A functional H3/H4 histone chaperone mediates abiotic stress adaptation via transcriptional regulation of diverse stress-related genes in rice.
Abstract
Modulation of gene expression is one of the most significant molecular mechanisms of abiotic stress response in plants. Via altering DNA accessibility, histone chaperones affect the transcriptional competence of genomic loci. However, in contrast to other factors affecting chromatin dynamics, the role of plant histone chaperones in abiotic stress response and adaptation remains elusive. Here, we studied the physiological function of a stress-responsive putative rice (Oryza sativa) histone chaperone of the NAP superfamily: OsNAPL6. We show that OsNAPL6 is a nuclear-localized H3/H4 histone chaperone capable of assembling a nucleosome-like structure. Utilizing overexpression and knockdown approaches, we found a positive correlation between OsNAPL6 expression levels and adaptation to multiple abiotic stresses. Results of comparative transcriptome profiling and promoter-recruitment studies indicate that OsNAPL6 functions during stress response via modulation of expression of various genes involved in diverse functions. For instance, we show that OsNAPL6 is recruited to OsRad51 promoter, activating its expression and leading to more efficient DNA repair and abrogation of programmed cell death under salinity and genotoxic stress conditions. These results suggest that the histone chaperone OsNAPL6 may serve a regulatory role in abiotic stress physiology possibly via modulating nucleosome dynamics at various stress-associated genomic loci. Taken together, our findings establish a hitherto unknown link between histone chaperones and abiotic stress response in plants.
Expression of the genome is largely influenced by chromatin structure that, in turn, is majorly governed by DNA methylation machinery and histone-associated chromatin factors (Eitoku et al., 2008). Nucleosome assembly/disassembly factors also known as histone chaperones constitute one of the major classes of such histone-associated chromatin factors. Histone chaperones modulate DNA accessibility via facilitating nucleosome assembly and disassembly both in a replication-coupled and replication-independent manner (Campos and Reinberg, 2009). Furthermore, by associating with free histones, they prevent promiscuous histone-DNA interactions (De Koning et al., 2007). Thus, histone chaperones play a critical role in regulating histone supply and nucleosomal incorporation of histones and hence are involved in almost all the cellular processes involving DNA.
Abiotic stress adaptation in plants requires fine-tuning of diverse pathways such as water uptake/loss, photosynthesis, ion uptake/sequestration, antioxidant balance, and osmolyte biosynthesis (Hirayama and Shinozaki, 2010). Modulating these many pathways requires alteration of expression of a diverse array of genes, which takes place via the coordinated action of various stress-responsive transcription factors as well as chromatin-associated factors. While there is some understanding regarding the role of transcription factors, enzymes catalyzing covalent histone modifications and chromatin remodeling complexes in responses to various abiotic stresses in plants (Sahoo et al., 2013; Kim et al., 2015), the role of histone chaperones in stress response remains enigmatic. Since the exchange of canonical histones with histone variants (e.g. H2A with H2A.Z) or that of covalently modified histone with an unmodified (e.g. acetylated with unacetylated) or a differently modified one (e.g. acetylated with methylated), mediated by histone chaperones, can dramatically alter gene expression at various loci (De Koning et al., 2007), histone chaperones can potentially contribute to stress adaptation in plants. Furthermore, most of the stresses lead to DNA damage that must be repaired (Bray and West, 2005). Histone chaperones promote the access of DNA repair machinery to the site of DNA damage (Polo et al., 2006; Gurard-Levin et al., 2014). Because histone chaperones possess the capacity to modulate gene expression and DNA repair, they may potentially play a pivotal role in plant adaptation to various stresses, which remains to be studied in detail. Indicative findings of a few previous studies (Liu et al., 2009; Zhu et al., 2011, 2013; Weng et al., 2014; Tripathi et al., 2015) suggest that a few histone chaperones, such as ASF1 and NAP1;3, may function in abiotic stress response in plants. However, evidence showing their precise function while a plant responds and adapts to various abiotic stresses is lacking.
In a previous study (Tripathi et al., 2015), we identified 25 rice (Oryza sativa) genes encoding putative histone chaperones belonging to the seven major families viz. CAF1 (Chromatin assembly factor I), FACT (Facilitates chromatin transcription), ASF1 (Antisilencing factor 1), HIRA (Histone regulatory homolog A), NASP (Nuclear Autoantigenic Sperm Protein), SPT6 (Suppressor of Ty element 6), and NAP (Nucleosome Assembly Protein). Via expression profiling, we found eight putative rice histone chaperones (two each from NAP and CAF1C and one each from CAF1A, CAF1B, SPT16, and SSRP families/subfamilies) to be responsive to multiple abiotic stress conditions. Apart from the members of the NAP family of proteins, all other abiotic stress-responsive rice histone chaperones are known to execute nucleosome assembly/disassembly function as multimeric complexes formed by two or more polypeptides (Eitoku et al., 2008; Tripathi et al., 2015).
Here, we investigated the role of a multiple abiotic stress-responsive putative NAP-family histone chaperone, OsNAPL6, vis-à-vis abiotic stress response in rice plants. We report that OsNAPL6 is a nuclear histone chaperone capable of assembling nucleosome-like structure in vitro. To study its physiological function, we generated two sets of transgenic rice plants, overexpressing and underexpressing OsNAPL6, and studied the response of these plants toward various abiotic stress conditions. Overexpression of OsNAPL6 enhanced the stress tolerance ability, while its underexpression showed the opposite effect. We then attempted to gain mechanistic insights into the observed positive correlation of OsNAPL6 expression and stress tolerance potential via high-throughput transcriptome analysis followed by qRT-PCR and chromatin immunoprecipitation-qPCR assays. The results suggested that OsNAPL6 may serve a regulatory role in stress physiology by affecting the expression of several stress-related genes. Together, our results shed light onto a novel function of plant histone chaperones in stress physiology.
RESULTS
OsNAPL6 Is a Nuclear-Localized Protein Possessing a Functional NLS Near Its N Terminus
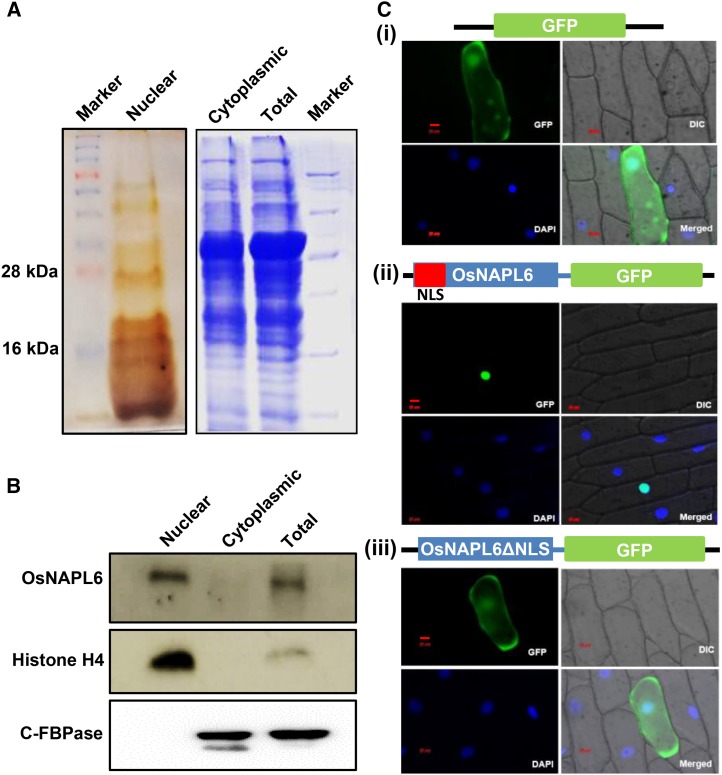
The subcellular localization of a histone chaperone can help in studying its cellular function (De Koning et al., 2007). We therefore examined the subcellular localization of OsNAPL6. The amino acid sequence of OsNAPL6 was first searched for the presence of putative nuclear localization signal (NLS) using cNLS mapper (Kosugi et al., 2009). cNLS mapper predicted a bipartite NLS near the N terminus of OsNAPL6 (Supplemental Fig. S1). To validate the in silico prediction of nuclear localization of OsNAPL6, subcellular fractionation followed by immunoblotting with anti-OsNAPL6 antibody was carried out. OsNAPL6 was found to be present in the nuclear fraction (Fig. 1, A and B). To further confirm its localization in planta, the full-length coding sequence of OsNAPL6 was cloned downstream to CaMV35S promoter in plant localization vector pMBPII so as to generate a fusion protein with a C-terminal GFP (OsNAPL6-GFP). The resultant plasmid (pMBPII-OsNAPL6) and the vector plasmid (pMBPII) were individually bombarded onto onion peel epidermal cells using biolistic method (see “Materials and Methods”), and the localization was examined under a fluorescence microscope (Axio Observer; Carl Zeiss). OsNAPL6-GFP fusion protein (in cells transformed with recombinant plasmid pMBPII-OsNAPL6) localized exclusively to the nucleus in contrast to GFP alone (pMBPII-GFP), which localized in the cytoplasm (Fig. 1, Ci and Cii). Besides, the predicted NLS was deleted to determine whether it is required for nuclear localization of OsNAPL6 protein. The NLS-deleted mutant (OsNAPL6ΔNLS) was expressed as a fusion protein with a C-terminal GFP (OsNAPL6ΔNLS-GFP) and was found to localize predominantly in the cytoplasm (Fig. 1, Ciii).
Figure 1.
