] [Google Scholar]
- Oelmüller R, Herrmann RG, Pakrasi HB (1996) Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J Biol Chem 271: 21848–21852 [DOI] [PubMed] [Google Scholar]
- Ohnishi N, Allakhverdiev SI, Takahashi S, Higashi S, Watanabe M, Nishiyama Y, Murata N (2005) Two-step mechanism of photodamage to photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 44: 8494–8499 [DOI] [PubMed] [Google Scholar]
- Olinares PDB, Kim J, Davis JI, van Wijk KJ (2011) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O, Kato Y, Adam Z, Sakamoto W (2007) Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol 48: 881–885 [DOI] [PubMed] [Google Scholar]
- Paila YD, Richardson LGL, Schnell DJ (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol 427: 1038–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rodermel SR (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc Natl Acad Sci USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy J, van Wijk KJ (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Powikrowska M, Khrouchtchova A, Martens HJ, Zygadlo-Nielsen A, Melonek J, Schulz A, Krupinska K, Rodermel S, Jensen PE (2014) SVR4 (suppressor of variegation 4) and SVR4-like: two proteins with a role in proper organization of the chloroplast genetic machinery. Physiol Plant 150: 477–492 [DOI] [PubMed] [Google Scholar]
- Pulido P, Llamas E, Llorente B, Ventura S, Wright LP, Rodríguez-Concepción M (2016) Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genet 12: e1005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Toledo-Ortiz G, Phillips MA, Wright LP, Rodríguez-Concepción M (2013) Arabidopsis J-protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. Plant Cell 25: 4183–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kirchhoff H (2013) A phosphorylation map of the photosystem II supercomplex C2S2M2. Front Plant Sci 4: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhao J, An R, Zhang J, Liang S, Shao J, Liu X, An L, Yu F (2016) Mutations in circularly permuted GTPase family genes AtNOA1/RIF1/SVR10 and BPG2 suppress var2-mediated leaf variegation in Arabidopsis thaliana. Photosynth Res 127: 355–367 [DOI] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (1998) A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc Natl Acad Sci USA 95: 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (1999) Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol 147: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (2002) Determinants for removal and degradation of transit peptides of chloroplast precursor proteins. J Biol Chem 277: 43888–43894 [DOI] [PubMed] [Google Scholar]
- Rivera-Rivera I, Román-Hernández G, Sauer RT, Baker TA (2014) Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates. Proc Natl Acad Sci USA 111: E3853–E3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RAO, Silva-Filho MC, Cline K (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65: 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Bruch EM, Ceccarelli EA (2011) Insights into the Clp/HSP100 chaperone system from chloroplasts of Arabidopsis thaliana. J Biol Chem 286: 29671–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotanova TV, Melnikov EE, Khalatova AG, Makhovskaya OV, Botos I, Wlodawer A, Gustchina A (2004) Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur J Biochem 271: 4865–4871 [DOI] [PubMed] [Google Scholar]
- Rowland E, Kim J, Bhuiyan NH, van Wijk KJ (2015) The Arabidopsis chloroplast stromal N-terminome: complexities of amino-terminal protein maturation and stability. Plant Physiol 169: 1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Tanaka R, Yamasato A, Tanaka A (2009) Determination of a chloroplast degron in the regulatory domain of chlorophyllide a oxygenase. J Biol Chem 284: 36689–36699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Yamasato A, Tanaka R, Tanaka A (2007) Functional analysis of N-terminal domains of Arabidopsis chlorophyllide a oxygenase. Plant Physiol Biochem 45: 740–749 [DOI] [PubMed] [Google Scholar]
- Samol I, Shapiguzov A, Ingelsson B, Fucile G, Crèvecoeur M, Vener AV, Rochaix JD, Goldschmidt-Clermont M (2012) Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell 24: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Yamamoto Y (2007) The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth Res 94: 203–215 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Adamska I (2012) Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol Plant 145: 224–234 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Mogg U, Adamska I (2011) A new principle of oligomerization of plant DEG7 protease based on interactions of degenerated protease domains. Biochem J 435: 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton JB, Robinson C (1991) Transport of proteins into chloroplasts: the thylakoidal processing peptidase is a signal-type peptidase with stringent substrate requirements at the −3 and −1 positions. J Biol Chem 266: 12152–12156 [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman RL, Inoue K (2009) Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett 583: 938–942 [DOI] [PubMed] [Google Scholar]
- Shipman-Roston RL, Ruppel NJ, Damoc C, Phinney BS, Inoue K (2010) The significance of protein maturation by plastidic type I signal peptidase 1 for thylakoid development in Arabidopsis chloroplasts. Plant Physiol 152: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Tanabe N, Lymperopoulos P, Khan NZ, Rodermel SR, Aronsson H, Clarke AK (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J Biol Chem 289: 11318–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LLE, Clarke AK (2011) Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell 23: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A, Moberg P, Ytterberg J, Panfilov O, Brockenhuus Von Lowenhielm H, Nilsson F, Glaser E (2002) Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J Biol Chem 277: 41931–41939 [DOI] [PubMed] [Google Scholar]
- Ståhl A, Nilsson S, Lundberg P, Bhushan S, Biverståhl H, Moberg P, Morisset M, Vener A, Mäler L, Langel U, et al. (2005) Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J Mol Biol 349: 847–860 [DOI] [PubMed] [Google Scholar]
- Sun R, Fan H, Gao F, Lin Y, Zhang L, Gong W, Liu L (2012) Crystal structure of Arabidopsis Deg2 protein reveals an internal PDZ ligand locking the hexameric resting state. J Biol Chem 287: 37564–37569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Fu T, Chen N, Guo J, Ma J, Zou M, Lu C, Zhang L (2010) The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XW, Wang LY, Zhang LX (2007) Involvement of DEG5 and DEG8 proteases in the turnover of the photosystem II reaction center D1 protein under heat stress in Arabidopsis thaliana. Chin Sci Bull 52: 1742–1745 [Google Scholar]
- Tapken W, Kim J, Nishimura K, van Wijk KJ, Pilon M (2015) The Clp protease system is required for copper ion-dependent turnover of the PAA2/HMA8 copper transporter in chloroplasts. New Phytol 205: 511–517 [DOI] [PubMed] [Google Scholar]
- Tapken W, Ravet K, Pilon M (2012) Plastocyanin controls the stabilization of the thylakoid Cu-transporting P-type ATPase PAA2/HMA8 in response to low copper in Arabidopsis. J Biol Chem 287: 18544–18550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PF, Glaser E (2013) Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta 1833: 360–370 [DOI] [PubMed] [Google Scholar]
- Urantowka A, Knorpp C, Olczak T, Kolodziejczak M, Janska H (2005) Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol Biol 59: 239–252 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ. (2015) Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol 66: 75–111 [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20: 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Funk C (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145: 203–214 [DOI] [PubMed] [Google Scholar]
- Wei L, Derrien B, Gautier A, Houille-Vernes L, Boulouis A, Saint-Marcoux D, Malnoë A, Rappaport F, de Vitry C, Vallon O, et al. (2014) Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal K, Langklotz S, Thomanek N, Narberhaus F (2012) A trapping approach reveals novel substrates and physiological functions of the essential protease FtsH in Escherichia coli. J Biol Chem 287: 42962–42971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Zhu Y, Ma Z, Sun Y, Quan Q, Li P, Hu P, Shi T, Lo C, Chu IK, et al. (2013) Proteomic evidence for genetic epistasis: ClpR4 mutations switch leaf variegation to virescence in Arabidopsis. Plant J 76: 943–956 [DOI] [PubMed] [Google Scholar]
- Yamasato A, Nagata N, Tanaka R, Tanaka A (2005) The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll B accumulation in Arabidopsis. Plant Cell 17: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka-Nishimura M, Yamamoto Y (2014) Quality control of photosystem II: the molecular basis for the action of FtsH protease and the dynamics of the thylakoid membranes. J Photochem Photobiol B 137: 100–106 [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S (2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park SS, Liu X, Foudree A, Fu A, Powikrowska M, Khrouchtchova A, Jensen PE, Kriger JN, Gray GR, et al. (2011) SUPPRESSOR OF VARIEGATION4, a new var2 suppressor locus, encodes a pioneer protein that is required for chloroplast biogenesis. Mol Plant 4: 229–240 [DOI] [PubMed] [Google Scholar]
- Zaltsman A, Ori N, Adam Z (2005) Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA (2002) Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol 9: 906–911 [DOI] [PubMed] [Google Scholar]
- Zhang D, Kato Y, Zhang L, Fujimoto M, Tsutsumi N, Sodmergen, Sakamoto W (2010) The FtsH protease heterocomplex in Arabidopsis: dispensability of type-B protease activity for proper chloroplast development. Plant Cell 22: 3710–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]