nana plant2 encodes an enzyme in the brassinosteroid biosynthetic pathway, and brassinosteroid and gibberellin interact in a developmentally specific manner to control maize architecture.
Abstract
A small number of phytohormones dictate the pattern of plant form affecting fitness via reproductive architecture and the plant’s ability to forage for light, water, and nutrients. Individual phytohormone contributions to plant architecture have been studied extensively, often following a single component of plant architecture, such as plant height or branching. Both brassinosteroid (BR) and gibberellin (GA) affect plant height, branching, and sexual organ development in maize (Zea mays). We identified the molecular basis of the nana plant2 (na2) phenotype as a loss-of-function mutation in one of the two maize paralogs of the Arabidopsis (Arabidopsis thaliana) BR biosynthetic gene DWARF1 (DWF1). These mutants accumulate the DWF1 substrate 24-methylenecholesterol and exhibit decreased levels of downstream BR metabolites. We utilized this mutant and known GA biosynthetic mutants to investigate the genetic interactions between BR and GA. Double mutants exhibited additivity for some phenotypes and epistasis for others with no unifying pattern, indicating that BR and GA interact to affect development but in a context-dependent manner. Similar results were observed in double mutant analyses using additional BR and GA biosynthetic mutant loci. Thus, the BR and GA interactions were neither locus nor allele specific. Exogenous application of GA3 to na2 and d5, a GA biosynthetic mutant, also resulted in a diverse pattern of growth responses, including BR-dependent GA responses. These findings demonstrate that BR and GA do not interact via a single inclusive pathway in maize but rather suggest that differential signal transduction and downstream responses are affected dependent upon the developmental context.
Aboveground plant form is achieved by the progressive addition of organs, which guarantees plant success through the acquisition of light and gases to support photosynthesis and reproduction. Multiple phytohormones regulate the architecture of organs, the rate at which they grow, and their placement on the growing axes of plants (Sussex and Kerk, 2001; Jaillais and Chory, 2010; Depuydt and Hardtke, 2011). This regulation not only contributes to the diversity of plant form and the adaptation of plants to their environments (Orshan, 1986; Chapin et al., 1987) but has been central to major gains in crop yield in the 20th and 21st centuries (Salamini, 2003; Salas Fernandez et al., 2009). Phytohormone regulation of plant architecture and components such as plant height, branching, and floral organ development has been studied extensively (Evans and Poethig, 1995; Choe et al., 2001; Hong et al., 2004; Sakamoto et al., 2006). The simultaneous presence of multiple phytohormones in all plant tissues raises the question: how do interactions between these developmental determinants influence the plant form? Multiple studies of gene expression demonstrated shared and distinct gene expression responses for various combinations of phytohormones in Arabidopsis (Arabidopsis thaliana; Nemhauser et al., 2006; Jaillais and Chory, 2010; Wang et al., 2011; Vanstraelen and Benková, 2012). However, the developmental consequences and changes in gene expression within these interactions resulting in their interdependency on plant development are largely unknown.
Brassinosteroid (BR) and gibberellin (GA) are two phytohormones that have similar effects on anisotropic cell expansion. As a result, both contribute to the directional expansion of plant organs and are involved in similar developmental processes (Santner et al., 2009; Santner and Estelle, 2009; Wolters and Jürgens, 2009; Hamant and Traas, 2010). BRs are polyhydroxylated steroidal compounds that control plant height, skotomorphogenesis and photomorphogenesis, branching, reproductive organ development, stomatal development, and disease resistance (Choe et al., 2001; Nakashita et al., 2003; Hartwig et al., 2011; Kim et al., 2012; Oh et al., 2012; Makarevitch et al., 2012; Wang et al., 2013). BR at concentrations of N × 10−10 is biologically active and sensitizes plants to auxin applications (Wada et al., 1981; Vert et al., 2008). BR and auxin positively regulate each other’s biosynthesis (Li et al., 2005; Kim et al., 2014), resulting in a positive regulatory loop. GA also influences plant height, branching, reproductive organ development, stomatal development, and disease resistance (Evans and Poethig, 1995; Eckardt, 2002; Saibo et al., 2003; Elfving et al., 2011).
Maize (Zea mays) is a monoecious species, which produces two types of imperfect flowers in two distinct locations. Staminate flowers are produced at the apex of the shoot in the tassel florets, and pistillate flowers are present on lateral branches in the ear florets. In the two inflorescences, organs are arrested or aborted postinitiation to prevent their manifestation in mature florets. BR-deficient mutants in maize exhibit severe dwarfism, altered leaf morphology, and pistil production in the tassel (Hartwig et al., 2011; Makarevitch et al., 2012). Other tasselseed (ts) mutants in maize are not dwarf. The genes responsible for some of these mutants are molecularly defined. Mutants such as ts1 and ts2 have a proposed function in jasmonic acid (JA) metabolism (DeLong et al., 1993; Acosta et al., 2009), while other mutants are involved in encoding an APETELA2-like transcription factor, Ts6, as well as its corresponding microRNA, ts4 (Chuck et al., 2007). As a result, the ability of JA to inhibit pistil production in the maize tassel has been a leading hypothesis for tassel seed production. However, work by Nickerson (1959) demonstrated that sustained GA3 treatments of the maize growing point induced tassel seeds in multiple wild-type maize inbred backgrounds. Consonant with a role for GA in the regulation of maize floral organ persistence, mutants deficient in GA biosynthesis were unable to suppress stamen production in the ear (Emerson et al., 1935). The JA and GA effects appear to be independent, as pistil persistence in the tassel florets of the ts1 and ts2 mutants was not suppressed by the loss of GA synthesis in double mutants between ts1 or ts2 and the GA-deficient mutant dwarf1 (d1; Irish et al., 1994). Thus, an overproduction of GA or a deficiency of JA in BR-deficient mutants could result in tassel seed production.
Much of what we currently know about the interactions between BR and GA is from experiments in Arabidopsis and a single seedling organ, the hypocotyl (Bai et al., 2012; Li et al., 2012; Stewart Lilley et al., 2013; Oh et al., 2014). In monocots, a study on rice (Oryza sativa) seedlings showed that BR and GA cross talk is dependent upon the tissue studied and the hormone concentration, exhibiting different results from Arabidopsis (Tong et al., 2014). If plants utilize nonoverlapping mechanisms to integrate signals, single-tissue studies may oversimplify their interactions and fail to explain the roles of these molecules in generating the diversity of plant adaptations and forms.
We report the characterization and cloning of nana plant2 (na2), first identified by H.S. Perry in the 1930s (Emerson et al., 1935). The na2 mutants exhibited severe dwarfism, suppression of tillers, altered leaf morphology, and a failure to suppress pistils in the tassel florets. We demonstrated that na2 was defective in the reduction of 24-methylenecholesterol to campesterol, caused by a mutation of Δ24-sterol reductase involved in BR biosynthesis. To investigate the hormonal regulation and cross talk of maize architecture, we created BR- and GA-deficient double mutants in maize. Through the analysis of these double mutants, we identified distinct genetic interactions between BR and GA mutants that were developmentally specific. GA was required for pistil production in the tassels of BR mutants, and BR was required for the increased branching observed in GA mutants, while BR and GA additively influenced plant height. Thus, the control of maize architecture by phytohormones, made visible by blocking their biosynthesis, is the result of interdependent signaling events that identify GA and BR cross talk as a critical determinant of maize development.
