SOB3 modulates seedling elongation by repressing the expression of genes associated with auxin signaling.
Abstract
Developing seedlings are well equipped to alter their growth in response to external factors in order to maximize their chances of survival. SUPPRESSOR OF PHYTOCHROME B4-#3 (SOB3) and other members of the AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) family of transcription factors modulate the development of Arabidopsis (Arabidopsis thaliana) by repressing hypocotyl elongation in young seedlings growing in light. However, the molecular mechanism behind how AHLs influence seedling development is largely unknown. We have identified genes associated with auxin-mediated hypocotyl elongation as downstream targets of SOB3. We found that YUCCA8 (YUC8) as well as members of the SMALL AUXIN UP-REGULATED RNA19 (SAUR19) subfamily were down-regulated in the short-hypocotyl, gain-of-function SOB3-D mutant and up-regulated in the dominant-negative, tall-hypocotyl sob3-6 mutant. SOB3-D and sob3-6 hypocotyls also exhibited altered sensitivity to the polar auxin transport inhibitor N-1-napthylphthalamic acid, suggesting a critical connection between auxin and the modulation of seedling elongation by SOB3. Finally, we found that overexpression of GREEN FLUORESCENT PROTEIN-SAUR19 in the SOB3-D line partially rescued defects in hypocotyl elongation, and SOB3 bound directly to the promoters of YUC8 and SAUR19 subfamily members. Taken together, these data indicate that SOB3 modulates hypocotyl elongation in young seedlings by directly repressing the transcription of genes associated with auxin signaling.
Strict regulation of development is critical for the survival of plants and other living organisms. However, rather than blindly following an endogenous road map for development, an organism must be able to integrate external signals with internal ones. This enables growth to be altered based on environmental conditions, maximizing chances of survival for the species. In particular, plants, as sessile organisms, are well equipped to alter their growth in response to external cues (for review, see Braidwood et al., 2014). For example, seedlings have the ability to substantially alter their developmental program in response to changing external conditions, such as light intensity, making them especially appropriate for studying how plants modulate their growth in response to various environmental signals (for review, see Arsovski et al., 2012; Boron and Vissenberg, 2014). Seedlings growing in darkness, such as those that have not yet emerged from soil following germination, devote most of their stored energy reserves to stem elongation in an attempt to rapidly reach sunlight at the soil surface, which is necessary for photosynthesis. This developmental program is known as skotomorphogenesis. In contrast, upon light exposure, seedlings switch to photomorphogenesis, diverting most of their energy to other processes, including chloroplast development, root growth, cotyledon expansion, and the production of true leaves. Skotomorphogenesis, photomorphogenesis, and other developmental processes are directed to a large degree by transcription factors, which connect hormone and other signal transduction pathways with the regulation of specific genes important for directly altering plant growth and architecture (for review, see Neff et al., 2000; Kaufmann et al., 2010; Arsovski et al., 2012). Research exploring the connections between transcription factors and development in plants is important because it unlocks the potential to directly manipulate these genes in order to improve crop productivity.
One group of genes coding for transcription factors that influence development specifically in plants is the AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) family (Fujimoto et al., 2004; Zhao et al., 2014). These transcription factors are characterized by the presence of two conserved regions, which are both essential for function: one or two DNA-binding AT-hook motifs, and a single plant and prokaryotic conserved (PPC) domain that is important for protein-protein interactions and nuclear localization (Fujimoto et al., 2004; Street et al., 2008; Zhao et al., 2013, 2014). Previous studies on AHLs, specifically those in the 29-member Arabidopsis (Arabidopsis thaliana) family (Fujimoto et al., 2004; Matsushita et al., 2007; Street et al., 2008; Zhao et al., 2013, 2014), have implicated some of these proteins in the direct regulation of downstream targets that influence various developmental processes. AHL25/AT-HOOK PROTEIN OF GIBBERELLIN FEEDBACK REGULATION1 binds to the promoter of the GA biosynthesis gene AtGA3Ox1 and helps maintain proper expression of the gene in response to feedback signals (Matsushita et al., 2007). The development of floral organs is influenced by AHL21/GIANT KILLER, which binds to and directly represses the expression of genes important for these processes, including AUXIN RESPONSE FACTOR3 (ARF3), JAGGED, KNUCKLES, and CRABS CLAW (Ng et al., 2009). AHL22 binds to an intragenic AT-rich region in FLOWERING LOCUS T, a gene that promotes flowering, and represses its expression (Yun et al., 2012). AHL16/TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK binds to an AT-rich region located in an intron of FLOWERING LOCUS C and the promoter of FLOWERING WAGENINGEN, both inhibitors of flowering, and represses the expression of both genes (Xu et al., 2013). Additional work on AHL16 has demonstrated that it can act not only as a repressor but also as an activator of transcription. AHL16 binds to AT-rich promoters of genes encoding arabinogalactan proteins, including AGP6, AGP11, AGP23, and AGP40, and activates their expression during the development of the pollen wall (Lou et al., 2014; Jia et al., 2015).
However, despite these recent findings, the downstream targets important for most developmental processes known to be influenced by AHLs have yet to be investigated. One notable example is their well-established role in modulating hypocotyl growth (Street et al., 2008; Xiao et al., 2009; Zhao et al., 2013). AHL29 was initially identified as SUPPRESSOR OF PHYTOCHROME B4-#3 DOMINANT (SOB3-D) in an activation-tagging screen, where it was found that enhanced expression of this gene suppresses the tall-hypocotyl phenotype characteristic of the photoreceptor mutant phyB-4 (Street et al., 2008). A role for SOB3, its closest homolog ESCAROLA (ESC), and other AHLs in repressing hypocotyl elongation in light-grown seedlings was confirmed subsequently through rigorous loss-of-function studies (Street et al., 2008; Xiao et al., 2009; Zhao et al., 2013). Additionally, three lines of evidence indicate that SOB3 represses hypocotyl elongation through its action as a transcription factor. First, SOB3 mutant alleles that either lack the AT-hook motif entirely or fail to bind DNA due to a single missense mutation (sob3-6 allele) behave in a dominant-negative fashion when expressed in Arabidopsis, producing an extremely tall-hypocotyl phenotype (Street et al., 2008; Zhao et al., 2013). Additionally, the TCP family of plant-specific transcription factors plays important roles in a variety of developmental processes (Cubas et al., 1999; Palatnik et al., 2003; Koyama et al., 2010; Danisman et al., 2012; Uberti-Manassero et al., 2012; Tao et al., 2013; Lucero et al., 2015; for review, see Manassero et al., 2013) and interacts with SOB3, which is essential for the repression of hypocotyl growth (Zhao et al., 2013). Finally, the extremely tall-hypocotyl phenotype characteristic of sob3-6 is completely abolished in the jaw-D background, where the expression levels of several TCP transcription factors are reduced due to enhanced expression of a microRNA important for posttranscriptional regulation of these genes (Palatnik et al., 2003; Zhao et al., 2013). These data strongly indicate that SOB3 represses hypocotyl elongation via its activity as a transcription factor.
Despite the well-established role of SOB3 in hypocotyl growth, downstream targets of SOB3 are unknown. Thus, in order to begin to understand the mechanism by which SOB3 modulates hypocotyl growth in light-grown seedlings, we sought to identify downstream targets important for this developmental process. We report the identification of several genes associated with auxin signaling and temperature-induced hypocotyl elongation that are strong candidates for being directly repressed at the transcriptional level by SOB3.
RESULTS
Genes Associated with Auxin Signaling Are Misregulated in SOB3-D and sob3-6
To identify candidate downstream targets of SOB3 important for its role in modulating hypocotyl elongation, we used RNA sequencing (RNA-seq) as a screen for genes misexpressed in SOB3 mutants. For this experiment, cDNA was generated from 4-d-old seedlings of Columbia-0 (Col-0), SOB3-D, and sob3-6, as we observed that hypocotyl phenotypes began to be apparent for both mutants around this time (Supplemental Fig. S1, A and B). Hierarchical clustering of the RNA-seq data indicated that perturbations in gene expression were much greater in SOB3-D than in sob3-6, as compared with the wild type (Supplemental Fig. S2). This was in agreement with our observation that SOB3-D had a much greater overall impact on plant development than sob3-6 (Supplemental Figs. S1, 3A, and S4). In order to screen for genes exhibiting expression changes correlating with hypocotyl growth, we calculated Pearson correlation coefficient (r) values for every gene using RPKM (reads per kilobase of exon model per million mapped reads) values and hypocotyl lengths for each genotype. For this analysis, we excluded any genes that exhibited less than a 2-fold change in expression between SOB3-D and sob3-6, as well as any genes for which transcript was not detected in the wild type. We then used PLAZA 3.0 software to perform Gene Ontology enrichment analysis on all genes for which an r value of greater than 0.8 or less than −0.8 was calculated (Proost et al., 2015). One group of enriched genes in this screen, annotated Response to Auxin, caught our interest (Supplemental Table S1).
This enriched group of genes associated with the auxin response seemed likely to be relevant to our study, since auxin signaling plays an important role in hypocotyl elongation, particularly in light-grown seedlings (Jensen et al., 1998), and SOB3 seems to be important for modulating hypocotyl elongation, specifically in light-grown seedlings (Street et al., 2008; Zhao et al., 2013). Within the group of auxin response genes identified here, we noticed that many members of the SMALL AUXIN UP-REGULATED (SAUR) gene family were included (Table I). SAUR genes, of which there are 79 functional members in Arabidopsis, are generally induced in response to auxin (Hagen and Guilfoyle, 2002; Spartz et al., 2012; for review, see Ren and Gray, 2015). SAURs were generally repressed in SOB3-D but up-regulated in sob3-6, as indicated by the positive r values for 21 of the 26 SAUR genes in this list. In addition, we also performed an RNA-seq screen using cDNA generated from 6-d-old Col-0 and SOB3-D seedlings. Among the genes exhibiting at least a 5-fold difference in expression between the two genotypes, Response to Auxin was again enriched as a Gene Ontology term (Supplemental Table S2). Also in agreement with the first RNA-seq screen, many of the genes included in this group were SAUR genes, and most of them (25 of 29) were expressed at lower levels in SOB3-D as compared with Col-0 (Supplemental Table S3).
