Abscisic acid signaling and biosynthesis are feedback-regulated by transcription factor OsbZIP23 that targets diverse genes involved in drought resilience.
Abstract
The OsbZIP23 transcription factor has been characterized for its essential role in drought resistance in rice (Oryza sativa), but the mechanism is unknown. In this study, we first investigated the transcriptional activation of OsbZIP23. A homolog of SnRK2 protein kinase (SAPK2) was found to interact with and phosphorylate OsbZIP23 for its transcriptional activation. SAPK2 also interacted with OsPP2C49, an ABI1 homolog, which deactivated the SAPK2 to inhibit the transcriptional activation activity of OsbZIP23. Next, we performed genome-wide identification of OsbZIP23 targets by immunoprecipitation sequencing and RNA sequencing analyses in the OsbZIP23-overexpression, osbzip23 mutant, and wild-type rice under normal and drought stress conditions. OsbZIP23 directly regulates a large number of reported genes that function in stress response, hormone signaling, and developmental processes. Among these targets, we found that OsbZIP23 could positively regulate OsPP2C49, and overexpression of OsPP2C49 in rice resulted in significantly decreased sensitivity of the abscisic acid (ABA) response and rapid dehydration. Moreover, OsNCED4 (9-cis-epoxycarotenoid dioxygenase4), a key gene in ABA biosynthesis, was also positively regulated by OsbZIP23. Together, our results suggest that OsbZIP23 acts as a central regulator in ABA signaling and biosynthesis, and drought resistance in rice.
Drought has become one of the most serious problems restricting the worldwide productivity of rice (Oryza sativa) due to global climate change, fresh water shortage, and increasing population. To improve the drought resistance of rice, many approaches such as marker-assisted selection, genome-wide association, and reverse genetics screening have been applied and significant progress has been achieved (Hu and Xiong, 2014).
Abscisic acid (ABA) signaling plays critical roles in the drought response, and it has been well characterized after the discovery of the ABA receptor in Arabidopsis (Arabidopsis thaliana; Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). Under drought stress, ABA accumulates in plant cells and is then perceived by an ABA receptor. Pyrabactin resistance1 (PYR1)/PYR1-like (PYL)/regulatory components of the ABA receptor (RCAR) inhibits the phosphatase activities of clade A protein phosphatase 2Cs (PP2Cs), and inactivation of PP2Cs causes the activation of Suc nonfermenting-1-related protein kinase2 (SnRK2s), which further activates the downstream substrates such as transcription factors and membrane ion channels (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Zhao et al., 2013).
Through bioinformatics analysis, homologs of ABA signaling components in rice have been identified. The rice genome contains 12 OsPYLs (Kim et al., 2012; He et al., 2014), 10 clade A PP2Cs (Xue et al., 2008), and 10 osmotic stress/ABA-activated protein kinases (SAPKs; Kobayashi et al., 2004). The molecular functions and interactions between some of these components were investigated in recent years. Most OsPYLs (OsPYL1-3, OsPYL6, and OsPYL10-11) can significantly inhibit the phosphatase activity of OsPP2C06 and OsPP2C09 in the presence of ABA, respectively (He et al., 2014). However, OsPYL12, which cannot bind to ABA, acts as a dimer form to constitutively inhibit OsPP2C06 and OsPP2C09 (He et al., 2014). OsPYL/RCAR5 was found to interact with OsPP2C30 and OsPP2C49 in yeast with the presence of ABA, and OsPP2C30 could interact with SAPK2 in a bimolecular fluorescence complementation (BiFC) assay that ultimately negatively regulates the transcriptional activity of OsREB1 (Kim et al., 2012).
A genome-wide and expression analysis of clade A PP2Cs in rice and Arabidopsis indicates that most of them were induced by ABA and drought stress or mannitol treatment, and the ABA response-related element (ABRE) motif was significantly enriched in the promoter region of them (Xue et al., 2008; Singh et al., 2010). Overexpression of the rice clade A PP2C gene OsPP2C108 in Arabidopsis enhanced the tolerance to salt, mannitol, and drought stresses, and sensitivity to ABA treatment (Singh et al., 2015). Recently, another clade A PP2C gene OsABIL2 was demonstrated as a negative regulator in ABA signaling (Li et al., 2015a, 2015b). In the presence of ABA, the OsPYL1-ABA-OsABIL2 complex causes a subcellular redistribution of OsABL2 and releases the inhibition on SAPK8 and SAPK10 in the nucleus (Li et al., 2015a, 2015b).
All of the 10 SAPKs were activated by hyperosmotic stress and SAPK8, 9, and 10 were also activated by ABA (Kobayashi et al., 2004). The bZIP transcription factor, TRAB1 (Transcription factor responsible for ABA regulation), was phosphorylated by SAPK8, 9, and 10, and the Ser-102 of TRAB1 was critical for its activation function (Hobo et al., 1999; Kagaya et al., 2002; Kobayashi et al., 2005). Recently, Tiller Enhancer (TE) was found to be another substrate of SAPK8, 9, and 10. TE interacts with ABA receptors (OsPYL2, 9, and 10) and promotes their degradation by the proteasome, while phosphorylated TE could inhibit its interaction with OsPYL10 and prevent the degradation of OsPYL10 (Lin et al., 2015). The OSRK1 (SAPK6) can phosphorylate and activate the bZIP transcription factor OREB1, and ectopic expression of OSRK1 in tobacco (Nicotiana tabacum) reduced the sensitivity to ABA in germinating seeds as well as root elongation (Chae et al., 2007). A more recent work using an in vitro phosphorylation assay and in vivo dual-luciferase assay indicated that SAPK2, SAPK6, and SAPK9 can phosphorylate and activate OsbZIP46, a subgroup A bZIP transcription factor (Tang et al., 2012; Kim et al., 2015).
Subgroup A bZIP transcription factors play important roles in ABA and drought stress response in plants, and their activation mechanisms are conserved between Arabidopsis and rice (Lu et al., 2009; E et al., 2014). Generally, subgroup A bZIP transcription factors need SnRKs to fully activate their transcriptional activation activity (Kagaya et al., 2002; Kobayashi et al., 2005; Chae et al., 2007; Fujii et al., 2007; Fujii and Zhu, 2009; Yoshida et al., 2010; Kim et al., 2015). In rice, 11 subgroup A bZIP transcription factors have been identified, and 6 of them have been associated with diverse biological functions (Hobo et al., 1999; Zou et al., 2007; Xiang et al., 2008; Zou et al., 2008; Lu et al., 2009; Amir Hossain et al., 2010; Hossain et al., 2010; Yang et al., 2011; Tang et al., 2012). Transgenic plants with overexpressed OsbZIP23 show significantly improved drought and salt tolerance and sensitivity to ABA (Xiang et al., 2008). OsbZIP46 (ABL1) has a high sequence identity to OsbZIP23. Unlike OsbZIP23, overexpression of intact OsbZIP46 showed increased ABA sensitivity but a slightly decreased drought tolerance at the seedling and reproductive stages. In contrast, overexpression of a constitutively active form of OsbZIP46 (OsbZIP46CA1) could activate downstream genes and enhance drought tolerance (Tang et al., 2012). However, despite their importance to drought tolerance in rice, little is known about the underlying molecular mechanisms of the subfamily A bZIP transcription factors, and their target genes have seldom been systematically investigated in plants.
In this study, we identified the upstream regulators of OsbZIP23, SAPK2, and OsPP2C49, and investigated their roles in the transcriptional activation of OsbZIP23 in rice. SAPK2 could directly phosphorylate and activate OsbZIP23, while OsPP2C49 negatively regulates SAPK2 and further inhibits the transcription activity of OsbZIP23. Genome-wide identification of the direct target genes of OsbZIP23 indicates that OsbZIP23 regulates numerous processes including stress response, hormone signaling, and development. Interestingly, we found that OsbZIP23 could directly and positively regulate OsPP2C49, and thus regulate ABA signaling. In addition, OsbZIP23 also regulated the ABA level by directly targeting the ABA synthesis gene OsNCED4. These results establish a complete regulatory network of OsbZIP23 in the ABA signaling pathway and the activation of downstream drought-related genes in rice.
RESULTS
Phosphorylated OsbZIP23 Activates Gene Expression
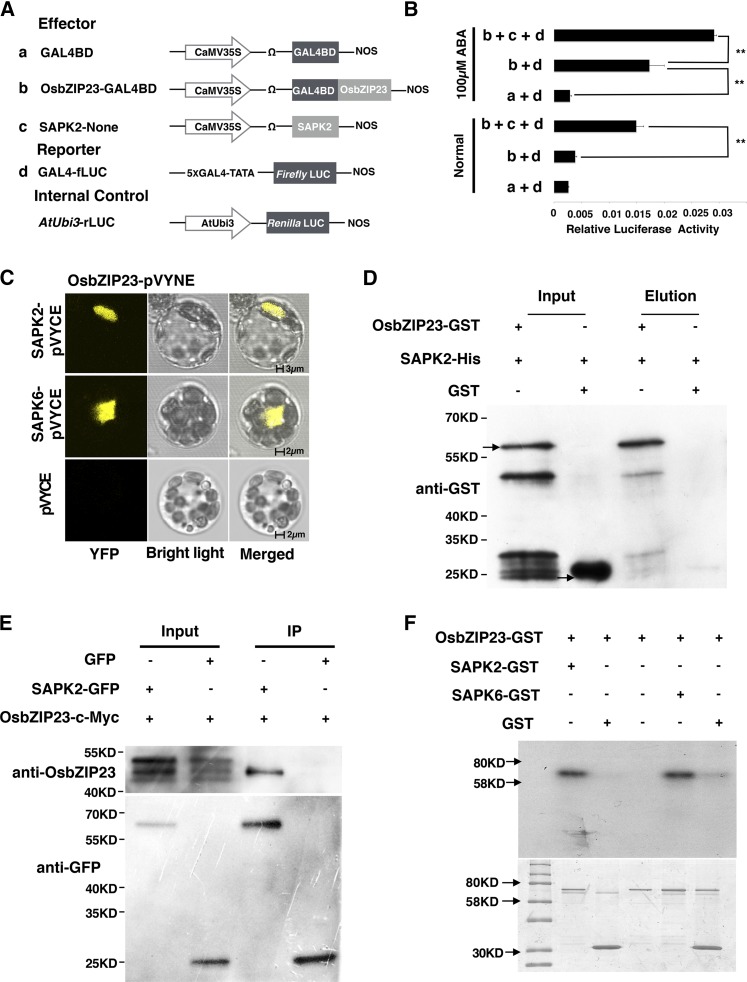
OsbZIP23 has been characterized as a positive transcriptional regulator of drought resistance in rice (Xiang et al., 2008). To further understand the transcriptional activity of OsbZIP23 in rice, we performed a dual-luciferase transient transcriptional activity assay in rice protoplasts. OsbZIP23 was fused with the yeast GAL4 binding domain (GAL4BD) as an effector plasmid and cotransformed into rice protoplasts with a firefly luciferase reporter gene driven by a promoter consisted of five copies of the GAL4 binding site (GAL4-fLUC; Fig. 1A). We only detected a slight increase in the transcriptional activation activity of OsbZIP23, but it was not statistically significant under normal conditions (Fig. 1B). However, after ABA treatment the transcriptional activation activity of OsbZIP23 was significantly induced (Fig. 1B). This result suggested that OsbZIP23 acts depending on the ABA signaling pathway, and that an upstream activator may be required for its activation under ABA treatment.