Putative histone chaperone OsNAPL6 is a nuclear-localized protein possessing a functional NLS near its N terminus. Nuclear and cytoplasmic protein fractions were resolved on SDS-PAGE (12%) followed either by Coomassie/silver staining (A) or immunoblotting (B) for OsNAPL6, histone H4 (a nuclear marker), and c-FBPase (a cytosolic marker). Immunoblotting for H4 and c-FBPase confirmed the purity of the nuclear and cytoplasmic fractions, respectively. C, Fluorescence micrographs show the intracellular localization of (i) GFP, (ii) OsNAPL6-GFP (OsNAPL6 fused with GFP at its C terminus), and (iii) OsNAPL6ΔNLS-GFP (NLS-deleted OsNAPL6 fused with GFP at its C terminus) in transiently transformed onion peel epidermal cells. Bar = 20 µm. See also Supplemental Figure S1.
OsNAPL6 Is a Functional Histone Chaperone Capable of Assembling Nucleosomes in Vitro
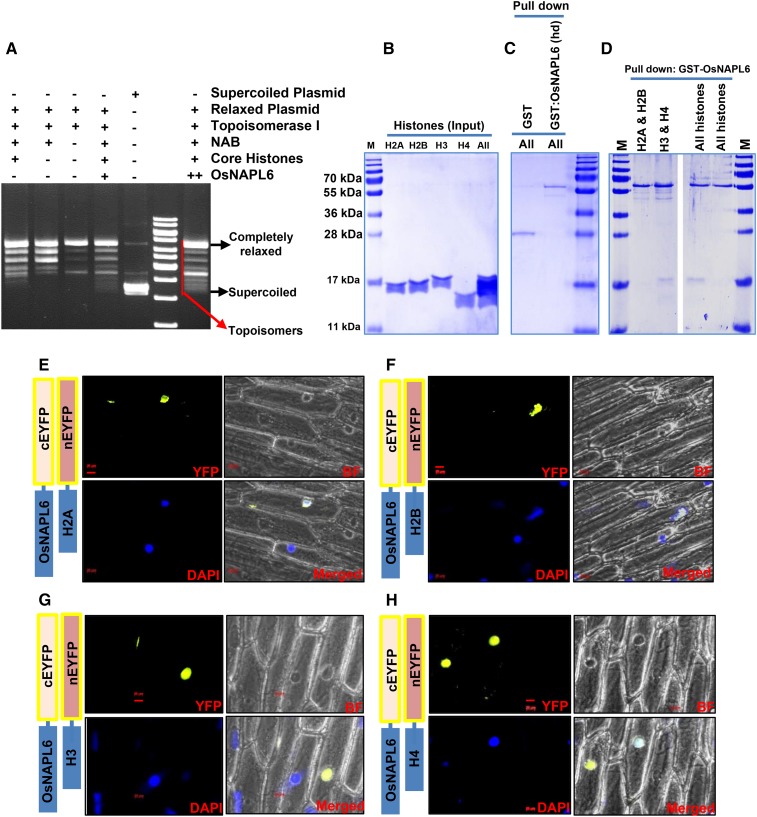
Having established that OsNAPL6 is a nuclear protein, we studied its ability to assemble nucleosomes. For this, we utilized a widely used in vitro nucleosome assembly assay, which is based on the supercoiling of prerelaxed plasmid DNA due to the formation of nucleosome-like structure (Umehara et al., 2002; Sato et al., 2012). The extent of nucleosome formation was measured by analyzing the superhelicity of plasmid DNA after incubation with recombinant OsNAPL6-histone octamer complex in the presence of Topoisomerase I. We found that OsNAPL6 possesses nucleosome assembly capacity as evidenced by the presence of topoisomers with a higher degree of supercoiling in the sample incubated with OsNAPL6-histone octamer compared to the histone control (without OsNAPL6; Fig. 2A). Besides, the observed supercoiling was further higher when double the amount of OsNAPL6 was used in the assay (Fig. 2A, lane marked as “++”), confirming the role of OsNAPL6 in the supercoiling of the prerelaxed plasmid.
Figure 2.
OsNAPL6 is a functional H3/H4 histone chaperone capable of assembling nucleosomes in vitro. A, A plasmid supercoiling assay was utilized for analyzing the nucleosome assembly activity of OsNAPL6 where supercoiled plasmid DNA was first relaxed with Topoisomerase I and the preincubated histone octamer-OsNAPL6 complex was added to the prerelaxed plasmid. Nucleosome-like structures were allowed to form on the relaxed plasmid template in the presence of Topoisomerase I. The proteins were digested and the DNA extracted. Supercoiling of the prerelaxed plasmid DNA resulted in topoisomers with different linking numbers. These topoisomers were separated on a 1.2% agarose gel and the gel was stained with ethidium bromide, postrunning. The positions of completely supercoiled and relaxed forms are indicated. NAB, Nucleosome assembly buffer. B to D, GST pull-down assay for analyzing the histone binding specificity of OsNAPL6. GST-OsNAPL6 fusion protein was immobilized onto glutathione-sepharose beads. Four histones in different combinations (H2A/H2B-H3/H4 [All], H3/H4 [H3 & H4], and H2A/H2B [H2A & H2B]) were added to the bead bound GST-OsNAPL6. Nonspecific binding was removed by multiple washing and the specifically bound histones were eluted. B, Input of histones used for each pull-down. C and D, The pull-down fractions were analyzed on a 15% SDS-PAGE gel. As a negative control, GST was checked for its activity to pull down the histones (C, first lane). To ensure specificity, heat denatured (hd) GST-OsNAPL6 fusion protein was analyzed for its ability to pull down the histones (C, second lane). “All” indicates all four core histones. E to H, BiFC assay was carried out to check the interaction of OsNAPL6 with various histones in planta. OsNAPL6 and H2A (E), H2B (F), H3 (G), or H4 (H) cloned in complementary split-YFP vectors were cotransformed into onion peel epidermal cells using biolistic bombardment method, as indicated. After 16 h of incubation at 28°C, the peels were stained with nuclear marker 4′,6-diamino-phenylindole (DAPI) and observed under a fluorescence microscope. Bars placed at the bottom of the micrographs represents 20 μm. BF, Bright field. See also Supplemental Figure S2.
OsNAPL6 Is an H3/H4 Histone Chaperone
Most of the histone chaperones are known to be specific to either the H3/H4 or H2A/H2B class of histones (Gurard-Levin et al., 2014). Nucleosome assembly activity of OsNAPL6 prompted us to investigate whether it is an H3/H4 chaperone or H2A/H2B chaperone. The NAP family is generally considered to comprise H2A-H2B histone chaperones (De Koning et al., 2007). In our previous study, the phylogenetic analysis of NAP family proteins from yeast, humans, Arabidopsis (Arabidopsis thaliana), and rice showed that OsNAPL6 is evolutionarily closer to HsSET than to other NAP proteins from yeast and humans (Tripathi et al., 2015). HsSET, in spite of being a NAP family histone chaperone, is much different from other NAP family proteins showing binding preference toward the H3-H4 histones (Karetsou et al., 2009), while ScNAP1 and other NAP proteins from humans are considered to be H2A-H2B histone chaperones (De Koning et al., 2007; Park et al., 2005). We attempted the validation of the inferences derived from the phylogenetic study by determining the histone preference of OsNAPL6. For this, a GST pull-down assay (Fig. 2, B–D) was carried out in which GST-NAPL6 fusion protein was incubated with H2A and H2B, with H3 and H4, or with all four core histones (H2A, H2B, H3, and H4). OsNAPL6 interacted directly only with H3/H4 histones and showed a higher binding preference toward histone H3 (Fig. 2D).
Since OsNAPL6 showed considerably higher affinity toward H3/H4 class of histones in vitro, we wondered if a similar interaction pattern exists in planta. For this, a bimolecular fluorescence complementation (BiFC) assay was performed (Supplemental Methods). As apparent from the micrographs (Fig. 2, E–H), we found fluorescence complementation in all the four sets when OsNAPL6 was coexpressed with each of the core histones with complementary YFP tags (c-EYFP-OsNAPL6 + n-EYFP-H2A; c-EYFP-OsNAPL6 + n-EYFP-H2B; c-EYFP-OsNAPL6 + n-EYFP-H3.1; c-EYFP-OsNAPL6 + n-EYFP-H4), suggesting that OsNAPL6 interacts with all the four histones in planta. From in vitro pull-down assays, we already know that the direct interaction of OsNAPL6 with histones H2A and H2B is, at most, weak. Therefore, the interaction of OsNAPL6 with histones H2A and H2B observed in the BiFC assay might be a result of their indirect interaction as explained (Supplemental Fig. S2). Nonetheless, our findings related to OsNAPL6-histone interaction indicate that OsNAPL6 is an H3-H4 histone chaperone but is present in plants in a complex (protein-protein, protein-DNA, or both) that also contains H2A-H2B histones.
Overexpression of OsNAPL6 Improves Tolerance of Rice Plants to Multiple Abiotic Stresses, While Plants Underexpressing OsNAPL6 Show Increased Sensitivity to Various Abiotic Stresses
To study the physiological functions of OsNAPL6, we used the overexpression and the knockdown approach. For overexpression, OsNAPL6 coding sequence was cloned downstream to a CaMV35S constitutive promoter (Supplemental Fig. S3A). For OsNAPL6 knockdown, a specific region from the 3′ untranslated region of OsNAPL6 was cloned both in the sense and antisense direction in the plant RNAi vector pFGC1008 (Supplemental Fig. S3B). pCAMBIA1301-OsNAPL6 overexpression (Ox) and pFGC1008-OsNAPL6-knockdown (KD) constructs were then used for Agrobacterium tumefaciens-mediated transformation of rice (Sahoo et al., 2011) followed by molecular confirmation of the plants for their transgenic status (Supplemental Figs. S4 and S5). Two lines (T2 generation) each showing the maximum overexpression (Ox2.4 and Ox3.2) and underexpression (KD1.2 and KD2.1) were then selected for further functional characterization vis-à-vis abiotic stress response of plants.