Table I. SAUR genes for which expression levels at 4 d correlate with hypocotyl phenotypes in SOB3 mutants.
Pearson correlation coefficient (r) values were calculated based on hypocotyl phenotypes at 6 d (Supplemental Fig. S1A) and RNA-seq RPKM values at 4 d (Supplemental Data Set S1) for SOB3-D, sob3-6, and Col-0 in seedlings grown under constant dim white light. Fold changes were calculated based on RPKM values from the same data set. SAUR genes for which an r value of greater than 0.8 or less than −0.8 are shown. Asterisks indicate genes that were repressed in SOB3-D based on the RNA-seq screen from 6-d-old seedlings and/or qRT-PCR data from 6-d-old seedlings (Supplemental Table S3; Supplemental Fig. S5).
| Name | r | Fold Change (sob3-6/SOB3-D) |
|---|---|---|
| SAUR45 | −1.00 | 0.17 |
| SAUR12* | −0.98 | 0.12 |
| SAUR34 | −0.95 | Not expressed in sob3-6 |
| SAUR69 | −0.93 | 0.04 |
| SAUR71* | −0.82 | 0.35 |
| SAUR20* | 0.81 | 2.48 |
| SAUR19* | 0.84 | 4.84 |
| SAUR61 | 0.88 | 6.28 |
| SAUR42* | 0.91 | Not expressed in SOB3-D |
| SAUR64 | 0.92 | 18.84 |
| SAUR6 | 0.92 | 2.19 |
| SAUR21* | 0.95 | 2.68 |
| SAUR41 | 0.96 | Not expressed in SOB3-D |
| SAUR68* | 0.97 | Not expressed in SOB3-D |
| SAUR28* | 0.97 | 17.79 |
| SAUR1* | 0.97 | 8.37 |
| SAUR7* | 0.97 | 8.37 |
| SAUR23* | 0.99 | 8.11 |
| SAUR29 | 0.99 | 12.56 |
| SAUR65 | 1.00 | 3.37 |
| SAUR22* | 1.00 | 28.78 |
| SAUR13* | 1.00 | Not expressed in SOB3-D |
| SAUR66* | 1.00 | 4.67 |
| SAUR24 | 1.00 | 17.27 |
| SAUR76 | 1.00 | 7.32 |
| SAUR63* | 1.00 | 3.78 |
In particular, one subclade of SAURs, the SAUR19 subfamily, which includes six members, SAUR19 to SAUR24 (Jain et al., 2006), was of particular interest for further analysis. All six of these genes showed a strong positive correlation with hypocotyl phenotypes based on the RNA-seq data from 4-d-old seedlings (Table I), and all but one of them, SAUR24, also were identified in the second RNA-seq screen using 6-d-old seedlings (Supplemental Table S3). Another reason SAUR19 to SAUR24 were of particular interest is because they have documented roles in promoting hypocotyl elongation (Franklin et al., 2011; Spartz et al., 2012, 2014). Additionally, it has been demonstrated that members of this subfamily are misregulated in tcp3 and tcp4 mutants (Koyama et al., 2010; Sarvepalli and Nath, 2011), and physical interactions with TCP4 have been linked to SOB3’s role in modulating hypocotyl growth (Zhao et al., 2013).
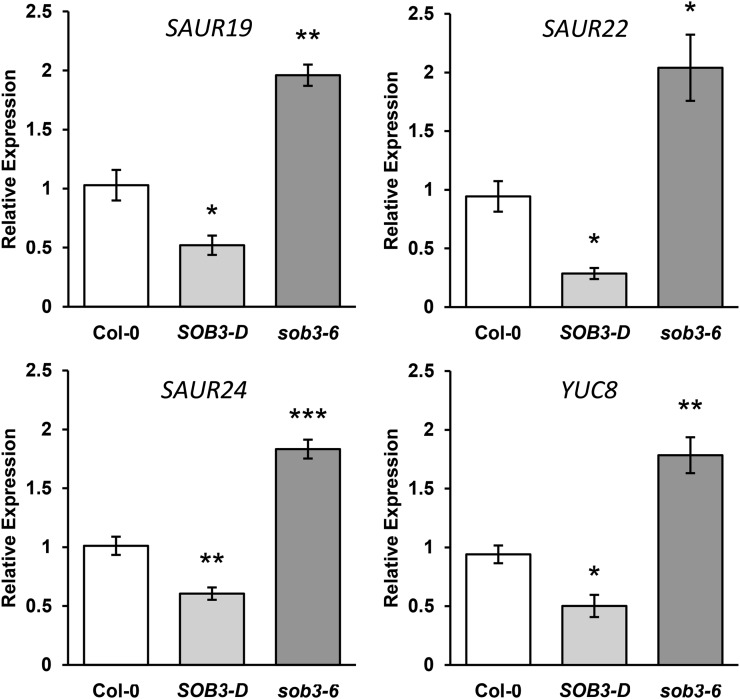
To further evaluate expression levels in the SOB3 mutants for three members of the SAUR19 subfamily, SAUR19, SAUR22, and SAUR24, we performed quantitative reverse transcription (qRT)-PCR using an independent set of cDNA preparations from 4-d-old seedlings grown in dim white light. These results confirmed that transcripts of SAUR19 subfamily members are depleted in SOB3-D and elevated in sob3-6 (Fig. 1). Also based on qRT-PCR analysis, the expression of several SAUR genes, including members of the SAUR19 subfamily, were down-regulated in SOB3-D seedlings grown for 6 d in white light or for 7 d in far-red light, further suggesting that SOB3 represses the transcription of these genes (Supplemental Figs. S5 and S6). However, we did not observe increased SAUR expression in sob3-6 seedlings at these later time points, which is not surprising considering that this mutant only exhibited enhanced hypocotyl growth compared with the wild type prior to day 6 (Supplemental Fig. S1). Therefore, the reduction of SAUR levels observed in 6-d-old SOB3-D seedlings likely results from ectopic expression of SOB3 artificially lengthening the time frame within which this protein functions as a repressor, mimicking the prolonged manner in which this allele impacted Arabidopsis development as compared with sob3-6 (Supplemental Figs. S1, 3A, and S4).
Figure 1.
Auxin-associated genes are repressed in SOB3-D but induced in sob3-6 at 4 d. Relative expression is shown for genes associated with auxin signaling in Col-0 and homozygous SOB3-D and sob3-6 seedlings, as determined by qRT-PCR. Transcript levels are normalized based on the expression of the MDAR4 housekeeping gene. PCR was performed in triplicate, and average expression values were calculated and used for analysis. All values are shown as fold change compared with the wild type. Error bars represent se from four biological replicates. In a Welch’s t test (unpaired two-tailed t test with unequal variance) compared with the wild type, *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Changes in SAUR gene expression in SOB3-D and sob3-6 could be the result of either direct or indirect repression by SOB3. One possibility is that changes in SAUR levels could result from altered auxin signaling in the mutants, perhaps due to differences in levels of the hormone. This is especially possible considering that SAUR19 to SAUR24 are all known to be induced by auxin (Spartz et al., 2012). The rate-limiting step in what is thought to be the main auxin biosynthetic pathway in plants is catalyzed by a class of flavin monooxygenases coded for by the 11 members of the YUCCA (YUC) family of genes (Zhao et al., 2001; Hofmann, 2011; Mashiguchi et al., 2011; Phillips et al., 2011; Stepanova et al., 2011; Won et al., 2011; Dai et al., 2013; for review, see Zhao, 2014). With this in mind, we examined data from both RNA-seq screens to evaluate if any YUC genes are misexpressed in the SOB3 mutants. Only one YUC gene, YUC8, fit our screening criteria for both RNA-seq data sets, correlating with hypocotyl phenotypes based on the expression values from 4-d-old seedlings (r = 0.99) and being repressed more than 5-fold in SOB3-D at 6 d (Supplemental Fig. S1A; Supplemental Data Sets S1 and S2). This indicated that YUC8 also could be a downstream target of SOB3. Indeed, when we examined transcript levels for YUC8 in 4-d-old Col-0, SOB3-D, and sob3-6 seedlings using qRT-PCR, we found that its expression pattern was very similar to that of SAUR19, SAUR22, and SAUR24 (Fig. 1). We also checked the expression patterns of both SAUR22 and YUC8 using cDNA generated from 5-d-old seedlings and observed similar results, providing further evidence that SOB3 causes downstream repression of both SAUR19 family members and YUC8 in seedlings less than 6 d old (Supplemental Fig. S7).
Auxin Signaling Is Important for SOB3 Mutant Hypocotyl Phenotypes
Since SAUR and YUC genes are both associated with auxin, we sought to test the hypothesis that SOB3 impacts hypocotyl growth by acting on this signaling pathway. With this aim in mind, we generated dose-response curves for seedlings grown in the presence of exogenous indole-3-acetic acid (IAA). Although endogenous auxin is generally thought to promote hypocotyl growth, exogenous IAA usually inhibits hypocotyl growth in light-grown wild-type seedlings, perhaps because the auxin response becomes saturated (Collett et al., 2000). However, mutant seedlings with alterations in auxin levels or auxin perception can exhibit resistance to such treatment or even display the complete opposite response, elongating in the presence of exogenous IAA. When we examined the responses of the SOB3 mutants to exogenous IAA, we found that SOB3-D responded similarly to the wild type (Supplemental Fig. S8). On the other hand, sob3-6 seemed to be slightly more sensitive than the wild type at higher concentrations of IAA.