Figure 1.
OsbZIP23 is a transcriptional activator. A, Scheme of the constructs used in the rice protoplast cotransfection assay. B, Phosphorylated OsbZIP23 but not its native status activation gene expression. OsbZIP23-GAL4BD, SAPK2-None, OsPP2C49-None, and the empty vector were used as the effector. The activity of GAL4-fLUC was used as the reporter and rLUC activity was used as an internal control. The fLUC/rLUC ratio represents the relative activity of the promoter. After transfection, one-half of the protoplasts were incubated with 100 μM ABA for 4 h. The value in each column represents the means of three independent replicates, and the error bars indicate the sd. The asterisks represent significant difference determined by the Student’s t-test; the double asterisks indicate P < 0.01. C, OsbZIP23 interacts with SAPK2 and SAPK6 in a BiFC assay. OsbZIP23-pVYNE and SAPK2-pVYCE or SAPK6-pVYCE were combined and transformed into rice protoplast. D, Pull-down assay confirmation of the interaction between OsbZIP23 and SAPK2 in vitro. SAPK2 (SAPK2-His) was used as a bait and the pull-down of OsbZIP23 was detected by anti-GST antibody. Arrows show the GST-tag-fused OsbZIP23 and the GST tag. E, Co-IP assay confirmation of the interaction between OsbZIP23 and SAPK2 in vivo. GFP-tagged SAPK2 was immunoprecipitated using anti-GFP antibody, and coimmunoprecipitated OsbZIP23-cMyc was detected by an anti-OsbZIP23 antibody. F, OsbZIP23 was phosphorylated by SAPK2 and SAPK6 in vitro.
The ABA signaling component SnRK2 protein kinases have been proven to activate bZIP transcription factors via phosphorylation in plants, and two SnRK homologs SAPK2 and SAPK6 in rice can phosphorylate OsbZIP46, which has a high sequence identity with OsbZIP23 (Tang et al., 2012). We examined whether OsbZIP23 is also activated by SAPKs. BiFC assay suggested that OsbZIP23 could interact with SAPK2 and SAPK6 in rice protoplasts (Fig. 1C). The interaction between SAPK2 and OsbZIP23 was further confirmed by an in vitro pull-down assay and an in vivo coimmunoprecipitation (Co-IP) assay (Fig. 1, D and E). Furthermore, the phosphorylation activity of SAPK2 and SAPK6 toward OsbZIP23 were demonstrated by an in vitro phosphorylation assay (Fig. 1F). To further investigate whether the phosphorylated OsbZIP23 could activate gene expression in rice, SAPK2 was cotransformed with OsbZIP23 into rice protoplasts. We found the transcriptional activity of OsbZIP23 was significantly enhanced even without ABA (Fig. 1B), suggesting that SAPK2 can activate OsbZIP23, which is consistent with a previous study (Kim et al., 2015).
Conserved Ser/Thr Is Required for Full Activation of OsbZIP23
OsbZIP23 exhibits high sequence similarity with TRAB1 and OsbZIP46 (Supplemental Fig. S1), whose activation has been previously reported (Kagaya et al., 2002; Tang et al., 2012). The phosphorylation site Ser-102 in TRAB1 is conserved in OsbZIP23 (Ser-106) and OsbZIP46 (Ser-103; Supplemental Fig. S1). Moreover, a previous study has shown that a conserved R-Q-X-S/T sequence in OREB1 can be recognized and phosphorylated by OSRK1 (Chae et al., 2007). There are three R-Q-X-S/T motifs in OsbZIP23, OsbZIP46, and TRAB1 (Supplemental Fig. S1). To further investigate the putative phosphorylation sites of OsbZIP23, we generated multiple amino acid substitutions in OsbZIP23 and checked their transcriptional activation activities in rice protoplasts (Supplemental Fig. S2). We found that single amino acid substitutions (S/T to A) could significantly decrease the transcriptional activation activity of OsbZIP23 under ABA treatment, but no significant differences were observed among the mutants of the four putative phosphorylation sites (Supplemental Fig, S2). Multiple (two to four) amino acid substitutions in the putative phosphorylation sites almost abolished the transcriptional activity of OsbZIP23 under ABA treatment (Supplemental Fig, S2). To further examine the activation sites of OsbZIP23 by SAPK2, the mutated forms of OsbZIP23 with multiple putative phosphorylation sites substituted were cotransformed with SAPK2 into rice protoplasts, and the results showed that the transcriptional activity was also significantly decreased (Supplemental Fig, S2). These results suggest that the four Ser/Thr sites are required for the full activation of OsbZIP23 under ABA treatment and through activation by SAPK2.
OsPP2C49 Negatively Regulates OsbZIP23 Transcriptional Activity through Deactivation of SAPK2
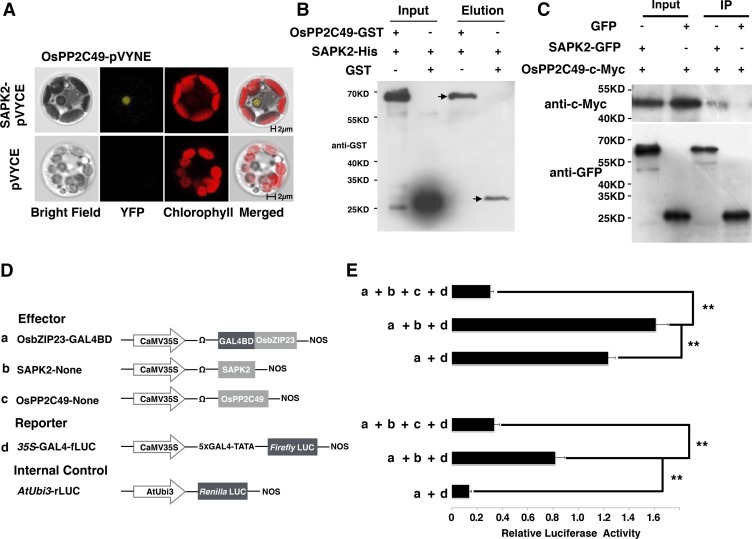
A previous study has shown that OsPP2C49, a clade A protein phosphatase 2C, could interact with a putative ABA receptor OsPYL/RCAR5 in an ABA-dependent fashion (Kim et al., 2012). We asked whether OsPP2C49 acts in the ABA signaling pathway through SAPK2 and OsbZIP23. To validate this, we performed in vitro pull-down, BiFC, and Co-IP assays, and the results suggested that OsPP2C49 could interact with SAPK2 (Fig. 2, A–C). Moreover, when OsPP2C49, SAPK2, and OsbZIP23 were cotransformed into the rice protoplasts, we found the transcriptional activation activity of OsbZIP23 was almost abolished (Fig. 2, D and E), suggesting that OsPP2C49 indeed acts upstream of SAPK2 and negatively regulates OsbZIP23 activation. The results above together with our previous genetic study (Xiang et al., 2008) suggest that OsbZIP23 functions in the classical ABA signaling pathway, and its transcriptional activation is regulated through phosphorylation.
Figure 2.
SAPK2 Interacts with OsPP2C49 in vitro and in vivo. A, SAPK2 interacts with OsPP2C49 in a BiFC assay. B, SAPK2 interacts with OsPP2C49 in the in vitro pull-down assay. His-tagged SAPK2 (SAPK2-His) was used as a bait, and pull-down of OsPP2C49 was detected by anti-GST antibody. Arrows show the GST-tagged OsPP2C49 and the GST tag. C, SAPK2 interacts with OsPP2C49 in the in vivo Co-IP assay. GFP-tagged SAPK2 was immunoprecipitated using an anti-GFP antibody, and coimmunoprecipitated OsPP2C49-cMyc was detected by antic-Myc antibody. D, Scheme of the constructs used in the rice protoplast cotransfection assay. E, OsPP2C49 negatively affects the transcriptional activation activity of OsbZIP23. The fLUC/rLUC ratio represents the relative activity of 35S promoter. After transfection, half of the protoplasts were incubated with 100 μM ABA for 4 h. The values in each column are the means of three independent replicates and error bars indicate the SD. The asterisks represent significant difference determined by the Student’s t test; double asterisks indicate P < 0.01.