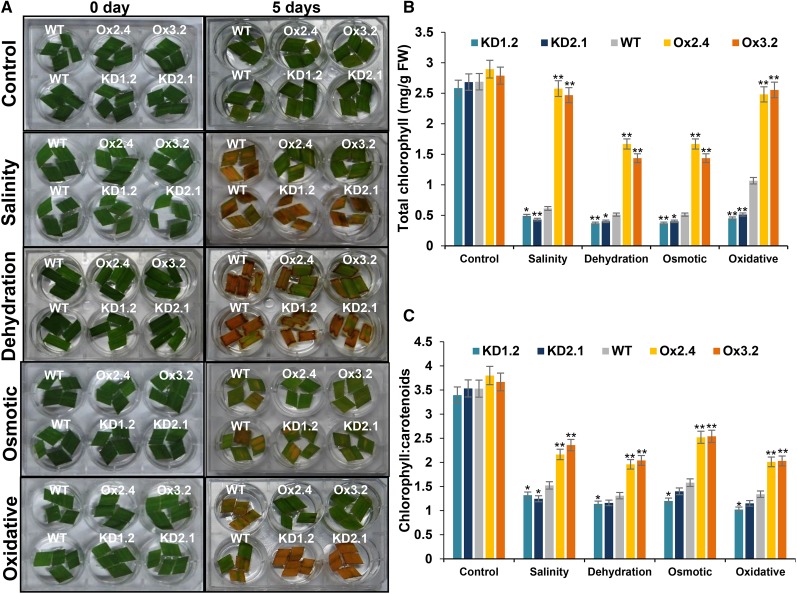
For initial assessment of the involvement of OsNAPL6 in stress tolerance of rice plants, leaf strip senescence assay was carried out. For this, leaf strips were excised and floated on water (as control) or on solutions containing inducing agents of various abiotic stresses viz. salinity, dehydration, osmotic, and oxidative. Leaf strips from Ox plants showed reduced senescence, reduced loss of total chlorophyll, and a higher chlorophyll-to-carotenoid ratio compared to the wild-type plants under all the stress conditions (Fig. 3). KD plants seemed to be more sensitive toward these stresses as evident by greater and faster senescence in leaf strips from these plants (Fig. 3A) and reduced chlorophyll content as well as chlorophyll-to-carotenoid ratio (Fig. 3, B and C) compared to the wild-type plants under most of the stress conditions.
Figure 3.
OsNAPL6 expression improves tolerance toward multiple abiotic stresses. A, Leaf strip senescence assay to analyze the response of OsNAPL6-overexpression and -knockdown toward salinity, dehydration, osmotic, and oxidative stresses. The assay was carried out to test the ability of OsNAPL6-Ox and -KD plants to resist loss in chlorophyll under salinity (200 mm NaCl), dehydration (5% PEG), osmotic (500 mm mannitol), and oxidative (5 mm H2O2) stress conditions. Leaf strips were floated on either water (control) or solutions containing stress agents as indicated. Numbers at the top indicate transgenic line number. Senescence was assessed until 120 h of stress. Leaf strips from wild-type, OsNAPL6-KD, and -Ox plants subjected to leaf strip senescence assay were used for estimation of either total chlorophyll (B) or chlorophyll-to-carotenoid ratio (C). FW, Fresh weight. Data shown are mean ± sd; n = 3. The * and ** represent statistically significant difference (compared to the wild type under the same conditions) at P < 0.05 and P < 0.01, respectively. Statistical significance was tested by one-way ANOVA followed by posthoc comparisons using Tukey-Kramer test.
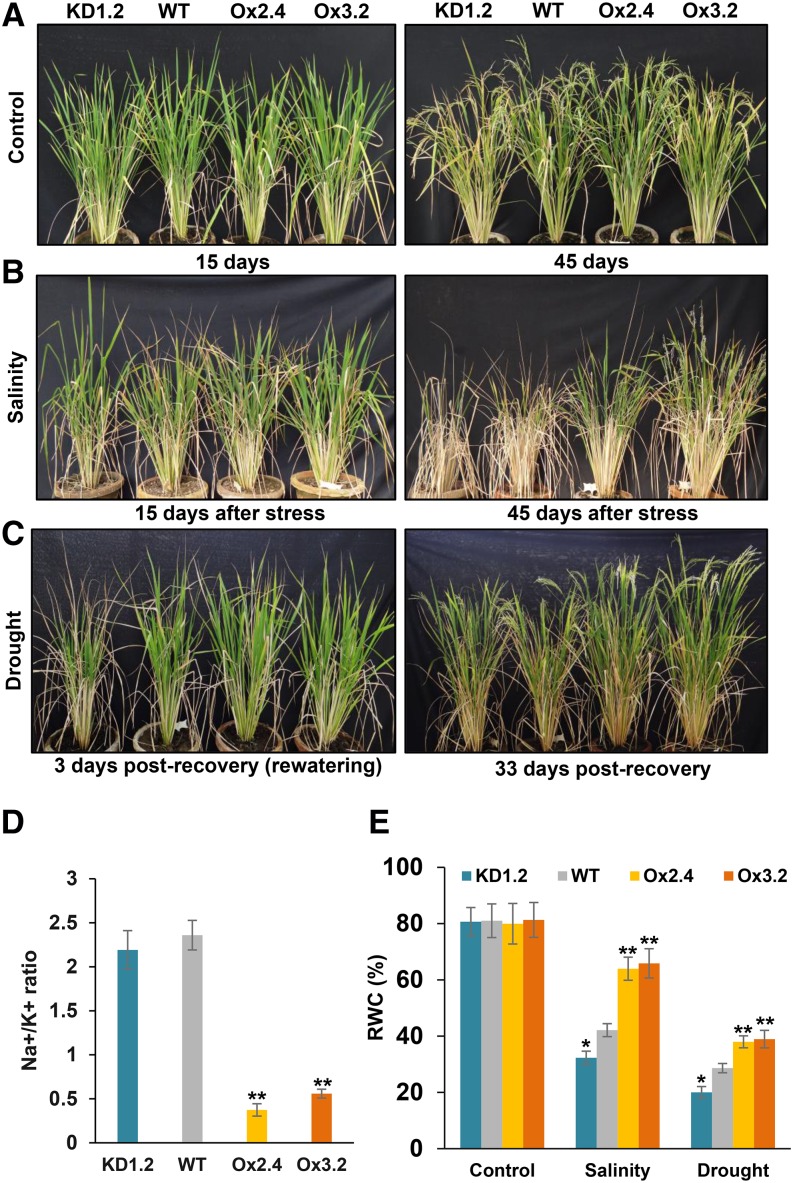
The stress tolerance potential of these plants was further assessed at the whole-plant level for salinity and drought stress at the late vegetative stage (Fig. 4; see “Materials and Methods”). Morphological observations (such as overall shoot growth, plant height, and root growth) indicated that plants overexpressing OsNAPL6 outperformed the wild-type plants under both salinity and drought stress conditions, while the knockdown plants were found to be more sensitive to both the stresses (Fig. 4, A–C; Supplemental Fig. S6). As one of the major mechanisms of adapting to ion toxicity during salinity stress is via maintaining a physiologically favorable Na+/K+ ratio (Zhu, 2001), we measured this ratio in leaf tissues from Ox, wild-type, and KD plants under both control and salinity stress conditions. Comparable Na+/K+ ratio was found under control conditions in all the plants, while under salinity stress conditions, Ox plants showed lesser accumulation of Na+ (relative to K+) compared to wild-type and KD plants as reflected by a lower Na+/K+ ratio (Fig. 4D). Furthermore, to evaluate the effect of osmotic component of salinity, we measured the relative water content (RWC) in these plants. Under salinity stress conditions, RWC of KD and wild-type plants decreased by ∼60 and ∼48%, respectively, compared to the control, whereas in case of Ox plants, the reduction in RWC was only up to 20% (Fig. 4E). Similarly, we found that RWC of KD and wild-type plants grown under drought conditions (measured at 3 d postrecovery) decreased by ∼75 and ∼65%, respectively, compared to the control, whereas in case of Ox plants, the reduction in RWC was around 52% (Fig. 4E). RWC is a comprehensive parameter providing an estimate of leaf water potential as well as osmotic adjustment, both of which play a pivotal role in maintaining turgor-related processes such as growth and stomatal conduction under stress conditions (Turner and Jones, 1980). Hence, the maintenance of a significantly higher RWC in Ox plants with respect to the wild type under both salinity and drought stress conditions indicates that this may be one of the possible physiological mechanisms of stress adaptation observed upon OsNAPL6 overexpression.
Figure 4.
Morphology of wild-type, OsNAPL6-Ox, and OsNAPL6-KD rice plants under salinity stress and after recovery from drought stress conditions. A, Approximately 2-month-old wild-type, OsNAPL6-Ox (lines Ox2.4 and Ox3.2), and OsNAPL6-KD (KD1.2) rice plants were maintained under control conditions by irrigating with water. Similar set of plants were subjected to salinity stress by fortnightly irrigation with a mixture of salt solutions leading to soil electrical conductivity (EC) of 10 dS/m (B) and drought stress (C). For drought stress, water was withheld for 12 d followed by rewatering. Growth of plants was monitored and pictures were taken on days as indicated. D, Leaves from salinity treated plants were harvested 15 d after stress and Na+/K+ ratio was determined by atomic absorption spectroscopy. E, RWC as measured 15 d postsalinity stress and 3 d postrecovery after drought stress. Data shown are mean ± sd; n = 3. The * and ** represent statistically significant difference (compared to the wild type under the same condition) at P < 0.05 and P < 0.01, respectively (one-way ANOVA followed by posthoc comparisons using Tukey-Kramer test).
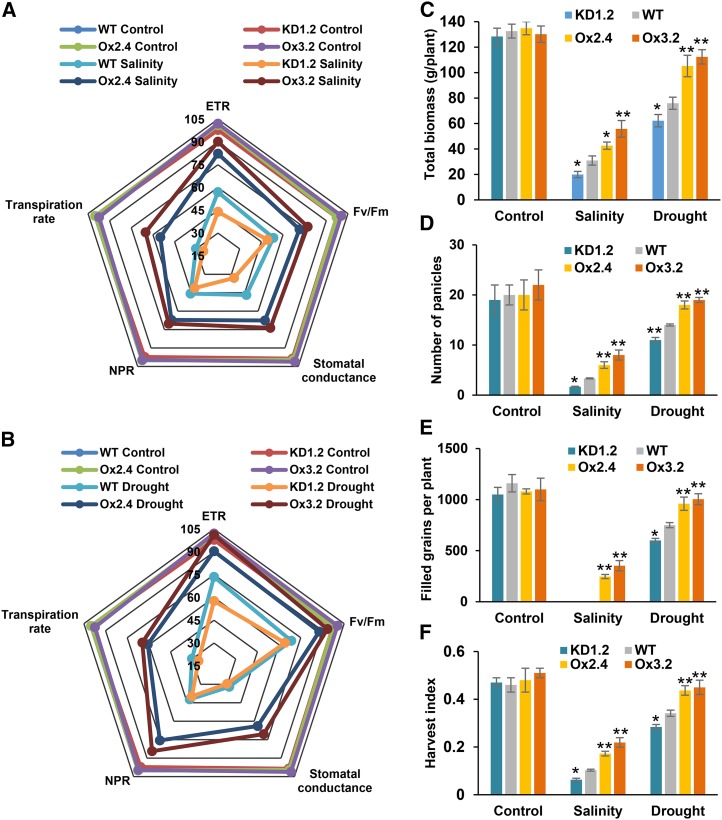
In sensitive plants, abiotic stresses such as salinity and drought lead to reduction in growth, senescence, and cell death apart from the direct effects on various metabolic processes (Munns and Tester, 2008; Sahoo et al., 2013). All these can lead to severe reduction in the efficiency of photosynthetic machinery. Because we found a positive correlation between OsNAPL6 expression and maintained growth (along with reduced senescence) even under salinity and drought stress conditions, we wondered if the plants adapting well to these stresses maintain better photosynthetic efficiency as well. Different photosynthetic parameters viz. net photosynthetic rate, Fv/Fm (PSII efficiency), transpiration rate, electron transport rate, and stomatal conductance were measured. Under control conditions, no appreciable difference was observed in each of the photosynthetic parameters (Fig. 5, A and B; Supplemental Fig. S7). However, under salinity and drought stress conditions, the Ox lines outperformed both wild-type and KD lines, while in most cases, evaluation of the physiological parameters indicated that KD plants are either more or equally sensitive to salinity and drought stresses as compared to the wild-type plants (Fig. 5, A and B; Supplemental Fig. S7). Thus, it appears that better growth and reduced senescence in Ox plants correlated well with improved photosynthetic efficiency under these stress conditions. These physiological as well as morphological observations corroborated well with the total biomass (at maturity), which was found to be in the order Ox > wild type > KD (one-way ANOVA followed by Tukey-Kramer test) under both salinity and drought stress conditions (Fig. 5C).
Figure 5.
Evaluation of various physiological and agronomic parameters shows positive correlation between OsNAPL6 expression and adaptation to salinity and drought stress. Approximately 2-month-old wild-type, OsNAPL6-Ox (lines Ox2.4 and Ox3.2), and OsNAPL6-KD (KD1.2) rice plants were subjected to salinity and drought stress. After 15 d of salinity stress (A) and 3 d postrecovery from drought stress (B), various physiological parameters, viz. net photosynthetic rate (NPR), Fv/Fm, stomatal conductance, transpiration rate, and electron transport rate (ETR), were measured and the relative values (wild-type control, taken as 100%) were plotted as a web diagram. Plants irrigated with water served as controls. After maturity, total biomass (C) and various yield-associated parameters viz. number of panicles (D), filled grains per plant (E), and harvest index (F) were determined and plotted as bar graphs. Data shown are mean ± sd; n = 3. The * and ** represent statistically significant difference (compared to the wild type under the same condition) at P < 0.05 and P < 0.01, respectively. Statistical significance was tested by one-way ANOVA followed by posthoc comparisons using Tukey-Kramer test.
OsNAPL6 Overexpression Reduces the Yield Gap under Salinity and Drought Stress Conditions
All stresses generally impose a “yield penalty” on plants (Cramer et al., 2011; Tripathi et al., 2012). We next addressed if reduced senescence and better photosynthetic efficiency in OsNAPL6 overexpression plants (compared to wild-type plants) under salinity and drought stress conditions can decrease the yield penalty. For this, grain yield of OsNAPL6-Ox, OsNAPL6-KD and wild-type plants was assessed by determining yield-related parameters, such as panicle number, grain number per plant, and harvest index. Under control conditions, no significant difference was observed in the number of panicles. However, under salinity stress conditions, substantially higher number of panicles was observed in Ox plants compared to KD and wild-type plants even though at the same time it was quite low as compared to plants grown under control conditions (Fig. 5D). Probably, the continuous maintenance of an EC of 10 dS/m led to this effect. Generally, salinity stress of this severity is not found in natural rice growing regions (Munns and Tester, 2008). In the plants recovered from drought stress, the decrease in panicle number was, though significant, not as drastic as compared to salinity stress. Under these conditions as well, the number of panicles in Ox plants was considerably higher compared to the KD and wild-type plants (Fig. 5D). The higher number of panicles in Ox plants compared to KD and wild-type plants under both salinity and drought stress conditions correlated well with the grain yield per plant. Under salinity stress conditions, properly filled grains could not develop in KD and wild-type plants whereas, in contrast, Ox plants could develop filled grains to some extent (Fig. 5E).
To further substantiate these results, the harvest index of Ox, KD, and wild-type plants was determined under these conditions. We found that under control conditions the harvest index (HI) varied from 0.46 to 0.51 for all the plants (wild-type, KD, and Ox; Fig. 5F). However, under salinity and drought stress conditions, considerable differences were found in the HI of Ox, KD, and wild-type plants. Under salinity stress conditions, HI reduced by ∼87% in KD plants and ∼78% in wild-type plants. However, in Ox plants, the corresponding decrease was only 57 to 64% (Fig. 5F). In plants recovered from drought stress conditions, the HI reduced by ∼40, 26, and 9 to 12% in KD, wild type, and Ox, respectively (Fig. 5F). This indicates that OsNAPL6 expression correlates well with stress tolerance of rice plants in terms of both vegetative and reproductive growth.
High-Throughput Comparative Transcriptome Analysis of Plants Overexpressing and Underexpressing OsNAPL6 Suggests That OsNAPL6 Expression Effects Large-Scale Changes in the Rice Transcriptome
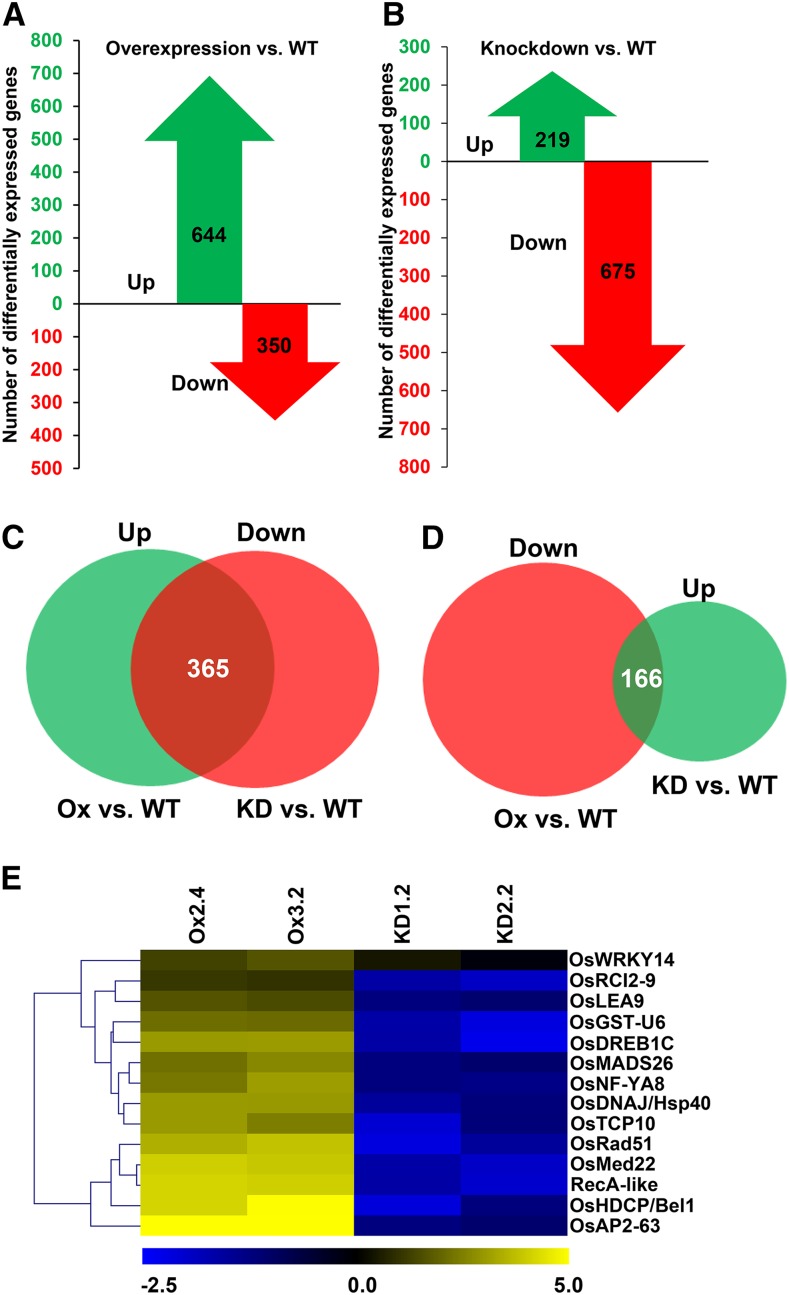
Since OsNAPL6, being a histone chaperone, is capable of assembling nucleosomes, we tested if OsNAPL6-mediated stress adaptation involves alteration in expression of stress-associated genes possibly via modulation of chromatin dynamics at the corresponding genetic loci. To test this hypothesis, microarray-based transcriptome analysis of OsNAPL6-Ox and OsNAPL6-KD as well as wild-type IR64 plants was carried out (see “Materials and Methods” and Supplemental Methods). Large-scale changes were observed both upon overexpression (Fig. 6A; Supplemental Fig. S8A) and underexpression (Fig. 6B; Supplemental Fig. S8B) of OsNAPL6. A total of 644 probe sets showed significantly higher expression (relative to the wild type), while 350 probe sets were downregulated (Fig. 6A) upon OsNAPL6 overexpression. On the other hand, comparison of transcriptome of KD plants with wild-type plants revealed down-regulation of 675 probe sets, while 219 probe sets were found to be upregulated (Fig. 6B). To gain further insights into genes whose expression gets altered upon OsNAPL6 expression, we selected those probe sets that showed anticorrelation of fold difference in expression (reciprocal expression pattern) between Ox and KD plants, i.e. probe sets expressed at a higher level in Ox (relative to the wild type) and showing lower expression in KD (with respect to the wild type) and vice versa were selected (Fig. 6, C and D). A total of 365 probe sets that were expressed at a higher level in Ox plants showed lower expression in KD plants (with respect to the wild type). On the other hand, compared to the wild type, 166 probe sets that showed lower expression in Ox plants were expressed at a higher level in KD plants, relative to the wild type.
Figure 6.
Manipulating OsNAPL6 expression by overexpression and knockdown causes large-scale changes in the rice transcriptome. A, Number of genes up-regulated (green) and down-regulated (red) in Ox plants with respect to wild-type plants. B, Number of genes up-regulated (green) and down-regulated (red) in KD plants with respect to wild-type plants. C, Venn diagram of genes showing up-regulation in Ox plants and down-regulation in KD plants with respect to wild-type plants. Genes common to both the categories and thus showing reciprocal expression pattern are represented by the intersection. D, Venn diagram of genes getting downregulated in Ox plants and upregulated in KD plants with respect to wild-type plants. Genes common to both the categories and thus showing reciprocal expression pattern are represented by the intersection. E, qRT-PCR based validation of the microarray-based expression profile of 14 genes functioning in DNA recombination or repair, transcription, or stress response. Heat maps represent fold change in expression (in log2 scale) of OsNAPL6-Ox and -KD seedlings over wild-type seedlings and were generated using Euclidean distance-based hierarchical clustering. For this, seeds of wild-type, Ox, and KD were germinated and 12-d-old seedlings were used to isolate RNA followed by qRT-PCR. Locus ids of the genes are given in Supplemental Table S1. See also Supplemental Figure S8.
We then attempted to validate the microarray-based expression pattern of 14 genes, selected on the basis of their observed expression pattern in the high-throughput analysis and their known molecular function, via qRT-PCR. These genes encode for proteins involved in transcription, translation, DNA repair, and for some proteins known to mediate stress response in plants. Many of the transcription factors and other proteins that were chosen for further study have been shown to play a role during abiotic stress adaptation in plants (Supplemental Table S1). qRT-PCR-based expression analysis showed that most of the genes showed expression pattern qualitatively similar to that observed in the microarray-based analysis (Fig. 6E; Supplemental Table S1) and their expression pattern indicated coexpression with OsNAPL6, i.e. expression at a higher level in Ox and a lower level in KD vis-à-vis the wild-type (Fig. 6E).
Recruitment of OsNAPL6 at the Respective Promoter Regions of OsRad51 and OsDREB1C
Coexpression of some genes with OsNAPL6 (Fig. 6E) can be attributed to a variety of possible mechanisms involving direct or indirect role of OsNAPL6 in modulating the expression of such genes. For this study, we were more interested in delineating the direct targets of OsNAPL6, i.e. those genes whose expression is regulated by OsNAPL6. Therefore, we used ChIP (chromatin immunoprecipitation)-based assays to study the recruitment of OsNAPL6 to the promoter regions of three genes (OsDREB1C, OsRad51, and OsLEA9) chosen on the basis of their cellular/physiological role, relevance in abiotic stress adaptation, and the results of our studies. OsDREB1C belongs to the DREB (Dehydration Response Element Binding) family of transcription factors, which generally functions in abscisic acid-independent osmotic stress response (Lata and Prasad, 2011). The LEA (Late Embryogenesis Abundant) class of proteins comprises hydrophilic proteins functioning in the prevention of protein aggregation during water stress (Hand et al., 2011) and hence has been shown to play an important role in adaptation to osmotic stresses (Goyal et al., 2005). OsRad51 belongs to a class of eukaryotic recombinases that play a pivotal role during recombination-associated with meiosis and, more importantly, during DNA repair (primarily, homologous recombination-mediated DNA repair; Wang et al., 2014).
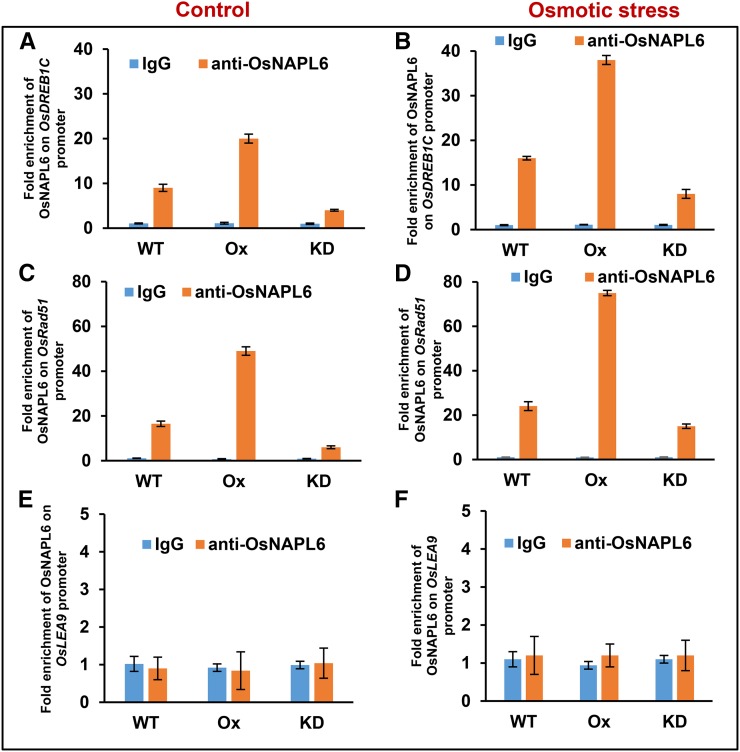
Results of ChIP-qPCR assay showed that while OsNAPL6 was recruited to the promoter regions of OsDREB1C and OsRad51 (Fig. 7, A–D), no significant binding of OsNAPL6 on OsLEA9 promoter could be detected over mock (immunoprecipitate pulled down with IgG; Fig. 7, E and F). The recruitment was more in Ox plants than in wild-type plants, whereas it was less in KD plants compared to the wild-type plants, which correlates well with the cellular levels of OsNAPL6 in each of the plant types (highest in Ox and lowest in KD). Furthermore, under osmotic stress conditions, the recruitment of OsNAPL6 was more on both OsDREB1C (Fig. 7, A and B) and OsRad51 promoter regions (Fig. 7, C and D), suggesting a potential link between higher expression of these under abiotic stress conditions (Lata and Prasad, 2011; Sahoo et al., 2013) and OsNAPL6 function. These results show that OsNAPL6 is recruited to the promoter regions of OsDREB1C and OsRad51, and the stimulation of their expression upon OsNAPL6 overexpression (Fig. 6E) may possibly be linked to the recruitment of OsNAPL6 onto their respective promoters (Fig. 7, A–D), though other mechanisms cannot be ruled out.
Figure 7.
ChIP-qPCR to analyze the recruitment of OsNAPL6 onto the promoter regions of OsDREB1C, OsRad51, and OsLEA9. Recruitment of OsNAPL6 onto the promoter regions of OsDREB1C, OsRad51, and OsLEA9 was analyzed by ChIP-qPCR. For this, seeds of wild-type, OsNAPL6-Ox, and OsNAPL6-KD plants were germinated. Twelve-day-old seedlings were then subjected to osmotic stress (500 mm mannitol) for 8 h. Both stressed and unstressed (control) whole seedlings were harvested and subjected to ChIP (using anti-OsNAPL6 antibody). The presence of DNA regions of interest (part of the promoter region [up to 400–500 bp upstream]) of OsDREB1C (A and B), OsRad51 (C and D), and OsLEA9 (E and F) was analyzed by qPCR using the immunoprecipitated DNA after elution. The recruitment is expressed as fold enrichment over IgG (mock). Data shown are mean ± se; n = 3.