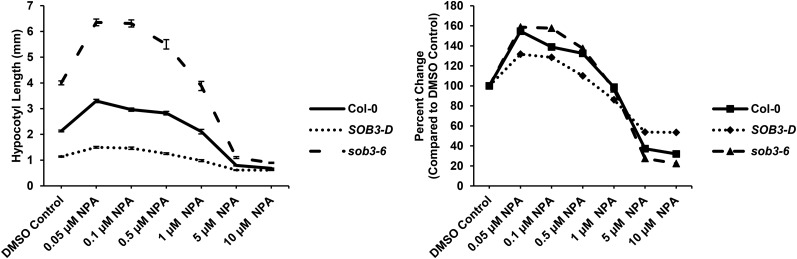
We also examined the effect of N-1-napthylphthalamic acid (NPA) on SOB3-D and sob3-6. NPA inhibits polar auxin transport, leading to altered hypocotyl elongation in light-grown seedlings while having very little effect on hypocotyl growth in dark-grown seedlings (Jensen et al., 1998). In our conditions, NPA promoted hypocotyl elongation at low concentrations, while at high concentrations, it inhibited elongation (Fig. 2). Furthermore, in contrast to the situation observed for IAA, both mutants clearly had different responses to NPA treatment as compared with the wild type. SOB3-D was less sensitive to both the inhibition and promotion of elongation by NPA when compared with the wild type, whereas, sob3-6 exhibited the opposite trend, showing greater overall sensitivity to NPA than the wild type. Furthermore, high concentrations of NPA almost completely eliminated differences in hypocotyl elongation between the three genotypes. These results indicate that there is a critical link between auxin signaling and the hypocotyl phenotypes observed in SOB3 mutants.
Figure 2.
SOB3 mutants exhibit altered sensitivity to NPA. Hypocotyl dose-response curves are shown for 6-d-old seedlings grown in dim white light on LS medium. Values represent means of either the actual measured hypocotyl length (left) or the sensitivity to NPA treatment (right) calculated as the percentage change in length compared with the same genotype on the DMSO control plates. Error bars represent se. For DMSO, Col-0, n = 34; SOB3-D, n = 35; and sob3-6, n = 41. For 0.05 µm NPA, Col-0, n = 35; SOB3-D, n = 31; and sob3-6, n = 37. For 0.1 µm NPA, Col-0, n = 38; SOB3-D, n = 38; and sob3-6, n = 44. For 0.5 µm NPA, Col-0, n = 41; SOB3-D, n = 37; and sob3-6, n = 43. For 1 µm NPA, Col-0, n = 40; SOB3-D, n = 33; and sob3-6, n = 38. For 5 µm NPA, Col-0, n = 38; SOB3-D, n = 34; and sob3-6, n = 24. And for 10 µm NPA, Col-0, n = 32; SOB3-D, n = 38; and sob3-6, n = 36.
Overexpressing SAUR19 Partially Rescues Hypocotyl Elongation Defects in SOB3-D
Since members of the SAUR19 subfamily exhibit altered expression in SOB3 mutants and the phenotypes of these mutants are heavily influenced by auxin signaling, we hypothesized that alterations in SAUR levels might be responsible for the altered hypocotyl growth in SOB3-D and sob3-6. Specifically, we suspected that the short-hypocotyl phenotype in SOB3-D may be caused by the repression of SAUR19 subfamily members, which are known to promote cell expansion and hypocotyl elongation (Franklin et al., 2011; Spartz et al., 2012, 2014). To test this hypothesis, we transformed the homozygous SOB3-D line with a construct expressing GFP-SAUR19 under the control of the cauliflower mosaic virus 35S promoter. The GFP epitope tag was translationally fused upstream of the SAUR19 coding sequence in order to stabilize the SAUR19 protein upon expression in the plant. The proteins coded for by SAUR19 subfamily members are unstable to the point that overexpressing them without an epitope tag produces no visible phenotype (Spartz et al., 2012). However, it has been demonstrated that attaching an N-terminal epitope tag stabilizes these proteins, resulting in enhanced hypocotyl elongation for lines expressing such constructs (Franklin et al., 2011; Spartz et al., 2012, 2014).
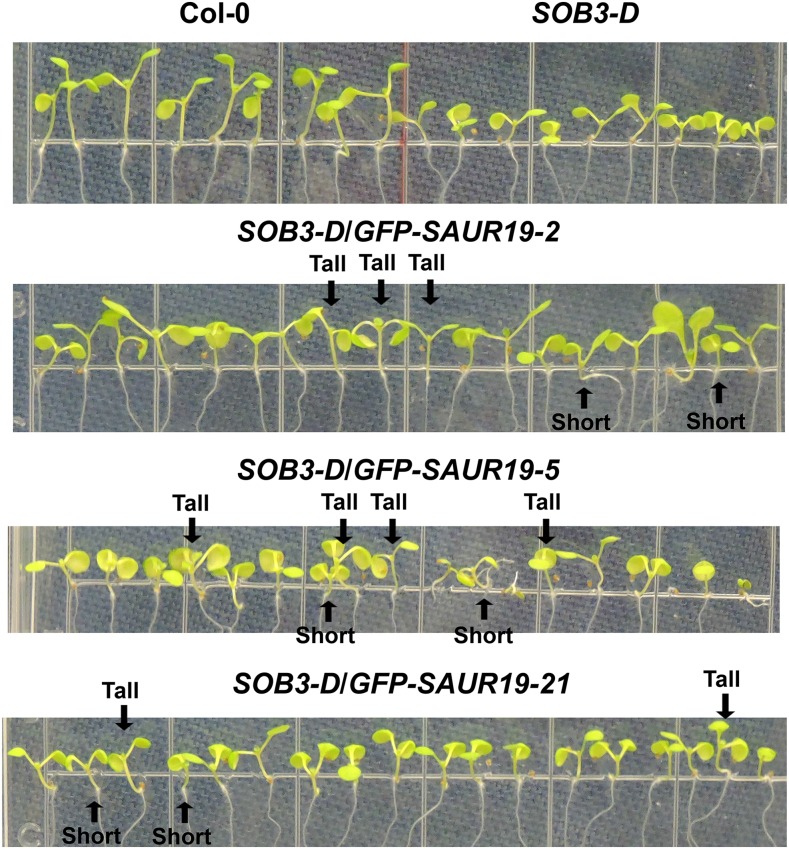
We examined both hypocotyl phenotypes and gene expression in T2 seedlings to evaluate if enhanced SAUR19 expression could rescue defects in elongation caused by SOB3-D. SOB3-D/GFP-SAUR19 seedlings were grown vertically on LS plates in dim white light for 6 d. At 6 d, seedlings were quickly photographed and then immediately harvested for RNA extraction. In multiple lines, the seedlings appeared to be segregating for hypocotyl length (Fig. 3). Within individual lines, some seedlings still resembled SOB3-D, having short hypocotyls, while others were noticeably taller.
Figure 3.
SOB3-D/GFP-SAUR19 lines segregate for hypocotyl phenotypes. Seedlings were grown on vertically oriented LS plates for 6 d in dim white light. All seedlings, except for Col-0, are homozygous at the SOB3-D locus. With regard to lines containing the GFP-SAUR19 transgene, these are segregating T2 generation seedlings. Similar results were obtained on at least two different plates for all lines, with one representative plate shown for each. For the double transgenic lines, the indicated tall and short seedlings were harvested for RNA extraction and cDNA synthesis, with these samples being used for the qRT-PCR analysis in Figure 4.
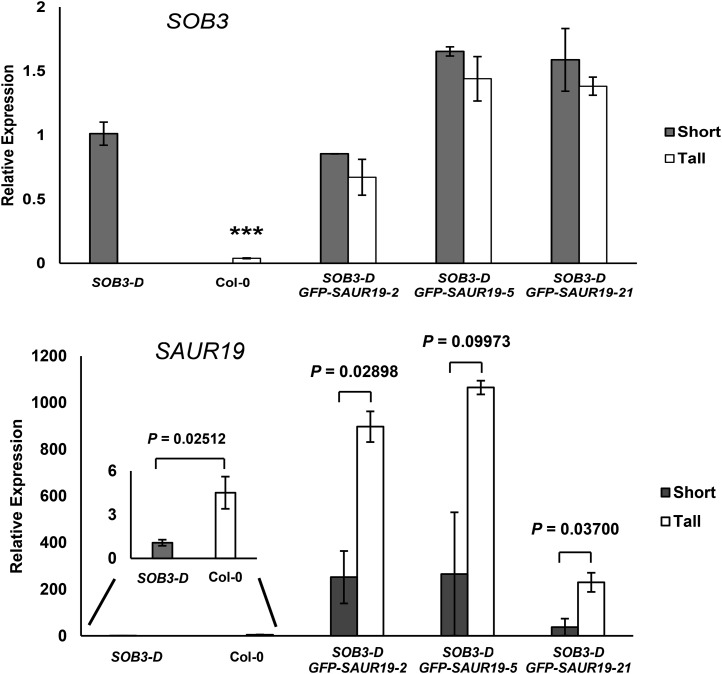
To test the hypothesis that enhanced expression of SAUR19 rescued defects in hypocotyl elongation for some T2 GFP-SAUR19 seedlings in the SOB3-D background, we compared gene expression between tall and short 6-d-old seedlings for each double transgenic line pictured in Figure 3. Examination of SOB3 transcript levels revealed no significant decrease in expression of this gene in tall seedlings, as compared with short individuals, when seedlings from the same line were compared (Fig. 4). In addition, expression of SOB3 in these three and other double transgenic lines was not substantially different as compared with the original SOB3-D line in 6-d-old seedlings (Fig. 4; Supplemental Fig. S9).
Figure 4.
Tall SOB3-D/GFP-SAUR19 seedlings have higher SAUR19 levels. Transcript levels are shown for the indicated genes as measured by qRT-PCR in short and tall seedlings harvested from vertically oriented LS plates. A representative plate for each sample is shown in Figure 3. All seedlings were grown in dim white light for 6 d. In a Welch’s t test (unpaired one-tailed t test with unequal variance), ***, P < 0.001 compared with SOB3-D (top) or is indicated for comparisons between specific pairs of samples (bottom). PCR was performed at least in triplicate, and average expression values were calculated and used for analysis. Transcript levels are normalized based on the expression of the MDAR4 housekeeping gene. All values are shown as fold change compared with SOB3-D. Error bars represent se for at least two biological replicates (when multiple short or tall seedlings were selected from the same plate, individuals were pooled together to constitute a single replicate).