RESULTS
na2 Mutants Are Disrupted in the Δ24-Sterol Reductase of Maize
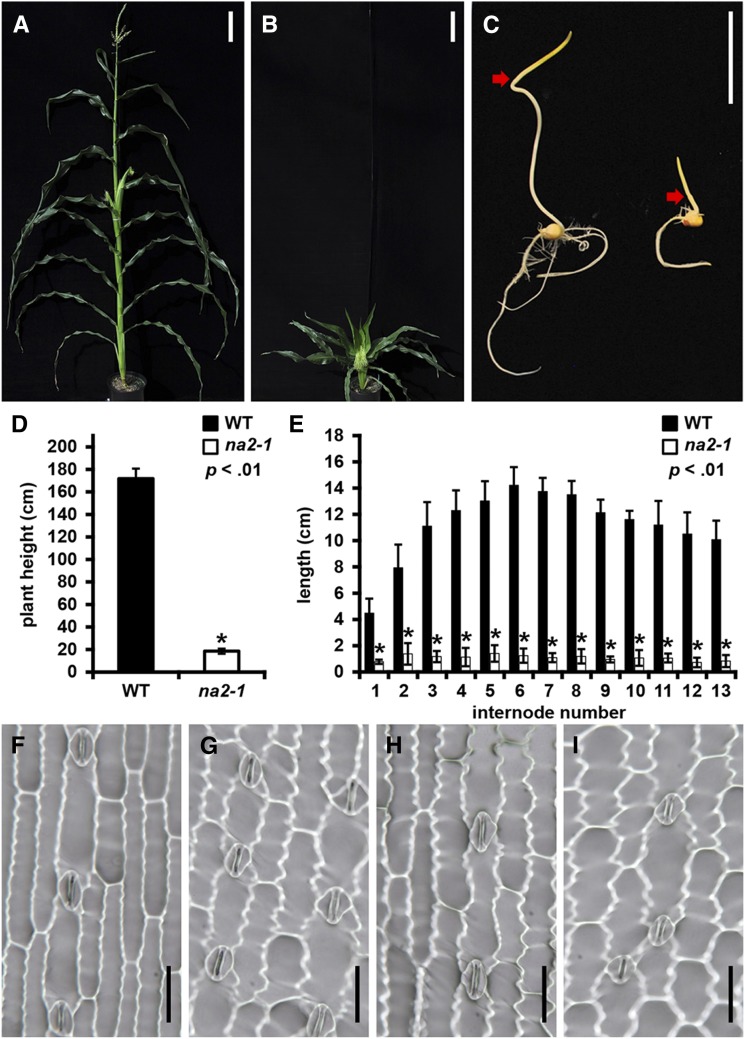
The maize line 4407D d*-N2374 was identified as an extreme dwarf among images of maize mutants (http://maizegdb.org/cgi-bin/displayvarrecord.cgi?id=75364). Figure 1 shows the phenotypes displayed by greenhouse-grown plants from this line, including dwarfism, feminized tassels, reduced branching, and upright leaves. Many of these are shared by the BR biosynthetic mutant na1 (Hartwig et al., 2011). Crosses between na1-1 and line 4407D d*-N2374 complemented these phenotypes to the wild type in the F1 generation, demonstrating that the 4407D d*-N2374 mutant defined a distinct locus. To test allelism between 4407D d*-N2374 and the classical mutant na2 (Emerson et al., 1935), homozygous mutant pollen parents were crossed to heterozygous ear parents. 4407D d*-N2374 failed to complement the na2 phenotypes in two stocks bearing mutations in na2, the na2-506D line and na2-518C introgressed into Hi27, demonstrating that line 4407D d*-N2374 is a new allele of na2 (wild type:na2, 73:57 [χ2 P = 0.22] and 16:15 [χ2 P = 0.86], respectively; for data from complementation crosses, see Supplemental Table S1). The accessions in the stock center will retain the nomenclature they have carried to date, and line 4407D d*-N2374 was designated na2-1. The na2-1 mutants were similar to na1 in the effects on plant height, tiller formation repression, and the suppression of floral organ growth and development. The na2-1 mutant was approximately 10% of the height of wild-type siblings at maturity and exhibited a tassel seed phenotype, displaying pistil development in the normally staminate tassel (Fig. 1, A, B, and D). The tassel seed phenotype of na2-1 was not completely penetrant and was environmentally influenced. In the summer of 2014, 74 of 123 plants (60.2%) grown in the field exhibited some degree of tassel seed, whereas only three of 13 plants (23.1%) grown in the greenhouse exhibited tassel seed (Fisher’s exact test P = 0.018). The na2-1 mutant also showed a deetiolated phenotype when grown in the dark as compared with its wild-type siblings (Fig. 1C). Reduced height at maturity resulted from the compression of all internodes in na2-1 as compared with wild-type siblings (Fig. 1, D and E). Stomatal imprints showed a slight difference in epidermal cell morphology between na2 mutants and wild-type siblings (Fig. 1, F–I). No significant differences were identified in stomatal index or stomatal density for either the abaxial or adaxial surface of na2-1 or na2-2 mutants compared with wild-type siblings (Supplemental Table S2).
Figure 1.
Morphological features of na2 mutants. A, Mature wild-type plant. B, na2-1 plant. C, Wild-type (left) and na2-1 seedlings (right) grown in the dark for 8 d. Red arrows indicate the distal end of the mesocotyls. D, Plant height of the wild type (WT) and na2-1 measured from the soil line to the highest leaf collar. E, Internode lengths of the wild type and the na2-1 mutant. Error bars are sd, and asterisks indicate Student’s t test (P < 0.01) for comparisons between the wild type and na2-1. F to I, Photomicrographs of imprints from wild-type abaxial surface (F), na2-1 abaxial surface (G), wild-type adaxial surface (H), or na2-1 adaxial surface (I). Bars = 20 cm in A and B, 5 cm in C, and 50 µm in F to I.
Due to the similarity in phenotypes of na2 and na1, we measured endogenous BR intermediates to determine if the BR pathway was blocked. Table I summarizes the BR pathway intermediates measured in na2-1 mutants and wild-type siblings. The na2-1 mutants accumulated 6-fold greater levels of the BR intermediate 24-methylenecholesterol than wild-type plants. Consistent with a block in the BR pathway at this step, the level of the downstream intermediate campesterol was reduced by 130-fold in the mutant as compared with the wild type. The proposed pathway for BR biosynthesis in maize is presented in Supplemental Figure S1, and the block in campesterol production suggests that na2 is defective in the C-24 reduction of 24-methylenechloesterol (Supplemental Fig. S1). This step in BR biosynthesis is encoded by the DWARF1 (DWF1) gene in Arabidopsis (Choe et al., 1999).
Table I. BR intermediates in wild-type and na2-1 plants.
Values shown are ng g−1 fresh weight from plants at 28 d after germination (DAG). Data are means of two independent experiments with 10 plants each. n.d., Not detected.
| Compound | +/na2-1 | na2-1/na2-1 |
|---|---|---|
| 24-Methylenecholesterol | 1,950 | 12,550 |
| Campesterol | 21,650 | 167 |
| Campestanol | 442 | 3.62 |
| 6-Oxocampestanol | 12.1 | 0.50 |
| 6-Deoxocathasterone | 0.21 | n.d. |
| 6-Deoxoteasterone | 0.04 | n.d. |
| 3-Dehydro-6-deoxoteasterone | 0.41 | n.d. |
| 6-Deoxotyphasterol | 2.71 | n.d. |
| 6-Deoxocastasterone | 6.78 | n.d. |
| Cathasterone | n.d. | n.d. |
| Teasterone | n.d. | n.d. |
| 3-Dehydroteasterone | n.d. | n.d. |
| Typhasterol | 0.04 | n.d. |
| Castasterone | 0.51 | n.d. |
| Brassinolide | n.d. | n.d. |
The protein sequence encoded by DWF1 was used as a query for a sequence similarity search by the BLAST algorithm (Tatusova and Madden, 1999) of the maize reference genome (AGPv3.21). This identified two predicted protein sequences in the maize genome as highly similar to DWF1 encoded by GRMZM2G057000 and GRMZM2G455658, both with greater than 79% identity to the DWF1 protein sequence. The conceptual translations of protein-coding sequences similar to DWF1 were obtained from whole-genome assemblies of plants available via Phytozome (Goodstein et al., 2012). A misannotation of the genes of maize, sorghum (Sorghum bicolor), and Selaginella (Selaginella moellendorffii) contains an N-terminal deletion, but closer inspection indicates conservation of these coding sequences within the regions annotated as 5′ untranslated region within GRZM2G455658, Sobic.004G355600, and Sm_144134, respectively. To investigate the phylogenetic relationships between the maize paralogs, DWF1, and the similar genes encoded in other plant genomes, we used a Bayesian Markov chain Monte Carlo approach implemented by MrBayes (Geyer, 1991; Ronquist et al., 2012) to construct the consensus tree depicted in Supplemental Figure S2. The tree is consistent with the two maize paralogs arising from a gene duplication that is older than the split between maize and sorghum. Analysis of the genomic contexts of both maize and sorghum genes using COGE GEVO (Lyons and Freeling, 2008) indicated extended regions of retained gene duplicates in comparisons between the sorghum paralogs, the maize paralogs, and the sorghum and maize paralog pairs. The extent of synteny was greater for paralogs that grouped together in the consensus tree (Supplemental Fig. S2; data not shown). Enzymes in this family are known to act on multiple substrates as Δ24-sterol reductases (Klahre et al., 1998) and, thus, are not specific to BR biosynthesis. Consistent with their role in forming structural sterols, such as sitosterol, genes encoding presumptive 24-sterol reductase activity are widely phylogenetically distributed in plants. In agreement with previous studies, we identified similar sequences in both Physcomitrella (Physcomitrella patens) and Selaginella (Mizutani and Ohta, 2010; Cheon et al., 2013; Hamberger and Bak, 2013). Physcomitrella and Selaginella each encode two paralogs that derive from duplication after the divergence of those lineages from each other and the remainder of the taxa (Supplemental Fig. S2). Supplemental Figure S3 shows a multiple sequence alignment of the na2 locus with Δ24-sterol reductases from sorghum, Setaria italica, rice, Arabidopsis, Glycine max, Manihot esculenta, Selaginella, and Physcomitrella. Inspection of the two maize paralogs from the B73 reference genotype identified two frame-shift mutations in GRMZM2G455658 that break and then restore the reading frame, resulting in more than 30 consecutive nonconserved amino acids disrupting a well-conserved region of the protein-coding sequence (Supplemental Fig. S3). Examination of the two sorghum paralogs also identified multiple frame-shift mutations accumulated in the sorghum gene Sobic.004G355600 that disrupt extensive regions of conserved protein-coding sequences (Supplemental Fig. S3).