Identification of OsbZIP23 Binding Sites by Chromatin Immunoprecipitation Sequencing
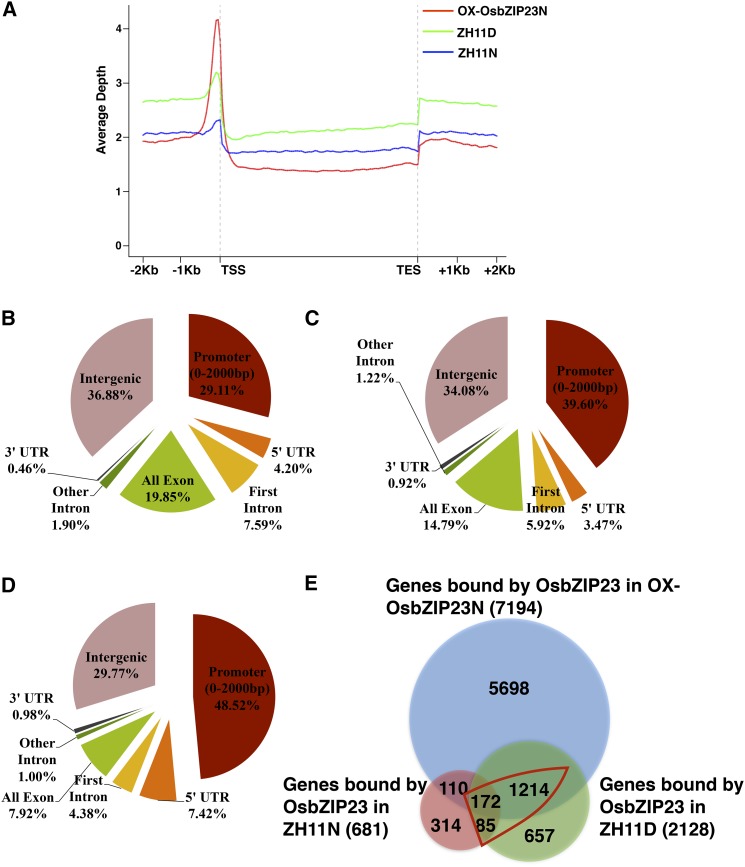
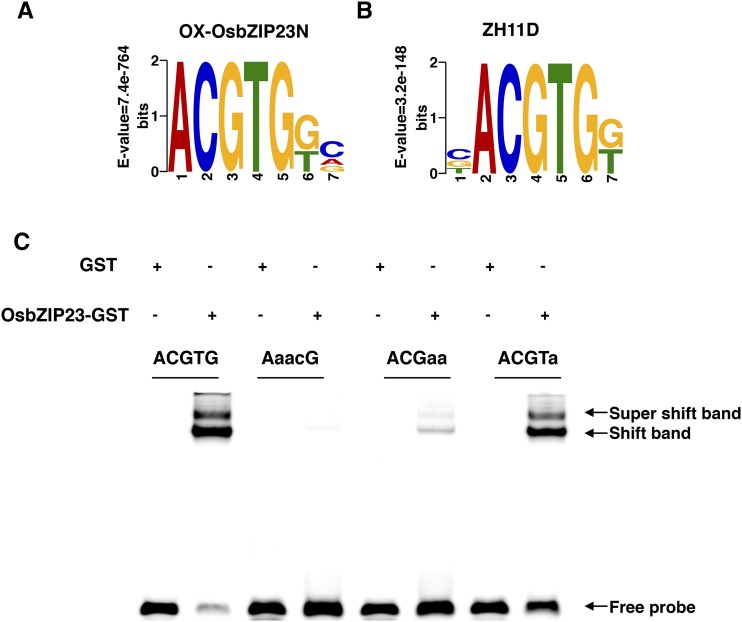
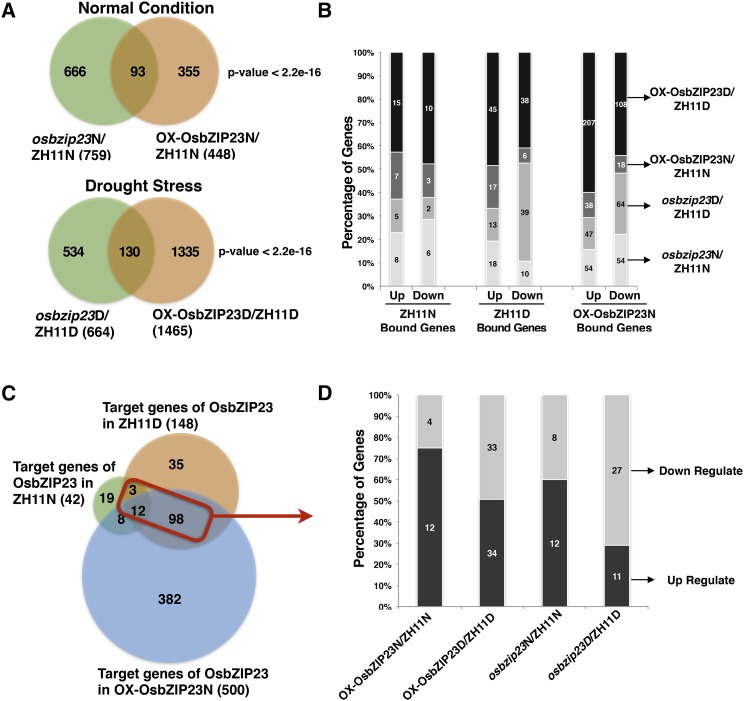
To further understand how OsbZIP23 regulates ABA signaling and confers drought resistance, we applied chromatin immunoprecipitation sequencing (ChIP-Seq) to identify the genome-wide binding sites of OsbZIP23. The ChIP assay was performed by using a specific antibody against OsbZIP23 (Supplemental Fig. S3) to pull down the putative OsbZIP23-bound DNA from the leaf tissues of wild-type rice Zhonghua 11 under normal growth conditions (ZH11N), and under moderate drought stress treatment (ZH11D), and OsbZIP23-overexpression rice under normal growth conditions (OX-OsbZIP23N). Three biological replicates for each tissue were used to generate ChIP-Seq libraries. After sequencing, we obtained 7,797,925, 9,927,059, and 6,963,151 unique mapped reads in ZH11N, ZH11D, and OX-OsbZIP23N, respectively (Supplemental Table S1). The unique mapped reads from the three samples were mainly located within 500 bp upstream of the transcription start site (TSS; Fig. 3A). By using model-based Analysis of ChIP-Seq software (Zhang et al., 2008), we identified 1738, 4260, and 11,611 peaks in ZH11N, ZH11D, and OX-OsbZIP23N, respectively (Supplemental Fig. S4). Further genomic distribution analysis revealed that a large proportion of OsbZIP23-associated peaks (29.11% in ZH11N, 39.6% in ZH11D, and 48.52% in OX-OsbZIP23N) were located within 2000 bp upstream of the TSS region (Fig. 3, B–D). Among the peaks, we identified 681, 2128, and 7194 putative OsbZIP23-bound genes in ZH11N, ZH11D, and OX-OsbZIP23N, respectively (Fig. 3E). By counting the overlapped OsbZIP23-bound genes between the ZH11N-ZH11D and ZH11D-OX-OsbZIP23N data sets (Fig. 3E, red triangle), 1471 genes were defined as OsbZIP23-bound genes involved in drought stress (OsbZIP23BGD; Supplemental File S1). Gene ontology analysis of OsbZIP23BGD showed that the number of genes in response to abiotic stress stimulus, regulation of transcription, and plant hormone signaling transduction were significantly enriched (Supplemental Fig. S5). Furthermore, we found that 152 (10.3%, P value < 2.2e-16) OsbZIP23BGD genes had known or suggestive functions (extract from RicENcode, http://ricencode.ncpgr.cn), and 48.7% (74 out of 152) of them were regulated by drought stress (Supplemental File S1). These OsbZIP23BGD genes with known or suggestive functions provide direct evidence and molecular mechanisms for OsbZIP23 in the regulation of a wide range of stress and hormone responses (Supplemental Fig. S6). In addition, 115 (7.8% of OsbZIP23BGD, P value = 2.75e-4) transcription factors were enriched and 35.7% (41 out of 115) of them were responsive to drought stress treatment (Supplemental File S1). The quality of the ChIP-Seq result was validated by a ChIP-quantitative PCR (qPCR) assay for 15 randomly selected genes (Supplemental Fig. S7). To investigate the binding motifs of OsbZIP23, ± 100 bp flanking sequences around the peaks were submitted to MEME-ChIP (http://meme-suite.org/tools/meme-chip) to search for enriched motifs. The most frequently captured motifs belonged to the ABA-responsive elements (ABREs; Fig. 4, A and B). An EMSA with point mutation verified that ACGTG is the core sequence of OsbZIP23 binding in the promoters of the target genes (Fig. 4C).
Figure 3.
Genome-wide distribution of OsbZIP23 binding sites in the rice genome. A, OsbZIP23 target sites are mainly enriched within the 500-bp upstream region of the TSS. All unique mapped reads (Supplemental Table S1) from each library were anchored to annotated genes in rice genome. B to D, Distribution of OsbZIP23 binding peaks within rice-genic regions: in ZH11N, wild-type rice under normal condition (B); ZH11D, wild-type rice under drought stress (C); and OX-OsbZIP23N, OsbZIP23 overexpressed rice under normal condition (D). E, Venn diagram showing the overlap of the OsbZIP23-bound genes in ZH11N, ZH11D, and OX-OsbZIP23N samples. The red triangle represents OsbZIP23BGDs.
Figure 4.
Motif analysis of OsbZIP23-bound promotors. A and B, The most significant motif identified in the OsbZIP23 binding peaks by discriminative regular expression motif elicitation in OX-OsbZIP23N and ZH11D, respectively. In both data sets, 200 bps around the top of the peak summit were subjected to discriminative regular expression motif elicitation. No significant motifs were found in OsbZIP23-bound promoters in ZH11N. C, EMSA showing ACGTG is required by OsbZIP23 binding to targets. GST-tagged OsbZIP23 was used in the EMSA, and the GST tag was used as a control. ACGTG probe sequence: CACGCGGCGACGCCACGTGTCCCCACCGATCCCGCCGCTCGGCCGACAGGTGGGCCCCA. ACGTG was substituted by AaacG, ACGaa, and ACGTa in the mutant probe and the substitute nucleotide acids were marked with lowercase characters.