We also examined SAUR19 expression levels in tall and short T2 GFP-SAUR19 individuals. Even in the short seedlings, higher levels of SAUR19 were observed, as compared with Col-0, which was attributed to the subtle nature of the phenotype combined with methodological constraints preventing us from choosing wild-type segregants at the GFP-SAUR19 locus with 100% accuracy. Since the seedlings did not always grow straight along the surface of the vertically oriented plates, quantification of individual hypocotyl lengths was impossible without the addition of another step, such as transferring the seedlings to acetate sheets and scanning them. This would have substantially delayed the time until seedlings could have been harvested for RNA extraction, potentially causing major changes in gene expression and making the qRT-PCR results unreliable. Hence, when choosing seedlings for RNA extraction, we relied entirely on visual identification of tall and short seedlings. Despite these difficulties, we were still able to observe 3- to 6-fold higher levels of SAUR19 expression in the selected tall individuals as compared with short siblings from the same line (Fig. 4). This indicates that enhanced levels of SAUR19 in the tall, double transgenic seedlings were responsible for partially rescuing hypocotyl elongation in these individuals.
For several segregating T2 GFP-SAUR19 single-locus insertion lines in the SOB3-D background, defects in rosette development and flowering disappeared for many individuals. Initially, this led us to think that enhanced expression of the SAURs could be compensating for low SAUR levels normally present in SOB3-D, thereby rescuing these plants (Supplemental Fig. S3A). However, when we examined transcript levels in SOB3-D rosette leaves using qRT-PCR, we found that SAUR19, SAUR22, and SAUR24 all were expressed at similar levels as compared with the wild type (Supplemental Fig. S10). Based on these results, we suspected that suppression of developmental defects for GFP-SAUR19 T2 plants in the SOB3-D background could be a result of reduced SOB3 expression due to trans-interactions between similar T-DNA insertions, leading to silencing (Daxinger et al., 2008; Gao and Zhao, 2013; Sandhu et al., 2013). Enhanced expression of SOB3 in the SOB3-D background is due to the presence of an activation-tagged construct containing enhancer elements from the cauliflower mosaic virus 35S promoter (Weigel et al., 2000; Street et al., 2008), which is the same promoter that was used in this experiment to drive the constitutive expression of GFP-SAUR19. Indeed, when we examined SOB3 transcript levels in rosette leaves of the wild-type-looking GFP-SAUR19 T2 individuals, we found that expression of the gene was significantly lower than in the original SOB3-D line (Supplemental Fig. S3B). These data strongly indicate that the suppression of developmental defects observed in some double transgenic plants was due to silencing of the enhancer elements that are responsible for increasing SOB3 expression in the SOB3-D background rather than an increase in the level of SAUR19.
SOB3 Binds to the Promoters of Genes Associated with Auxin Signaling
To test the hypothesis that SOB3 directly represses the expression of SAUR genes, we decided to investigate promoter binding using chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR). We generated transgenic lines expressing a SOB3-GFP translational fusion under the control of the endogenous SOB3 promoter. To avoid interference with endogenous SOB3 protein, we transformed the ProSOB3:SOB3-GFP construct into the null sob3-4 background, which was described previously (Street et al., 2008). We identified a homozygous single-locus insertion T3 line that displayed phenotypes reminiscent of SOB3-D, only more mild, including short hypocotyls, curled rosette leaves, and delayed flowering and senescence (Supplemental Figs. S4 and S11). These phenotypes indicate that the SOB3-GFP protein is functional and expressed at somewhat higher levels in this line than in Col-0. We chose to use this line, subsequently referred to simply as SOB3-GFP, for ChIP-qPCR analysis (Fig. 5).
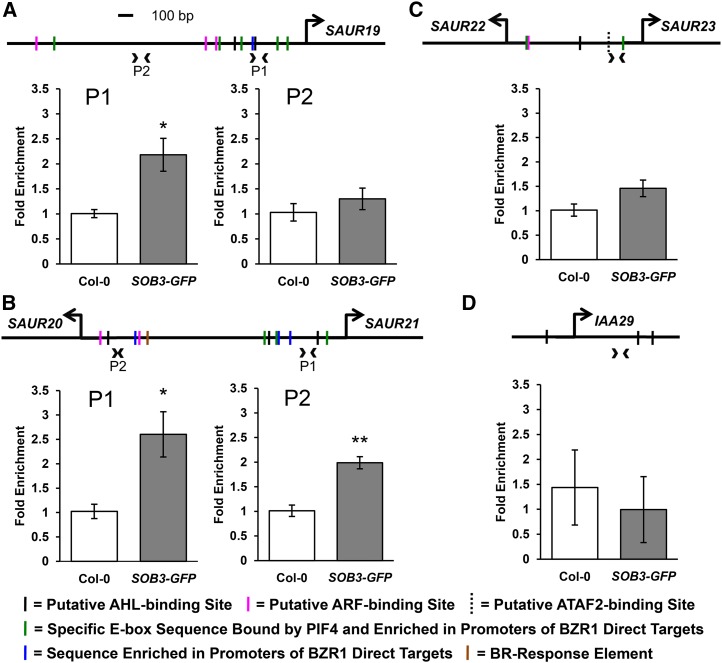
Figure 5.
SOB3 binds to SAUR promoters. A, Relative enrichment within the SAUR19 promoter in samples prepared from wild-type or transgenic SOB3-GFP lines using a ChIP-qPCR procedure employing an antibody against the GFP epitope. B, Relative enrichment within a dual promoter region located between SAUR20 and SAUR21. C, Enrichment within a dual promoter region located between SAUR22 and SAUR23. D, ChIP-qPCR results for a control primer set that amplifies an intragenic region of the IAA29 gene. Arrowheads on the promoter diagrams indicate primer pairs used for quantitative PCR (qPCR), and large arrows indicate the transcription start sites. Solid black vertical lines on the diagrams indicate the presence of one of five consensus sequences bound by AHLs in the same clade as SOB3 (AATTAAAT and ATTATAAT bound by AHL20; AATATATT bound by both AHL20 and AHL25; and AATTAATT and AATAAAT bound by AHL25; Verkest et al., 2014). Magenta lines indicate motifs bound by ARFs (TGTCTC and TGTCGG; Ulmasov et al., 1999b; Boer et al., 2014). Green lines indicate the presence of a specific E-box motif (CACATG), which is bound by PIF4 (Hornitschek et al., 2012). This motif also is enriched in promoters of genes that are both activated in response to brassinosteroids and directly bound by BZR1 (Sun et al., 2010) as well as in regions of the genome commonly bound by BZR1 and PIF4 (Oh et al., 2012). Blue lines indicate a 5-bp motif (GGTCC) enriched in the promoters of brassinosteroid-regulated genes also bound by BZR1 (Sun et al., 2010). The brown line indicates the presence of a brassinosteroid response element (CGTGTG), which is bound by BZR1 (He et al., 2005; Sun et al., 2010). The dashed vertical line indicates the presence of a CCA1-binding site (AAAAATCT), a potential ATAF2-binding site (Peng et al., 2015). The same set of chromatin immunoprecipitation (ChIP) samples was used for all PCRs shown. All values are shown as fold change compared with the Col-0 control samples. Error bars represent se from three different ChIP preparations. In a Welch’s t test (unpaired one-tailed t test with unequal variance), *, P < 0.05 and **, P < 0.01.
In turn, ChIP-qPCR revealed significant enrichment, in SOB3-GFP samples, as compared with Col-0, for a region amplified by a primer set landing approximately 250 bp upstream of the transcription start site in the SAUR19 promoter (Fig. 5A). This suggests that SOB3 binds directly to the SAUR19 promoter. Interestingly, contained within the region amplified by this primer set is a consensus sequence known to be bound by another AHL in the same clade as SOB3 (Verkest et al., 2014). Another clade A AHL consensus-binding sequence also is located slightly farther upstream. We also examined enrichment in the SOB3-GFP samples at a site located farther upstream, approximately 1,000 bp away from the SAUR19 transcription start site, in an area lacking any known binding sites for clade A AHLs. Only very slight enrichment at this locus was observed in SOB3-GFP, indicating little or no binding by SOB3.
We also evaluated other SAUR promoters for SOB3 binding. The six members of the SAUR19 subfamily are all located on chromosome 5 in Arabidopsis within a region spanning approximately 21 kb (Spartz et al., 2012). Within this region, two pairs of genes, SAUR20/21 and SAUR22/23, are positioned head to head, with less than 2 kb separating their transcription start sites. These intervening regions likely act as dual promoters, controlling the expression of both adjacent SAUR genes. Our ChIP-qPCR results indicated that SOB3 binds to at least two sites in the dual promoter region between SAUR20 and SAUR21 (Fig. 5B). This promoter region contains three known clade A AHL consensus-binding sequences, two of which are located in the immediate vicinity of the regions where enrichment was observed in the SOB3-GFP samples. Slight enrichment also was observed for a region located within the dual promoter separating SAUR22 and SAUR23, although the increase was not quite significant (Fig. 5C). However, in a second set of ChIP samples, which were prepared using the same procedure except that sonication of nuclei was increased from 20 to 30 min, a slightly higher level of binding was observed at this locus, with significant enrichment observed this time in SOB3-GFP samples, as compared with Col-0 (Supplemental Fig. S12).
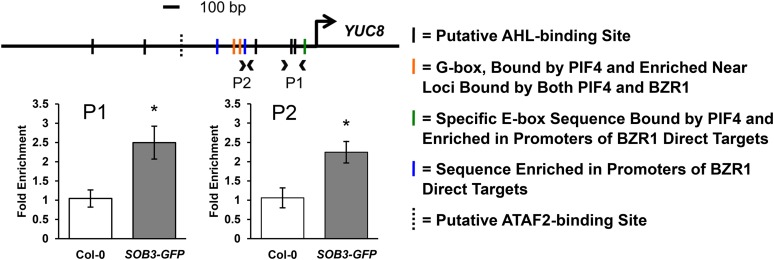
Additionally, we used ChIP-qPCR to test if SOB3 binds to the promoter of the auxin biosynthesis gene YUC8. We observed enrichment at two different loci located within the YUC8 promoter in the SOB3-GFP samples as compared with the Col-0 control, indicating that SOB3 also binds to this promoter (Fig. 6). Similar to the results for the SAURs, the two sites in the YUC8 promoter where enrichment was observed either contain, or are located close to, consensus clade A AHL-binding sequences. Although enrichment for SAUR and YUC8 promoter regions in the ChIP-qPCR assays was small, we checked for enrichment at several of these loci in a second, independent set of ChIP preparations and obtained similar results (Supplemental Fig. S12). Taken together, these results suggest that SOB3 directly binds to and regulates the expression of YUC8 and SAUR19 family members, both associated with auxin signaling. Furthermore, a lack of enrichment observed in the SOB3-GFP line for an intragenic region located within another auxin-response gene, IAA29, indicates that the enrichment observed for YUC8 and SAUR promoters is region specific and not simply an artifact arising from differences in ChIP preparations between the two lines (Fig. 5D; Supplemental Fig. S12).
Figure 6.
SOB3 binds to the YUC8 promoter. Relative enrichment is shown within the YUC8 promoter in samples prepared from Col-0 or transgenic SOB3-GFP lines using a ChIP-qPCR procedure employing an antibody against the GFP epitope. Arrowheads on the promoter diagram indicate primer pairs used for qPCR, and the large arrow indicates the transcription start site. Solid black vertical lines on the diagram indicate the presence of one of the five consensus sequences bound by AHLs in the same clade as SOB3 (AATTAAAT and ATTATAAT bound by AHL20; AATATATT bound by both AHL20 and AHL25; and AATTAATT and AATAAAT bound by AHL25; Verkest et al., 2014). Orange lines indicate a G-box motif (CACGTG), which is bound by PIF4 (Huq and Quail, 2002; Hornitschek et al., 2012; Sun et al., 2012) and also enriched in regions of the genome commonly bound by BZR1 and PIF4 (Oh et al., 2012). The green line indicates the presence of a specific E-box motif (CACATG), which is bound by PIF4 (Hornitschek et al., 2012). This motif also is enriched in promoters of genes that are both activated in response to brassinosteroids and directly bound by BZR1 (Sun et al., 2010), as well as in regions of the genome commonly bound by BZR1 and PIF4 (Oh et al., 2012). Blue lines indicate a 5-bp motif (GGTCC) enriched in the promoters of brassinosteroid-regulated genes also bound by BZR1 (Sun et al., 2010). The dashed vertical line indicates the presence of an evening element (AAAATATCT), a potential ATAF2 binding site (Peng et al., 2015). The same set of ChIP samples was used as for Figure 5. All values are shown as fold change compared with the Col-0 control sample. Error bars represent se from three different ChIP preparations. In a Welch’s t test (unpaired two-tailed t test with unequal variance), *, P < 0.05.
Phenotypes of SOB3 Mutants Are Influenced by Temperature
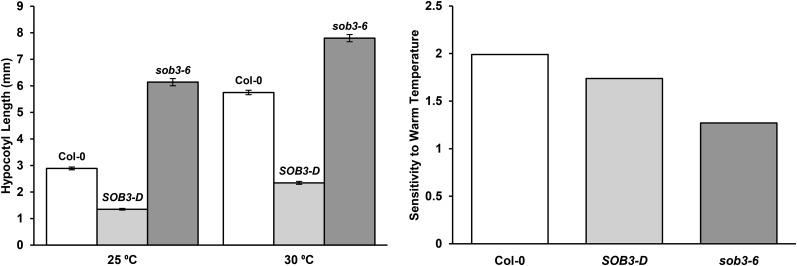
Since YUC8 and SAUR19 subfamily members are both thought to function as components of a signaling pathway that promotes elongation growth in response to elevated temperatures (Franklin et al., 2011; Sun et al., 2012; for review, see Proveniers and van Zanten, 2013), we investigated if the SOB3-D and sob3-6 phenotypes were affected by temperature. We compared hypocotyl elongation at 25°C and 30°C. Hypocotyl phenotypes in SOB3-D and sob3-6 were apparent at both temperatures, but the impact of both alleles on seedling elongation clearly changed based on the temperature (Fig. 7). SOB3-D exhibited lower sensitivity to enhanced hypocotyl elongation promoted by growth at 30°C. This indicates that the repressive effect of the SOB3-D allele on hypocotyl growth was more severe at 30°C than at 25°C. The sob3-6 mutant also exhibited lower sensitivity to warm temperature-induced hypocotyl elongation. As a result, the tall-hypocotyl phenotype characteristic of sob3-6 was severely attenuated at 30°C as compared with 25°C. This indicates that a connection exists between temperature-induced hypocotyl elongation and SOB3. Along these lines, we also tested if the expression of one SAUR, SAUR22, differs based on temperature in our growth conditions. Indeed, we found that the expression of SAUR22 was higher at 25°C than at 18°C (Supplemental Fig. S13), which is consistent with previous reports indicating that SAUR19 subfamily members are induced by warm temperatures (Franklin et al., 2011; Delker et al., 2014; Johansson et al., 2014; Bours et al., 2015). This indicates that SAUR19 subfamily genes are regulated by temperature in our growth conditions. Taken together with the phenotypic data for SOB3-D and sob3-6 at 25°C and 30°C, this lends further support to the hypothesis that SOB3 represses hypocotyl growth by acting on components important for mediating hypocotyl elongation in response to warm temperatures.
Figure 7.
Temperature impacts hypocotyl phenotypes in SOB3-D and sob3-6. Hypocotyl growth is shown for homozygous mutants grown for 6 d in the same intensity of dim white light at either 25°C or 30°C. Values represent either the mean of the actual measured hypocotyl length (left) or the sensitivity to warm temperature-induced hypocotyl elongation (right), presented for each genotype as hypocotyl length at 30°C divided by hypocotyl length at 25°C. Error bars represent se. For 25°C, Col-0, n = 89; SOB3-D, n = 71; and sob3-6, n = 91. For 30°C, Col-0, n = 55; SOB3-D, n = 43; and sob3-6, n = 65.
DISCUSSION
A Model for the Repression of Hypocotyl Growth by SOB3
Seedlings fine-tune their rate of elongation in response to changing environmental conditions. Transcription factors functioning as components of signaling pathways are essential to alter the expression of downstream target genes, which are directly responsible for imparting changes in growth. We have generated several pieces of evidence indicating that SOB3 represses hypocotyl elongation in light-grown seedlings by suppressing the expression of YUC8 and members of the SAUR19 subfamily. First, data from our RNA-seq screen and qRT-PCR analyses indicated that transcription of these genes is robustly down-regulated in the SOB3-D gain-of-function mutant, including in seedlings grown for 4, 5, or 6 d in white light as well as in far-red light for 7 d (Fig. 1; Table I; Supplemental Table S3; Supplemental Figs. S5–S7). Our data also indicated that transcripts of these genes are elevated in the loss-of-function sob3-6 mutant in seedlings less than 6 d old, which is the stage of development when this allele confers enhanced hypocotyl elongation (Table I; Fig. 1; Supplemental Figs. S1 and S7). Additionally, overexpression of SAUR19 partially rescued the short-hypocotyl phenotype caused by enhanced expression of SOB3 in the SOB3-D mutant (Figs. 3 and 4). Although the results from this experiment do not exclude the possibility that SOB3 acts in a pathway parallel to auxin signaling, when considered in light of the aforementioned expression data obtained from the SOB3-D and sob3-6 mutants, it seems much more likely that YUC8 and SAUR19 to SAUR24 function downstream of SOB3. These results, combined with our ChIP-qPCR data, which indicated that SOB3 binds directly to the promoters of YUC8 and multiple SAUR19 subfamily members (Figs. 5 and 6; Supplemental Fig. S12), strongly suggest that SOB3 inhibits hypocotyl elongation by directly repressing the transcription of these auxin signaling pathway components. That said, cross talk frequently occurs between various plant hormone signaling pathways (for review, see Depuydt and Hardtke, 2011). Therefore, it is possible that SOB3 acts on other hormone pathways in addition to that of auxin. A rigorous transcriptomics study in which multiple replicates and time points are included would be informative to determine if SOB3 exerts its effect on hypocotyl growth mainly by acting on auxin signaling or if it also acts on other pathways, such as those associated with brassinosteroids or GAs.
YUC8 and SAURs function at opposite ends of the auxin signaling pathway (Fig. 8). YUC8 functions in biosynthesis of the hormone and is essential for increasing endogenous auxin levels in response to warm temperatures, which in turn leads to hypocotyl elongation (Gray et al., 1998; Stavang et al., 2009; Franklin et al., 2011; Sun et al., 2012; Ma et al., 2016). Eleven YUC genes in Arabidopsis code for flavin monooxygenases, which function in the biosynthesis of auxin from Trp in what seems to be the major pathway for the synthesis of this hormone in plants (Zhao et al., 2001; Hofmann, 2011; Mashiguchi et al., 2011; Phillips et al., 2011; Stepanova et al., 2011; Won et al., 2011; Dai et al., 2013; for review, see Zhao, 2014). Specifically, these enzymes catalyze the rate-limiting conversion of indole-3-pyruvic acid to the active hormone IAA, the second of two steps in this short biosynthetic pathway. The presence of multiple YUC genes in plants seems to be important for the precise regulation of auxin biosynthesis based on tissue, stage of development, and external stimuli (Cheng et al., 2006, 2007; Yamamoto et al., 2007; Hornitschek et al., 2012; Kriechbaumer et al., 2012; Li et al., 2012; Sun et al., 2012; Hersch et al., 2014; Ma et al., 2016; for review, see Zhao, 2010, 2014).