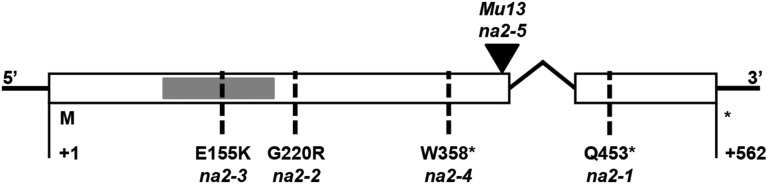
We determined the sequences of GRMZM2G057000 in multiple na2 mutants. A summary of these data is presented in Figure 2, and the genetic material used is described in Supplemental Table S3. Sequencing of this locus in na2-1 identified a C-to-T nucleotide transition at position 1,357 resulting in a premature stop codon and truncation of the last 110 amino acids (Fig. 2). Three additional alleles, na2-2, na2-3, and na2-4, were identified among plants with reduced stature in an ethyl methanesulfonate (EMS)-treated population derived from B73 (Weil, 2009). The na2-2 mutant had a G-to-A transition at nucleotide position 658 resulting in a missense mutation that converted the Gly codon to Arg. Similarly, na2-3 had a G-to-A transition at nucleotide position 463 resulting in a missense mutation that converted a Glu codon to Lys. These two missense mutations were drastic amino acid replacements that change codons that are 100% conserved between all angiosperms analyzed, as well as Selaginella and Physcomitrella, except for the missense mutation in na2-2 in the sorghum paralog Sobic.004G355600 (Supplemental Fig. S3). The na2-4 allele was a G-to-A mutation at nucleotide position 1,074 resulting in a nonsense codon and truncation of the last 206 amino acids. We also generated an insertion allele, na2-5, in a targeted mutagenesis experiment among approximately 100,000 F1 plants from crosses using na2-1 pollen onto Mu-active ears. The single novel allele, na2-5, contains a Mu13 insertion at the codon for amino acid 407. Two additional alleles without molecular information were identified by allelism tests: na2-6, also taken from the maize B73 EMS population (Weil, 2009), and na2-7, which was in the 4405B d*-N1074C stock at the MaizeCOOP. All seven novel alleles of na2 displayed similar dwarf phenotypes and rates of tassel seed production similar to the original mutant (Emerson et al., 1935).
Figure 2.
Structure of the na2 locus and alleles. Exons are depicted as white boxes, the intron as a bent black line, and the sequence encoding the putative FAD-binding domain as a small gray box. Alleles are indicated with dashed lines for single-nucleotide polymorphism variations in na2-1, na2-2, na2-3, and na2-4 and a triangle at the site of the Mu13 transposable element insertion in na2-5. The amino acid substitution that results from each single-nucleotide polymorphism is specified above each allele designation, with an asterisk indicating the induction of a stop codon.
NA2 mRNA Is Accumulated in Rapidly Growing Tissues of B73
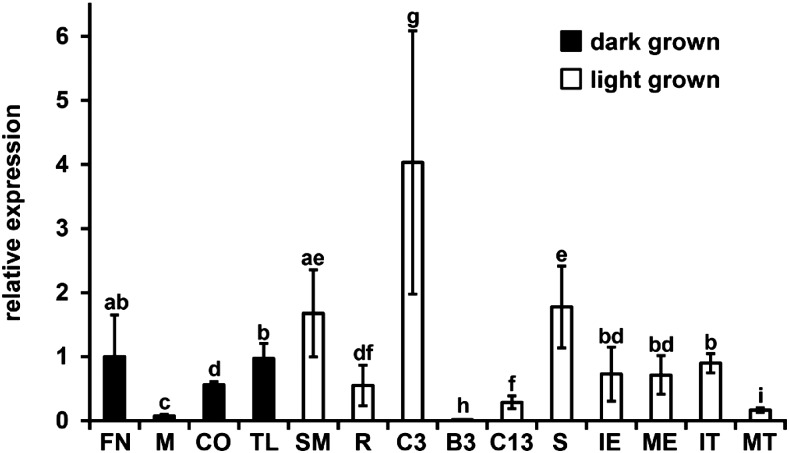
We performed quantitative reverse transcription-PCR (qRT-PCR) to analyze the organ-specific expression of NA2. Figure 3 summarizes the means of relative expression normalized to controls for NA2 in mRNA extract from different organs and treatments of wild-type B73 plants. The NA2 transcript was detected in all tissues analyzed. The highest expression of NA2 was in mRNA from the collar (comprising the intercalary meristem, auricle, and ligule) of leaf 3 in plants at 9 DAG. The accumulation of NA2 was generally lower in older tissues. For example, the collar of leaf 13 taken from mature plants had less NA2 mRNA than the third leaf from 9-DAG plants. Interestingly, NA2 expression was low in the mesocotyl of dark-grown tissues despite the dependence of NA2 for elongation of this tissue, as indicated by the deetiolated phenotype of na2-1 seedlings (Fig. 1C). Tissue age was not the only factor controlling NA2 expression. NA2 accumulation was strongly influenced by the linear axis of leaf expansion. The lowest NA2 mRNA expression was found in the blade of leaf 3 at 9 DAG, just distal to the collar that provided the highest observed NA2 accumulation.
Figure 3.
Comparison of relative NA2 mRNA accumulation measurement by qRT-PCR. qRT-PCR was carried out on mRNA extracted from multiple organs. Relative expression was obtained by normalization to the expression level of molybdenum cofactor (GRMZM2G067176) as an internal standard. Samples are indicated on the x axis: FN, 0.25-cm segment containing the first stem node of etiolated seedlings at 5 DAG; M, mesocotyl tissue of 5-DAG etiolated seedlings; CO, coleoptile of 5-DAG etiolated seedlings; TL, true leaves of 5-DAG etiolated seedlings; SM, shoot meristem and surrounding tissue of 9-DAG plants; R, roots of 9-DAG plants; C3, collar of the third leaf from 9-DAG plants; B3, blade from the third leaf of 9-DAG plants; C13, collar of the 13th leaf at maturity; S, stem segment from V7-stage plants; IE, immature ears 0.5 to 2.5 cm in length at the time of harvest; ME, mature ear sampled at silk emergence; IT, immature tassel 0.5 to 2.5 cm in length at the time of harvest; MT, mature tassel sampled at anthesis. Histogram height is proportional to the mean, and error bars indicate sd. Three biological replicates were run in three technical replications each for each sample. Lowercase letters indicate connecting letter report as determined by ANOVA with a posthoc analysis using the Holm-Sidak algorithm with P < 0.05. Samples that share the same letter do not differ statistically, whereas samples that are not connected by the same letter are statistically different.
Much like the frame-shift mutations that distinguished na2 from na2-like (GRMZM2G455658; Supplemental Fig. S3), expression level differences between the paralogs also were visible. The accumulation of mRNA from na2 was greater than that from the na2-like gene, as recorded in summaries of RNA sequencing experiments in the qTeller database (http://www.qteller.com). Shown in Supplemental Figure S4 are RNA fragments per kilobase of transcript per million mapped reads (FPKM) values for the two maize and two sorghum genes from the qTeller database. The maize na2 gene (GRMZM2G057000) had an average FPKM value of 179.2 across all tissues and treatments, with the highest expression in the developing leaf (617; Supplemental Fig. S4A). The maize na2-like gene (GRMZM2G455658) had an average FPKM value of 0.1 across all tissues and treatments, with the highest expression in one of the pollen samples (2.9), but was inconsistently found in pollen RNA sequencing libraries (Supplemental Fig. S4B). The expression levels of the sorghum na2 (Sobic.010G277300) and na2-like ortholog Sobic.004G355600 (Supplemental Fig. S2) had average FPKM values of 167.8 and 0.04, respectively (Supplemental Fig, S4, C and D). Thus, relative to their paralogs, the mRNA encoded by the na2 genes was accumulated to 3 and 4 orders of magnitude greater levels and the na2-like paralogs had accumulated numerous frame-shift mutations after the divergence of maize and sorghum. This is consistent with the na2-like paralogs encoding pseudogenes that have lost expression and function. The observation that nonsense and missense mutations of GRMZM2G057000 were sufficient to dwarf maize plants favor this interpretation.
BR and GA Biosynthetic Mutants Exhibit Complex Genetic Interactions Controlling Organ Development and Growth
BR and GA biosynthetic mutants exhibit defects in floral development and severe dwarfism (Suttle, 1924; Hartwig et al., 2011). BR interacts with GA metabolism in rice (Schmitz et al., 2013; Tong et al., 2014) and at the signal transduction level in Arabidopsis (Bai et al., 2012; Li et al., 2012; Stewart Lilley et al., 2013). To investigate the BR and GA interaction in maize, we generated double mutants between BR and GA biosynthetic mutants. The BR mutants na2-1 and na1-1, a 5α-steroid reductase, and the GA mutants d1, a GA 3-oxidase, and d5, an ent-kaurene synthase, were used in this study (Hedden and Phinney, 1979; Hartwig et al., 2011; Chen et al., 2014). F2 populations from crosses between na2-1 and d5, na2-1 and d1, and na1-1 and d1 segregated for the mutants and effects of the genetic backgrounds. Some parental lines were of unknown backgrounds (Supplemental Table S3). To control for segregating background mutation effects, wild-type plants, single mutants, and double mutants were phenotyped and compared between F2 siblings to test BR and GA mutant effects without confounding the genetic background. The F2 generation of each cross was phenotyped for plant height, tiller number, anther ear formation, and production of tassel seeds in the summers of 2013 (na2-1/d5) and 2014 (na2-1/d5, na2-1/d1, and na1-1/d1). These results are summarized in Table II, and images of the resulting phenotypes are presented in Figure 4. Segregation ratios of phenotypes in the F2 generation were different from 9:3:3:1 for the na2/d5 population (χ2 P = 5.43E-16) and for the na2-1/d1 population (χ2 P = 0.011); however, the na1-1/d1 population was not different from a 9:3:3:1 segregation (χ2 P = 0.400). The na2-1/d5 and na2-1/d1 populations had an underrepresentation of single and double mutants as compared with wild-type plants. These results are consistent with weak seedling establishment in BR and GA mutants, as had been noted previously (Emerson et al., 1935).
Table II. Morphometric analysis of BR and GA double mutant combinations.
| Genotype | n | Height (cm)a | Tillers per Planta | Plants with Tassel Seedsb | Plants with Anthers in Ear Floretsb |
|---|---|---|---|---|---|
| na2-1 × d5; F2 | 949 | – | – | – | – |
| Wild type | 658 | 180.2 ± 21.2 a | 0.8 ± 1.0 a | 2 (0.3%) a | 0 (0%) a |
| na2-1 | 123 | 21.5 ± 5.0 b | 0 ± 0 b | 74 (60%) b | 0 (0%) a |
| d5 | 145 | 30.3 ± 5.7 c | 2.1 ± 1.2 c | 0 (0%) ac | 145 (100%) b |
| na2-1/d5 | 23 | 7.2 ± 2.8 d | 0.09 ± 0.3 d | 2 (9%) c | 22 (96%) b |
| na2-1 × d1; F2 | 576 | – | – | – | – |
| Wild-type | 349 | 193.9 ± 23.1 a | 0.3 ± 0.6 a | 3 (0.9%) a | 0 (0%) a |
| na2-1 | 111 | 23.0 ± 6.1 b | 0.0 ± 0.0 b | 55 (50%) b | 0 (0%) a |
| d1 | 97 | 37.6 ± 9.1 c | 1.7 ± 1.2 c | 0 (0%) a | 97 (100%) b |
| na2-1/d1 | 19 | 7.3 ± 1.9 d | 0.0 ± 0.0 d | 1 (5%) a | 19 (100%) b |
| na1-1 × d1; F2 | 172 | – | – | – | – |
| Wild type | 102 | 216.3 ± 22.9 a | 0.7 ± 0.8 a | 0 (0%) a | 0 (0%) a |
| na1-1 | 35 | 51.7 ± 22.7 b | 0.0 ± 0.0 b | 6 (17%) b | 0 (0%) a |
| d1 | 29 | 35.3 ± 7.8 c | 2.0 ± 1.1 c | 0 (0%) ab | 29 (100% ) b |
| na1-1/d1 | 6 | 14.2 ± 3.9 d | 0.0 ± 0.0 d | 0 (0%) ab | 6 (100%) b |
Data are presented as means with sd. Lowercase letters indicate connecting letter report as determined by ANOVA with posthoc analysis using the Holm-Sidak algorithm with P < 0.05.
Number of plants with tassel seeds or anthers in ear florets, with the percentage of plants in parentheses. Lowercase letters indicate connecting letter report as determined by Fisher’s exact test with P < 0.01.
Figure 4.
Morphological features of na2-1, d5, and double mutant combinations. All images were taken at maturity. A to D, Wild-type plant (A), na2-1 plant (B), d5 plant (C), and na2-1/d5 plant (D). E to H, Tassel phenotypes of wild-type (E), na2-1 (F), d5 (G), and na2-1/d5 (H) plants. I to L, Ear phenotypes of wild-type (I), na2 -1 (J), d5 (K), and na2-1/d5 (L) plants. Silks have been partially removed from K and L. Red arrows indicate stamens on ears in K and L. Bars = 10 cm (A–H) and 2 cm (I–L).
The BR and GA biosynthetic mutations displayed multiple genetic interactions in the double mutants as compared with the single mutants. These interactions occurred in a development- and organ-specific manner. Wild-type plants on average were 180.2 cm, na2-1 plants were 21.5 cm, d5 plants were 30.3 cm, and na2-1/d5 plants were 7.2 cm, with overall plant height being measured from the soil level to the leaf collar of the top leaf (Fig. 4, A–D; Table II). The na2-1 and d5 plants showed 88.1% and 83.2% reductions in plant height as compared with wild-type plants, respectively. The na2-1/d5 double mutants were 96% reduced in plant height as compared with the wild type and 66.5% or 76.2% shorter than na2-1 or d5, respectively. This demonstrates additivity and, thus, no interaction between na2-1 and d5 for overall plant height.
Unlike plant height, tillering displayed epistasis. A lack of endogenous GA causes maize plants to branch more and produce multiple tillers. By contrast, loss of BR biosynthesis suppressed tiller production. The na2-1/d5 double mutants resembled the na2-1 single mutants and did not display profuse tillers. The wild-type plants from the na2-1 × d5 F2 generation had 0.8 tillers per plant, no na2-1 plants had tillers, d5 mutants had an average of 2.1 tillers, and na2-1 d5 had an average of 0.09 tillers, as only a single double mutant produced a single tiller (Fig. 4, A–D). These results demonstrate that na2-1 is epistatic to d5 for tiller production. This demonstrates that BR biosynthesis is required for a loss of GA to increase branch numbers.
BR and GA are required for the developmental failure of floral organs required to produce imperfect flowers in maize. BR mutants failed to suppress pistil development in the tassel, resulting in the tassel seed reported here (Fig. 1) and previously (Hartwig et al., 2011; Makarevitch et al., 2012). GA mutants failed to suppress anther development in the ear resulting in perfect flowers on the ear, also called anther ear (Emerson, 1912; Suttle, 1924; Evans and Poethig, 1995). To determine if these phenotypes were additive like plant height or displayed epistasis like tillering, the inflorescences of all F2 siblings in the cross between na2-1 and d5 were carefully observed for floral organ persistence. When tassel seeds were carefully phenotyped in the summers of 2013 and 2014 in the field, 0.3% (n = 658) of wild-type plants showed some degree of pistil production in the tassel. In the single mutants, 60% of na2-1 plants (n = 123) and 0% of d5 plants (n = 145) had tassel seeds. Double mutants were substantially suppressed as compared with na2-1 single mutants, with 9% of na2-1/d5 double mutant plants (n = 23) exhibiting some tassel seed formation (Fig. 4, E–H; Table II). This was greater than the wild type but far less than na2-1 single mutants, suggesting that d5 is required for the induction of tassel seeds mediated by the loss of na2-1. The ears of the same plants were scored for anther production. Wild-type plants showed no development of anthers in ears (n = 658). The single mutants were differentially affected, with 0% of the na2-1 plants displaying anther ear (n = 123) and 100% of d5 plants developing anther ear (n = 145). Unlike tillering, where na2-1 was epistatic to d5, 96% of na2-1/d5 plants exhibited anther ear (n = 23; Fig. 4, I–L; Table II). This demonstrates that the persistence of anthers in the ears of d5 mutants was not influenced by na2-1 and the loss of BR.
The observations in all three double mutant combinations of the different BR and GA loci were mainly similar. The phenotyping results for all three F2 families can be found in Table II. The na2-1/d1 double mutant combinations resulted in the same genetic interactions as described above for na2-1/d5 double mutants. In addition, the phenotyping of the na1-1/d1 double mutants showed identical genetic interactions between na1-1 and d1 as seen with na2-1 and d1 or d5. There were notable differences between the F2 populations, none of which altered the interaction between the BR and GA mutants. Most obvious among these are the increased height of na1-1 single mutants and a lower percentage of tassel seed production (17%; n = 35) as compared with the na2-1 mutants in the crosses to the same d1 background. As a result of observing multiple phenotypes expressed in different developmental contexts, we find that there is no single mode of interaction between GA and BR in maize. In summary, BR and GA additively affected plant height, GA was required for the production of tassel seeds in the absence of BR, BR was required for the production of tillers in the absence of GA, but BR was not involved in the persistence of anthers in the ears that occurs in GA-deficient mutants.
Exogenous Application of na2 and d5 with GA3
The double mutant analysis indicated multiple interactions between BR and GA affecting plant morphology. To investigate the effect of GA3 on BR-dependent plant architecture, na2-1, d5, and their wild-type siblings were treated with exogenous GA3. GA3 at 866 µm was applied directly into the leaf whorl as 1 mL every third day from 25 DAG until tassel emergence, similar to the approach taken by Nickerson (1959). Multiple phenotypes were measured, including plant height, tillers per plant, leaf angle, width and length of the leaf below the flag leaf and the leaf subtending the ear, node number, days to tassel, tassel branch number, tassel length, presence or absence of tassel seeds, and presence or absence of anthers in ear florets. The means and sd values of these measurements are presented in Table III. GA3 application increased total plant height of both na2-1 and the wild type (Table III). Plant height increased by similar amounts of 60% and 62% in wild-type and na2-1 plants, respectively. This increase in height resulted in significant increases in all internodes except internode 14 of wild-type plants, whereas only internodes 1 to 4 and 7 were increased significantly in na2-1 plants upon GA3 application (Supplemental Fig. S5). These results are consistent with our observation of additivity between BR and GA disruption for plant height and indicate that BR-deficient mutants in maize respond to GA3 treatment, resulting in internode elongation no different from that in the wild-type siblings.
Table III. Morphometric analysis of na2-1 treated with GA3.
| Parameter | Mock |
+GA3 |
||
|---|---|---|---|---|
| +/na2-1 | na2-1/na2-1 | +/na2-1 | na2-1/na2-1 | |
| n | 16 | 13 | 16 | 14 |
| Plant heighta | 192.9 ± 39.1 a | 32.7 ± 5.7 b | 309.6 ± 39.2 c | 52.9 ± 16.8 d |
| Days to tassela | 63.8 ± 3.6 a | 73.5 ± 8.6 b | 60.0 ± 3.2 c | 73.5 ± 5.3 b |
| Tillers per planta | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a |
| Plants with tassel seedsb | 0 (0%) a | 3 (23%) a,b | 7 (44%) b | 13 (93%) c |
| Plants with anthers in ear floretsb | 0 (0%) a | 0 (0%) a | 0 (0%) a | 0 (0%) a |
| Tassel branchesa | 14.3 ± 5 a | 11.7 ± 5.6 a | 13.6 ± 4.2 a | 11.9 ± 6.0 a |
| Total nodesa | 15.1 ± 0.9 a | 14.4 ± 1.3 a | 15.5 ± 1.3 a | 13.1 ± 1.1 b |
| Node of top eara | 10.4 ± 0.9 a | 10.5 ± 1.1 a | 9.9 ± 1.0 a,b | 9.1 ± 1.0 b |
| Tassel lengtha | 50.6 ± 4.2 a | 28.2 ± 6.5 b | 54.0 ± 13.5 a | 31.7 ± 10.7 b |
| Angle of upper leafa | 25.9 ± 16.3 a | 49.6 ± 19.9 b | 23.8 ± 17.3 a | 56.4 ± 17.7 b |
| Length of upper leafa | 48.8 ± 7.1 a,c | 23.7 ± 8.5 b | 42.0 ± 15.9 a | 34.9 ± 10.1 c |
| Width of upper leafa | 5.4 ± 0.8 a | 4.3 ± 1.0 b | 3.5 ± 0.9 c | 3.2 ± 0.9 c |
| Angle of lower leafa | 45.9 ± 14.7 a | 61.2 ± 12.8 b | 50.6 ± 9.8 a | 74.5 ± 10.2 c |
| Length of lower leafa | 95.5 ± 9.6 a | 61.4 ± 5.5 b | 95.6 ± 14.8 a | 77.8 ± 11.3 c |
| Width of lower leafa | 8.0 ± 0.9 a | 8.1 ± 1.4 a | 4.9 ± 1.2 b | 4.3 ± 1.3 b |
Data are presented as means with sd. Lowercase letters indicate connecting letter report as determined by ANOVA with posthoc analysis using the Holm-Sidak algorithm with P < 0.05.
Number of plants with tassel seeds or anthers in ear florets, with the percentage of plants in parentheses. Lowercase letters indicate connecting letter report as determined by Fisher’s exact test with P < 0.01.
Treatment with GA3 caused the development of tassel seeds in wild-type plants and enhanced this phenotype in na2-1 as compared with mock treatments (Table III). Thus, just as a loss of GA biosynthesis via the d1 and d5 mutants inhibited tassel seed production, exogenous GA3 was sufficient to increase tassel seeds in both na2-1 and the wild type. GA3 application also accelerated tassel emergence in wild-type plants as compared with controls. This application had no effect on na2-1 tassel emergence, indicating a BR dependence on GA3-induced flowering time acceleration. The node carrying the top ear in na2-1 plants was lower in GA3-treated plants, which also produced fewer internodes overall. GA3 treatment had no effect on internode number or ear placement, as detected in wild-type plants. This indicates that a loss of BR was necessary for GA3 to influence internode number.
GA3 caused na2-1 upper (leaf below the top leaf) and lower (leaf above the node containing the uppermost ear) leaves to become narrower and longer. Wild-type leaf lengths were not affected by GA3 treatment, but both upper and lower leaves were narrower (Table III). Applying GA3 also resulted in an increase in the angle of the lower leaf for na2-1 (more upright). Again, GA3 treatment had no effect on leaf angles for wild-type plants. Thus, the loss of BR was required for this treatment to control leaf length and leaf angle. All other parameters measured showed no significant difference between GA3 and mock treatment or na2-1 and the wild type, respectively.
Some phenotypes were measured in these materials that had not been measured in our initial characterization of the na2-1 mutant. Among these, significant differences were identified between na2-1 and wild-type siblings. The angles of both the upper and lower leaves were significantly greater for na2-1 plants as compared with wild-type plants in the untreated controls. In addition, the na2-1 mutants exhibited shorter tassels than the wild-type plants under mock conditions. Although tassel branch numbers were lower in the na2-1 mutants than in wild-type plants, the difference was not statistically significant, likely due to a remarkably high sd in tassel branch number in our conditions.
GA3 effects on the single mutant d5 also were scored, and means and sd values are summarized in Table IV. Applying GA3 to d5 and wild-type siblings resulted in a complementation of total plant height (Table IV). The GA3 application increased all internode lengths in d5 (Supplemental Fig. S5). The d5 mutants took longer than the wild type to reach tassel emergence; however, GA3 application resulted in d5 reaching tassel emergence faster than mock treatment, although it did not completely recover delayed flowering time (Table IV). The excess tiller formation in d5 mutants also was complemented by treatment with GA3. Application of GA3 resulted in the development of tassel seeds in 100% of d5 plants, just as it had in wild-type plants (Table IV). This result provides a further test that the suppression of tassel seed production in the na2-1/d5 double mutants was not due to a genetic background effect suppressing tassel seed formation. The GA3 application failed to suppress the expression of anther ear in d5 mutants, perhaps owing to the application to the whorl not delivering GA3 to the ear branch at the time of floral organ specification and arrest. Tassel branches and tassel length were fewer and shorter, respectively, in d5 compared with the wild type with mock treatment. Only tassel length was complemented in d5 mutants upon GA3 treatment, resulting in normal length tassels with fewer branches. The d5 mutants had shorter and wider leaves as compared with the wild type under mock conditions. The leaf lengths were complemented upon GA3 treatment, whereas d5 leaves were narrower than both untreated d5 and wild-type plants. In addition to the GA3-affected phenotypes, the angle of the lower leaf of d5 was significantly more upright than that of the wild type under control conditions and was unaffected by GA3 treatment. All other parameters measured showed no significant difference between GA3 and mock treatment or d5 and wild-type contrasts.
Table IV. Morphometric analysis of d5 treated with GA3.
| Parameter | Mock |
+GA3 |
||
|---|---|---|---|---|
| +/d5 | d5/d5 | +/d5 | d5/d5 | |
| n | 17 | 15 | 16 | 15 |
| Plant heighta | 242.6 ± 28.9 a | 38.8 ± 12.4 b | 301.9 ± 40.9 c | 313.3 ± 56.6 c |
| Days to tassela | 58.4 ± 3.6 a | 71.7 ± 6.7 b | 55.5 ± 6.9 a | 61.6 ± 3.8 c |
| Tillers per planta | 0 ± 0 a | 0.9 ± 0.9 b | 0 ± 0 a,c | 0.2 ± 0.4 c |
| Plants with tassel seedsb | 0 (0%) a | 0 (0%) a | 16 (100%) b | 15 (100%) b |
| Plants with anthers in ear floretsb | 0 (0%) a | 15 (100%) b | 0 (0%) a | 15 (100%) b |
| Tassel branchesa | 21.8 ± 6.1 a | 7.9 ± 5.7 b,c | 16.3 ± 12.0 a,c | 4.6 ± 3.2 b |
| Total nodesa | 14.1 ± 1.1 a | 14.5 ± 1.8 a | 14.3 ± 1.1 a | 13.5 ± 1.0 a |
| Node of top eara | 9.1 ± 0.6 a | 8.7 ± 1.0 a | 9.4 ± 0.7 a | 9.1 ± 1.0 a |
| Tassel lengtha | 61.2 ± 5.1 a | 29.7 ± 3.5 b | 56.3 ± 16.6 a,c | 46.1 ± 11.9 c |
| Angle of upper leafa | 47.1 ± 17.1 a | 47.0 ± 21.1 a | 48.4 ± 13.1 a | 50.7 ± 22.2 a |
| Length of upper leafa | 45.4 ± 5.5 a | 28.4 ± 9.2 b | 49.8 ± 10.8 a | 50.3 ± 10.9 a |
| Width of upper leafa | 5.7 ± 0.6 a | 7.3 ± 1.6 b | 4.4 ± 1.6 c | 3.6 ± 0.8 c |
| Angle of lower leafa | 47.9 ± 8.8 a | 59.3 ± 11.6 b | 47.8 ± 13.3 ab | 55.7 ± 11.0 a,b |
| Length of lower leafa | 105.9 ± 7.6 a | 50.7 ± 7.7 b | 100.0 ± 10.3 a | 92.5 ± 7.7 a |
| Width of lower leafa | 7.1 ± 0.7 a | 9.6 ± 1.2 b | 5.1 ± 1.2 a | 3.8 ± 0.7 c |
Data are presented as means with sd. Lowercase letters indicate connecting letter report as determined by ANOVA with posthoc analysis using the Holm-Sidak algorithm with P < 0.05.