Target Genes Are Directly Regulated by OsbZIP23 under Drought Stress
To identify the direct target genes regulated by OsbZIP23 at the transcript level, the same rice materials as those used for ChIP-Seq were used for RNA-seq. We used both the overexpression and mutant seedlings of OsbZIP23 under normal and drought stress conditions to generate RNA-seq libraries. A total of 758, 448, 664, and 1465 differentially expressed genes were found in osbzip23 (compared to wild type) under normal conditions (osbzip23N), OsbZIP23-overexpresssion plants under normal conditions (OX-OsbZIP23N), osbzip23 under drought stress (osbzip23D), and OsbZIP23-overexpression plants under moderate drought conditions (OX-OsbZIP23D), respectively (Fig. 5A; Supplemental File S2). The high quality of the RNA-seq results was validated by reverse transcription-quantitative PCR (RT-qPCR) assay for 29 randomly selected genes (Supplemental Fig. S8). The direct target genes of OsbZIP23 were identified by combining the data sets of differentially expressed genes with the OsbZIP23-bound genes from the ChIP-Seq data sets (Fig. 5B). A total of 42, 148, and 500 direct target genes of OsbZIP23 were identified in the ZH11N, ZH11D, and OX-OsbZIP23N samples, respectively (Fig. 5C). Among them, 113 direct target genes involved in drought stress (defined as OsbZIP23TGD, indicated by a red square in Fig. 5C; Supplemental File S3) were further analyzed for their differential expression patterns (Fig. 5D). We found that 51.3% (58 out of 113) of OsbZIP23TGDs were induced by drought stress treatment, while only 13.2% (15 out of 113) of OsbZIP23TGDs were suppressed by drought stress treatment (Supplemental File S3). Moreover, we identified 14 OsbZIP23TGD genes with known function in drought response or drought resistance such as OsbZIP23 itself (Xiang et al., 2008), OCPI1 (Huang et al., 2007), OsLEA3-2 (Duan and Cai, 2012), OsSRO1c (You et al., 2013, 2014), and OsETOL1 (Du et al., 2014; Supplemental File S3). These results further demonstrate that OsbZIP23 plays an important role in regulating drought resistance through direct binding to the promoters of drought stress-related genes.
Figure 5.
Identification of OsbZIP23 direct target genes by comparing ChIP-Seq and RNA-seq data. A, Venn diagram showing the number of genes regulated by OsbZIP23 based on the RNA-seq analysis. Differentially expressed genes (fold change ≥ 2 with P value < 0.05) in overexpressed OsbZIP23 and osbzip23 samples in both normal and drought stress conditions were used for comparing. P values were calculated using the Fisher’s exact test. B, Overlapped genes between the OsbZIP23-bound genes (ChIP-Seq data) in ZH11N, ZH11D, and OX-OsbZIP23N, respectively, and the OsbZIP23-regulated genes (RNA-seq data). C, Venn diagram showing the number of OsbZIP23 direct target genes in ZH11N, ZH11D, and OX-OsbZIP23N samples. The red square indicates OsbZIP23TGDs. D, Differentially expressed OsbZIP23TGDs in four RNA-seq classes.
OsbZIP23 Directly and Positively Regulates the Expression of OsPP2C49
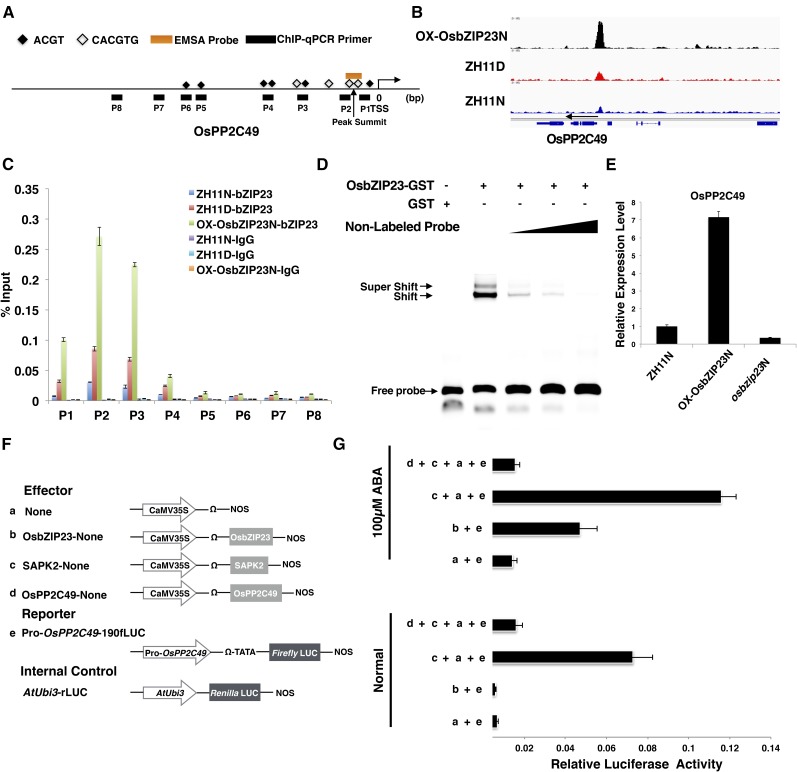
By carefully checking the gene identities of the OsbZIP23TGDs, we were surprised to find that OsPP2C49, which is mentioned above, is also a target gene of OsbZIP23. A distinctive peak was detected approximately 1 Kb upstream of the TSS of OsPP2C49 in all three ChIP-Seq libraries (Fig. 6, A and B). ChIP-qPCR showed that OsbZIP23 specifically binds to the 500-bp region upstream of the TSS (Fig. 6C). We selected a 59-bp sequence around the peak summit as a probe that contained a typical G-box (CACGTG) to conduct EMSA, and the results confirmed the specific binding by OsbZIP23 to the OsPP2C49 promoter (Fig. 6D). RNA-seq data suggested that the transcript level of OsPP2C49 was positively regulated by OsbZIP23, and this was confirmed by RT-qPCR (Fig. 6E). Furthermore, we performed a dual-luciferase transient transcriptional activity assay to further confirm the regulation of OsPP2C49 by OsbZIP23 (Fig. 6F). The OsPP2C49 promoter-driven reporter was up-regulated only when SAPK2 or ABA was added (Fig. 6G). In addition, we also observed that OsPP2C49 had a negative effect toward the activation of its own promoter through the deactivation of SAPK2 (Fig. 6G). These results together suggest that OsbZIP23 can directly and positively regulate the expression level of OsPP2C49.
Figure 6.
OsbZIP23 directly and positively regulates the expression of OsPP2C49. A, Scheme showing the structure of the OsPP2C49 promoter region. Short lines indicate the region detected by ChIP-qPCR in C. The black squares indicate the ABREs. The empty squares indicate the G-box (CACGTG). The orange line indicates the location of the probe used for EMSA in D. B, ChIP-Seq data showing OsbZIP23 specifically binding to the promoter region of OsPP2C49. C, Validation of the direct binding of OsbZIP23 to the promoter of OsPP2C49 by ChIP-qPCR. The enrichment values were normalized to Input. D, qRT-PCR showing that OsbZIP23 positively affects the expression levels of OsPP2C49 in rice. Actin was used as a reference gene. E, EMSA showing that OsbZIP23 could directly bind to the promoter of OsPP2C49. The 5-, 10-, and 30-fold excess nonlabeled probes were used for competition. F, Scheme of the constructs used in the rice protoplast cotransfection assay. G, OsbZIP23 activates OsPP2C49 expression in a phosphorylation-dependent manner. The fLUC/rLUC ratio represents the relative activity of the OsPP2C49 promoter. After transfection, half of the protoplasts were incubated with 100 μm ABA for 4 h. The values in each column are the mean of three independent replicates and the error bars indicate the sd.
OsPP2C49 Is a Negative Regulator of ABA Signaling in Rice
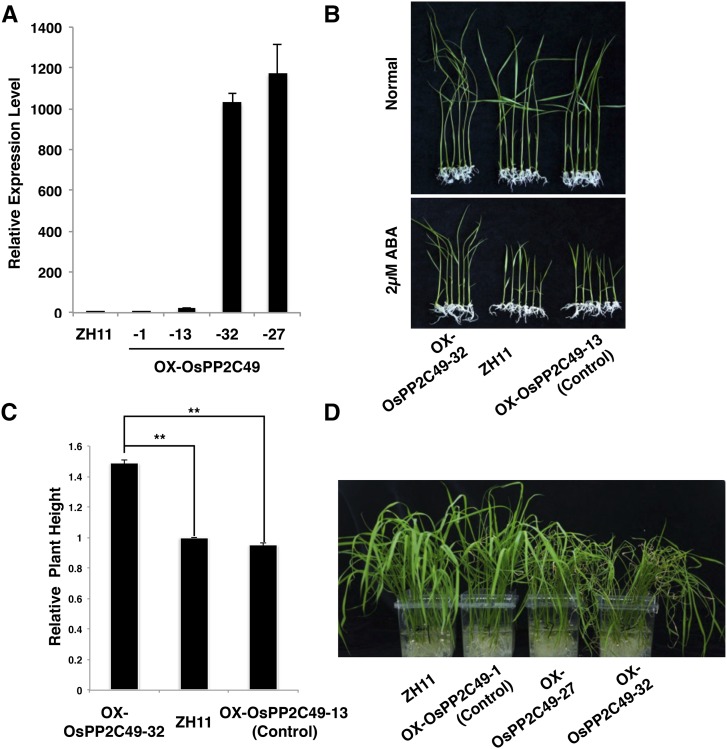
Because OsPP2C49 is a close homolog of ABI1, which has been well characterized as a negative regulator of ABA signaling in Arabidopsis, we further investigated the biological function of OsPP2C49 in ABA signaling in rice by generating OsPP2C49-overexpression plants (OX-OsPP2C49) to examine the ABA sensitivity (Fig. 7A). As shown in Figure 7B, the relative plant height of the overexpression line (OX-OsPP2C49-32) was significantly higher than the wild-type (ZH11) and the negative transgenic line (OX-OsPP2C49-13) under ABA treatment (Fig. 7C). Moreover, we observed a rapid dehydration phenotype of the OsPP2C49 over-expression lines when compared to the wild type and the negative transgenic controls (Fig. 7D), which is similar to the phenotype observed in the overexpression plants of another ABI1-like OsPP2C gene (Li et al., 2015a). These results together indicate that OsPP2C49 acts as an important negative regulator in ABA signaling and drought response in rice. By using CRISPR-CAS9 technology, we generated a mutant of OsPP2C49, which contains a point mutation in the coding region. However, this mutant exhibited ABA sensitivity similar to wild type (Supplemental Fig. S9), which may be due to functional redundancy of the clade A PP2C subfamily.