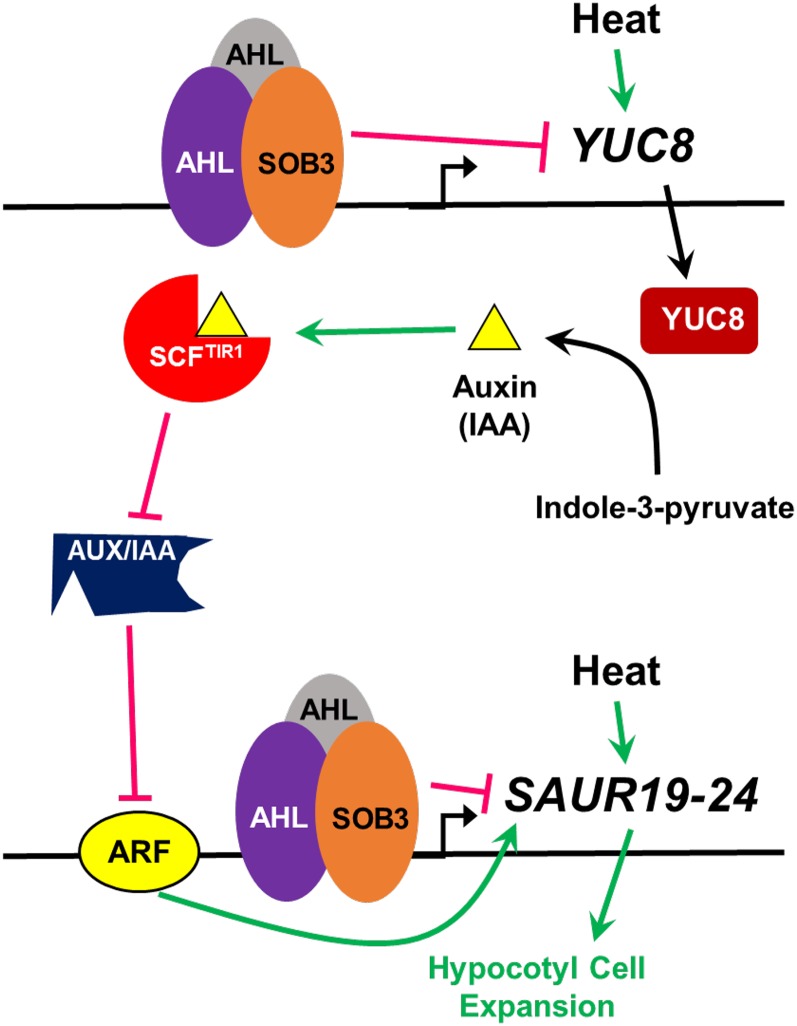
Figure 8.
Model for the modulation of hypocotyl growth by SOB3. Increased temperature leads to hypocotyl elongation through a pathway that is at least partially mediated through YUC8 and SAUR19 subfamily members. Heat induces the expression of YUC8, which leads to enhanced auxin production. Auxin promotes physical interactions between SCFTIR1 and AUX/IAA repressors, inhibiting the activity of AUX/IAAs by promoting their degradation. This leads to the attenuation of posttranslational repression of ARF activity by AUX/IAAs, which in turn promotes the transcription of growth-promoting SAUR19 subfamily members. Based on the results of this study, we propose that SOB3 inhibits hypocotyl elongation by some combination of direct and indirect repression of SAUR19 subfamily member expression. SOB3 inhibits production of the hormone signal, auxin, by binding to the promoter of YUC8 and repressing its expression, which in turn leads to the inhibition of SAUR19 to SAUR24 transcription via the SCFTIR1-mediated auxin signaling pathway. Additionally, SAUR19 to SAUR24 are repressed directly at the transcriptional level by SOB3.
In turn, auxin influences the transcription of at least a portion of its downstream targets through a short signaling pathway mediated by the receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) (Fig. 8; Gray et al., 1999; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; for review, see Enders and Strader, 2015; Salehin et al., 2015; Li et al., 2016). Binding of auxin to TIR1 stabilizes interactions between the SCFTIR1 E3 ubiquitin ligase complex and AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressors, the latter of which are ubiquitinated and subsequently degraded via the 26S proteasome (Gray et al., 1999, 2001; Tiwari et al., 2001; Tian et al., 2003; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007; Maraschin et al., 2009). AUX/IAA proteins are potent repressors of transcription, and in the absence of auxin promoting their degradation, they complex with and inhibit the function of activating ARFs (Tiwari et al., 2001, 2003; Szemenyei et al., 2008; Korasick et al., 2014; for review, see Guilfoyle and Hagen, 2007; Wright and Nemhauser, 2015). Auxin response elements located in the promoters of genes targeted by this signaling pathway are bound by ARFs, which always seem to function as transcriptional activators in the case of targets antagonized by AUX/IAAs (Ulmasov et al., 1997a, 1997b, 1999a, 1999b; Tiwari et al., 2003; Tatematsu et al., 2004; Oh et al., 2014; for review, see Enders and Strader, 2015; Salehin et al., 2015; Li et al., 2016). SAUR19 to SAUR24 transcripts are all induced by auxin but severely depleted in a mutant exhibiting enhanced stability of an AUX/IAA protein, while their promoters also are bound by ARF6, indicating that this subfamily is regulated by the SCFTIR1-AUX/IAA-ARF pathway (Spartz et al., 2012; Oh et al., 2014; for review, see Proveniers and van Zanten, 2013). It is also known that SAUR19 subfamily members directly interact with and inhibit the function of PP2C-D phosphatases, which in turn leads to the activation of plasma membrane H+-ATPases, causing a decrease in apoplastic pH and, finally, an increase in cell expansion due to the enhanced activity of expansins and other cell wall-modifying enzymes (Spartz et al., 2012, 2014; Takahashi et al., 2012; for review, see Dünser and Kleine-Vehn, 2015). Therefore, it is clear that SAUR19 to SAUR24 function at the end of the auxin signaling pathway to convert the hormonal signal into a specific developmental output, cell expansion.
Considering our data in light of this information, we propose a model whereby SOB3 represses auxin signals involved in promoting hypocotyl cell expansion in response to elevated temperatures at two different points, both at the beginning of the signaling pathway, via the repression of YUC8 and the synthesis of the hormone, and at the end of it, through the repression of SAUR19 to SAUR24 (Fig. 8). Although this seems to be the most likely scenario given that SOB3 bound directly to the promoters of both YUC8 and SAURs in our study (Figs. 5 and 6; Supplemental Fig. S12), additional work is needed to conclusively rule out the possibility that SOB3 directly represses the transcription of only YUC8 or SAURs. For instance, direct repression of YUC8 transcription alone by SOB3 could reduce auxin levels and signaling through the SCFTIR1-mediated signaling pathway, indirectly leading to a reduction in the abundance of SAUR19 to SAUR24 transcripts. Conversely, it is also conceivable that SOB3 may only directly repress the transcription of SAUR19 subfamily members, in turn leading to the misregulation of YUC8 via a feedback loop. Transgenic rice plants overexpressing the auxin-inducible OsSAUR39 gene exhibit reduced levels of the hormone (Kant et al., 2009), likely indicating that SAUR genes are involved in the feedback regulation of auxin biosynthetic genes. Interestingly, in this study, SOB3-D lacked any changes in sensitivity to exogenous IAA (Supplemental Fig. S8), which are characteristic of at least some mutants exhibiting either global changes in auxin levels or defects in transmission of the hormone signal (Collett et al., 2000). Hence, this may indicate that any changes in auxin levels as a result of altered YUC8 expression in SOB3-D either are minimal or are simply restricted to sites where exogenous IAA cannot be efficiently transported (Cheng et al., 2006). On the other hand, overall, sob3-6 was somewhat more sensitive to IAA application. This could indicate that YUC8-mediated changes in auxin levels are more important for producing the defects in hypocotyl growth observed in sob3-6 than those in SOB3-D.
Does SOB3 Repress Additional SAUR Genes?
A question of interest for future studies is whether SOB3 regulates the expression of other SAUR genes outside the SAUR19 subfamily. This is a distinct possibility, considering that, in addition to SAUR19 to SAUR24, many other SAURs were identified, via the RNA-seq screens performed in this study, as potential downstream targets of SOB3 (Table I; Supplemental Table S3). Additionally, this seems very likely considering that only small changes in expression were observed for SAUR19, SAUR22, and SAUR24 in SOB3-D and sob3-6 (Fig. 1), yet high GFP-SAUR19 expression seems to only partially rescue hypocotyl growth in the SOB3-D background (Figs. 3 and 4). Members of another subclade, consisting of SAUR61 to SAUR68, as well as SAUR75, are excellent candidates for repression by SOB3, as they also promote hypocotyl elongation (Chae et al., 2012; Sun et al., 2016), and our RNA-seq screen at 4 d indicated that SAUR61, SAUR63 to SAUR66, and SAUR68 are all down-regulated in SOB3-D and up-regulated in sob3-6 (Table I). Additionally, we also saw evidence for the repression of SAUR62, SAUR63, and SAUR66 to SAUR68 in SOB3-D at later time points (Supplemental Table S3; Supplemental Figs. S5 and S6). Furthermore, SAUR65 is regulated directly by TCP3 and TCP15 (Koyama et al., 2010; Uberti-Manassero et al., 2012; Lucero et al., 2015), while the expression of SAUR63, SAUR66, and SAUR67 is influenced by TCP10 (Danisman et al., 2013). TCP3 and TCP10 may be important for SOB3 function due to the fact that they are two of five TCP family members exhibiting reduced expression in the jaw-D background (Palatnik et al., 2003), where the extremely tall-hypocotyl phenotype typically characteristic of sob3-6 is abolished (Zhao et al., 2013).
The Seedling-Specific Impact of SOB3 Is a Point of Interest for Future Studies
Another point of interest for future studies is the relatively narrow time frame in which the sob3-6 allele impacts plant development as well as the transcription of downstream targets (Fig. 1; Table I; Supplemental Figs. S1 and S5–S7). These results suggest that endogenous SOB3 only impacts hypocotyl elongation in seedlings less than 6 d old, which is surprising, considering that this gene is still expressed in 7-d-old seedlings (Street et al., 2008). One possible explanation for this is that SOB3 requires a cofactor, missing in older seedlings, that is necessary for it to bind target promoters and repress gene transcription. One interesting possible cofactor is the transcription factor ATAF2, which interacts physically with SOB3’s closest homolog, ESC, as well as with AHL12 (Zhao et al., 2013), and is known to function as a direct transcriptional repressor of two brassinosteroid-inactivating enzymes that influence hypocotyl growth, BAS1 and SOB7 (Neff et al., 1999; Turk et al., 2005; Peng et al., 2015). Interestingly, we found an evening element and CCA1-binding site in the promoters of YUC8 and SAUR22/SAUR23, respectively, which also are sequences bound by ATAF2. TCPs also are good candidate cofactors important for the activity of SOB3 as a transcriptional repressor, given that they are indispensable for manifestation of the sob3-6 phenotype (Zhao et al., 2013), and they also are known to regulate the expression of SAUR genes (Koyama et al., 2010; Uberti-Manassero et al., 2012; Danisman et al., 2013; Lucero et al., 2015).
SOB3’s function as a repressor of SAURs also could be dependent on interactions with transcription factors responsible for activating these genes. For example, transcriptional repression by SOB3 could be dependent on binding to and antagonizing the transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4), which is important for directly activating both YUC8 and SAUR19 subfamily members in response to warm temperatures, promoting hypocotyl elongation (Koini et al., 2009; Franklin et al., 2011; Oh et al., 2012, 2014; Sun et al., 2012; Johansson et al., 2014; Ma et al., 2016; for review, see Proveniers and van Zanten, 2013). PIF4 also plays a major role in light signaling (Huq and Quail, 2002; Lorrain et al., 2008; for review, see Leivar and Quail, 2011), and some evidence indicates that it activates YUC8 in response to low light intensities (Hornitschek et al., 2012). This makes PIF4 a particularly interesting candidate, considering that previous studies revealed that defects in hypocotyl elongation observed in SOB3 mutants are apparent mainly at low light intensities (Street et al., 2008; Zhao et al., 2013). Along these same lines, SOB3’s function as a repressor of SAURs could be dependent on interactions with ARFs, which activate these genes (Fig. 8; Oh et al., 2014; for review, see Proveniers and van Zanten, 2013). Interestingly, cis-elements known to be bound by PIF4 and ARFs are found in the promoter regions where binding of SOB3 was observed in this study (Figs. 5 and 6). This further suggests that SOB3 may repress transcription by antagonizing the functions of PIF4 and ARFs. Motifs bound by the transcription factor BZR1, which is a key component of the brassinosteroid signaling pathway, also are present in the promoter regions where SOB3 binding was observed. Since BZR1, PIF4, and ARF6 all interact physically and frequently function together as coactivators of transcription (Oh et al., 2012, 2014) it is possible that SOB3 represses gene transcription by antagonizing complexes composed of these three transcription factors.
The sob3-6 allele also is unique in that it confers a tall-hypocotyl phenotype in seedlings without producing other visible phenotypes at the seedling, juvenile, or adult stage (Supplemental Figs. S1, S3A, and S4). This is consistent with SOB3 repressing the expression of SAUR19 to SAUR24, direct promoters of cell growth in hypocotyls (Spartz et al., 2012, 2014), rather than affecting components involved generally in light signaling or photomorphogenesis (for review, see Neff et al., 2000; Arsovski et al., 2012; Boron and Vissenberg, 2014). Many mutations conferring defects in hypocotyl elongation are in components of light-signaling pathways; hence, they produce pleiotropic phenotypes, as in the case of loss-of-function phyB mutants, which exhibit constitutive shade avoidance, reduced levels of chlorophyll, and early flowering (Reed et al., 1993; Lorrain et al., 2008; Leivar et al., 2009; for review, see Neff et al., 2000; Arsovski et al., 2012). Manipulating these types of genes for agricultural purposes is largely impractical, because a number of undesirable phenotypes are likely to accompany a desired characteristic conferred by a mutant allele. However, since the effect of SOB3 on development is restricted to repressing elongation in young seedlings, this makes its homologs in crop plants ideal candidates to manipulate in order to improve stand establishment. Since AHLs seem to be found ubiquitously as large gene families in higher plants, it is likely that homologs possessing similar functions to SOB3 are widespread in crop plants (Zhao et al., 2014). Cultivars mutated in ways that mimic the sob3-6 allele presumably would have enhanced hypocotyl elongation, helping them to reach the soil surface rapidly, yet otherwise be unaffected developmentally. Therefore, off-target, negative effects on other agronomically important traits could be avoided. This could be particularly helpful for improving yields in dry climates, as it would enable seeds to be planted deeper in the soil, facilitating access to moisture.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study are in the Col-0 ecotype. The SOB3-D, sob3-6, and sob3-4 mutants were all described previously (Street et al., 2008; Zhao et al., 2013). Except for soil-grown plants, seeds were sown on one-half-strength LS medium with 1.5% Suc and 1% phytagel (Sigma). For assays with IAA or NPA, the compound was added in solution to the medium to achieve the specified concentration, with the amount of solvent held constant among all plates used for the same experiment. In order to synchronize germination, seeds were incubated in the dark at 4°C for about 4 d, then transferred to red light and 25°C for 12 h, prior to being grown in a chamber. For the dim white light condition, seedlings were grown in an E-30B growth chamber (Percival), where both fluorescent and incandescent bulbs were used to supply continuous light with a red to far-red light ratio of approximately 1:1 and a fluence rate of 23 μmol m−2 s−1. Except where other growth temperatures are specified, seedlings were grown at 25°C.
For soil-grown plants, seeds were sown on moist soil, then pots were placed in 4°C for about 1 week to synchronize germination. Subsequently, plants were grown in a CMP4030 Controller (Conviron) at 21°C constant temperature in long-day conditions (16-h days/8-h nights), irradiating with 200 μmol m−2 s−1 white light supplied by a mixture of fluorescent and incandescent bulbs.
RNA-Seq Screen
RNA was extracted from seedlings using the RNeasy Plant Mini Kit (Qiagen). During extraction, the On-Column DNase I Digestion Set (Sigma) was used to eliminate genomic DNA contamination. Following RNA extraction, the MicroPoly(A) Purist Kit (Applied Biosystems) was used to enrich for mRNA. Since the purpose of this experiment was to simply identify candidate genes misregulated in SOB3 mutants and not to perform rigorous transcriptomic analysis, at this point, mRNA samples from the same genotype and condition were pooled together, effectively resulting in single replicates for sequencing. A total of 50 ng of poly(A) purified mRNA was used for Ion Proton sequencing library construction using the Ion Total RNA-seq kit version 2, with the exception that all purifications and size selections were performed using AMPureXP beads (Beckmann-Coulter Genomics). Libraries were quantified by qPCR. An Ion One Touch 2 was used for emulsion PCR and sequencing bead preparation with Ion P1 OT2 200 V3 reagents. Sequencing beads were characterized and quantified by flow cytometry (Guava EasyCyte; Millipore) and sequenced at three samples per P1 chip using 520 flows on an Ion Proton. Signal processing, base calling, and barcode separation were done using Torrent Suite version 4.2 software. Reads were mapped to the Arabidopsis genome (TAIR 10), and RPKM values were calculated using CLC Genomics Workbench 7.5 software (www.clcbio.com). Hierarchical clustering analysis was performed using the settings Euclidian Distance for distance measure and Single Linkage for cluster linkage. Fold changes in RPKM values, as well as Pearson correlation coefficients, were calculated using Microsoft Excel software.
Hypocotyl Measurements
Seedlings were harvested for hypocotyl measurements at the time points specified. A ScanJet3500 (Hewlett-Packard) or an Epson Perfection V600 Photo flat-bed scanner was used to generate TIFF or JPEG format images of the seedlings. NIH ImageJ version 1.48 (Schneider et al., 2012) was used to measure hypocotyl lengths, and data were analyzed and graphs generated using Microsoft Excel software.
Photography and Image Processing
All photographs were taken using a Nikon Coolpix P600 camera. Images were cropped and assembled using Inkscape software 0.91.
qRT-PCR
RNA was extracted from seedlings using the RNeasy Plant Mini Kit (Qiagen). Genomic DNA contamination was eliminated either with the On-Column DNase I Digestion Set (Sigma) or the gDNA Eraser (Takara). First-strand cDNA synthesis was conducted using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad) or the PrimeScript RT Reagent Kit (Takara). qPCR was carried out using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) or Thunderbird SYBR qPCR Mix (Toyobo) in a 7500 Fast Real Time PCR System (Applied Biosystem), a CFX96 Touch Real-Time PCR Detection System (Bio-Rad), or an Mx3000p qPCR System (Agilent). Primers used for amplification are listed in Supplemental Table S4. Microsoft Excel software was used to analyze and compare data using the ΔΔCT method.
Generation of GFP-SAUR19 Transgenic Lines
The GFP coding sequence was amplified from the pEarleyGate 103 vector (Earley et al., 2006) using primers 5′-GGTACTCCATGGCTCGAGATGGTAGATCTGACTAGTAAAGG-3′ and 5′-GGTACTGAGCTCCCTAGGCTGCAGTATGCATATGCAGGTACCGCTAGCTTTGTATAGTTCATCC-3′. The resulting PCR product and the pGEM T-Easy vector (Promega) were digested with NcoI and SacI, and the two fragments were ligated together to generate pGEM-GFP. The SAUR19 coding sequence was then amplified from cDNA using primers 5′-CGGGCAGGTACCAAAATGGCTTTCGTGAGAAGTC-3′ and 5′-ATTAAATCTGCAGTGTTGGATCATCTTCATTGGAG-3′. The resulting PCR product and the pGEM-GFP plasmid were then both cleaved with KpnI and PstI. This enabled subsequent ligation of the SAUR19 coding sequence into the pGEM-GFP plasmid, 3′ of GFP. The resulting GFP-SAUR19 translational fusion construct was then sequenced and confirmed to be free of errors. GFP-SAUR19 was cut out of the resulting vector using XhoI and XbaI and cloned into pEarleyGate 103 at the XhoI and AvrII sites. This plasmid was transformed into Agrobacterium tumefaciens, which was subsequently used to transform SOB3-D plants, using the floral dip method (Clough and Bent, 1998). Successful transformants were identified by screening on LS plates containing 30 ng mL−1 Basta (Bayer CropScience). Single-locus insertion T2 lines were identified by growing plants on selection plates, scoring the number of resistant and susceptible individuals, and selecting lines that segregated at a 3:1 ratio based on χ2 analysis. Kanamycin (30 ng mL−1) also was included on all plates used for screening SOB3-D/GFP-SAUR19 lines, in an attempt to increase the chances of identifying lines where SOB3 was not being silenced.
ProSOB3:SOB3-GFP Plasmid Construction
Around 1.2 kb of the promoter region of SOB3 together with its coding sequence (without stop codon) was amplified with the primer pair 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCTTCACCTTTAGAATATATGAC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTACTTAAAGGCTGGTCTTGGTGGTGCG-3′ and cloned into the entry pENTR/D-TOPO plasmid using the pENTR/D-TOPO cloning kit (Thermo Fisher Scientific). The resulting plasmid was sequenced to confirm the absence of errors. Subsequently, ProSOB3-SOB3 was recombined into the pMDC107 binary vector (Curtis and Grossniklaus, 2003), 5′ of and in frame with GFP, via a Gateway LR reaction (Invitrogen). sob3-4 plants were transformed, and transgenic plants were isolated as described above.
ChIP
ChIP was carried out essentially as described previously (Ikeuchi et al., 2015) except for the following modifications. Four-day-old whole seedlings grown in dim white light were used in this study, and the cross-linking step with 1% formaldehyde was only performed for 4 min. Sonication was performed in a Bioruptor Plus UCD-300 (Diagenode) for 20 or 30 cycles (30 s on/30 s off) on the H power setting, which resulted in an average fragment size of approximately 150 bp. Anti-GFP antibody (ab290; Abcam) was used for immunoprecipitation. RNase A (Thermo) at a final concentration of 1 mg mL−1 at 37°C for 30 min was used to degrade RNA.
Accession Numbers
Arabidopsis Genome Initiative numbers for the sequences used in this study are as follows: IAA29 (AT4G32280), MDAR4 (AT3G27820), PMP (AT3G24160), PP2AA3 (AT1G13320), SAUR1 (AT4G34770), SAUR6 (AT2G21210), SAUR7 (AT2G21200), SAUR12 (AT2G21220), SAUR13 (AT4G38825), SAUR19 (AT5G18010), SAUR20 (AT5G18020), SAUR21 (AT5G18030), SAUR22 (AT5G18050), SAUR23 (AT5G18060), SAUR24 (AT5G18080), SAUR28 (AT3G03830), SAUR29 (AT3G03820), SAUR34 (AT4G22620), SAUR41 (AT1G16510), SAUR42 (AT2G28085), SAUR45 (AT2G36210), SAUR57 (AT3G53250), SAUR61 (AT1G29420), SAUR62 (AT1G29430), SAUR63 (AT1G29440), SAUR64 (AT1G29450), SAUR65 (AT1G29460), SAUR66 (AT1G29500), SAUR68 (AT1G29490), SAUR69 (AT5G10990), SAUR71 (AT1G56150), SAUR76 (AT5G20820), SOB3 (AT1G76500), UBQ10 (AT4G05320), and YUC8 (AT4G28720).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. sob3-6 does not affect development after day 6.
Supplemental Figure S2. Hierarchical clustering of RNA-seq data.
Supplemental Figure S3. Rosette phenotypes in SOB3 mutants and SOB3-D/GFP-SAUR19 double transgenic lines.
Supplemental Figure S4. Phenotypes in 55-d-old SOB3 mutants.
Supplemental Figure S5. SAUR genes are repressed in SOB3-D at 6 d.
Supplemental Figure S6. SAUR genes are down-regulated in SOB3-D seedlings grown in far-red light.
Supplemental Figure S7. SAUR22 and YUC8 expression patterns in SOB3 mutants at 5 d.
Supplemental Figure S8. Responses of SOB3 mutants to exogenous IAA.
Supplemental Figure S9. SOB3 expression in SOB3-D/GFP-SAUR19 seedlings.
Supplemental Figure S10. SAUR expression in rosettes leaves.
Supplemental Figure S11. SOB3-GFP phenotypes are reminiscent of SOB3-D.
Supplemental Figure S12. SOB3 binding to SAUR and YUC8 promoters.
Supplemental Figure S13. Comparison of SAUR22 expression at 25°C and 18°C.
Supplemental Table S1. Expression of auxin-response genes correlating with hypocotyl lengths in SOB3 mutants.
Supplemental Table S2. Auxin-response genes misregulated in SOB3-D at 6 d.
Supplemental Table S3. Misregulated SAUR genes in SOB3-D based on RNA-seq at 6 d.
Supplemental Table S4. Primers used for qRT-PCR and ChIP-qPCR.
Supplemental Data Set S1. RNA-seq data from 4-d-old seedlings.
Supplemental Data Set S2. RNA-seq data from 6-d-old seedlings.
Supplementary Material
Acknowledgments
We thank Derek Pouchnik and Mark Wildung (Washington State University) for help generating the RNA-seq data and for the use of the Bioruptor Plus UCD-300 (Diagenode) used in the ChIP procedure, Dr. Minami Matsui (RIKEN) for providing growth space for the seedlings grown in far-red light, and Mariko Ohnuma (RIKEN) for expertise regarding the ChIP procedure.
Glossary
- RNA-seq
RNA sequencing
- IAA
indole-3-acetic acid
- NPA
N-1-napthylphthalamic acid
- ChIP-qPCR
chromatin immunoprecipitation followed by quantitative PCR
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
Footnotes
This work was supported by the U.S. Department of Agriculture National Institute of Food and Agriculture (Agriculture and Food Research Initiative grant no. 2013–67013–21666 to M.M.N. and HATCH grant no. 1007178 to M.M.N.), the Brubbaken and Reinbold Monocot Breeding Fund (to M.M.N.), the Global Plant Sciences Initiative Research Fellowship from Washington State University (to D.S.F.), and the East Asia and Pacific Summer Institutes for U.S. Graduate Students Fellowship (National Science Foundation and Japan Society for the Promotion of Science award to D.S.F.).
Articles can be viewed without a subscription.
References
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL (2012) Photomorphogenesis. The Arabidopsis Book 10: e0147, doi/10.1199/tab.0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589 [DOI] [PubMed] [Google Scholar]
- Boron AK, Vissenberg K (2014) The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep 33: 697–706 [DOI] [PubMed] [Google Scholar]
- Bours R, Kohlen W, Bouwmeester HJ, van der Krol A (2015) Thermoperiodic control of hypocotyl elongation depends on auxin-induced ethylene signaling that controls downstream PHYTOCHROME INTERACTING FACTOR3 activity. Plant Physiol 167: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidwood L, Breuer C, Sugimoto K (2014) My body is a cage: mechanisms and modulation of plant cell growth. New Phytol 201: 388–402 [DOI] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O (2000) Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol 124: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao Y (2013) The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288: 1448–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van Dijk AD, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent GC, Immink RG (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot 64: 5673–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I (2008) Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci 13: 4–6 [DOI] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–R373 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dünser K, Kleine-Vehn J (2015) Differential growth regulation in plants: the acid growth balloon theory. Curr Opin Plant Biol 28: 55–59 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Enders TA, Strader LC (2015) Auxin activity: past, present, and future. Am J Bot 102: 180–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Matsunaga S, Yonemura M, Uchiyama S, Azuma T, Fukui K (2004) Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol Biol 56: 225–239 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y (2013) Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol Plant 6: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C (2014) Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci USA 111: 6515–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR. (2011) YUC and TAA1/TAR proteins function in the same pathway for auxin biosynthesis. Plant Cell 23: 3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, Veylder LD et al. (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1: 15089. [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88: 360–371 [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia QS, Zhu J, Xu XF, Lou Y, Zhang ZL, Zhang ZP, Yang ZN (2015) Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for nexine formation. Mol Plant 8: 251–260 [DOI] [PubMed] [Google Scholar]
- Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R, Halliday KJ (2014) Arabidopsis cell expansion is controlled by a photothermal switch. Nat Commun 5: 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA 111: 5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Wang P, Hawes C, Abell BM (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70: 292–302 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ (2016) A review of auxin response factors (ARFs) in plants. Front Plant Sci 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lou Y, Xu XF, Zhu J, Gu JN, Blackmore S, Yang ZN (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat Commun 5: 3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manassero NG, Viola IL, Welchen E, Gonzalez DH (2013) TCP transcription factors: architectures of plant form. Biomol Concepts 4: 111–127 [DOI] [PubMed] [Google Scholar]
- Maraschin FdS, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KH, Yu H, Ito T (2009) AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol 7: e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Peng H, Zhao J, Neff MM (2015) ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142: 4129–4138 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proveniers MC, van Zanten M (2013) High temperature acclimation through PIF4 signaling. Trends Plant Sci 18:59–64 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. ( 1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Koirala PS, Neff MM (2013) The ben1-1 brassinosteroid-catabolism mutation is unstable due to epigenetic modifications of the intronic T-DNA insertion. G3 (Bethesda) 3: 1587–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Interaction of TCP4-mediated growth module with phytohormones. Plant Signal Behav 6: 1440–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR Inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolome J, Gomez MD, Yoshida S, Asami T, Olsen JE, Garcia-Martinez JL, Alabadi D, Blazquez MA (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Shah PK, Smith AM, Avery N, Neff MM (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J 54: 1–14 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW (2003) Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J 36: 643–651 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999a) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999b) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Abeel T, Heyndrickx KS, Van Leene J, Lanz C, Van De Slijke E, De Winne N, Eeckhout D, Persiau G, Van Breusegem F, et al. (2014) A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol 164: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RC, Nemhauser JL (2015) New tangles in the auxin signaling web. F1000Prime Rep 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71: 39–50 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Stroud H, Gu X, Sun B, Gan ES, Ng KH, Jacobsen SE, He Y, Ito T (2013) A matrix protein silences transposons and repeats through interaction with retinoblastoma-associated proteins. Curr Biol 23: 345–350 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Kim YS, Jung JH, Seo PJ, Park CM (2012) The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J Biol Chem 287: 15307–15316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Peng H, Neff MM (2013) Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci USA 110: E4688–E4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Qiu J, Roalson EH, Neff MM (2014) Insights into the evolution and diversification of the AT-hook motif nuclear localized gene family in land plants. BMC Plant Biol 14: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2014) Auxin biosynthesis. The Arabidopsis Book 12: e0173, doi/10.1199/tab.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.