OsNAPL6 Mediates Efficient DNA Repair and Aids in Abrogation of Programmed Cell Death under Salinity and Genotoxic Stress Conditions
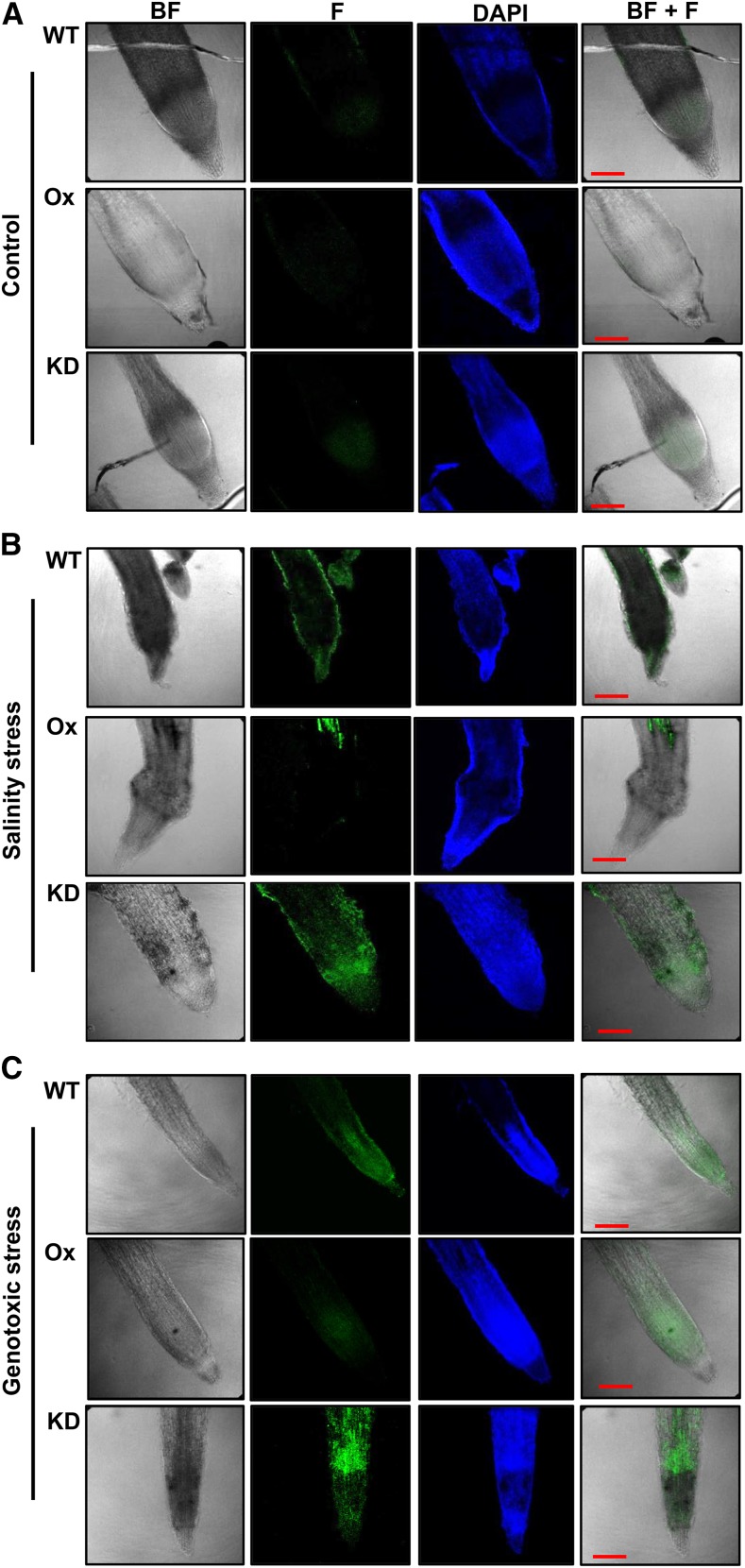
All stresses generally lead to at least some degree of DNA damage either directly or indirectly (via reactive oxygen species) or even both (Bray and West, 2005). Our results that OsNAPL6 may stimulate the expression of OsRad51 suggested an intriguing mechanism vis-à-vis the role of OsNAPL6 in abiotic stress response. That is, apart from other mechanisms (for example, transcriptional stimulation of OsDREB1C, other transcription factors, and stress-related proteins), efficient DNA repair mediated by OsNAPL6 may be an important stress defense mechanism. Therefore, we addressed as to if there exists a correlation between OsNAPL6 expression and efficiency of DNA repair. For this, we assessed DNA damage (primarily nicks or breaks) in wild-type, OsNAPL6-Ox, and OsNAPL6-KD plants under both control and stress conditions. An in situ TUNEL (terminal deoxynucleotidyl transferase dUTP nick-end labeling) assay was carried out to detect DNA fragmentation. Since breaks in internucleosomal DNA almost invariably lead to programmed cell death (PCD), TUNEL assay can also provide some information about PCD. Results of TUNEL assay showed that differences in the extent of DNA fragmentation in meristematic regions of roots overexpressing (Ox) and underexpressing OsNAPL6 (KD) compared to the wild type were more pronounced under stress conditions (Fig. 8; Supplemental Fig. S9). Nonetheless, even under control conditions, the extent of DNA fragmentation was found to be more in KD plants compared to the wild type (Fig. 8A; Supplemental Fig. S9A). Under salinity stress conditions, confocal microscopy revealed lesser incorporation of fluorescein-12-dUTP in meristematic regions of roots of Ox plants compared to the wild type (Fig. 8B; Supplemental Fig. S9B). On the contrary, the meristematic regions of roots of KD plants showed higher incorporation of fluorescein-12-dUTP compared to that of the wild-type roots (Fig. 8B; Supplemental Fig. S9B). As the incorporation of fluorescein-12-dUTP in the TUNEL reaction utilized here is directly proportional to the number of free DNA termini (usually due to breaks), these results indicate that roots of Ox plants have reduced DNA damage and, hence, possibly lower PCD as compared to the wild-type plants under salinity stress conditions. In contrast, meristematic regions of roots from KD plants showed more fluorescein-12-dUTP fluorescence indicating more free DNA termini, which implies enhanced breakage and, hence, exacerbated PCD under salinity stress conditions (Fig. 8B; Supplemental Fig. S9B).
Figure 8.
TUNEL assay to detect in situ DNA fragmentation in root tips of wild-type, OsNAPL6-Ox, and OsNAPL6-KD rice seedlings under control, salinity, and genotoxic stress conditions. To detect in situ DNA fragmentation in root tips, seeds from wild-type, OsNAPL6-Ox, and OsNAPL6-KD plants were germinated and grown on 0.5× MS + solid agar medium for 12 d and then subjected to 200 mm NaCl (for salinity stress) for 48 h (B) or to genotoxic stress by using 40 mg/L aphidicolin for 48 h (C) followed by fixing, permeabilization, and TUNEL reaction. Water-treated seedlings served as controls (A). Green color represents fluorescein-12-dUTP, while blue color shows DAPI staining. For microscopy, samples were mounted on slides in ProLong Gold Antifade mountant with DAPI (Life Technologies) and the slides were viewed under a confocal microscope (Nikon A1R) using a 20× objective. Images were acquired and processed using NIS Elements software (Nikon). Bar = 100 µm. BF, Bright field; F, fluorescein (green).
Because salt stress conditions cause DNA damage indirectly (for example, via reactive oxygen species generation; Mittler, 2002), the observations (Fig. 8B; Supplemental Fig. S9B) can be explained by considering three possibilities: there is lesser damage in the Ox plants, the repair mechanism is more efficient in these plants, or both. To comment upon the exact cause of apparent lesser DNA damage in overexpression plants and to test the direct correlation between DNA repair efficiency and observed extent of DNA fragmentation possibly coupled with PCD, the seedlings were exposed to a DNA damaging agent aphidicolin. A considerably higher DNA fragmentation was detected in the roots of aphidicolin-treated KD seedlings compared to the wild-type plants, while fluorescein-12-dUTP labeling and, hence, DNA fragmentation was found to be lowest in the root cells of Ox seedlings (Fig. 8C; Supplemental Fig. S9C). These observations indicate that OsNAPL6 expression is crucial in providing resistance to PCD and the associated DNA fragmentation under stress conditions, possibly due to improved DNA repair mediated via OsRad51, among other probable mechanisms.
DISCUSSION
Histone chaperones, owing to their nucleosome assembly/disassembly activity in conjunction with other chromatin-associated factors, can substantially change the transcriptional competence of a region of chromatin (Akey and Luger, 2003). Owing to this, histone chaperones can potentially play a significant role in modulation of responses to various stresses in plants as transcription is a major point of regulation of most of the stress-associated physiological processes. However, there is a paucity of reports elucidating the role of plant histone chaperones in stress response and adaptation. Previously, through expression profiling of histone chaperones from rice and Arabidopsis, we had shown that a few putative histone chaperones show differential expression under various abiotic stress conditions (Tripathi et al., 2015). Out of the eight stress-regulated putative histone chaperones in rice, except the NAP family of proteins, all others are known to execute histone chaperone function as multimeric complexes formed by two or more polypeptides (De Koning et al., 2007; Tripathi et al., 2015); hence, altering the expression of a single member of such complexes might not show a phenotype. For studying the function of histone chaperones during abiotic stress response and adaptation in plants, we therefore selected a multiple stress-responsive histone chaperone OsNAPL6 (a member of the rice NAP family of histone chaperones) primarily because of its potential ability to function as a histone chaperone without its association with other proteins.
Subcellular localization of a histone chaperone defines its exact function. For example, a nuclear-localized histone chaperone may be involved in deposition/eviction of histones onto/from the nucleosomes, whereas a cytosolic histone chaperone may buffer histones at their site of biosynthesis (Cook et al., 2011; Gurard-Levin et al., 2014). In this context, exclusive nuclear localization of OsNAPL6 (Fig. 1) suggests that it may play a role in replication-coupled or replication-independent nucleosome assembly/disassembly. Once OsNAPL6 was found to be a nuclear-localized protein, we tested its ability to assemble nucleosomes. Though there have been some reports that have classified a plant protein as histone chaperone utilizing various bioinformatics tools (Zhu et al., 2013), not many studies have shown the nucleosome assembly/disassembly activity of such histone chaperones. Therefore, we utilized the plasmid supercoiling assay and verified that the putative histone chaperone OsNAPL6 is capable of assembling nucleosomes and hence is indeed a histone chaperone (Fig. 2A). To further test the specificity, we also used a higher amount of OsNAPL6 in the supercoiling assay and expectedly there was an increase in the proportion of topoisomers with a higher superhelicity (in magnitude terms; Fig. 2A, lane marked as ++).
During chromatin assembly, (H3-H4)2 tetramers are believed to be deposited prior to the deposition of H2A-H2B dimers (Burgess and Zhang, 2013). Therefore, H3-H4 deposition is thought to be a crucial step during chromatin assembly. It is in this context that the histone specificity of OsNAPL6 toward H3/H4 histones gains significance (Fig. 2). Being an H3/H4 histone chaperone, it is possibly involved in the first step of chromatin assembly and hence might play a greater role (as compared to an H2A/H2B histone chaperone) in determining the chromatin dynamics and hence DNA accessibility of particular loci. Because DNA accessibility during transcription as well as during DNA repair is a key point of regulation of these processes and considering the fact that both transcription and DNA repair are altered under various stress conditions to which plants are exposed (Bray and West, 2005; Sahoo et al., 2013), it is plausible that the H3-H4 histone chaperone OsNAPL6 plays a role during stress response and adaptation.
In yeast, HIRA histone chaperone has been shown to be involved in transcriptional stimulation in response to osmotic and oxidative stresses (Chujo et al., 2012). Previous studies, including ours (Zhu et al., 2013; Tripathi et al., 2015), have suggested that expression of a few plant histone chaperones is perturbed during biotic and abiotic stress conditions. However, the lack of functional characterization studies has precluded a detailed understanding of the function of histone chaperones during abiotic stress response and adaptation in plants. It is against this background that the findings of our study gain significance as we report the role of a histone chaperone during abiotic stresses. Our findings that alteration in levels of OsNAPL6 positively correlates with adaptation to various abiotic stresses both in terms of vegetative growth and yield (Figs. 3–5) provide evidence for the role of OsNAPL6 in response and adaptation to abiotic stresses.
Owing to the complexity of the means through which plants respond to different abiotic stresses, numerous cellular processes and the network of genes involved therein together determine the outcome of stress response (Urano et al., 2010; Hirayama and Shinozaki, 2010). Taking clues from some previous studies that have shown genome-wide transcriptional changes attributed to the nucleosome assembly/disassembly activity of histone chaperones (Zhu et al., 2006; Chujo et al., 2012), we hypothesized that manipulating the expression of OsNAPL6 may alter the expression of several genes, many of which might be associated with abiotic stress response. Consistent to our hypothesis, we observed large-scale changes in the rice transcriptome when the expression of OsNAPL6 was manipulated by overexpression or knockdown (Fig. 6, A–D). The coexpression of a histone chaperone (Fig. 6E) with its putative target gene can be attributed either to direct or indirect effects of histone chaperone or even it may have no cause-and-effect relation. When we examined these possibilities with respect to the observed coexpression of OsNAPL6 with OsDREB1C, OsLEA9, and OsRad51 (Fig. 6E; Supplemental Table S1), we found that OsNAPL6 is recruited to the promoter regions of OsDREB1C and OsRad51 (Fig. 7, A–D); hence, the coexpression of OsNAPL6 with these two genes might possibly be a result of transcriptional stimulation-mediated by the histone chaperone activity of OsNAPL6. Histone chaperones possess the capacity to stimulate gene expression via taking part in transient nucleosome disassembly or via histone exchange (e.g. exchange of H2A with H2A.Z; De Koning et al., 2007). We cannot, at present, comment upon the exact mode explaining our results, i.e. whether it is nucleosome disassembly or histone exchange or both. Nevertheless, our results show that a plant histone chaperone is recruited to genomic loci and affects gene expression. These finding have huge implications toward the role of OsNAPL6 in abiotic stress adaptation. This is because DREB1C belongs to a class of dehydration-responsive transcription factors (DREB), which has previously been found to mediate tolerance to osmotic and other abiotic stresses (Lata and Prasad, 2011), and Rad51 is a crucial factor in repair of double-stranded breaks in DNA (Wang et al., 2014), which often occur during severe abiotic stress conditions like salinity, drought, and exposure to heavy metals (Bray and West, 2005).
Assessment of DNA damage in seedlings subjected to salinity and genotoxic stress led us to conclude that DNA damage is more efficiently repaired in overexpression plants, while the repair mechanism is somewhat compromised in the knockdown plants (Fig. 8). These findings suggest that OsNAPL6 promotes efficient DNA damage repair and helps in abrogating programmed cell death. Previously, it has been shown that some histone chaperones such as ASF1 in humans mediate the recognition step of DNA repair by facilitating histone exchange and nucleosome disassembly (Polo et al., 2006). Furthermore, it has also been found that during the restoration step as well, histone chaperones like HIRA and CAF-I aid the reassembly of nucleosomes (Soria et al., 2012; Adam et al., 2013). In case of plants, it has been shown that double mutation of NRP1 and NRP2 (histone chaperones from the NAP family) in Arabidopsis increases sensitivity to DNA damage (Zhu et al., 2006). However, the mechanistic link between DNA damage repair and plant histone chaperones still remains elusive. Our results help in bridging some missing links as we have shown that a rice histone chaperone (OsNAPL6) stimulates OsRad51 expression by getting recruited to its promoter region and the enhanced levels of OsRad51 (Figs. 6E, 7C, 7D, and 8) may be instrumental in the efficient repair of DNA damage. These results are also unique insofar as we show that OsNAPL6 confers tolerance to abiotic stresses via promoting DNA repair apart from other possible means. Furthermore, this mechanism explains the similar response of OsNAPL6-Ox plants to multiple abiotic stress conditions (Figs. 3–5) as DNA damage is a common consequence of most of the abiotic stresses.
CONCLUSION
Taken together, our findings implicate histone chaperones in abiotic stress response and adaptation in plants. The results of this study have together unraveled one of the major mechanisms of modulation of stress response by the rice histone chaperone OsNAPL6. We have shown the recruitment of a plant histone chaperone to genomic loci, which, to our knowledge, was hitherto unreported. The finding that OsNAPL6 is recruited to the promoter region of OsDREB1C, a transcription factor, which can then drive the expression of various osmotic stress-associated genes, is significant in the context of stress response and adaptation. Furthermore, the involvement of efficient DNA repair in stress adaptation mediated by OsNAPL6, possibly via OsRad51, provides a common mechanism of tolerance to multiple abiotic stresses. While we have examined the transcriptional modulation of two of the predicted target genes (OsDREB1C and OsRad51) by OsNAPL6, many more such putative targets remain to be studied in detail. Furthermore, precisely how a plant histone chaperone, in general, and OsNAPL6, in particular, is recruited to specific genomic loci is hitherto unanswered. It is plausible that specific DNA-binding factors may aid the recruitment of OsNAPL6 to particular loci, experimental proof for which remains to be generated. Future studies on these lines would improve our understanding regarding the role of plant histone chaperones in abiotic stress response.
MATERIALS AND METHODS
Plant Material
Rice (Oryza sativa sp. indica cv IR64) has been used throughout the study including for the generation of transgenic plants. T2 generation transgenic plants overexpressing (Ox) or underexpressing (KD) OsNAPL6 were used to assess the effect of OsNAPL6 expression on stress response of rice plants.
Origin of the Gene Sequences
OsNAPL6 sequence used throughout the study, including for cloning and in silico analysis, is from O. sativa sp. indica cv IR64 (GenBank ID: EEC73468.1). To design primers for the other genes/promoter fragments, sequence information corresponding to Nipponbare (a japonica rice cultivar) was obtained from Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). For cloning of genes, cDNA prepared from RNA isolated from leaf tissue of rice plants of the IR64 genotype was used as template.
Subcellular Localization by Creating a C-Terminal GFP Fusion and Transient Expression in Onion Peel Epidermal Cells
Subcellular localization of OsNAPL6 was studied by gold particle bombardment-mediated transient expression in onion peel epidermal cells as detailed in Supplemental Methods.
Subcellular Fractionation
Nuclear and cytoplasmic fractions from leaves of 14-d-old rice seedlings were isolated using plant nuclei isolation/extraction kit (CelLytic PN; Sigma-Aldrich) following manufacturer’s instructions. Polyclonal anti-OsNAPL6 antiserum was generated by immunizing rabbit with homogeneously purified recombinant OsNAPL6 following the standard procedure.
In Vitro Plasmid Supercoiling Assay
The assay was carried out using prerelaxed pUC19 as the template and purified 6x-His-OsNAPL6 recombinant protein as the putative histone chaperone following a procedure described previously (Sato et al., 2012) with some modifications. To obtain recombinant OsNAPL6 protein, its coding sequence was cloned in the bacterial expression vector pET28a (EMD Millipore) at BamHI and EcoRI restriction sites. Primers used for PCR amplification have been listed in Supplemental Methods. The construct was transformed in Escherichia coli BL-21 (DE3) strain and the expression was induced using 0.1 mm IPTG followed by cell lysis. Recombinant His-tagged OsNAPL6 (6xHis-OsNAPL6) was purified via Ni-NTA affinity chromatography following standard procedure. Purity of the recombinant protein was determined by resolving it on a 12% SDS-PAGE gel followed by staining with Coomassie Brilliant Blue.