Figure 7.
OsPP2C49 is a negative regulator of ABA signaling. A, Transcript levels of overexpressed OsPP2C49 and control plants. B, Decreased sensitivity of OX-OsPP2C49 plants to ABA treatment. Three-day-germinated seeds were transplanted in either half-strength MS medium (Normal) or half-strength MS medium supplemented with 2 μm ABA for 7 d. The wild-type (ZH11) and negative transgenic line (OX-OsPP2C49-13) were used as controls. C, Relative plant height of the ZH11, OX-OsPP2C49-13, and OX-OsPP2C49-32 plants under ABA treatment. The bars represent the sd (n > 6) and the double asterisks represent a significant difference determined by the Student’s t test at P < 0.01. D, OX-OsPP2C49 seedlings are shown rapidly dehydrated after exposure to air. Three-day-germinated seeds were transplanted into a rooting tube with half-strength MS medium and grown for 11 d. The sealing film was partially open to investigate the dehydration performance.
OsbZIP23 Affects ABA Levels through the Positive Regulation of OsNCED4
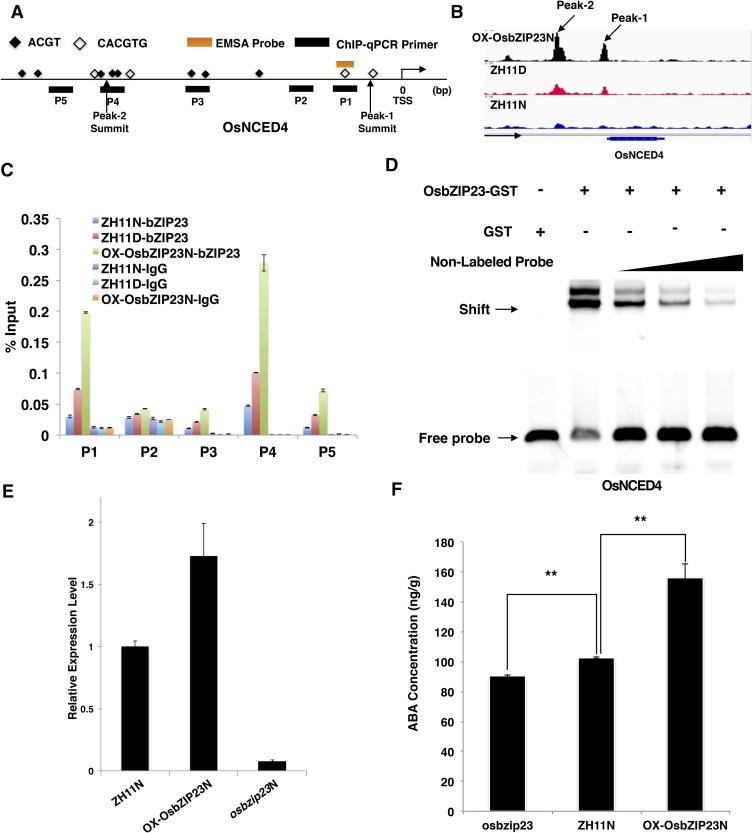
Interestingly, we noticed that OsNCED4, encoding a key enzyme in ABA synthesis, is also a target gene of OsbZIP23 (Fig. 8, A and B). A previous study showed that the OsNCED4 expression level is correlated with the ABA level in rice (Liang et al., 2014). In the ChIP-Seq analysis, the OsNCED4 promoter sequence was highly enriched in the ZH11D and OX-OsbZIP23N samples, which was further confirmed by ChIP-qPCR (Fig. 8C). EMSA with a 51-bp probe around the peak summit suggested that OsbZIP23 could directly bind to the promoter of OsNCED4 (Fig. 8D). In addition, OsbZIP23 also positively affected the expression level of OsNCED4. In the OsbZIP23-overexpression seedlings, OsNCED4 was significantly up-regulated, while OsNCED4 was down-regulated in osbzip23 (Fig. 8E). Because OsNCED4 is a key enzyme in ABA synthesis, we further measured the ABA levels in the OsbZIP23 overexpression and osbzip23 plants. As shown in Figure 8F, the ABA level was significantly increased in the overexpression plants but decreased in the osbzip23 mutants. In addition, we found another four genes, MHZ5, PSY1, LCYe and DSM1, that are involved in ABA biosynthesis and that could be also directly bound by OsbZIP23 (Supplemental Fig. S10). These results suggest that OsbZIP23 also functions as a positive regulator of ABA biosynthesis in rice.
Figure 8.
OsbZIP23 affects the ABA level through directly and positively regulating the expression of the OsNCED4 gene. A, Scheme showing the structure of the OsNCED4 promoter region. The black bars indicate the region detected by ChIP-qPCR in C. The orange bar indicates the probe used for EMSA in E. The black squares indicate the ABREs. The empty squares indicate the G-box (CACGTG). B, ChIP-Seq data showing OsbZIP23 specifically binding to the promoter region of OsNCED4. C, Validation of the direct binding of OsbZIP23 to the promoter of OsNCED4 by ChIP-qPCR. The enrichment values were normalized to Input. D, EMSA showing that OsbZIP23 could directly bind to the promoter of OsNCED4. The 5-, 10-, and 30-fold excess nonlabeled probes were used for competition. E, qRT-PCR showing that OsbZIP23 positively affects the expression levels of OsNCED4 in rice. Actin was used as a reference gene. F, OsbZIP23 positively affects the ABA levels in rice. The values in each column are the mean of three independent replicates, and the error bars indicate the sd. The double asterisks represent a significant difference determined by the Student’s t test at P < 0.01.
DISCUSSION
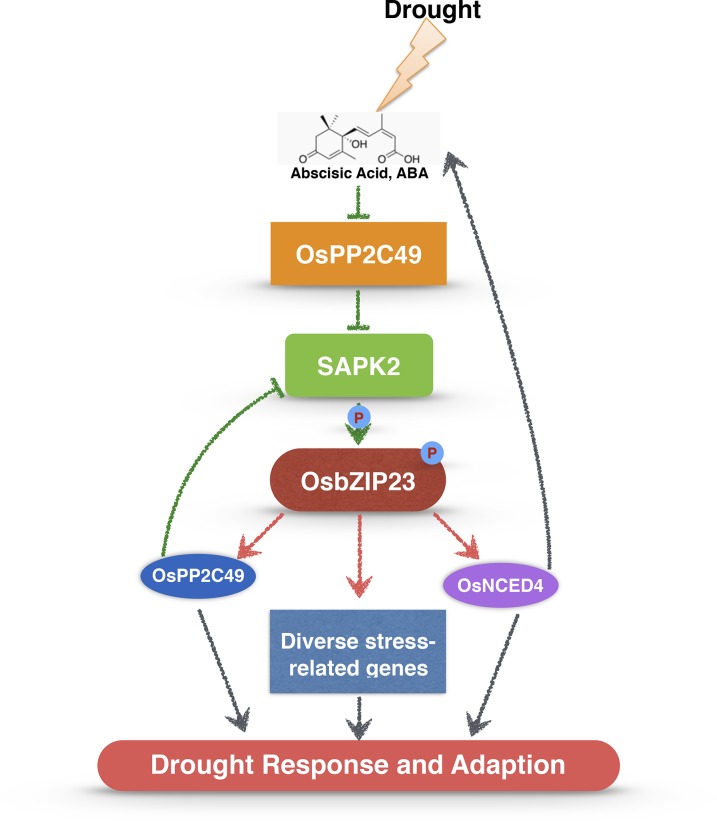
Our previous study found that OsbZIP23 confers ABA-dependent drought resistance in rice. In this study, we further revealed the role of this transcription factor in the regulation of ABA signaling and the expression of downstream genes. We provided strong evidence that SAPK2 and OsPP2C49 act as positive and negative regulators, respectively, of OsbZIP23. We also identified the genome-wide target genes of OsbZIP23 under normal and drought stress conditions. Most significantly, we found that OsbZIP23 can feedback-regulate the ABA signaling components and biosynthesis. These findings, with a working model of OsbZIP23 function presented in Figure 9 and discussed below, have enriched our understanding of the molecular mechanisms underlying ABA and drought stress responses.
Figure 9.
Working model of OsbZIP23-mediated regulation of ABA signaling pathway under drought stress in rice. ABA, which is induced by drought stress, inhibits the activity of OsPP2C49 to release the kinase activity of SAPK2 for further activation of OsbZIP23 through phosphorylation. OsbZIP23 could directly bind to the promoters of OsPP2C49 and positively regulates the expression of OsPP2C49, which in turn negatively regulates the ABA signaling. Meanwhile, OsbZIP23 could positively affect the ABA levels through direct regulation of OsNCED4. In addition, OsbZIP23 also directly regulates diverse stress-related genes involved in the drought response and adaptation in rice (see details in Supplemental Fig. S6). Green arrows indicate protein-protein interaction, red arrows indicate direct binding by OsbZIP23, and gray arrows point to putative functions. P, Phosphorylation.
OsbZIP23 Needs Phosphorylation for Its Transcriptional Activation
The subgroup A bZIP transcription factors such as TRAB1, OsbZIP46, and OsbZIP12 in rice and ABI5, ABRE1, ABRE2, and ABF1 in Arabidopsis generally require ABA for transcriptional activation (Gampala et al., 2002; Kobayashi et al., 2005; Yoshida et al., 2010; Kim et al., 2015). Recently, OsbZIP46 was found to be phosphorylated by SAPK2 and SAPK6 in vitro, and cotransformation of OsbZIP46 with SAPK2, SAPK6, or SAPK9 into rice protoplasts can significantly induce the transcriptional activation activity of OsbZIP46 (Kim et al., 2015). OsbZIP23 shows high similarity with OsbZIP46 (Nijhawan et al., 2008). We found that SAPK2 and SAPK6 could also interact and phosphorylate OsbZIP23 and significantly induce the transcriptional activity of OsbZIP23 in vivo (Fig. 1), which may indicate that both SAPK2 and SAPK6 phosphorylate the conserved Ser/Thr sites in OsbZIP23 and OsbZIP46 such as the R-Q-X-S/T sequence (Chae et al., 2007). This hypothesis was confirmed in our further experiments when we found four Ser/Thr substitutions in OsbZIP23 that almost abolish SAPK2-activated transcriptional activation of OsbZIP23 (Supplemental Fig. S2). However, further experiment is needed to examine which sites could be phosphorylated by SAPK2 in vivo.
Without ABA, clade A PP2Cs deactivate SnRKs through dephosphorylation. Both OsPP2C30 and OsABIL2 could deactivate SAPK2 and then negatively regulate the expression of downstream genes mainly through bZIP transcription factors (Kim et al., 2012; Li et al., 2015a, 2015b). In this work, we found that SAPK2 could interact with OsPP2C49. In a dual luciferase assay, OsPP2C49 almost abolished the SAPK2-activated transcriptional activation of OsbZIP23. Moreover, OsPP2C49 was shown to interact with OsPYL5 in an ABA-dependent manner, which further supports that OsbZIP23 participates in a classical ABA signaling pathway in rice (Fig. 9). The transcriptional activity of OsbZIP46 differed in the activation by SAPK2, SAPK6, and SAPK9; SAPK2 showed the highest activation (Kim et al., 2015). Whether such difference exists for OsbZIP23 needs to be further tested.
OsbZIP23 Regulates Diverse Functional Genes Mainly Involved in Stress Responses
In this study, we applied ChIP-Seq and RNA-seq to investigate the downstream genes of OsbZIP23. Our results provide, to our knowledge, new insights into the gene network regulated by OsbZIP23 underlying the drought resistance of rice. OsbZIP23 mainly binds to the regions 500 bp upstream of the TSSs in all three samples (Fig. 3A), which is consistent with the transcription factor role of OsbZIP23 and with the previous ChIP-Seq results of other transcription factors in Arabidopsis and maize (Zea mays L.; Zhang et al., 2011; Ram et al., 2014; Li et al., 2015a, 2015b). Moreover, we found that the conserved ABREs are significantly enriched in the OsbZIP23-bound regions (Fig. 4), indicating that OsbZIP23 may mainly target the ABA-responsive genes.
The genes bound by OsbZIP23 in the three samples differ from each other. A small number of genes were detected in the wild-type sample under normal conditions (ZH11N), while over 7000 genes were detected in the OsbZIP23-overexpression lines. Notably, more than 2000 genes were detected in the wild type under drought stress conditions (ZH11D), which is consistent with the expression induction of OsbZIP23 under drought stress conditions (Nijhawan et al., 2008; Xiang et al., 2008). To identify OsbZIP23-bound genes with high confidence, we chose the overlapping genes among the three samples, defined as OsbZIP23BGD (Fig. 3E), for further analysis. The molecular functions of the OsbZIP23BGD genes are very diverse (Supplemental Fig. S5). Almost half of the function-known genes in the OsbZIP23BGD are involved in stress response or hormone signaling including SNAC1, OsbZIP72, DWA1, OsGARS3, and OsBZR1, which are important components in the plant stress response and hormone pathways (Supplemental Fig. S6). Moreover, we also detected several developmental process-related genes such as OsCO3, COPT1, PDIL1, and NAL1 (Supplemental Fig. S6), which are directly targeted by OsbZIP23. This suggested that OsbZIP23 may also regulate developmental processes in addition to its critical role in stress response regulation. At least, we observed that the osbzip23 mutant showed slightly reduced plant height and a change in the heading date. How OsbZIP23 regulates developmental processes in rice may be an interesting project for further study.
Genome-wide identification of target genes including LONG HYPOCOTYL5, BRASSINAZOLE RESISTANT1 (BZR1), LEAFY, and FAR-RED ELONGATED HYPOCOTYL3 in Arabidopsis (Sun et al., 2010; Ouyang et al., 2011; Winter et al., 2011; Zhang et al., 2011), KNOTTED1 in maize (Zea mays L.; Bolduc et al., 2012), and rice homeobox1 and IDEAL PLANT ARCHITECTURE1 in rice (Lu et al., 2013; Tsuda et al., 2014), have demonstrated that transcription factors are significantly enriched. In this work, 115 transcription factors were identified in OsbZIP23BGDs, and over 35% of them were regulated by drought stress (Supplemental File S1). OsbZIP23BGDs include several transcription factors previously shown to play an important role in drought stress, such as OsBZ8, OSVP1, SNAC1, OsGRAS23, OsWRKY45, OsBZR1, TRAB1, OsbZIP72, and OsNAC5 (Supplemental Fig. S6), which further implies that OsbZIP23 acts as a central regulator of drought resistance in rice.
OsbZIP23 Feedback Regulates ABA Signaling by Directly Targeting OsPP2C Genes
A recent study showed that clade A PP2C-mediated ABA signaling has evolved in ancestral land plants to control preexisting mechanisms for desiccation tolerance and facilitate rapid propagation on land (Komatsu et al., 2013). As negative regulators in ABA signaling, the expression levels of clade A PP2Cs are also regulated by stresses (Xue et al., 2008). In our RNA-seq analysis, 7 out of 10 clade A PP2Cs in rice were up-regulated by drought stress (Supplemental Table S2), which may imply that the induced expression of clade A PP2Cs may be needed to keep a reasonable scale of the ABA signaling in cells. Investigation of the cis-elements within the 1 Kb promoter region of clade A PP2Cs in rice and Arabidopsis suggests that the frequency of ABREs in the promoter region of clade A PP2Cs is much higher than that in the whole genome (Xue et al., 2008; Singh et al., 2010). The high enrichment of ABREs in the promoter region of PP2Cs indicates that AREB bZIP transcription factors may be involved in the regulation of these PP2C genes. Several studies have reported that clade A PP2Cs were differentially regulated by subgroup-A bZIP transcription factors (Xiang et al., 2008; Yoshida et al., 2010; Tang et al., 2012; Liu et al., 2014), but it has not been proven whether the bZIP transcription factors can directly regulate their expression levels. Our data clearly suggests that OsbZIP23 can directly and positively regulate the expression of OsPP2C49, which acts as a negative regulator of OsbZIP23 by deactivating SAPK2. We checked the ChIP-Seq data for the other clade A OsPP2Cs. To our surprise, OsbZIP23 could directly bind to the promoters of eight other OsPP2Cs under drought stress conditions (Supplemental Table S2). However, only OsPP2C49 and OsPP2C51 were differentially regulated by OsbZIP23 in the RNA-seq results (Supplemental Table S2), which indicates that the regulation of different clade A PP2Cs may need different transcription factors such as NACs and MYCs to coordinate with bZIP transcription factors for the differential regulation (Singh et al., 2012; Xu et al., 2013; Maurya et al., 2015).
In agreement with ABI1 and ABI2 in Arabidopsis and OsABIL2 in rice (Leung et al., 1997; Gosti et al., 1999; Komatsu et al., 2013; Li et al., 2015a, 2015b), overexpression of OsPP2C49 in rice also caused reduced ABA insensitivity and a drought hypersensitivity phenotype. However, a frame-shift mutation in OsPP2C49, which was generated by CRISPR-CAS9 technology, did not show an obvious change in the ABA sensitivity (Supplemental Fig. S9). In Arabidopsis, only the mutants with knockouts of multiple clade A PP2Cs can lead to ABA hypersensitivity, and osabil1 and osabil2 single mutations in rice also showed no phenotypic changes (Saez et al., 2006; Rubio et al., 2009; Li et al., 2015a, 2015b). Therefore, function redundancy may exist among the 10 clade A OsPP2Cs, and mutations of multiple genes may be effective in enhancing the ABA sensitivity and/or drought resistance of rice.
OsbZIP23 Positively Regulates the ABA Level in Rice
Because OsbZIP23 could directly regulate OsPP2C49, an important component of ABA signaling, we wondered whether OsbZIP23 could affect the expression of other upstream components in the ABA signaling pathways. However, except for eight clade A-OsPP2Cs and SAPK3, all of the OsPYLs and the other SAPKs were not found in the OsbZIP23BGDs (Supplemental Table S3). Interestingly, five ABA synthesis-related genes, DSM2, MHZ5, PSY1, LCY-e, and OsNCED4 (Du et al., 2010a, 2010b; Liang et al., 2014; Li et al., 2015a, 2015b; Yin et al., 2015) were found to be directly targeted by OsbZIP23 (Supplemental Fig. S10), and the ABA level was significantly increased in the OsbZIP23-overexpression line (Fig. 9). Such feedback regulation of ABA signaling and biosynthesis is similar to the BZR1 transcription factor, which is a central regulator in brassinosteroid (BR) signaling in Arabidopsis, and the downstream target genes of BZR1 include several BR biosynthesis enzymes and BR signaling components (Sun et al., 2010).
Plants have evolved multiple pathways to fine-tune ABA signaling including cross talks with other hormone signaling. For example, BRASSINOSTEROID INSENSITIVE2 was found to positively modulate ABA signaling through phosphorylation of subgroup III SnRKs in Arabidopsis (Cai et al., 2014), and ARK plays an essential role in the activation of SnRK2 in the moss Physcomitrella patens (Cai et al., 2014; Saruhashi et al., 2015). Recently, ABI1 was found to be degraded through ubiquitination by PUB12 and PUB13 in the presence of ABA and an ABA receptor in Arabidopsis, which may be regarded, to our knowledge, as a new positive regulation pathway of ABA signaling (Kong et al., 2015).
In this study, we found that ABA signaling as well as ABA biosynthesis can be feedback-regulated by OsbZIP23, which has significantly increased our understanding of ABA signaling regulation (Fig. 9). The feedback up-regulation of both ABA synthesis and PP2C49 is quite interesting in terms of fine control of the amplification of stress signaling because PP2C49 is a negative regulator of ABA signaling. We propose that feedback up-regulation of ABA synthesis may help amplify ABA signaling especially under the continuous drought stress; however, plants have evolved other mechanisms to avoid over-amplification of the signaling, for example through up-regulation OsPP2C49 to attenuate the ABA signaling. Moreover, OsbZIP23 acts as a central transcriptional regulator in drought responses based on the fact that a large number of drought stress-related genes are directly regulated by OsbZIP23 (Supplemental Fig. S6). Therefore, OsbZIP23 may be a promising candidate gene for engineering drought resistance in rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice Zhonghua 11 (ZH11; Oryza sativa ssp. japonica) was used as the wild-type. The OsbZIP23-overexpression line and the osbzip23 mutant were from our previous study (Xiang et al., 2008). Seedlings were grown in sand/paddy (1:3) soil under natural long-d conditions (approximately 14 h light/10 h dark) in July at Wuhan. Seedlings at the four-leaf stage were used for drought stress treatments. The stressed plants were harvested until all of the leaves were rolled and the nonstressed samples were collected at the same time. For all of the samples, the aboveground parts of the seedlings were harvested and frozen in liquid nitrogen for RNA isolation or immediately into 1% formaldehyde for chromatin isolation.
For generating overexpressed OsPP2C49 transgenic rice plants, the genome fragment that contains the entire OsPP2C49 cDNA fragment amplified with the primer set PP49FK/RB (Supplemental Table S4) was inserted into the KpnI and BamHI sites of the binary expression vector pCAMBIA1301U (driven by a maize ubiquitin promoter). The construct was introduced into rice ZH11 by Agrobacterium tumefaciens (EH105)-mediated transformation. For generating ospp2c49 mutants, specific sgRNA targeting OsPP2C49 (Supplemental Table S4) was designed and assembled into the binary expression vector pCXUN-CAS9 (sgRNA was driven by the U3 promoter). The construct was introduced into rice ZH11 by A. tumefaciens (EH105)-mediated transformation.
Protein Extraction and Western Blot
For nuclear protein extraction, leaves from 4-leaf-old rice plants were ground to a fine powder in liquid nitrogen and then protein was extracted with extraction buffers 1, 2, and 3, which is the same as the nuclear protein extraction method in ChIP assay (Bowler et al., 2004). Nuclear protein was separated with a 12% SDS-PAGE gel, and transferred to a PVDF membrane. The membrane was blocked and incubated with the OsbZIP23 polyclonal antibody (1:5,000) that was generated by the Abmart Company and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody. The signals were detected with x-ray films (Fujifim).
ChIP Sequencing
Wild-type rice under normal conditions, wild-type rice under drought stress conditions, and overexpressed OsbZIP23 rice under normal conditions were used for ChIP-Seq analysis. ChIP was performed as described previously with some modifications (Zong et al., 2013). Briefly, leaf tissues from four-leaf-old seedling were immediately fixed after harvest by 1% formaldehyde under vacuum for 30 min and 3 g tissues for each sample were used for chromatin isolation. Isolated chromatin was sheared to approximately 100 to 500 bp by Diagenode Bioruptor. For ChIP-Seq, the DNA was immunoprecipitated by anti-OsbZIP23 antibody as described previously, and the precipitated DNA was purified and dissolved in distilled water. For each library, three independent replicated samples were prepared and then an equal weight of purified DNA from each replicate was mixed together to generate the sequencing library, which was processed by the Beijing Genomics Institute.
ChIP-Seq data processing and analysis were performed as described previously in Zong et al. (2013). Briefly, raw sequencing reads from each library were mapped to the rice genome (RGAP ver. 7.0, http://rice.plantbiology.msu.edu) using SOAP2 (Li et al., 2009), and only uniquely mapped reads were used for peak identification. The Model-based Analysis of ChIP-Seq (MACS) software was used to identify OsbZIP23-associated regions with default parameters. The .wig files of the MACS output were visualized using the Integrated Genomics Viewer (V.2.3.26; Robinson et al., 2011). A gene was regarded as an OsbZIP23-bound gene if the promoter region of the gene (including 1 kb upstream of the transcription start site) had at least 1 bp overlapping with the peaks. To identify enriched motifs, we extracted 200 bp around the peak summits (100 bp upstream and 100 bp downstream) of each library and then subjected them to MEME-ChIP (Machanick and Bailey, 2011; Ma et al., 2014; http://meme-suite.org/tools/meme-chip). For functional category analysis, KEGG pathway information was collected from the KEGG database (Kanehisa et al., 2012) and the functional category (Rice_japonica_mapping_merged_08 download) was collected from the Mapman Web site (Thimm et al., 2004). The enriched functions of genes bound by OsbZIP23 under drought stress were determined by using Fisher’s exact test. The gene ontology of the OsbZIP23-bound genes under drought stress was also performed by the AgriGO software with default parameters (Du et al., 2010b).
RNA-Seq
RNA-seq was used to identify target genes of OsbZIP23 by integrating analysis with ChIP-Seq data. Total RNAs were extracted from 4-leaf-old rice seedlings using TRIzol (Invitrogen) reagent according to the user manual. For each library, three independent replicated RNA samples were prepared and 100 μg RNA of each replicate was mixed to construct the sequencing library. The libraries were then sequenced with Illumina HiSeq 2000, which was processed by the BerryGenomics Company. Clean reads were mapped to the reference genome of rice (RGAP v. 7.0) using TopHat (v. 2.0.8). To identify differentially expressed genes between the libraries, edgeR (Robinson et al., 2010) was used. The gene expression levels were calculated by using the RPKM method. Genes with an expression change ≥2-fold with P value < 0.05 were defined as differentially expressed genes.
ChIP-Quantitative PCR
The product of ChIP was analyzed via quantitative real-time PCR (the primer sequences are listed in Supplemental Table S4) with a StepOnePlus Real-Time PCR System (Life Tech). Three replicates of each sample were performed and the enrichment values were normalized to the input sample.
Real-Time PCR
Total RNAs were isolated from rice seedlings using TRIzol (Invitrogen) reagent according to the manufacturer's instructions. RNAs (3 µg) were first treated with DNase I (Invitrogen) and then used for cDNA synthesis with the SuperScript III First Strand cDNA Synthesis Kit (Invitrogen). FastStart Universal SYBR Green Master (Roche) was used in real-time PCR analysis with the StepOnePlus Real-Time PCR System (Life Tech). Three technical replicates were performed for each sample, and the expression levels were normalized to Actin for expression quantification. The primer sequences are listed in Supplemental Table S4.
Dual Luciferase Transcriptional Activity Assay in Rice Protoplasts
Rice protoplasts were isolated from 14-d-old seedlings of ZH11 (O. sativa spp. japonica) as described previously in Xie and Yang (2013) with some modifications. Briefly, the rice protoplasts were isolated by digesting rice sheath strips in digestion solution (10 mm MES, pH 5.7, 0.6 m Mannitol, 1 mm CaCl2, 5 mm β-mercaptoethanol, 0.1% BSA, 0.3% Cellulase RS, and 0.75% Macerozume R10) for 4 h with gentle agitation. The protoplasts were then incubated in W5 solution for 30 min, collected by centrifugation at 100g for 8 min, and finally resuspended in MMG solution. For transformation, 3 μg of each plasmid was added together and gently mixed with 100 μL of protoplasts and 110 μL of PEG-CaCl2 solution, which was then incubated at room temperature for 15 min in the dark. Two volumes of W5 solution were added to stop the transformation. The transformed protoplasts were collected by centrifugation and then resuspended in WI solution. The transformed protoplasts were then divided into two equal samples and incubated for 12 h in the dark. After 12 h of incubation, one of the transformed protoplasts was treated with 100 μm ABA in the dark for 4 h. Finally, the protoplasts were collected by centrifugation at 100g for 8 min and immediately utilized in the luciferase assay.
The Dual-Luciferase Reporter Assay System (Promega) was used to measure the luciferase activity. Briefly, the transformed protoplasts were resuspended in 60 μL of 1× Passive Lysis buffer, and 40 μL of lysate was used to measure the luciferase activity. Forty microliters of luciferase assay substrate buffer was added into the lysate and the firefly luciferase (fLUC) activity was measured with the Infinite M200 System (Tecan). Forty microliters of Stop & Glo substrate buffer was then added to the reaction and the renilla luciferase (rLUC) activity was measured. Three independent transformations for each sample were performed, and the relative luciferase activity was calculated as the ratio between fLUC and rLUC (fLUC/rLUC). “None” and yeast GAL4 binding domain vectors (GAL4BD) were used as effectors; 35S-GAL4-fLUC, GAL4-fLUC, and 190LUC were used as reporters; and AtUbi::rLUC was used as an internal control. Primers used for the vector construction are listed in Supplemental Table S4. All of the plasmids used in this assay were purified with the Plasmid Midi Kit (Qiagen).
BiFC Assay
The rice SAPK2 and SAPK6 genes were cloned into the pVYCE vector, and OsbZIP23 and OsPP2C49 were cloned into the pVYNE vector (Waadt et al., 2008). Primers used for the vector construction are listed in Supplemental Table S4. Five micrograms of each vector was mixed and transformed into the rice protoplasts, as described above. After incubation in the dark for 16 h, the fluorescence was observed by confocal microscopy (Leica Microsystems).
In Vitro Pull-Down Assay
For GST-tagged OsbZIP23 or OsPP2C49 protein expression, the pGEX4T-1-OsbZIP23 or OsPP2C49 was constructed and expressed in the Escherichia coli BL21 (DE3) strain (primers are listed in Supplemental Table S4). The expressed protein was purified using the Glutathione Sepharose 4 Fast Flow (GE Healthcare) according to the manufacturer’s instructions. For His-tagged SAPK2 protein expression, the pET28a-SAPK2 was constructed and expressed in the E. coli BL21 (DE3) strain and purified using the Ni-NTA Superflow resin (Qiagen; primers are listed in Supplemental Table S4). Equal volumes of GST or SAPK2-His; OsbZIP23-GST or SAPK2-His; and OsPP2C49-GST or SAPK2-His recombinant proteins were incubated in 1 mL of His pull-down buffer (20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm MgCl2, 0.5% Lgepal CA-630, and 20 mm imidazole) for 6 h at 4°C. One-hundred microliters of Ni-NTA Superflow resin (Qiagen) was added into the protein mixture and it was incubated for another 2 h at 4°C. The resin was washed five times with His pull-down buffer and the pulled proteins were eluted by boiling and further analyzed by immunoblotting using anti-GST (Abcam).
Co-IP Assay in Rice Protoplasts
The Co-IP assay was performed in the rice protoplast system. Rice protoplast isolation and transformation procedures were the same as mentioned above. For each Co-IP assay, the transformation system was 10-fold-amplified. After transformation, the protoplasts were incubated in the 8 mL WI solution in the dark for 16 h at room temperature. After incubation, the protoplasts were collected and then resuspended in 600 μL Co-IP buffer (50 mm Tris-HCl, pH 8.0, 150 mm KCl, 1 mm EDTA, 0.5% Triton X-100, 1 mm DTT, 1 mm PMSF, and 1× Protease Inhibitor Cocktail [Roche]). For thorough lysis of the protoplasts, the Co-IP buffer-protoplast mix was incubated on ice for 10 min. The total protoplast protein was extracted by centrifugation at 12,000 rpm for 15 min and the supernatant was collected. Ten percent of the supernatant was used as an input sample. Forty microliters of GFP-Trap M beads (Chromotek) was mixed with the supernatant and incubated with gentle end-over-end rolling at 4°C for 4 h. The beads were washed five times with the Co-IP buffer and resuspended in 100 μL 2× Laemmli buffer. Finally, they were heated to 95°C for 10 min to dissociate the immunocomplexes and then analyzed by western blot with anti-GFP (ab290; Abcam), anti-OsbZIP23, and anti-cMyc (AE308; ABclonal) antibodies.
In Vitro Protein Phosphorylation Assay
The in vitro phosphorylation assay was performed as described previously by Tang et al. (2012). Briefly, purified GST-fused OsbZIP23 was incubated with SAPK2 or SAPK6 in the kinase buffer (50 mm Tris-HCl, 10 mm MgCl2, 1 mm DTT, 0.2 mm ATP, and 0.5 μCi of [γ-32P]ATP) at room temperature for 30 min and terminated by adding 6 μL of 4× Laemmli buffer and heating at 90°C for 10 min. The reaction product was separated on a 12% SDS-PAGE gel and further exposed to x-ray films (Fujifim) for detecting the signal.
Electrophoretic Mobility Shift Assay
GST-fused OsbZIP23 was expressed and purified as described above (see “In Vitro Pull-Down Assay”). 5′ FAM oligonucleotides were synthesized and labeled by the Shanghai Sangon Company (Supplemental Table S4). To generate double-stranded oligos, an equal amount of the complementary single-stranded oligos was mixed, heated to 95°C for 2 min, and annealed by gradually cooling down to 25°C. To perform the EMSA, purified GST-fused OsbZIP23 was preincubated with the Binding buffer [1 μg poly(dI-dC), 10 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm DTT, 2.5% glycerol, and 5 mm MgCl2] at room temperature for 20 min. For the competition assay, nonlabeled probe was incubated with protein and Binding buffer at room temperature for 20 min. After that, 1 μL 5′ FAM labeled probe (10 μmol/L) was added and incubated at room temperature for 20 min. The samples were then subjected to electrophoresis on 6% PAGE gels running with 0.5× Tris-borate-EDTA buffer at 4°C in the dark for 1 h. The fluorescence signaling was captured by FLA-5100 (Fuji).
ABA Treatment
To test the ABA sensitivity of OsPP2C49-overexpressed transgenic plants at the seedling stage, positive seeds were selected by germinating on half-strength MS medium containing 25 mg/L hygromycin, and the negative lines and the wild type were germinated on normal half-strength MS medium. After 3 d of germination in the dark, the germinated seeds were transplanted to normal half-strength MS medium or half-strength MS medium containing 2 μM ABA and grown for 7 d under 14-h light/10-h dark conditions. After 7 d of growth, shoot and root length were measured.
Quantification of ABA Levels in Rice
ABA extraction and quantification was performed as described previously in Liu et al. (2012). Briefly, 20 mg of freeze-dried four-leaf stage seedlings were extracted twice with 750 μL of plant hormone extraction buffer (methanol:water:glacial acetic acid, 80:19:1, v/v/v) supplemented with 20 ng/mL 2H6ABA internal standards. Quantification was performed in an ABI 4000 Q-Trap (Applied Biosystems).
Accession Numbers
The sequence data from this article can be found in the RGAP database (http://rice.plantbiology.msu.edu) under the following accession numbers: OsbZIP23, LOC_Os02g52780; SAPK2, LOC_Os07g42940; SAPK6, LOC_Os02g34600; OsPP2C49, LOC_Os05g38290; Rab16A, LOC_Os11g26790; OsNCED4, LOC_Os07g05940; DSM2, LOC_Os03g03370; PSY1, LOC_Os06g51290; LCYe, LOC_Os01g39960; and Actin, LOC_Os03g50885. The ChIP-Seq and RNA-seq raw data are deposited in NCBI’s Gene Expression Omnibus with accession code GSE81462.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignment of OsbZIP23, OsbZIP46, and TRAB1.
Supplemental Figure S2. Effects of Ser/Thr substitution on SAPK2-mediated transcriptional activity activation of OsbZIP23.
Supplemental Figure S3. Immunoblotting detects the specificity of the OsbZIP23 antibody.
Supplemental Figure S4. Statistical analysis of the length of the OsbZIP23-bound regions.
Supplemental Figure S5. Functional categories of the OsbZIP23 bound genes in drought stress conditions.
Supplemental Figure S6. OsbZIP23BGDs with known functions in various cellular processes and responses.
Supplemental Figure S7. ChIP-qPCR verification of the ChIP-Seq results.
Supplemental Figure S8. Correlation analysis between the RT-qPCR and the RNA-seq data.
Supplemental Figure S9. The ospp2c49 showed similar ABA sensitivity with the wild-type.
Supplemental Figure S10. OsbZIP23 directly binds to the promoters of the ABA synthesis genes.
Supplemental Table S1. Statistics summary for the ChIP-Seq libraries.
Supplemental Table S2. OsbZIP23 directly binds to the promoter of clade A PP2Cs in rice.
Supplemental Table S3. OsPYLs and SAPKs identified in OsbZIP23BGDs.
Supplemental Table S4. Primers used in this study.
Supplemental File S1. OsbZIP23 bound genes under drought stress conditions (OsbZIP23BGDs).
Supplemental File S2. RNA-seq results of differentially OsbZIP23-regulated genes under normal and drought stress conditions.
Supplemental File S3. OsbZIP23-target genes involved in drought stress (OsbZIP23TGDs).
Supplementary Material
Acknowledgments
We thank Dr. Honghong Hu (Huazhong Agricultural University, China) and Dr. Hao Du (Harvard University, Cambridge, MA) for critical reading of the article. We thank Dr. Hongbo Liu and Dr. Dongqin Li (Huazhong Agricultural University, China) for technical assistance in ABA level quantification analyses and Dr. Masaru Ohme-Takagi (National Institute of Advanced Industrial Science and Technology, Japan) for providing the plasmids 35S-GAL4-fLUC, GAL4-fLUC, and 190fLUC and Dr. Shouyi Chen (Institute of Genetics and Developmental Biology, China) for providing the plasmids GAL4BD, ‘None’, and AtUbi3-rLUC.
Footnotes
Articles can be viewed without a subscription.
References
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72: 557–566 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez JdeD, Fujii H, Wang X (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA 111: 9651–9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS (2007) A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol 63: 151–169 [DOI] [PubMed] [Google Scholar]
- Du H, Wang N, Cui F, Li X, Xiao J, Xiong L (2010a) Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol 154: 1304–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wu N, Cui F, You L, Li X, Xiong L (2014) A homolog of ETHYLENE OVERPRODUCER, OsETOL1, differentially modulates drought and submergence tolerance in rice. Plant J 78: 834–849 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010b) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Cai W (2012) OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 7: e45117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E ZG, Zhang YP, Zhou JH, Wang L (2014) Mini review roles of the bZIP gene family in rice. Genet Mol Res 13: 3025–3036 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in three abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Finkelstein RR, Sun SS, Rock CD (2002) ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J Biol Chem 277: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Hao Q, Li W, Yan C, Yan N, Yin P (2014) Identification and characterization of ABA receptors in Oryza sativa. PLoS One 9: e95246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB (2010) The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167: 1512–1520 [DOI] [PubMed] [Google Scholar]
- Hu H, Xiong L (2014) Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol 65: 715–741 [DOI] [PubMed] [Google Scholar]
- Huang Y, Xiao B, Xiong L (2007) Characterization of a stress responsive proteinase inhibitor gene with positive effect in improving drought resistance in rice. Planta 226: 73–85 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Kim N, Moon SJ, Min MK, Choi EH, Kim JA, Koh EY, Yoon I, Byun MO, Yoo SD, Kim BG (2015) Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front Plant Sci 6: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M, Taji T, Hayashi T, Sakata Y (2013) Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, Gong Z (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, Xiao H, Song R (2015a) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell 27: 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Shen H, Wang T, Wang X (2015b) ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol 56: 2396–2408 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, Chu C (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wu F, Sheng P, Zhang Z, Zhang X, Guo X, Wang J, Cheng Z, Wang J, Wang H, Wan J (2015) The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat Commun 6: 7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84: 19–36 [DOI] [PubMed] [Google Scholar]
- Liu H, Li X, Xiao J, Wang S (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615 [DOI] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, et al. (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Noble WS, Bailey TL (2014) Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc 9: 1428–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya JP, Sethi V, Gangappa SN, Gupta N, Chattopadhyay S (2015) Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J 83: 439–450 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram H, Priya P, Jain M, Chattopadhyay S (2014) Genome-wide DNA binding of GBF1 is modulated by its heterodimerizing protein partners, HY5 and HYH. Mol Plant 7: 448–451 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Saruhashi M, Kumar Ghosh T, Arai K, