Characterization of new constitutively active mutants of phytochromes reveals that both biologically active structure and nuclear localization are crucial for the light-independent function in plants.
Abstract
Plant phytochromes are photoreceptors that mediate a variety of photomorphogenic responses. There are two spectral photoisomers, the red light-absorbing Pr and far-red light-absorbing Pfr forms, and the photoreversible transformation between the two forms is important for the functioning of phytochromes. In this study, we isolated a Tyr-268-to-Val mutant of Avena sativa phytochrome A (AsYVA) that displayed little photoconversion. Interestingly, transgenic plants of AsYVA showed light-independent phytochrome signaling with a constitutive photomorphogenic (cop) phenotype that is characterized by shortened hypocotyls and open cotyledons in the dark. In addition, the corresponding Tyr-303-to-Val mutant of Arabidopsis (Arabidopsis thaliana) phytochrome B (AtYVB) exhibited nuclear localization and interaction with phytochrome-interacting factor 3 (PIF3) independently of light, conferring a constitutive photomorphogenic development to its transgenic plants, which is comparable to the first constitutively active version of phytochrome B (YHB; Tyr-276-to-His mutant). We also found that chromophore ligation was required for the light-independent interaction of AtYVB with PIF3. Moreover, we demonstrated that AtYVB did not exhibit phytochrome B activity when it was localized in the cytosol by fusion with the nuclear export signal and that AsYVA exhibited the full activity of phytochrome A when localized in the nucleus by fusion with the nuclear localization signal. Furthermore, the corresponding Tyr-269-to-Val mutant of Arabidopsis phytochrome A (AtYVA) exhibited similar cop phenotypes in transgenic plants to AsYVA. Collectively, these results suggest that the conserved Tyr residues in the chromophore-binding pocket play an important role during the Pr-to-Pfr photoconversion of phytochromes, providing new constitutively active alleles of phytochromes by the Tyr-to-Val mutation.
Phytochromes are photoreceptors that regulate many aspects of plant growth and development in response to red (R) and far-red (FR) light signals from the environment (Rockwell et al., 2006; Li et al., 2011; Wang and Wang, 2015). They are encoded in higher plants by small gene families. For example, Arabidopsis (Arabidopsis thaliana) contains a phytochrome gene family with five members, designated phytochrome A (phyA) to phyE (Clack et al., 1994; Mathews, 2010). Previous studies on phytochrome mutant alleles revealed that phyA is mainly responsible for sensing FR, whereas phyB to phyE play predominant roles under R or white light conditions (Rockwell et al., 2006; Franklin and Quail, 2010). As an example, during seedling de-etiolation, which involves hook opening, cotyledon expansion, and inhibition of hypocotyl elongation, the effects of continuous FR light are mediated exclusively by phyA and those of continuous R light are mediated predominantly by phyB.
Phytochromes are dimeric chromoproteins with each monomer possessing a covalently linked chromophore, and they exist in two photochromic species, R light-absorbing Pr and FR light-absorbing Pfr forms (Rockwell et al., 2006; Nagatani, 2010). They are biosynthesized as the Pr form in the dark, which can be phototransformed into the biologically active Pfr form upon exposure to R light. The absorption of R light by the Pr form triggers a Z-to-E isomerization of the chromophore and leads to reversible conformational changes throughout the protein moiety, resulting in the Pfr form (Vierstra and Zhang, 2011; Song et al., 2013; Burgie and Vierstra, 2014). Conversely, the Pfr form can be converted to the Pr form by the absorption of FR light. The photoconversion between the Pr and Pfr forms is a unique feature of phytochromes, in which the Pr-to-Pfr photoactivation is known as a critical step in the induction of a highly regulated signaling network for photomorphogenesis in plants (Quail, 2002; Jiao et al., 2007; Chory, 2010; Xu et al., 2015).
Once photoactivated, phytochromes are translocated from the cytosol to the nucleus, which has been suggested as a pivotal step in phytochrome signaling (Sakamoto and Nagatani, 1996; Kircher et al., 2002; Fankhauser and Chen, 2008). In the case of phyB, the nuclear import of phyB has been proposed to be accomplished by an intrinsic nuclear localization signal (NLS) in the C-terminal domain (Chen et al., 2005), and more recently, was shown to be facilitated by phytochrome-interacting factors (PIFs), such as PIF3 (Pfeiffer et al., 2012). On the other hand, the translocation of phyA requires FHY1 (for far-red elongated hypocotyl 1) and FHL (for FHY1-Like), in which the Pfr form of phyA utilizes the NLS of FHY1/FHL through direct physical interactions (Hiltbrunner et al., 2005, 2006; Rösler et al., 2007; Genoud et al., 2008; Rausenberger et al., 2011). In the nucleus, phytochromes interact with a wide array of downstream signaling components, among which PIFs, a small subset of basic helix-loop-helix transcription factors, have been suggested to be canonical components for the regulation of the transcriptional network that drives multiple facets of photomorphogenesis (Leivar and Quail, 2011). Among PIFs, PIF3 is the founding member that interacts with phytochromes in a Pfr-specific manner and negatively regulates phytochrome signaling (Ni et al., 1998; Kim et al., 2003). Moreover, the physical interaction of phytochromes with PIF3 is known to lead to the latter’s phosphorylation and subsequent degradation via the 26S proteasome (Al-Sady et al., 2006; Ni et al., 2014; Shin et al., 2016), which eventually regulates the transcription of various photoresponsive genes for photomorphogenesis in plants (Bae and Choi, 2008; Chen and Chory, 2011; Xu et al., 2015).
Constitutively active alleles of a gene are useful for studying its molecular function and the regulatory roles in its signal transduction. In the case of phytochromes, although many mutant alleles have been used to elucidate its molecular function, most of them are loss-of-function alleles (Rockwell et al., 2006; Franklin and Quail, 2010). So far, only one mutation site for constitutively active alleles of phyA and phyB has been reported (Su and Lagarias, 2007; Hu et al., 2009). The Tyr-276-to-His mutant of Arabidopsis phyB (Y276H-AtphyB; also known as YHB) has been shown to confer a constitutive photomorphogenic (cop) phenotype to transgenic plants (i.e. open cotyledons and shortened hypocotyls in the dark). In addition, the Tyr-242-to-His mutant of Arabidopsis phyA (Y242H-AtphyA; also known as YHA) has been suggested to adopt a signaling active state that is similar to that of YHB but confers a weak cop phenotype to transgenic plants (i.e. shortened hypocotyls in the dark but longer than light-grown seedlings). These constitutively active YHA and YHB mutants have been used to investigate phytochrome-mediated light signaling. For example, YHA was used to elucidate the FR light signaling mechanisms of phyA (Rausenberger et al., 2011), and YHB was used to investigate the molecular function of HEMERA, which is known as an essential component for both phyB localization to nuclear bodies and the degradation of PIFs (Chen et al., 2010; Galvão et al., 2012). In addition, YHB was used to prove that a noncovalently attached chromophore can mediate phyB signaling (Oka et al., 2011) and also to study the phyB function on the circadian clock (Jones et al., 2015). Therefore, the constitutively active mutants are useful tools for studying the molecular functions of phytochromes in plant light signaling.
In this study, we report a new mutation site for constitutively active alleles of phyA and phyB. Initially, we isolated the Tyr-268-to-Val mutant of Avena sativa phyA (Y268V-AsphyA) that showed poor photoconversion and found that transgenic phyA-201 plants with the mutant displayed a cop phenotype in the dark. In addition, the corresponding Arabidopsis mutants (Y303V-AtphyB and Y269V-AtphyA) to Y268V-AsphyA also showed the cop phenotype. Furthermore, we demonstrated that the nuclear localization of the mutants, as well as the biologically active structure, is crucial for their constitutive phytochrome function in plants. Therefore, this study suggests that the Tyr residues at Y268-AsphyA, Y269-AtphyA, and Y303-AsphyB play an important role in Pr-to-Pfr photoconversion, and the Tyr-to-Val mutants mediate the phytochrome signaling in a light-independent manner when they are localized in the nucleus, providing new constitutively active mutant alleles of phytochromes.
RESULTS
Isolation of New Constitutively Active Phytochrome Mutants
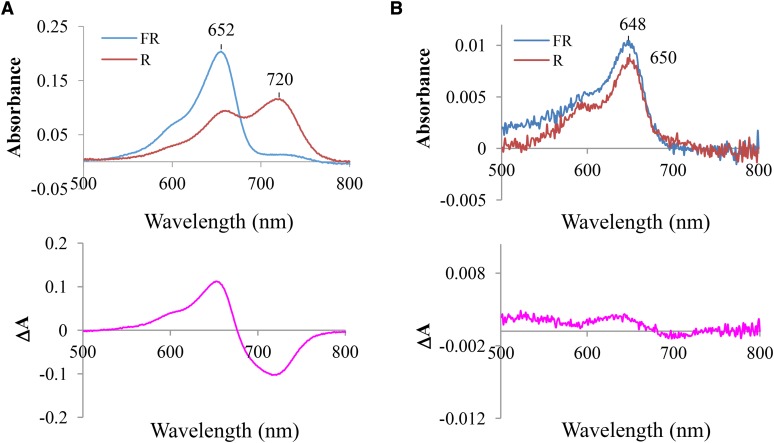
The photochemical properties of phytochromes are determined by interactions between a chromophore and the amino acid residues in its binding pocket (Vierstra and Zhang, 2011). These amino acid residues provide chemical environments for covalent attachment of the chromophore to a Cys residue via a thioether linkage and structural changes and stabilization during the photoconversion between the Pr and Pfr forms. From the photochemical analysis of site-specific mutants of AsphyA for the conserved amino acid residues in the GAF (for cGMP phosphodiesterase/Adenylate cyclase/FhlA) domain, we isolated a mutant that exhibited little photoconversion (Fig. 1). The A. sativa Tyr-268-to-Val mutant of phyA (hereafter, AsYVA) showed covalent chromophore attachment in vitro as confirmed by the zinc fluorescence assay but barely detectable Pr-to-Pfr photoconversion as shown in its difference spectrum (Supplemental Fig. S1; Fig. 1B). The absorption spectra of AsYVA after saturating irradiation by FR and R light were similar to that of the Pr form of wild-type AsphyA, except that its absorption wavelength maximum is slightly blue-shifted (648/650 nm versus 652 nm). On the other hand, the Tyr-268-to-Phe (Y268F) mutant showed a similar difference spectrum to wild-type AsphyA (Supplemental Fig. S2). Together, these results suggest an important role of the phenyl ring at the Y268 position during the photoconversion.
Figure 1.
AsYVA displays little photoconversion. Absorbance (top) and difference (bottom) spectra of wild-type AsphyA (A) and AsYVA (B) are shown. The absorbance spectra were obtained after saturating irradiation by FR (for Pr form) and R (for Pfr form) light, in which absorption maxima are indicated. The difference spectra were obtained by subtracting the Pfr spectrum (R) from the Pr spectrum (FR). The proteins were expressed in Pichia pastoris and purified from ammonium sulfate-precipitated extracts using streptavidin affinity chromatography (Supplemental Fig. S1).
Based on the structure of the photosensory module from AtphyB, three Tyr residues (Y276, Y303, and Y361) are located near the D-ring of the chromophore (Burgie et al., 2014). It is notable that Y303-AtphyB corresponds to Y268-AsphyA or Y269-AtphyA, and the Tyr residues are completely conserved in the GAF domain of plant phytochromes (Supplemental Fig. S3A). Since a Z-to-E isomerization in the C15-C16 double bond between the C- and D-rings of the chromophore has been known to be important for the photoconversion (Rockwell et al., 2006; Nagatani, 2010), these Tyr residues are expected to play a role for the photochemistry of phytochromes. Indeed, when we analyzed the Tyr-326-to-Val (Y326V) mutant of AsphyA, we found that the mutant did not show the ligation with the chromophore in vitro, while the Y326F mutant showed the chromophore ligation and displayed a similar difference spectrum to wild-type AsphyA (Supplemental Fig. S2). Since Y326-AsphyA corresponds to Y361 of AtphyB, these results suggest that Y361-AtphyB might play an important role for the chromophore ligation. Moreover, previous studies showed that YHB (i.e. Y276-to-His mutant of AtphyB) exhibited little photoconversion and conferred a cop phenotype in transgenic plants (Su and Lagarias, 2007; Hu et al., 2009). Thus, in this study, we investigated the biological function of Y303V-AtphyB (hereafter, AtYVB) as well as AsYVA.
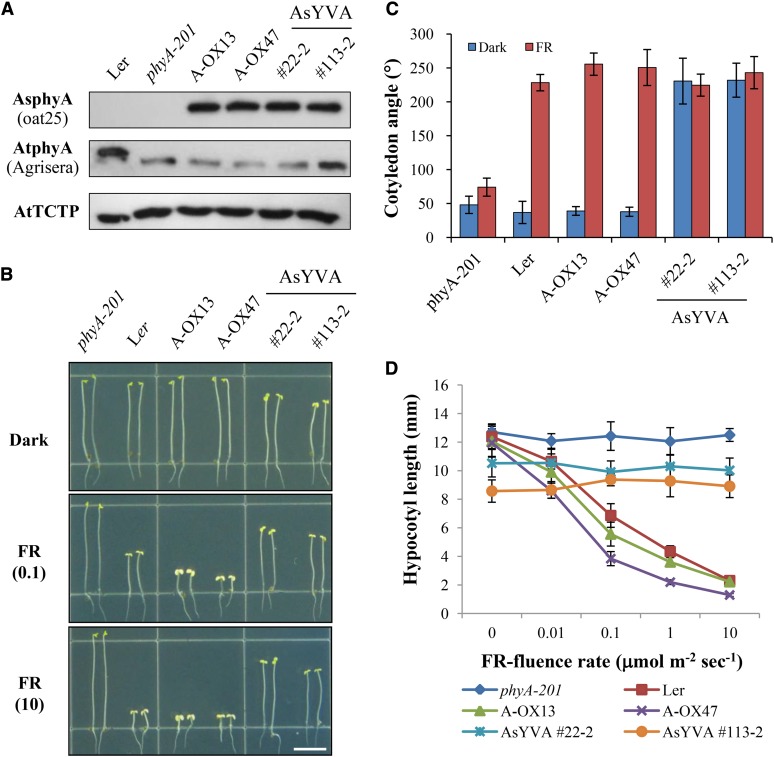
To investigate the in vivo function of AsYVA and AtYVB, we produced transgenic lines with phyA-201 or phyB-5 background (Landsberg erecta [Ler] ecotype) expressing AsYVA and AtYVB cDNAs under the control of the 35S promoter, respectively (Supplemental Fig. S4, A and B). As a control, we also produced transgenic phyB-5 lines expressing the YHB cDNA. We then selected transgenic lines showing comparable protein expression levels of phytochromes with transgenic plants overexpressing wild-type AsphyA (A-OX) or AtphyB (B-OX; Fig. 2A; Supplemental Fig. S5A), and investigated the seedling responses to light conditions. First, we found that the AsYVA plants showed a cop phenotype in the dark, which is characterized by open cotyledons and shortened hypocotyls compared with Ler or A-OXs (Fig. 2, B and C). However, the hypocotyl lengths of the AsYVA plants were much longer than those of the light-grown control plants (i.e. Ler or A-OXs), while the cotyledon openings in the dark were comparable to those of Ler and A-OXs under continuous FR light. More importantly, the hypocotyl lengths of the AsYVA plants were not shortened under FR light with increasing fluence rates (Fig. 2D). These results suggest that AsYVA is constitutively active but not fully functional in plants, resulting in a weak cop phenotype in the transgenic plants.
Figure 2.
Transgenic plants with AsYVA exhibit a weak cop phenotype. A, Immunoblot analysis to show the protein levels of AsphyA and AtphyA in transgenic plants. phyA-201, PhyA-deficient Arabidopsis (Ler ecotype); A-OX, transgenic phyA-201 lines overexpressing wild-type AsphyA; AsYVA, transgenic phyA-201 lines with Y268V-AsphyA. Numbers represent independent homozygous lines. AsphyA-specific (oat25), AtphyA-specific (Agrisera), or AtTCTP-specific antibody was used for the immunoblots. The lower bands in the immunoblot for AtphyA represent truncated AtphyA proteins in phyA-201 that were used for transformation. AtTCTP is shown as loading control. B, Hypocotyl de-etiolation of representative seedlings grown under continuous FR light conditions. The numbers 0.1 and 10 in parentheses are the fluence rates (µmol m−2 s−1) used in the analysis. Scale bar = 5.0 mm. C, Average cotyledon angles of seedlings grown in the dark or continuous FR light (10 µmol m−2 s−1). Data are the means ± sd (n ≥ 30). D, FR fluence rate response curves for the inhibition of hypocotyl growth. Data are the means ± sd (n ≥ 30).
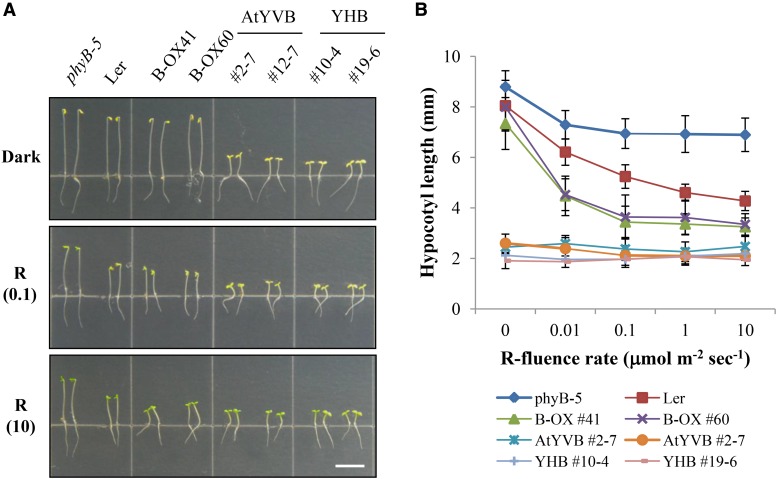
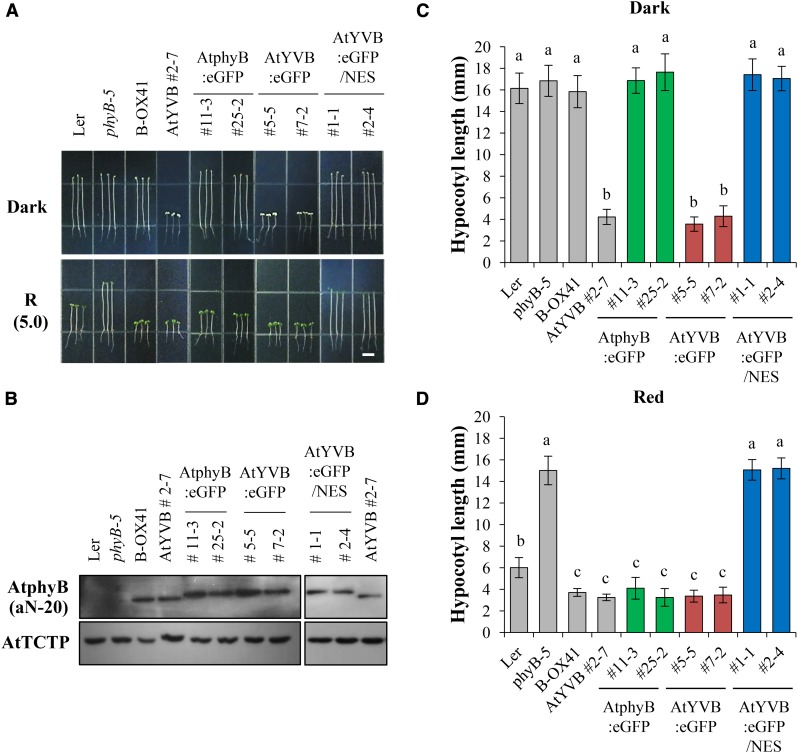
Next, we obtained representative transgenic phyB-5 lines with AtYVB or YHB (Supplemental Fig. S5) and investigated the seedling de-etiolation responses under continuous R light. The results showed that the AtYVB plants in the dark were fully de-etiolated by exhibiting shortened hypocotyls and open cotyledons, which were comparable to Ler and B-OXs under R light (Fig. 3A). Moreover, the shortened hypocotyl lengths of the AtYVB plants were similar under R light with different fluence rates (Fig. 3B). These results suggest that AtYVB is constitutively active and fully functional in plants, resulting in a strong cop phenotype. As a control, we also confirmed the strong cop phenotype of the YHB plants (Fig. 3). Since the seedling phenotypes of the AtYVB plants were similar to those of the YHB plants, this study provides a new allele for the constitutively active phyB.
Figure 3.
Transgenic plants with AtYVB exhibit a strong cop phenotype. A, Hypocotyl de-etiolation of representative seedlings grown under continuous R light conditions. phyB-5, PhyB-deficient Arabidopsis (Ler ecotype); B-OX, transgenic phyB-5 lines overexpressing wild-type AtphyB; AtYVB, transgenic phyB-5 lines with Y303V-AtphyB; YHB, transgenic phyB-5 lines with Y276H-AtphyB. The numbers 0.1 and 10 in parentheses are the fluence rates (µmol m−2 s−1) used in the analyses. Scale bar = 5.0 mm. B, R fluence rate response curves for the inhibition of hypocotyl growth. Data are the means ± sd (n ≥ 30).
AtYVB Shows Light-Independent Nuclear Localization and Interaction with PIF3
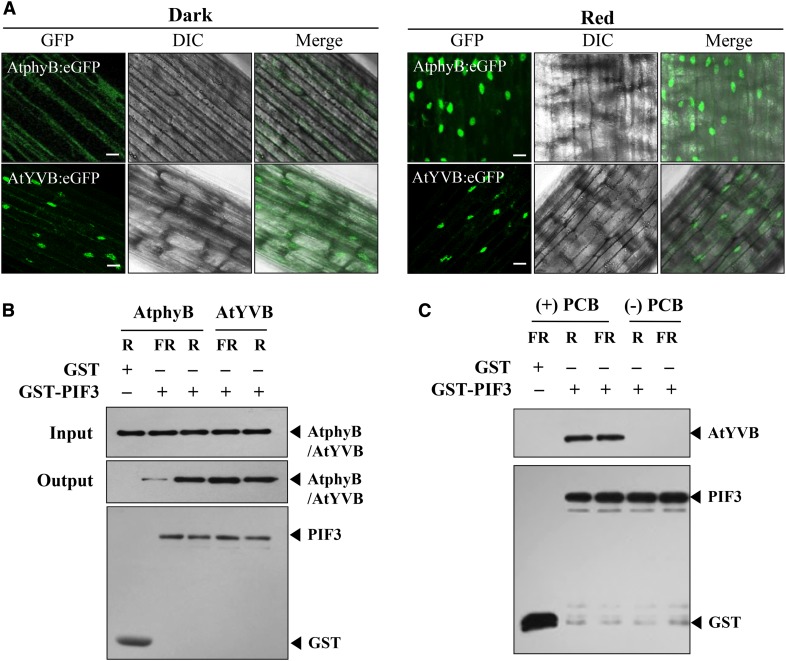
It is well known that the light-dependent translocation of phytochromes into the nucleus is one of the most critical steps for their signaling in plants (Fankhauser and Chen, 2008). In addition, YHB has been reported to localize in the nucleus in a light-independent manner (Su and Lagarias, 2007). Thus, we performed a nuclear localization analysis of AtYVB using the fusion of GFP. For this, we produced transgenic phyB-5 lines expressing wild-type AtphyB:eGFP or AtYVB:eGFP (eGFP, enhanced GFP; Supplemental Fig. S4B), and investigated their subcellular localization in the dark or R light. The results showed that AtYVB:eGFP was localized in the nucleus in both dark and R light conditions (Fig. 4A). As a control, we confirmed the light-dependent nuclear localization of wild-type AtphyB:eGFP. Combined with the results of the fluence rate-independent activity of AtYVB (Fig. 3B), these results suggest that AtYVB is a constitutively active mutant whose conformation is similar to the biologically active form.
Figure 4.
AtYVB is localized in the nucleus and interacts with PIF3 in a light-independent manner. A, Nuclear localization analysis of AtphyB:eGFP and AtYVB:eGFP. Three-day-old dark-grown seedlings were kept in the dark or exposed to R light (5.0 µmol m−2 s−1) for 1 h. DIC, Different interference contrast. Scale bar = 10 μm. B, In vitro protein-protein interaction analysis between AtphyB/AtYVB and PIF3. AtphyB-specific (aN-20) and GST-specific (sc-138) antibodies were used for the detection of AtphyB/AtYVB and PIF3 proteins, respectively. GST was included as a negative control. The phytochrome proteins were expressed in P. pastoris and purified using streptavidin affinity chromatography. PIF3 protein was expressed in E. coli and purified using glutathione affinity chromatography. C, Interaction analysis of PIF3 with apo- and holo-proteins of AtYVB. After preparing ammonium sulfate-precipitated extracts from P. pastoris cells, apo- and holo-proteins of AtYVB were purified without addition of chromophore [(−)PCB] or with the chromophore addition [(+)PCB], respectively.
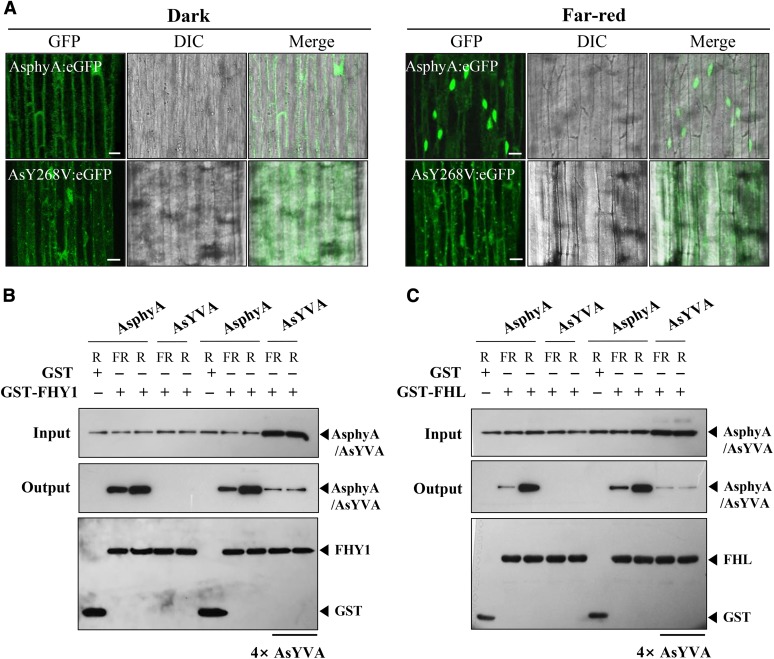
The phytochromes also show light-dependent interaction with downstream signaling components, such as PIFs (Matsushita et al., 2003; Toledo-Ortiz et al., 2010). Thus, to further examine the light-independent activity of AtYVB, we investigated the in vitro protein-protein interaction between AtYVB and PIF3, which is known to interact specifically with the Pfr form of phytochromes. The results showed that AtYVB interacted with PIF3 independently of light, whereas wild-type AtphyB interacted with PIF3 in a Pfr-specific manner (Fig. 4B). This light-independent interaction of AtYVB with PIF3 might be well correlated with its light-independent nuclear localization, if we consider the previous result that the nuclear import of phyB is facilitated by PIF3 (Pfeiffer et al., 2012). Moreover, we observed that apo-protein of AtYVB could not interact with PIF3 (Fig. 4C). This result suggests that chromophore binding is necessary for the interaction with PIF3 and probably for the function of AtYVB. Together, our results support the notion that chromophore-ligated AtYVB adopts a signaling-active structure, although it has one spectral form whose absorption spectrum is similar to the Pr form of wild-type AsphyA.
Nuclear Export Signal-Fused AtYVB Is Not Functional in Plants
To address the importance of nuclear localization for the constitutive phytochrome function, we further produced transgenic phyB-5 lines expressing nuclear export signal (NES)-fused AtYVB:eGFP (Supplemental Fig. S4B). As expected, AtYVB:eGFP/NES was localized in the cytosol in both dark and R light conditions (Supplemental Fig. S6). Then, to examine the in vivo function of AtYVB:eGFP/NES, we investigated the seedling de-etiolation responses in continuous R light after selecting transgenic lines showing comparable protein expression levels of AtphyB to the B-OX plant (Fig. 5, A and B). Results showed that the seedling de-etiolation was not observed in the AtYVB:eGFP/NES plants in either dark or R light condition, similar to phyB-5 (Fig. 5, C and D). On the other hand, the transgenic plants with AtYVB:eGFP exhibited a strong cop phenotype in the dark, as shown in the AtY303V plant (Fig. 5C). The seedling responses to R light were also indistinguishable between the AtYVB and AtYVB:eGFP plants (Fig. 5D). As controls, other plants, including Ler, B-OX, and transgenic plants with AtphyB:eGFP, showed typical etiolated phenotypes in the dark and de-etiolated phenotypes in R light. These results indicate that NES-fused AtYVB is not functional, demonstrating that the nuclear localization is crucial for the constitutive activity of AtYVB.
Figure 5.
Photoresponse analysis of transgenic plants with NES-fused AtYVB. A, Seedling de-etiolation responses under dark or continuous R light (5.0 μmol m−2 s−1). AtphyB:eGFP, Transgenic phyB-5 lines with eGFP-fused wild-type AtphyB; AtYVB:eGFP, transgenic phyB-5 lines with eGFP-fused Y303V-AtphyB; AtYVB:eGFP/NES, transgenic phyB-5 lines with NES-fused AtYVB:eGFP. Numbers represent independent homozygous lines. Scale bar = 5.0 mm. B, Immunoblot analysis to show AtphyB protein levels in transgenic plants. Loading controls (AtTCTP) are shown in the bottom panel. C and D, Average hypocotyl lengths of seedlings in A. Data are the means ± sd (n ≥ 25). Means with different letters are significantly different at P < 0.01, using Duncan’s multiple range test.
AsYVA Is Impaired in Nuclear Localization and Light-Induced Protein Degradation
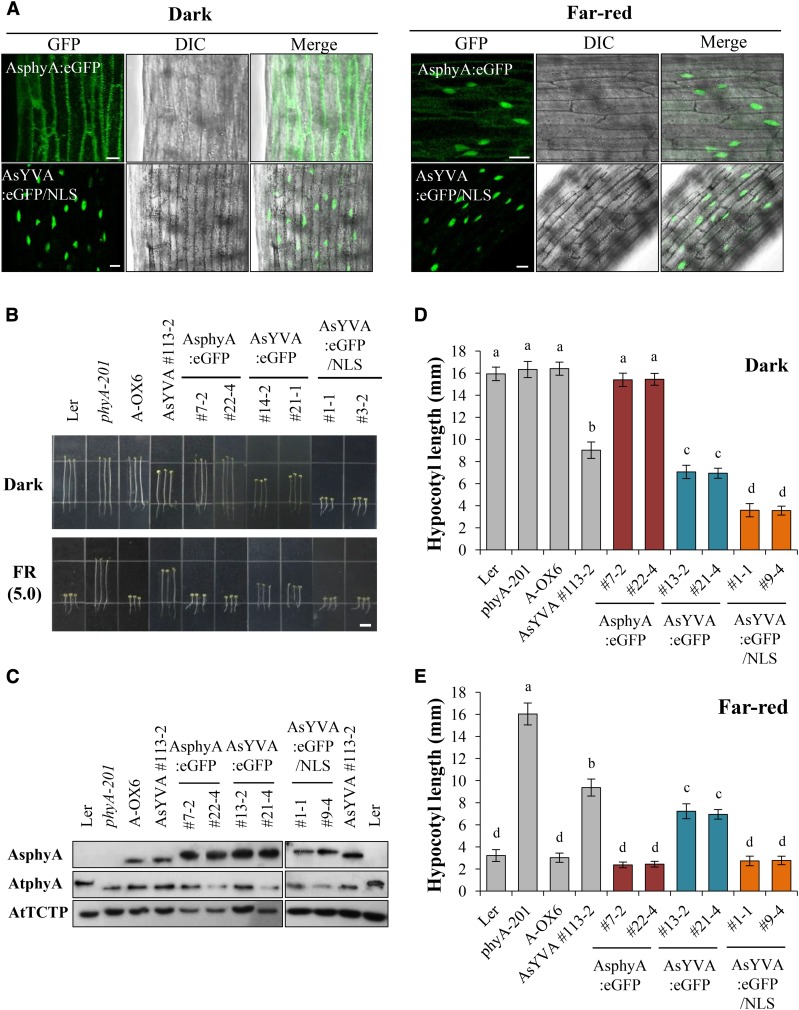
Our data show that the AsYVA plants exhibited a weak cop phenotype (Fig. 2) and that light-independent nuclear localization is required for the constitutive activity of AtYVB (Fig. 5). Accordingly, we investigated the nuclear localization of AsYVA. For this, we produced transgenic phyA-201 lines expressing wild-type AsphyA:eGFP or AsYVA:eGFP (Supplemental Fig. S4A). The results showed that AsYVA:eGFP did not show a clear nuclear localization in either dark or FR light condition (Fig. 6A; Supplemental Fig. S7). Although we found nucleus-localized AsYVA:eGFP occasionally, most of the AsYVA:eGFP was detected in the cytosol in speckles (Supplemental Fig. S7B). To analyze the nuclear localization of AsYVA more clearly, we further investigated the in vitro protein-protein interaction of AsYVA with FHY1 and FHL, i.e. the phytochrome-interacting proteins that participate in the nuclear localization of phyA. The results showed that the interaction of AsYVA with both FHY1 and FHL was significantly reduced, whereas wild-type AsphyA interacted with FHY1 and FHL in a Pfr-preferential manner (Fig. 6, B and C). To evaluate the strength of interactions between AsphyA/AsYVA and FHY1/FHL, we further determined dissociation constants (Kd) by measuring fluorescence polarization using IAEDANS-labeled AsphyA/AsYVA proteins with increasing concentrations of FHY1 or FHL. With the Pfr form of IAEDANS-labeled AsphyA protein, the Kd values were calculated to be 0.66 × 10−6 m for FHY1 and 0.54 × 10−6 m for FHL, indicating that the Pfr form of AsphyA has similar affinity to FHY1 and FHL (Supplemental Fig. S8A). In contrast, when the same experiment was repeated with the IAEDANS-labeled AsYVA (R-irradiated), the Kd values were calculated to be approximately 6.68 × 10−6 m for FHY1 and 8.34 × 10−6 m for FHL, indicating a significant reduction in affinity for the protein-protein interaction. The Pr form of AsphyA and FR-irradiated AsYVA also showed significantly reduced interactions with FHY1 and FHL (Supplemental Fig. S8B). These results are consistent with the in vitro protein-protein interaction results in which the interaction between AsYVA and FHY1/FHL could be detected only with a higher amount of AsYVA protein (Fig. 6C). Moreover, it is noted that AsYVA interacted with FHY1/FHL similarly under FR and R light conditions, indicating a light-independent interaction. Taken together, our results suggest that the nuclear localization of AsYVA is impaired due to reduced interaction affinity with FHY1 and FHL, which might be the reason for the weak cop phenotype in its transgenic plants.
Figure 6.
AsYVA is impaired in the nuclear localization. A, Nuclear localization analysis of AsphyA:eGFP and AsYVA:eGFP. Three-day-old dark-grown transgenic seedlings were kept in the dark or exposed to FR light (5 µmol m−2 s−1) for 1 h. DIC, Different interference contrast. Scale bar = 10 μm. B and C, In vitro protein-protein interaction analysis of AsphyA/AsYVA with FHY1 and FHL. The interaction assays of AsYVA with FHY1/FHL were conducted with the same (1 μg each) or 4-fold higher concentration (4×) of the AsYVA protein. AsphyA-specific (oat25) and GST-specific (sc-138) antibodies were used for the detection of AsphyA/AsYVA and FHY/FHL proteins, respectively. GST was included as a negative control.
Another possible reason for a weak cop phenotype in the AsYVA plants would be decreases in protein stability of phyA. It has been known that phyA accumulates in etiolated seedlings and degrades rapidly upon light exposure (Henriques et al., 2009). Thus, we further investigated the light-dependent protein degradation of phyA in transgenic plants under R or white light. When we investigated time-dependent degradation with R light irradiation, we found that the protein levels of AsYVA were not changed up to 8 h, whereas wild-type AsphyA protein degraded rapidly (Supplemental Fig. S9A). Furthermore, AsYVA protein was not degraded even after white light treatment for 24 h, whereas most of wild-type AsphyA proteins were degraded (Supplemental Fig. S9B). These results indicate that AsYVA is much more stable than wild-type AsphyA and suggest that the weak cop phenotype in the transgenic plants is not due to decreased protein stability of AsYVA.
NLS-Fused AsYVA Confers a Strong cop Phenotype to Plants
To prove that the weak cop phenotype in the AsYVA plants is due to impaired nuclear localization, we further investigated the in vivo function of NLS-fused AsYVA:eGFP. For this, we produced transgenic phyA-201 lines expressing AsYVA:eGFP/NLS (Supplemental Fig. S4A). Then, we investigated the subcellular localization and found that AsYVA:eGFP/NLS was localized in the nucleus in both dark and FR light conditions (Fig. 7A). As a control, we confirmed that wild-type AsphyA:eGFP showed light-dependent nuclear localization. Next, we investigated the seedling de-etiolation responses in continuous FR light after selecting transgenic lines showing comparable protein expression levels of phytochromes to the transgenic plants expressing AsphyA-eGFP (Fig. 7, B and C). Results showed that the AsYVA:eGFP/NLS plants exhibited a strong cop phenotype in the dark (Fig. 7, B and D), suggesting that the constitutively nucleus-localized AsYVA mutant is more functionally active. As a control, we confirmed that the AsYVA:eGFP plants showed a weak cop phenotype in the dark, which was similar to the AsYVA plant (Fig. 7D). It is also notable that the hypocotyl lengths of the AsYVA:eGFP plants were shorter than those of the AsYVA plants (Fig. 7, D and E), which might be due to a higher phytochrome expression level in the AsYVA:eGFP plants than that in the AsYVA plant (Fig. 7C). In addition, the AsYVA:eGFP/NLS plants also showed fully de-etiolated phenotypes in FR light, whereas partially de-etiolated phenotypes were observed in the AsYVA and AsYVA:eGFP plants (Fig. 7E). Together, these results indicate that AsYVA:eGFP/NLS is a constitutively active mutant of phyA adopting a signaling-active structure, and suggest that the weak cop phenotype in the AsYVA plants is caused by the impaired nuclear localization, probably due to significantly reduced interaction with FHY1 and FHL.
Figure 7.
Subcellular localization and photoresponse analyses of NLS-fused AsYVA. A, Subcellular localization analysis. Three-day-old dark-grown seedlings were kept in the dark or exposed to FR light (5.0 µmol m−2 s−1) for 1 h. DIC, Different interference contrast. Scale bar = 10 μm. B, Seedling de-etiolation responses of transgenic plants under dark or continuous FR light (5.0 µmol m−2 s−1). Scale bar = 5.0 mm. AsphyA:eGFP, Transgenic phyA-201 lines with eGFP-fused wild-type AsphyA; AsYVA:eGFP, transgenic phyA-201 lines with eGFP-fused Y268V-AsphyA; AsYVA:eGFP/NLS, transgenic phyA-201 lines with NLS-fused AsYVA:eGFP. C, Immunoblot analysis to show AsphyA protein levels in transgenic plants. Loading controls (AtTCTP) are shown in the bottom panel. D and E, Average hypocotyl lengths of seedlings in B. Data are the means ± sd (n ≥ 25). Means with different letters are significantly different at P < 0.01, using Duncan’s multiple range test.
AtYVA Is Also Impaired in Nuclear Localization, Exhibiting a Weak cop Phenotype in Transgenic Plants
In this study, we introduced a heterologous phyA into Arabidopsis plants for the functional analysis of the Tyr-to-Val mutant. Thus, we extended our work using the Arabidopsis Tyr-269-to-Val mutant of phyA (hereafter, AtYVA) that corresponds to AsYVA. For this, we produced transgenic phyA-201 lines expressing eGFP-fused wild-type AtphyA or AtYVA (Supplemental Fig. S4C). Then, we investigated its subcellular localization and found that AtYVA is mostly localized in the cytosol in both dark and FR light conditions (Supplemental Fig. S11A), which is a similar result to that of AsYVA. Next, we investigated the seedling de-etiolation responses in continuous FR light using representative transgenic plants showing comparable protein expression levels of phyA to the A-OX plant (Supplemental Fig. S11, B and C). The results showed that the AtYVA plants exhibited similar seedling de-etiolation responses in the dark and FR light to the AsYVA plants (Supplemental Fig. S11, D and ES). Therefore, our data suggest that Tyr-269 in AtphyA and Tyr-268 in AsphyA might have a conserved role in the photochemistry of phytochromes.
DISCUSSION
Previously, the mutations at the Y242-AtphyA/Y276-AtphyB residues have been reported to generate constitutively active mutants of phyA and phyB, such as YHA and YHB (Su and Lagarias, 2007). In this study, we found that the constitutively active phytochromes could also be obtained by the mutations at the Y268-AsphyA/Y269-AtphyA/Y303-AtphyB residues, which included AsYVA, AtYVA, and AtYVB (Figs. 2 and 3; Supplemental Fig. S11). The previous constitutively active mutants (i.e. YHA and YHB) were originally obtained from the study of a red fluorescent mutant of Synechocystis Cph1 (Fischer and Lagarias, 2004). It was the Tyr-176-to-His mutant of Cph1 that exhibited greatly reduced difference spectrum but enhanced fluorescence (Fischer et al., 2005). Thus, we further performed fluorescence analysis of the corresponding Cph1 mutant to AsYVA (i.e. Y203V-Cph1). As a result, we could not detect enhanced fluorescence with the Y203V and Y203H mutants of Cph1 (Supplemental Fig. S10). Therefore, although the transgenic plants with YHA/YHB and AsYVA/AtYVA/AtYVB exhibited similar cop phenotypes, the photochemical properties are not the same, suggesting possible differences in protein properties for their function (see below).
Early in this study, we found that the degree of the cop phenotype was different in the transgenic plants with AsYVA and AtYVB. While the AtYVB plants exhibited a strong cop phenotype in the dark (Fig. 3), the cop phenotype of the AsYVA plants was relatively weak; the hypocotyls were not as shortened as the light-grown control seedlings (Fig. 2). Therefore, it was expected that there was a defect in the phytochrome function of AsYVA. To this end, we found differences between AsYVA and AtYVB in their nuclear localization. Our results clearly demonstrated the light-independent nuclear localization of AtYVB (Fig. 4A), while AsYVA showed impaired nuclear localization (Fig. 6A). Subsequently, we further observed that the interactions of AsYVA with FHY1 and FHL were significantly reduced (Fig. 6B; Supplemental Fig. S8). These results suggest the possibility that only a small portion of AsYVA might be localized in the nucleus, which is correlated with the weak cop phenotype observed in the AsYVA plants. Previously, YHA was also shown to confer a weak cop phenotype to its transgenic plants, and it has been proposed that the partial activity of YHA might be due to reduced protein stability (Su and Lagarias, 2007) and/or reduced nuclear localization (Rausenberger et al., 2011). In this study, we confirmed no light-dependent protein degradation of AsYVA (Supplemental Fig. S9), but significantly reduced nuclear localization of AsYVA and AtYVA (Fig. 6A; Supplemental Fig. S11A). Therefore, our results are in good agreement with the hypothesis that the weak cop phenotype is caused by the reduced nuclear localization. However, the present results of AsYVA are different from the previous results of YHA because YHA was shown to interact strongly with FHY1 and FHL independent of light (Rausenberger et al., 2011). Since the expression of YHA in wild-type background resulted in a dominant-negative phenotype, it has been suggested that the constitutive interaction of YHA with FHY1/FHL could prevent nuclear import of phyA due to trapping of FHY1/FHL. Thus, the reduced nuclear import of phyA would be explained well with either constitutive or impaired interaction with FHY1/FHL. Therefore, further studies will be necessary to elucidate the structural differences between YHA and AsYVA/AtYVA for the differential interaction with FHY1/FHL.
The results in this study suggest that the structure of AsYVA/AtYVB is similar to the biologically active form, which has also been previously proposed for YHA and YHB (Su and Lagarias, 2007). However, our data of AsYVA protein stability suggest that the structure is not likely to be the same as the Pfr form. The Pfr form of phyA is known to degrade rapidly in both the cytosol and the nucleus (Sharrock and Clack, 2002; Toledo-Ortiz et al., 2010). However, we did not observe the light-induced degradation of AsYVA (Supplemental Fig. S9), although it is expected that AsYVA might adopt a signaling-active structure because of the cop phenotype observed in its transgenic plants. Rather, we could detect cytosolic speckles in the dark-grown AsYVA:eGFP plants (Supplemental Fig. S7). Previously, it has been found that phyA exhibits rapid light-dependent aggregation, forming sequestered areas of phytochrome before its degradation via the ubiquitin-26S proteasome pathway (Eichenberg et al., 1999). Thus, our results suggest that the structure of AsYVA is similar to the biologically active form, which can induce the formation of sequestered areas of phytochrome in the cytosol, but different from the Pfr form that might be necessary for induction of protein degradation. Further studies will be necessary to elucidate the structural differences between AsYVA and the Pfr form of wild-type AsphyA.
This study demonstrated that the NES-fused AtYVB did not exhibit phyB activity in plants, showing a phyB-deficient phenotype (Fig. 5). In contrast, the NLS-fused AsYVA exhibited the strong cop phenotype observed in the AtYVB plants (Fig. 7). These results suggest that nuclear localization is absolutely necessary for the function of both phyA and phyB, although it is likely that both AtYVB and AsYVA have a biologically active conformation. Regarding these data, it is notable that the transgenic fhy1/fhl plants expressing YHA remained fully etiolated in both dark and FR light conditions, and the nuclear accumulation of YHA was increased by overexpressing FHY1 (Rausenberger et al., 2011). Considering the very weak interaction of AsYVA with FHY1 and FHL (Fig. 6B; Supplemental Fig. S8), it would be expected that the level of nucleus-localized AsYVA is dependent on the available amounts of FHY1/FHL in plants, i.e. little nuclear localization in fhy1/fhl plants and increased nuclear localization in FHY1-overexpressing plants. Thus, the weak cop phenotype in the AsYVA plants might reflect the amounts of nucleus-localized AsYVA with a low binding affinity to FHY1/FHL. This is consistent with our results that the hypocotyls of the AsYVA:eGFP plants were more shortened in the dark than those of the AsYVA plants, in which higher expression of AsYVA:eGFP was detected than that of AsYVA (Fig. 7, B–E). Since the higher expression may increase the amounts of AsYVA that interact with FHY1 and FHL, more nucleus-localized AsYVA can be achieved in plants, resulting in a relatively stronger cop phenotype. In addition to the nuclear localization, active structural conformation is also necessary for the constitutive activity of phytochromes. Previously, etiolated phenotypes of transgenic plants with NLS-fused wild-type phyA have been shown in the dark, suggesting that nuclear localization is not enough to show phytochrome activity (Genoud et al., 2008; Toledo-Ortiz et al., 2010). In this study, we showed that only the NLS-fused AsYVA displayed the strong cop phenotype (Fig. 7). Therefore, we conclude that both constitutive nuclear localization and biologically active conformation are required for the constitutively active alleles of phytochromes.
Recently, the crystal structure of photosensory module of AtphyB in the Pr form (PDB ID: 4OUR) has been reported (Burgie et al., 2014). Thus, we obtained structural models for the Pr and Pfr forms of AtphyB using molecular modeling studies to predict the possible roles of Tyr residues near the D-ring of chromophore during the Pr-to-Pfr photoconversion (Supplemental Data Files S1 and S2). Our modeling data suggest that the cavity for the D-ring of the chromophore in the GAF domain mainly consisted of three Tyr residues and an Asp residue (Supplemental Fig. S12). In the Pr form, T-shaped π-π interaction was observed between Y361 and the D-ring (Supplemental Fig. S12A). In addition, the other two Tyr residues, Y276 and Y303, were involved in alkyl-π interactions. In the Pfr form, the T-shaped π-π interaction was observed between Y303 and D-ring, and Y276 and Y361 were involved in alkyl-π interactions (Supplemental Fig. S12B). Thus, our results propose that the D-ring of chromophore formed the T-shaped π-π interaction with Y361 in the Pr form, but the π-π interaction was moved toward Y303 in the Pfr form. Moreover, the distances between Tyr residues and the D-ring in the Pr and Pfr forms were calculated to be 1.00 and 0.26 nm for Y276, 0.85 and 0.25 nm for Y303, and 0.34 and 0.27 nm for Y361, respectively (Supplemental Fig. S12C). These data suggest that the interaction patterns of the D-ring with Tyr residues are different in the Pr and Pfr forms, in which the D-ring is more closely located to the Tyr residues in the Pfr form than in the Pr form. Overall, the results of photochemical analyses (Fig. 1; Supplemental Fig. S2) and the molecular dynamics (MD) simulations (Supplemental Fig. S12) suggest that the three Tyr residues are important for the chromophore ligation and the Pr-to-Pfr photoconversion, which might explain why the mutations at Y241-AtphyA/Y276-AtphyB and Y268-AsphyA/Y269-AtphyA/Y303-AtphyB can generate constitutively active alleles of phyA and phyB.
In conclusion, this study provides new constitutively active alleles of phyA and phyB. In the case of phyB, a Tyr-to-Val mutation in the GAF domain, i.e. Y303V-AtphyB, is sufficient for the light-independent activity in plants. In the case of phyA, the Tyr-to-Val mutation, i.e. Y268V-AsphyA and A269V-AtphyA, is not enough, but a constitutively active phyA allele is obtained by the fusion of NLS to the Tyr-to-Val mutant (i.e. AsYVA:eGFP/NLS). This study also suggests that the structure of the isolated constitutively active mutants is similar to the biologically active form of phytochromes, but not the same as the Pfr form, especially in the case of the AsYVA mutant that exhibits significantly reduced interaction with FHY1 and FHL. Overall, the light-independent and constitutively active phytochrome mutants isolated in this study would be useful for understanding the molecular function of phytochromes in plant light signaling.
MATERIALS AND METHODS
Spectroscopic Analysis of Recombinant Phytochrome Proteins
Full-length phytochrome cDNAs of wild-type AsphyA and the AsYVA mutant fused with a 10-amino acid streptavidin affinity-tag at the C terminus were cloned into pPIC3.5K (Invitrogen), and recombinant phytochrome proteins were prepared using the Pichia protein expression system (Invitrogen) and streptavidin affinity chromatography (IBA), as described previously (Kim et al., 2004; Shin et al., 2014, 2016). The QuickChange Site-Directed Mutagenesis System (Stratagene) was used to generate AsYVA with the mutagenic primers shown in Supplemental Table S1. Phycocyanobilin (PCB) prepared from lyophilized Spirulina cells by methanolysis, as described previously (Beale and Cornejo, 1991). The PCB sample in DMSO was added to phytochrome samples at a final concentration of 20 μm to generate chromophore-assembled holo-phytochrome proteins.
All of the spectroscopic experiments were carried out under green safety light conditions, which consisted of a white fluorescent lamp equipped with a plastic filter (Rosco) with a maximal transmittance at 500 nm. Absorption spectra were recorded by a diode array UV/VIS spectrophotometer (Cary) after R or FR light irradiation. A fiber-optic illuminator system (Cole-Parmer) equipped with 656- and 730-nm interference filters (Oriel) was used as a light source, and the samples were illuminated with R or FR light for 15 min. A differential spectrum was obtained by subtracting the Pfr spectrum from the Pr spectrum. To assess the chromophore ligation, zinc fluorescence assays were conducted as described previously (Berkelman and Lagarias, 1986). The phytochrome samples were analyzed on SDS-polyacrylamide gels, and the gels were then soaked in 20 mm zinc acetate/150 mm Tris-HCl, pH 7.0, for 10 min at room temperature with gentle shaking. Zinc fluorescence of holo-phytochromes was visualized under UV light (312 nm).
For the measurements of fluorescence emission spectra, Cph1 mutants including Y176H, Y203V, and Y203H were generated using the QuickChange Site-Directed Mutagenesis System with the mutagenic primers (Supplemental Table S1). Each Cph1 gene for the region corresponding to N-terminal 514 amino acids (N514) was then subcloned into pBAD-MycHisC (Invitrogen), as described previously (Gambetta and Lagarias, 2001). The vector was cotransformed into Escherichia coli strain LMG194 (Invitrogen) with pPL-PCB vector. Recombinant expression of PCB-assembled Cph1 (N514) protein was induced by adding 1 mm isopropyl-β-d-thiogalactopyranoside and 0.002% (w/v) arabinose, and purified by the His affinity chromatography. Fluorescence emission spectra were then measured at 30°C using a Shimadzu RF-5301PC spectrofluorometer with a temperature-controlled cuvette.
Phytochrome Constructs for Arabidopsis Transformation
The AsYVA cDNA was cloned into pBI121, as done previously with the wild-type AsphyA gene (Kim et al., 2004). AtYVB and a previously known constitutively active mutant, YHB (Su and Lagarias, 2007), were also generated by site-directed mutagenesis and cloned into pBI121 to generate transgenic Arabidopsis (Arabidopsis thaliana) plants (Supplemental Fig. S4). For eGFP-fused constructs, an eGFP expression cassette under the control of a cassava vein mosaic virus (CsVMV) promoter from pCsVMV-eGFP-N-999 vector (Kim et al., 2008) was cloned into pBI121 using HindIII and PvuII to generate a pBI121-eGFP vector. Then, the AsphyA and AtphyB cDNAs were subcloned into the pBI121-eGFP vector using SalI/BamHI and XbaI/KpnI, respectively (Supplemental Fig. S4, A and B). In addition, wild-type and Y269V mutant cDNAs of AtphyA were also cloned into the pBI121-eGFP vector using SalI/BamHI (Supplemental Fig. S4C). The primers used for these clonings are shown in Supplemental Table S1. For the fusion constructs of phytochromes with NLS (LQPKKKRKVGG) or NES (LQNELALKLAGLDINKTGG) derived from SV40 and PKI (Matsushita et al., 2003; Toledo-Ortiz et al., 2010), the NLS or NES was fused to the C-terminal end of eGFP by two consecutive PCR reactions using the eGFP expression cassette as a template and the primers listed in Supplemental Table S1, which generated the pBI121-eGFP/NLS or pBI121-eGFP/NES vector. Then, the AsYVA and AtYVB cDNAs were subcloned into the pBI121-eGFP/NLS and pBI121-eGFP/NES vectors, respectively (Supplemental Fig. S4, A and B).
Plant Materials
Arabidopsis plants were grown on soil in a culture room (22°C with a 16-h photoperiod). The constructs of AsphyA and AtphyA in pBI121, pBI121-eGFP, or pBI121-eGFP/NLS were each introduced into the phyA-201 mutant using the Agrobacterium tumefaciens (strain GV3101)-mediated floral dip method (Clough and Bent, 1998). In the case of the AtphyB constructs in pBI121, pBI121-eGFP, and pBI121-eGFP/NES, they were introduced into the phyB-5 mutant. Transgenic lines segregating approximately 3:1 for antibiotic resistance in the T2 generation were selected, and the homozygous generation was used for subsequent analyses. After obtaining the homozygous lines, western-blot analysis was carried out with crude extracts from 4-d-old dark-grown seedlings to assess the expression of phytochrome proteins using 1:5,000 AsphyA-specific oat25 monoclonal antibody (Cordonnier et al., 1983), 1:1,000 AtphyB-specific polyclonal antibody (aN-20; Santa Cruz Biotechnology), or 1:1,000 AtphyA-specific polyclonal antibody (Agrisera). For loading controls in this analysis, we used a 1:10,000 polyclonal antibody against Arabidopsis translationally controlled tumor protein (AtTCTP; At3g16640), which is known to be ubiquitously expressed and distributed (Berkowitz et al., 2008; Kim et al., 2012). We selected independent homozygous lines of each transgenic plant that showed a comparable level of phytochrome protein expression to the transgenic line with wild-type phytochrome for further analysis.
Photoresponse Analyses of Transgenic Arabidopsis Plants
Seeds were surface-sterilized, incubated at 4°C for 3 d in the dark, and sown on 0.8% (w/v) phytoagar plates containing half-strength Murashige and Skoog salts and vitamins. The seeds were then exposed to white light for 6 h to promote germination, returned to darkness (22°C) for 24 h, and grown for 4.5 d in darkness or under continuous FR or continuous R light with various fluence rates, by using an LED growth chamber with R (λmax = 654 nm, bandwidth = 25 nm) or FR (λmax = 738 nm, bandwidth = 42 nm) light (Vision Science). The seedlings were photographed with a digital camera (Nikon), and hypocotyl lengths and cotyledon angles of 30 seedlings were then measured with Scion image analysis software.
Confocal Microscopy Analysis
To investigate the subcellular localization of phytochromes, 3-d-old dark-grown seedlings were kept in the dark or exposed to R or FR light (5 μmol m−2 s−1) for 1 h just before the localization analysis. The seedlings were then transferred onto a microscope slide immersed under a cover slip. All confocal images were obtained using a laser confocal scanning microscope (Leica TCS SP5 AOBS/Tandem) with a 40× objective (HCX PL APO N.A. 1.25 Oil UV) and HyD detector at Korea Basic Science Institute. To visualize the GFP fluorescence, 488 nm of Ar laser was used for excitation and spectral bandwidths from 495 nm to 540 nm were used for the detection of emitted light.
In Vitro Protein-Protein Interaction Assays
For recombinant proteins of phytochrome-interacting proteins including PIF3, FHY1, and FHL, glutathione S-transferase and streptavidin affinity tags (GST/strep) were fused to their N and C termini, respectively. They were subcloned into the pGEX 4T vector (GE Healthcare) with primer sets (Supplemental Table S1) and transformed into E. coli strain BL21-CodonPlus (Invitrogen). The recombinant proteins in E. coli cells were induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mm, following further incubation for 3 to 4 h at 37°C. The proteins were then purified using streptavidin affinity chromatography. The concentrations of the purified recombinant proteins were determined using a Quant-iT Protein Assay Kit (Invitrogen).
To examine the protein-protein interaction of phytochromes with phytochrome-interacting proteins in vitro, pull-down experiments were performed. Phytochromes (0.5 or 1.0 μg) were incubated with GST/strep-fused PIF3, FHY1, or FHL (1.0 μg) for 60 min at 4°C in 500 μL of pull-down buffer (100 mm Tris-HCl, pH 7.6, 1 mm EDTA, 150 mm NaCl, and 100 μg/mL bovine serum albumin). Glutathione resin was then added and incubated for an additional 30 min. Phytochromes and GST/strep-fused phytochrome-interacting proteins in the supernatant and precipitate were detected using an ECL western-blotting analysis system (Thermo Scientific) with 1:5,000 AsphyA-specific (oat25), 1:1,000 AtphyB-specific (aN-20), and 1:2,000 GST-specific antibodies (sc-138; Santa Cruz Biotechnology).
Determination of Dissociation Constants
After saturating irradiation by FR and R light, wild-type AsphyA and AsYVA proteins were labeled with a thiol-reactive fluorogenic probe, IAEDANS [5-((((2-Iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid], in dark condition as described previously (Jeganathan et al., 2006). The amount of bound IAEDANS was determined by the absorption at 336 nm (Ɛ336 = 6,100 m−1 cm−1). The molar ratio (protein/IAEDANS) was 0.4 for the labeling, and it was determined that approximately 0.8 molecule of the probe was bound to AsphyA monomer after the reaction. The molar ratio for the bound IAEDANS in proteins was calculated to be 1.0 for AsphyA and 0.92 for AsYVA.
After mixing the IAEDANS-labeled AsphyA/AsYVA with increasing concentrations of FHY1 or FHL at 30°C for 30 min, fluorescence polarization (P) of the probe was measured at 30°C using a Shimadzu RF-5301PC spectrofluorometer with a temperature-controlled cuvette. The excitation wavelength of 336 nm and the emission wavelength of 490 nm were selected with monochromators, and the P values were calculated with P = (III − I⊥)/(III + I⊥), where III and I⊥ are intensities measured with polarizers parallel and perpendicular to the vertically polarized exiting beam, respectively. All fluorescent measurements were also undertaken in the dark condition. Dissociation constants (Kd values) for the protein-protein interaction were then calculated using single-ligand binding model and the following equation as described previously (Park and Raines, 2004): P = ∆P × F/(Kd + F) + Pmin, where ∆P (= Pmax − Pmin) is the total change in polarization and F is the concentration of FHY1 or FHL.
In Vivo Degradation Assay of Phytochrome
The transgenic seeds were germinated and grown for 3.5 d in the dark. Seedlings of each transgenic plant were exposed to R light (28 μmol m−2 s−1) or white light (150 μmol m−2 s−1), and harvested at the indicated times. The seedlings were ground in liquid nitrogen using TissueRuptor (Qiagen) and extracted with an extraction buffer [70 mm Tris-HCl, pH 8.3, 35% ethylene glycol, 98 mm (NH4)2SO4, 7 mm EDTA, 14 mm sodium metabisulfite, 0.07% polyethyleneimine, and protease inhibitors]. Then, 25 μg of total protein extracts was used for western-blot analysis to detect AsphyA using the oat25 antibody.
Computational Details for Molecular Modeling
The Pr and Pfr structures of AtphyB bound with phytochromobilin (PΦB) were constructed by molecular modeling studies. To predict high-quality model structures for the Pr and Pfr forms of AtphyB, homology modeling studies were performed using multitemplates as described previously (Larsson et al., 2008). The Pr form of crystal structure of AtphyB is available, but missing parts exist in the PHY domain (Burgie et al., 2014). Thus, in addition to the structure of AtphyB (PDB ID: 4OUR), the structure of Syn-Cph1 (PDB ID: 2VEA) was used as template to construct the homology-modeled structure for the Pr conformation of AtphyB. To predict the Pfr conformation, AtphyB (PDB ID: 4OUR) was used for the main template structure, and SyB-Cph1 (PDB ID: 2KLI) and DrBphP (PDB ID: 5C5K) were also selected as template structures. DrBphP structure in the photoactivated state (Burgie et al., 2016) was used for the conformation of tongue region in the PHY domain, and SyB-Cph1 structure (Rockwell et al., 2009) was used for the conformation of amino acid residues, including Tyr-54, Phe-86, Asp-86, Val-100, Arg-101, His-139, Tyr-142, and His-169, in the GAF domain. The PΦB molecule was also obtained from SyB-Cph1 (PDB ID: 2KLI). The MD snapshot structures of the homology-modeled 3D structures for the Pr and Pfr forms of AtphyB are available from our Web site (http://bio.gnu.ac.kr/pdb/AtphyB_Pr.pdb and AtphyB_Pfr.pdb).
To gain insight into chromophore binding modes at the atomic level, MD simulations were conducted with the modeled structures in the water solvent using the Groningen Machine for Chemical Simulation (GROMACS) 5.0.4 package. The topology parameters for PΦB were obtained by the SwissParam Web server, and a CHARMM27 force field was applied to gain a correct arrangement of the atoms and refined side-chain orientations. The transferable intermolecular potential 3P (TIP3P) water model surrounded the modeled systems. Then, the net charges of all systems were neutralized by adding Na+ counter-ions. To form the thioether bond between a Cys residue and PΦB, CAC atom (PΦB) and SG atom (Cys residue) were fixed during the simulations due to the lack of parameters for PΦB in GROMACS. The energy minimization was performed using the steepest descent algorithm to remove possible bad contacts from initial structures until energy convergence reached 1,000 kJ mol−1. NVT and NPT equilibration were carried out at 100 ps under 300 K. After equilibration, production runs were performed for the systems for 5 ns. From the trajectories of the last 1 ns, the representative structure was selected by clustering analysis.
Statistical Analysis
Experimental data were subjected to ANOVA using IBM SPSS Statistics 20 software. Significant difference of mean values were compared by the lsd at P < 0.01. All of the data were represented as the mean ± sd of at least three independent experiments.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Purified recombinant wild-type (AsphyA) and Y268V-AsphyA (AsYVA) proteins used in the photochemical analysis.
Supplemental Figure S2. Photochemical analyses of purified recombinant AsphyA mutant proteins.
Supplemental Figure S3. Amino acid sequence alignments of phytochromes.
Supplemental Figure S4. T-DNA regions of the binary vector constructs used for plant transformation.
Supplemental Figure S5. Transgenic phyB-5 plants expressing AtYVB and YHB.
Supplemental Figure S6. Nuclear localization of NES-fused AtYVB.
Supplemental Figure S7. Subcellular localization analyses of wild-type AsphyA (A) and AsYVA (B).
Supplemental Figure S8. Determination of dissociation constants for the protein-protein interaction between AsphyA/AsYVA and FHY1/FHL.
Supplemental Figure S9. Protein stability of AsYVA in plants.
Supplemental Figure S10. Photochemical analyses of purified recombinant Cph1 mutant proteins.
Supplemental Figure S11. Subcellular localization and photoresponse analyses of AtYVA.
Supplemental Figure S12. Prediction for the positions of Tyr residues near the d-ring of chromophore in the Pr and Pfr forms of AtphyB.
Supplemental Table S1. Primers used in this study.
Supplemental Data File S1. Homology model for the Pr form of AtphyB.
Supplemental Data File S2. Homology model for the Pfr form of AtphyB.
Supplementary Material
Acknowledgments
We thank the Kumho Life Science Laboratory in Chonnam National University for providing plant growth facilities.
Glossary
- NLS
nuclear localization signal
- NES
nuclear export signal
- MD
molecular dynamics
- PCB
phycocyanobilin
Footnotes
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant no. NRF-2014R1A1A2057739 to J.-I.K.), and in part by Next-Generation BioGreen 21 Program from the Rural Development Administration, Republic of Korea (grant nos. PJ01104001 to J.-I.K. and PJ01106202 to K.W.L.).
Articles can be viewed without a subscription.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Beale SI, Cornejo J (1991) Biosynthesis of phycobilins. 3(Z)-phycoerythrobilin and 3(Z)-phycocyanobilin are intermediates in the formation of 3(E)-phycocyanobilin from biliverdin IX alpha. J Biol Chem 266: 22333–22340 [PubMed] [Google Scholar]
- Berkelman TR, Lagarias JC (1986) Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal Biochem 156: 194–201 [DOI] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pollmann S, Masle J (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD (2014) Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci USA 111: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Vierstra RD (2014) Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26: 4568–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Zhang J, Vierstra RD (2016) Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24: 448–457 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J (2005) Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Chory J. (2010) Light signal transduction: an infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cordonnier MM, Smith C, Greppin H, Pratt LH (1983) Production and purification of monoclonal antibodies to Pisum and Avena phytochrome. Planta 158: 369–376 [DOI] [PubMed] [Google Scholar]
- Eichenberg K, Kunkel T, Kretsch T, Speth V, Schäfer E (1999) In vivo characterization of chimeric phytochromes in yeast. J Biol Chem 274: 354–359 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chen M (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Lagarias JC (2004) Harnessing phytochrome’s glowing potential. Proc Natl Acad Sci USA 101: 17334–17339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC (2005) Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry 44: 15203–15215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta GA, Lagarias JC (2001) Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA 98: 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T, Schweizer F, Tscheuschler A, Debrieux D, Casal JJ, Schäfer E, Hiltbrunner A, Fankhauser C (2008) FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan S, von Bergen M, Brutlach H, Steinhoff HJ, Mandelkow E (2006) Global hairpin folding of tau in solution. Biochemistry 45: 2283–2293 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jones MA, Hu W, Litthauer S, Lagarias JC, Harmer SL (2015) A constitutively active allele of phytochrome B maintains circadian robustness in the absence of light. Plant Physiol 169: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Shen Y, Han YJ, Park JE, Kirchenbauer D, Soh MS, Nagy F, Schäfer E, Song PS (2004) Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Han YJ, Hwang OJ, Lee SS, Shin AY, Kim SY, Kim JI (2012) Overexpression of Arabidopsis translationally controlled tumor protein gene AtTCTP enhances drought tolerance with rapid ABA-induced stomatal closure. Mol Cells 33: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Wallner B, Lindahl E, Elofsson A (2008) Using multiple templates to improve quality of homology models in automated homology modeling. Protein Sci 17: 990–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S. (2010) Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22: 4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Nagatani A. (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Kong SG, Matsushita T (2011) A non-covalently attached chromophore can mediate phytochrome B signaling in Arabidopsis. Plant Cell Physiol 52: 2088–2102 [DOI] [PubMed] [Google Scholar]
- Park SH, Raines RT (2004) Fluorescence polarization assay to quantify protein-protein interactions. Methods Mol Biol 261: 161–166 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Nagel MK, Popp C, Wüst F, Bindics J, Viczián A, Hiltbrunner A, Nagy F, Kunkel T, Schäfer E (2012) Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Rausenberger J, Tscheuschler A, Nordmeier W, Wüst F, Timmer J, Schäfer E, Fleck C, Hiltbrunner A (2011) Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Shang L, Martin SS, Lagarias JC (2009) Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proc Natl Acad Sci USA 106: 6123–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler J, Klein I, Zeidler M (2007) Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci USA 104: 10737–10742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin AY, Han YJ, Baek A, Ahn T, Kim SY, Nguyen TS, Son M, Lee KW, Shen Y, Song PS, Kim JI (2016) Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun 7: 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin AY, Han YJ, Song PS, Kim JI (2014) Expression of recombinant full-length plant phytochromes assembled with phytochromobilin in Pichia pastoris. FEBS Lett 588: 2964–2970 [DOI] [PubMed] [Google Scholar]
- Song C, Rohmer T, Tiersch M, Zaanen J, Hughes J, Matysik J (2013) Solid-state NMR spectroscopy to probe photoactivation in canonical phytochromes. Photochem Photobiol 89: 259–273 [DOI] [PubMed] [Google Scholar]
- Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Kiryu Y, Kobayashi J, Oka Y, Kim Y, Nam HG, Mochizuki N, Nagatani A (2010) Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol 51: 1648–1660 [DOI] [PubMed] [Google Scholar]
- Vierstra RD, Zhang J (2011) Phytochrome signaling: solving the Gordian knot with microbial relatives. Trends Plant Sci 16: 417–426 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H (2015) Phytochrome signaling: time to tighten up the loose ends. Mol Plant 8: 540–551 [DOI] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E (2015) Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci 20: 641–650 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.