For in vitro plasmid supercoiling assay, 200 ng supercoiled plasmid DNA (pUC19) was relaxed with one unit of Topoisomerase I (Promega) by incubating the mixture at 37°C for 1 h in a total volume of 9 µL (reaction mixture I). Histone octamer-histone chaperone complex was formed by incubating 600 ng of core histone octamer (core histones HeLa; Millipore) with 1 µg of recombinant OsNAPL6 (putative histone chaperone) in 1.5× nucleosome assembly buffer (15 mm Tris-HCl, pH 8.0, with 1.5 mm EDTA, 225 mm NaCl, and 150 µg/mL BSA in a total volume of 18 µL) for 30 min at 30°C (reaction mixture II). The two reaction mixtures were combined (core histone octamer-histone chaperone complex [18 µL] was added to the relaxed plasmid [9 µL]) and incubated at 37°C for 1 h. Nucleosome-like structures were allowed to form on the prerelaxed plasmid template (in the presence of Topoisomerase I present in the reaction mixture I). The reaction was stopped by digesting the proteins with Proteinase K (final concentration 0.1 mg/mL) in the presence of SDS (final concentration 0.2% w/v) at 37°C for 15 min. DNA was extracted using phenol-chloroform-isoamyl alcohol followed by precipitation using isopropanol in the presence of 0.2 m NaCl and 100 µg/mL glycogen. Supercoiling of the prerelaxed plasmid would result into a population of topoisomers with different linking numbers, which were separated on a 1.2% agarose gel in the absence of ethidium bromide and the gel was stained with ethidium bromide (0.5 µg/mL) postrunning for visualization.
GST Pull-Down Assay
Direct interaction of histones with recombinant GST-OsNAPL6 was analyzed using GST pull-down assay as described previously (Natsume et al., 2007) with modifications as described in Supplemental Methods.
In Planta Interaction of OsNAPL6 with Histones
In planta interaction of OsNAPL6 with histones was studied using BiFC assay. The detailed procedure has been provided in Supplemental Methods.
Generation of OsNAPL6-Ox and -KD Rice Transgenic Plants
Ox and KD plants were generated following standard procedures of cloning and plant transformation followed by selection, regeneration, hardening (Sahoo et al., 2011), and molecular confirmation of the transgenic status by various means as detailed in Supplemental Methods.
Stress Treatment and Evaluation of Various Physiological and Agronomical Parameters
Leaf strip senescence assay was carried out as detailed in Supplemental Methods. For salinity stress treatment at the late vegetative stage, 2-month-old plants (wild-type, overexpression lines Ox2.4 and 3.2, and knockdown line KD1.2) of comparable growth were subjected to irrigation every fortnight with a mixture of salts (50 mm NaCl, 3.75 mm MgCl2, 15 mm MgSO4, and 6.25 mm CaCl2) resulting in soil electrical conductivity (EC) of 10 dS/m, which was maintained until the completion of their life cycle. The growth was monitored regularly and various physiological parameters were evaluated 15 d after the first application of salinity stress whereas yield parameters and root length were determined after the plants attained maturity. For drought stress, approximately 2-month-old plants of comparable growth were subjected to water withdrawal for 12 d following which the plants were recovered by rewatering. Various physiological parameters were evaluated 3 d postrecovery (evaluation of physiological parameters could not be carried out during the period of water withdrawal due to leaf rolling), while yield parameters and root length were measured after the plants attained maturity. The physiological parameters were measured as described previously (Ghosh et al., 2014). HI was determined by calculating the ratio of dry mass of harvested grains and total shoot dry mass. Total biomass of plants was determined after the plants attained maturity (harvesting stage).
High-Throughput Comparative Transcriptome Profiling
Microarray-based high-throughput comparative transcriptome profiling was carried out using Ox, KD, and wild-type leaf tissue from 12-d-old seedlings of the T2 generation (grown under control conditions) as the starting material. Detailed methodology followed for high-throughput comparative transcriptome profiling including data analysis has been provided in the Supplemental Methods section.
qRT-PCR and ChIP-qPCR
qRT-PCR was carried out as described elsewhere (Tripathi et al., 2015). The sequences of various primers used for qRT-PCR has been provided in the Supplemental Methods. ChIP-qPCR was performed using leaf tissues from 12-d-old rice seedlings of the wild type as well as transgenic lines (grown under control and osmotic stress conditions) as the starting material following the procedure described in Supplemental Methods.
TUNEL Assay
In situ TUNEL assay was carried out using DeadEnd Fluorometric TUNEL system (Promega) following the manufacturer’s instructions with some modifications. Briefly, seeds from wild-type, OsNAPL6-Ox, and OsNAPL6-KD plants were germinated and grown on 0.5× MS (Murashige and Skoog medium) + solid agar medium for 12 d and then subjected either to 200 mm NaCl (for salinity stress) for 48 h or to genotoxic stress by using 40 mg/L aphidicolin for 48 h followed by fixing, permeabilization, and TUNEL reaction as detailed in Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EEC73468.1 (OsNAPL6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence analysis of OsNAPL6 predicts a bipartite NLS near its N terminus.
Supplemental Figure S2. Model showing various possibilities explaining the apparent in planta interaction of OsNAPL6 with histones H2A and H2B.
Supplemental Figure S3. Schematic representation of OsNAPL6-overexpression and -knockdown vector constructs used for generation of transgenic rice plants overexpressing and underexpressing OsNAPL6.
Supplemental Figure S4. Molecular confirmation of OsNAPL6 overexpression transgenic plants via Southern hybridization, qRT-PCR, and immunoblotting.
Supplemental Figure S5. Confirmation of siRNA-mediated knockdown of OsNAPL6 in OsNAPL6-knockdown plants.
Supplemental Figure S6. Root growth and plant height of wild-type, OsNAPL6-overexpression, and OsNAPL6-knockdown rice plants under salinity and drought stress conditions.
Supplemental Figure S7. Various photosynthetic parameters of OsNAPL6-overexpression and OsNAPL6-knockdown rice plants under salinity stress and 3 d postrecovery from drought stress conditions.
Supplemental Figure S8. Volcano plot showing differentially expressed genes in OsNAPL6-overexpression and OsNAPL6-knockdown plants as compared to the wild-type plants.
Supplemental Figure S9. Extent of DNA fragmentation in root tips of OsNAPL6-overexpression and OsNAPL6-knockdown plants relative to the wild-type plants as observed in TUNEL assay.
Supplemental Table S1. Details of a few genes that were found to be differentially expressed in the high-throughput comparative transcriptome analysis of OsNAPL6-overexpression, OsNAPL6-knockdown, and wild-type plants and were selected for further expression analysis by qRT-PCR.
Supplementary Material
Acknowledgments
A.K.T. thanks the Department of Biotechnology, Government of India, for providing a senior research fellowship.
Glossary
- BiFC
bimolecular fluorescence complementation
- RWC
relative water content
- HI
harvest index
- ChIP
chromatin immunoprecipitation
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- PCD
programmed cell death
Footnotes
Articles can be viewed without a subscription.
This work was supported by funds from the Department of Biotechnology, Government of India and internal grants of the International Centre for Genetic Engineering and Biotechnology.
References
- Adam S, Polo SE, Almouzni G (2013) Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106 [DOI] [PubMed] [Google Scholar]
- Akey CW, Luger K (2003) Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13: 6–14 [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE (2005) DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z (2013) Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Chujo M, Tarumoto Y, Miyatake K, Nishida E, Ishikawa F (2012) HIRA, a conserved histone chaperone, plays an essential role in low-dose stress response via transcriptional stimulation in fission yeast. J Biol Chem 287: 23440–23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G (2011) A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell 44: 918–927 [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G (2007) Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14: 997–1007 [DOI] [PubMed] [Google Scholar]
- Eitoku M, Sato L, Senda T, Horikoshi M (2008) Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci 65: 414–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Pareek A, Sopory SK, Singla-Pareek SL (2014) A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J 80: 93–105 [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin ZA, Quivy JP, Almouzni G (2014) Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem 83: 487–517 [DOI] [PubMed] [Google Scholar]
- Hand SC, Menze MA, Toner M, Boswell L, Moore D (2011) LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol 73: 115–134 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Karetsou Z, Emmanouilidou A, Sanidas I, Liokatis S, Nikolakaki E, Politou AS, Papamarcaki T (2009) Identification of distinct SET/TAF-Ibeta domains required for core histone binding and quantitative characterisation of the interaction. BMC Biochem 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H (2009) Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–4748 [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Gao J, Dong AW, Shen WH (2009) A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol Plant 2: 688–699 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446: 338–341 [DOI] [PubMed] [Google Scholar]
- Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K (2005) Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem 280: 1817–1825 [DOI] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G (2006) New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Sahoo KK, Tripathi AK, Pareek A, Singla-Pareek SL (2013) Taming drought stress in rice through genetic engineering of transcription factors and protein kinases. Plant Stress 7: 60–72 [Google Scholar]
- Sahoo KK, Tripathi AK, Pareek A, Sopory SK, Singla-Pareek SL (2011) An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 7: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ishiai M, Toda K, Furukoshi S, Osakabe A, Tachiwana H, Takizawa Y, Kagawa W, Kitao H, Dohmae N, et al. (2012) Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J 31: 3524–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G (2012) Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734 [DOI] [PubMed] [Google Scholar]
- Tripathi AK, Pareek A, Sopory SK, Singla-Pareek SL (2012) Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes. Rice (N Y) 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AK, Singh K, Pareek A, Singla-Pareek SL (2015) Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol 15: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Jones MM. 1980. Turgor maintenance by osmotic adjustment: a review and evaluation. In Turner NC, Kramer PJ eds, Adaptation of Plants to Water and High Temperature Stress. Wiley Interscience, New York, pp 87–103 [Google Scholar]
- Umehara T, Chimura T, Ichikawa N, Horikoshi M (2002) Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells 7: 59–73 [DOI] [PubMed] [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Wang H, Cheng Z, Li W, Zhu G, Wang Y, Ma H (2014) The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol 201: 292–304 [DOI] [PubMed] [Google Scholar]
- Weng M, Yang Y, Feng H, Pan Z, Shen WH, Zhu Y, Dong A (2014) Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ 37: 2128–2138 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Meyer D, Pichon O, Renou JP, Cao K, Shen WH (2006) Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18: 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Shen WH (2013) Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta 1819: 343–348 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, Dong A (2011) Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J 66: 443–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.