Number of plants with tassel seeds or anthers in ear florets, with the percentage of plants in parentheses. Lowercase letters indicate connecting letter report as determined by Fisher’s exact test with P < 0.01.
DISCUSSION
We report here the molecular cloning of na2, the maize ortholog of the DWF1 gene from Arabidopsis required for the conversion of 24-methylenecholesterol to campesterol in the BR biosynthesis pathway. The phenotype of na2 was largely similar to the previously identified BR biosynthetic mutants of maize na1 and brd1 (Fig. 1; Hartwig et al., 2011; Makarevitch et al., 2012). BR intermediates were measured in the na2-1 mutant and compared with controls (Table I). This identified a block in the conversion of 24-methylenecholesterol to campesterol in the mutant plants. Metabolite data served as an orthogonal data type that complemented a gene-based mapping and allele sequencing approach to confirm the na2 gene identity as one of two DWF1-like sequences in maize. A total of seven alleles of na2 were identified, five of which we also characterized molecularly with respect to the nature of the mutation (Fig. 2). All seven novel alleles of na2 exhibited a phenotype comparable to the original na2 allele identified by H.S. Perry in the 1930s (Emerson et al., 1935).
Many phenotypes observed in na2 and other maize BR-deficient mutants indicated some lack of conservation for the effects of this phytohormone on plant development. The severe dwarfism of na2 was similar to the effects of BR deficiency in other plants and resulted from a shortening of all internodes. The suppression of pistil development observed in maize has not been reported in the Arabidopsis mutants. Also in contrast to the Arabidopsis mutants, we found that the loss of BR biosynthesis downstream of na2 was not required for stomata development in maize (Supplemental Table S2), unlike Arabidopsis, where BR affects the stomatal density and index (Gudesblat et al., 2012; Kim et al., 2012; Wang et al., 2015). This may be because BRs are not required for stomata production in all plants or may be due to an na2-independent production of bioactive BR in maize.
The persistence of pistils in the tassel of na2 mutants required GA biosynthesis (Fig. 4; Table II). We also noted an environmental influence on this phenotype, with the expression of tassel seed production reduced in greenhouse conditions. Phenotype expression was strong in the winter nursery in PR (data not shown), indicating that it was not simply a daylength effect between our field and greenhouse experiments. Temperature, light quality and quantity, and rooting media influence all could be factors.
A phylogenetic analysis was performed using protein-coding sequences similar to Δ24-sterol reductases from 36 plant taxa with fully sequenced genomes. This identified two paralogs each in maize and sorghum. These paralogs are the result of gene duplication prior to the divergence of maize and sorghum (Supplemental Fig. S2). Analysis of mRNA accumulation using the qTeller database found that mRNA from na2 and the orthologous sorghum gene, Sobic.004G277300, were accumulated to at least 3 orders of magnitude greater counts than their paralogs, which showed very low or no detectable accumulation across all tissues and experiments (Supplemental Fig. S4). Analysis of the protein-coding sequences identified multiple frame-shift mutations that disrupted conserved regions of the maize (GRMZM2G455658) and sorghum (Sobic.004G355600) paralogs (Supplemental Fig. S3). These mutations were not shared between maize and sorghum but, rather, arose after the divergence of maize and sorghum. These frame-shift mutations resulted in poor multiple alignments (Supplemental Fig. S3). The presence of multiple independent frame-shift mutations that are likely to disrupt protein function and the observation of very low expression levels suggest that the na2-like subclade may have lost function already or be more prone to loss. In support of this possibility, this subclade is already absent in the genomes of S. italica, rice, and Brachypodium distachyon. It is also possible that the observed frame shifts resulted from poor genome assemblies. We were not able to test this with available EST sequences. The low expression level of the na2-like genes resulted in all EST data present in the National Center for Biotechnology Information being either derived from the na2 orthologs or failing to cover the regions of the frame-shift mutations.
Interactions between BR and GA were demonstrated previously in Arabidopsis and rice (Bai et al., 2012; Li et al., 2012; Stewart Lilley et al., 2013; Tong et al., 2014). We have extended these observations of cross talk affecting plant development to maize. In addition, we have demonstrated many plant organs affected by the interaction between these phytohormones (Table II). The lack of a single conserved mode for the interaction demonstrates that plant height, tiller formation, and reproductive organ development will integrate the information from these two signals in some distinct manner and require the existence of a complex interaction scheme between BR- and GA-deficient mutants.
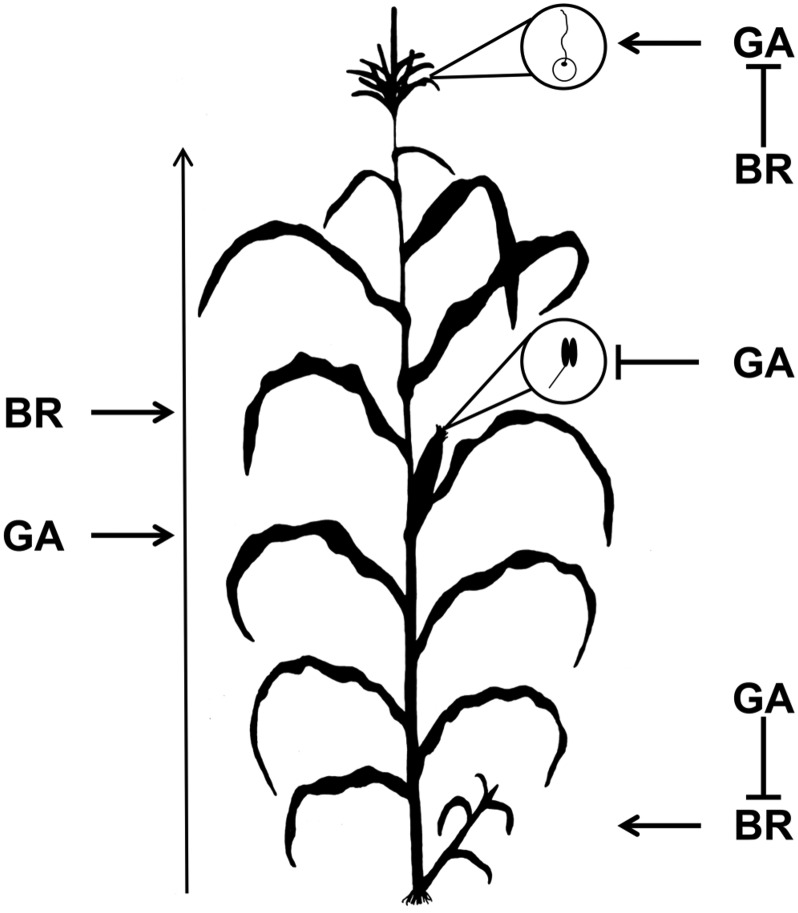
A model for the interactions between BR and GA that is consistent with our genetic interaction data is presented in Figure 5. Loss of d5 or d1 impacted apical dominance and increased tillering, which was suppressed by the loss of na2 or na1 (Table II). Thus, the tiller growth manifested in the absence of GA required BR production, and these experiments provide, to our knowledge, the first evidence in maize for BR tillering regulation. Tillering was shown previously to involve BR in other grasses such as barley (Hordeum vulgare) and rice (Tong et al., 2009; Hussien et al., 2014). Pistil production in the staminate tassel occurs in the na2 and na1 mutants, but mutation of d5 or d1 prevented tassel seed production (Fig. 4; Table II). This demonstrates that tassel seed production in the BR-deficient dwarves required GA. Furthermore, it suggested that the loss of BR results in tassel seed production by the overproduction of GA, similar to the tassel seed produced by sustained GA3 applications performed by Nickerson (1959). GA production loss in the d5 mutants resulted in anther ear development in maize, but unlike tillering, na2 had no effect on this phenotype (Fig. 4; Table II). Thus, the interactions between BR and GA that result in their myriad effects on plant architecture were made up of a diverse set of interaction types. No developmental stage or organ type was predictive of the interaction. For example, the two phytohormones interacted to suppress pistils in the tassel but showed no interaction in stamen arrest in the ear florets, demonstrating that even for reproductive organ suppression, no one mode of interaction governed the outcome. By investigating other BR and GA biosynthetic mutant combinations and showing that the observed phenotypes are consistent with na2/d5, we demonstrate that they are not locus-specific interactions but reflect cross talk between BR and GA deficiency. Also, consistent with our observed dependence of tassel seed formation in na2 and na1 on GA biosynthesis (Table II), GA3 application was able to induce tassel seed formation in d5 and wild-type siblings (Table IV), indicating that excess GA in the tassel primordia was sufficient to block pistil abortion in the tassel florets even in the d5 genetic background.
Figure 5.
Model of the distinct interactions between na2 and d5 in different organs. BR and GA promote stem elongation and overall plant height additively. BR is required for the increase in tillering when GA is lost. GA suppresses anther formation in the ear independent of BR levels. GA is required for the production of pistil development in tassel florets when BR is absent. The vertical arrow at left of the maize plant indicates total plant height; lines with arrows represent positive regulation; lines with blocked arrows represent negative regulation.
GA3 application to na2-1 and d5 allowed us to further elucidate the cross talk between BR and GA (Tables III and IV). GA3 application results in fewer total nodes and a lower terminal ear node in na2-1 but had no such effect on wild-type siblings. Thus, in the absence of BR, GA3 application reduced node number. This reduction in node number was not associated with a decrease in flowering time. The na2-1 mutants flowered later than the wild types in both mock and GA3 treatments. GA3 decreased flowering time for the wild type without changing node number (Table III). Thus, na2-1 treated with GA3 produced fewer nodes over a longer duration. This demonstrates a role for both BR and GA in determining the plastochron time, or time interval between each maize node initiation. The decrease in organ initiation rate by GA3 in the na2-1 plants is the opposite of what one would expect for GA, given the shorter flowering time in wild-type plants. This demonstrates that BR is required for GA to speed up flowering and increase the rate of new organ initiation. This is similar to the gating of GA effects by BR signaling in etiolating Arabidopsis hypocotyls (Bai et al., 2012). This may share a mechanistic relationship with tiller outgrowth, in which BR was required for the loss of GA to induce tiller outgrowth.
One maize BR-deficient phenotype demonstrated in this work is increased leaf angle, as reported previously of BR-deficient mutants in rice (Hong et al., 2004). Upright leaves are an agronomically important trait that contributed to increased planting densities and yield increases achieved by modern maize hybrids (Duvick, 2005). The na2-1 mutants leaves were more upright than those of wild-type controls. Potentially contributing to this phenotype, the highest expression of NA2 was in young developing leaf collars (Fig. 3). NA2 mRNA accumulation also was relatively high in the shoot meristem and developing stem, with lower accumulation in the leaf blade (Fig. 3). The abundant expression of this gene raises the possibility that manipulation of BR levels can be used for crop improvement. We demonstrated that we can simultaneously reduce plant height and increase leaf angle. This could both improve harvest index and allow for greater maize planting densities. GA3 treatment to na2-1 plants resulted in further increases to leaf angles (Table III). Thus, BR reduction sensitized the leaf collar to GA3 treatment. In contrast, it was shown in rice that GA inhibited lamina inclination with accumulated BR or enhanced BR signaling (Tong et al., 2014). Leaf length also was strongly dependent on BR and GA, but in this case, GA only increased leaf length in the absence of BR. This should be interpreted with caution, however, as treatment with GA3 was performed by application into the whorl, which may deliver GA3 to the site of leaf primordia initiation, young leaves during expansion, and meristems but not over the entire developmental phase of the leaf or post intercalary meristem expansion of the blade. The observations of elongated leaves in na2-1 suggest that defects in growth established early in leaf development by a loss of BR can be partially overcome by GA3 applications.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All of the maize (Zea mays) genetic stocks used in this study are summarized in Supplemental Table S3. The na2-506D and na2-518C stocks were obtained from the MaizeCOOP as lines 506D and 518C, respectively. The na2-1 and na2-7 alleles were obtained from the MaizeCOOP as lines 4407D d*-N2374 and 4405B d*-N1074C, respectively. na2-2 (PB03NM417), na2-3 (04INW22CW04351), na2-4 (04IAB73PS020G1), and na2-6 (04CAB73SH0365) were identified in an EMS population as M3 generation plants in the summer of 2011. na2-5 was identified as an F1 plant in a Mu-tagging screen of approximately 120,000 plants crossed with na2-1 in the summer of 2012. Seeds of the d5 mutant were obtained from the Carolina Biological Supply Company as item 177110. The d1 allele used was obtained from the MaizeCOOP as line 302A d1-6016. The na1-1 allele (Hartwig et al., 2011) corresponds to line 310G in the Maize COOP. The GRMZM2G057000 locus was amplified from the na2-1 to na2-5 alleles using primers described in Supplemental Table S4. PCR products were sequenced using the Sanger method (Sanger et al., 1977; Macrogen USA).
Double mutant analyses were conducted on plants grown in the field in West Lafayette, Indiana, at the Agronomy Center for Research and Education in the summer of 2013 (na2/d5) and 2014 (na2/d5, na2/d1, and na1/d1). All greenhouse experiments were conducted in the Purdue Horticulture Plant Growth Facility using temperature settings of 27°C (day) and 21°C (night) with a 16-h daylength provided by supplemental lighting, as needed. Plants were fertilized with 200 ppm Miracle-Gro Excel (Scotts) adjusted to pH 6 following the manufacturer’s recommendations.
The plant tissue used for qRT-PCR analysis of gene expression in dark-grown material was collected from 5-DAG etiolated plants of B73 grown in type 601 flat trays filled with Turface MVP (Profile Products) as the potting substrate. Plants were grown in complete darkness in a Conviron E8 growth chamber at 27°C. The material sampled included a 0.25-cm segment containing the first stem node, mesocotyl tissue, coleoptile tissue, and the true leaves. Light-grown tissues for qRT-PCR analysis of gene expression were sampled from 9-DAG seedlings in Turface incubated in the greenhouse conditions described above. Material collected included the shoot meristem and surrounding tissue, roots, the leaf collar of the third leaf, and the third leaf blade cut through the intercalary meristem. Nonseedling tissues collected from light-grown plants were collected from plants grown in the field. These included the uppermost stem segments from V7-stage plants that were 1 cm in length, immature ears 0.5 to 2.5 cm in length at the time of harvest, mature ears collected at silk emergence from the husks, immature tassels 0.5 to 2.5 cm in length at the time of harvest, and mature tassels harvested at anthesis.
Leaf Imprints for Microscopic Analysis of Stomata Development
Wild-type, na2-1, and na2-2 families were grown for 28 DAG (V5 stage) in the greenhouse under the conditions stated previously. Epidermal cell layer imprints were produced from the widest portion of the fifth leaf using ethyl 2-cyanoacrylate (Scotch SuperGlue; 3M) applied to the leaves and then pressed onto and recovered on glass microscope slides (Thermo Fisher Scientific). Imprints of the adaxial and abaxial leaf surfaces were taken and observed by light microscopy using a UNICO H606T microscope (United Products & Instruments). Stomatal indices and densities were calculated for each imprint by counting all the cells in five randomly selected viewable areas in each imprint at 40× magnification. Only areas between vasculature tissues from high-quality imprints where all cell boundaries were completely visible were considered. For each genotype, mean parameters were calculated from five viewable areas from each of six plants per genotype.
Quantification of BR Intermediates
For internal BR measurements, wild-type and na2-1 siblings were grown for 28 d in the greenhouse, and shoot tissue was harvested and immediately frozen in liquid nitrogen. Samples were then lyophilized using the VirTis 2KBTES-55 freeze dryer (SP Scientific) for 3 d at −78°C and 0.53 Pa. Purification and quantification of BR was performed as described (Fujioka et al., 2002). A summary of the two independent experiments is shown in Supplemental Table S5.
Phylogenetic Analyses and in Silico Analysis of Expression Data
GRMZM2G057000 protein homolog DNA-coding sequences were obtained using the Inparanoid method (O’Brien et al., 2005) through Phytozome version 10.2.2 (Goodstein et al., 2012) with a Dual Affine Smith-Waterman alignment score greater than 1,500, and a sequence similarity of greater than 50% resulted in a total of 57 sequences from 36 taxa. All sequences were downloaded using BioMart (Smedley et al., 2015) and loaded into Mesquite version 3.03 (Maddison and Maddison, 2015), where they were aligned using ClustalW version 2.1 (Larkin et al., 2007). FastTree version 2.1.7 (Price et al., 2010) was used to create an approximate maximum-likelihood phylogenetic tree from the aligned sequences. The maximum-likelihood tree was used as a starting tree for MrBayes version 3.2.5 (Ronquist et al., 2012) to infer a Markov chain Monte Carlo (Geyer, 1991) phylogenetic tree using a general time reversible model with a proportion of invariable sites and a γ-shaped distribution of rates across sites model with a variable partition rate (rates varied under a flat Dirichlet prior). The analysis was run for 10 million generations.
FPKM values and histograms were obtained from qteller.com for selected maize and sorghum (Sorghum bicolor) genes (Wang et al., 2009; Li et al., 2010; Davidson et al., 2011, 2012; Dugas et al., 2011; Schnable and Freeling, 2011; Waters et al., 2011; Bolduc et al., 2012; Chang et al., 2012; Kakumanu et al., 2012; Mizuno et al., 2012). Recreation of these histograms could be obtained at http://www.qteller.com.
qRT-PCR
Total RNA was isolated as described (Sekhon et al., 2011) from various B73 tissues and treated with DNaseI (Thermo Fisher Scientific). Complementary DNA was synthesized using reverse transcriptase (iScript; Bio-Rad) according to the manufacturer’s instructions. All primers used for qRT-PCR analysis are described in Supplemental Table S4 and were designed using PrimerExpress software (Applied Biosystems, Invitrogen). All primers used showed 90% to 110% efficiency at 500 nm concentration when conducting complementary DNA dilution standard curves using StepOne software (Applied Biosystems, Invitrogen) for amplification efficiency (Taylor et al., 2010). The molybdenum cofactor biosynthesis gene (GRMZM2G067176) was used as the internal control (Sekhon et al., 2011). The StepOnePlus (Applied Biosystems, Invitrogen) instrument was used as described (Orlova et al., 2009) to conduct qRT-PCR.
GA3 Experiments
Maize plants treated with GA3 were grown under greenhouse conditions as stated previously. Plants were grown in 2-gallon pots with a 2:1 mixture of Turface MVP (Profile Products) to peat germinating mix (Conrad Fafard). GA3 (Gold Biotechnology) at 866 µm in water with a final concentration of 0.02% ethanol and 0.005% Silwet L-77 (Sigma-Aldrich) or a mock preparation of ethanol and Silwet L-77 without GA was applied to the leaf whorl every third day, starting 25 d after planting until tassel emergence. All measurements were taken at maturity.
Statistical Analyses
Microsoft Excel and the add-in XL Toolbox (version 6.50; http://xltoolbox.sourceforge.net) was used for statistical analysis. ANOVA for sets of data groups was performed with multiple comparisons/post hoc testing within XL Toolbox. If a significant difference (P < 0.05) was detected, post hoc tests using a Holm-Sidak correction for multiple tests (Zar, 2010) were performed. The Microsoft Excel add-in Real Statistics Resource Pack (version 3.3.1; http://real-statistics.com) was used for hypothesis testing on binary data. The Real Statistics Resource Pack formula for Fisher’s exact test was used to determine significant differences (P < 0.01) between samples, and then multiple comparisons were conducted between data groups (Zar, 2010).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: KX443688 (na2-1), KX443689 (na2-2), KX443690 (na2-3), KX443691 (na2-4), and KX443692 (na2-5).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Proposed BR pathway and measured intermediates from na2-1 and wild-type plants.
Supplemental Figure S2. Phylogenetic tree of presumed Δ24-sterol reductases from 36 taxa.
Supplemental Figure S3. Multiple sequence alignment of maize NA2 protein sequence with orthologous sequences.
Supplemental Figure S4. RNA expression levels of maize or sorghum genes from the qTeller data set.
Supplemental Figure S5. Internode lengths of na2-1, d5, and wild-type siblings with and without GA3 application.
Supplemental Table S1. Segregation ratios of crosses.
Supplemental Table S2. Stomatal indices and densities of na2-1, na2-2, and wild-type plants.
Supplemental Table S3. Sources of maize genetic stocks.
Supplemental Table S4. Primers used in this study.
Supplemental Table S5. Two independent experiments showing BR intermediates in wild-type and na2-1 plants.
Supplementary Material
Acknowledgments
We thank Jim Beaty and the Purdue University Agronomy Farm for expert assistance as well as Rob Eddy and the greenhouse team for the Horticulture Greenhouse facility for the production of high-quality growth environments; Johal laboratory members A. DeLeon, R. Zhan, and K. Chu for help with chemical treatments; and Dilkes laboratory members E. Buescher, C. Addo-Quaye, and M. Silva Guzman for reading early drafts of the article and returning comments.
Glossary
- BR
brassinosteroid
- JA
jasmonic acid
- EMS
ethyl methanesulfonate
- DAG
days after germination
- FPKM
fragments per kilobase of transcript per million mapped reads
Footnotes
This work was supported by the National Science Foundation (EAGER grant no. 1450341 to B.P.D. and CAREER grant no. 1054918 to B.S.).
Articles can be viewed without a subscription.
References
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265 [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Liu WY, Shih AC, Shen MN, Lu CH, Lu MY, Yang HW, Wang TY, Chen SC, Chen SM, et al. (2012) Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Bloom AJ, Field CB, Waring RH (1987) Plant responses to multiple environmental factors. Bioscience 37: 49–57 [Google Scholar]
- Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, McCarty DR, Tan BC (2014) The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol 166: 2028–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon J, Fujioka S, Dilkes BP, Choe S (2013) Brassinosteroids regulate plant growth through distinct signaling pathways in Selaginella and Arabidopsis. PLoS ONE 8: e81938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al. (1999) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S (2007) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517–1521 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Buell CR (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J 71: 492–502 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. (2011) Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- DeLong A, Calderon-Urrea A, Dellaporta SL (1993) Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74: 757–768 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–R373 [DOI] [PubMed] [Google Scholar]
- Dugas DV, Monaco MK, Olsen A, Klein RR, Kumari S, Ware D, Klein PE (2011) Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 12: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. (2005) The contribution of breeding to yield advances in maize (Zea mays L.). Adv Agron 86: 83–145 [Google Scholar]
- Eckardt NA. (2002) Foolish seedlings and DELLA regulators: the functions of rice SLR1 and Arabidopsis RGL1 in GA signal transduction. Plant Cell 14: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving DC, Visser DB, Henry JL (2011) Gibberellins stimulate lateral branch development in young sweet cherry trees in the orchard. Int J Fruit Sci 11: 41–54 [Google Scholar]
- Emerson RA. (1912) The inheritance of certain “abnormalities” in maize. Am Breeders Assoc Annu Rep 7/8: 385–399 [Google Scholar]
- Emerson RA, Beadle GW, Fraser AC (1935) A summary of linkage studies in maize. Cornell Univ Agric Exp Stn Mem 1–83
- Evans MM, Poethig RS (1995) Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol 108: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CJ. (1991) Markov chain Monte Carlo maximum likelihood. In Keramidas M, ed, Computing Science and Statistics: Proceedings of the 23rd Symposium on the Interface. Interface Foundation of North America, Fairfax Station, VA, pp 156–163 [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Hamant O, Traas J (2010) The mechanics behind plant development. New Phytol 185: 369–385 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DPV, Choe S, Johal GS, Schulz B (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108: 19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phinney BO (1979) Comparison of ent-kaurene and ent-isokaurene synthesis in cell-free systems from etiolated shoots of normal and dwarf-5 maize seedlings. Phytochemistry 18: 1475–1479 [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Matsuoka M (2004) Brassinosteroids and rice architecture. J Pestic Sci 29: 184–188 [Google Scholar]
- Hussien A, Tavakol E, Horner DS, Muñoz-Amatriaín M, Muehlbauer GJ, Rossini L (2014) Genetics of tillering in rice and barley. Plant Genome 7: 1–20 [Google Scholar]
- Irish EE, Langdale JA, Nelson TM (1994) Interactions between tassel seed genes and other sex-determining genes in maize. Dev Genet 15: 155–171 [Google Scholar]
- Jaillais Y, Chory J (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kwon M, Jeon J, Schulz B, Corvalán C, Jeong YJ, Choe S (2014) The Arabidopsis gulliver2/phyB mutant exhibits reduced sensitivity to brassinazole. J Plant Biol 57: 20–27 [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH (1998) The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW (2005) Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Li QF, Wang C, Jiang L, Li S, Sun SSM, He JX (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal 5: ra72–ra72 [DOI] [PubMed] [Google Scholar]
- Lyons E, Freeling M (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J 53: 661–673 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (2015) Mesquite: a modular system for evolutionary analysis, version 3.03. http://mesquiteproject.org
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 7: e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Kawahigashi H, Kawahara Y, Kanamori H, Ogata J, Minami H, Itoh T, Matsumoto T (2012) Global transcriptome analysis reveals distinct expression among duplicated genes during sorghum-Bipolaris sorghicola interaction. BMC Plant Biol 12: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33: 887–898 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nickerson NH. (1959) Sustained treatment with gibberellic acid of five different kinds of maize. Ann Mo Bot Gard 46: 19–37 [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer EL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova I, Nagegowda DA, Kish CM, Gutensohn M, Maeda H, Varbanova M, Fridman E, Yamaguchi S, Hanada A, Kamiya Y, et al. (2009) The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta. Plant Cell 21: 4002–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orshan G. (1986) Plant form as describing vegetation and expressing adaptation to environment. Ann Bot (Lond) 44: 7–38 [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2010) FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo NJ, Vriezen WH, Beemster GT, Van Der Straeten D (2003) Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J 33: 989–1000 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Salamini F. (2003) Hormones and the green revolution. Science 302: 71–72 [DOI] [PubMed] [Google Scholar]
- Salas Fernandez MG, Becraft PW, Yin Y, Lübberstedt T (2009) From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14: 454–461 [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M (2009) Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 5: 301–307 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schmitz AJ, Folsom JJ, Jikamaru Y, Ronald P, Walia H (2013) SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol 198: 1060–1070 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. (2015) The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43: W589–W598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart Lilley JL, Gan Y, Graham IA, Nemhauser JL (2013) The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J 76: 165–173 [DOI] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM (2001) The evolution of plant architecture. Curr Opin Plant Biol 4: 33–37 [DOI] [PubMed] [Google Scholar]
- Suttle A. (1924) The genetic interrelations of different types of dwarf corn. PhD thesis. Cornell University, Ithaca, NY [Google Scholar]
- Tatusova TA, Madden TL (1999) BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M (2010) A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50: S1–S5 [DOI] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28: 463–487 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol 2: 323–325 [Google Scholar]
- Wang M, Yang K, Le J (2015) Organ-specific effects of brassinosteroids on stomatal production coordinate with the action of Too Many Mouths. J Integr Plant Biol 57: 247–255 [DOI] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng YQ, Wu Y (2011) Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68: 249–261 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27: 681–688 [DOI] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil CF. (2009) TILLING in grass species. Plant Physiol 149: 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H, Jürgens G (2009) Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Zar J. (2010) Biostatistical Analysis, Ed 5 Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.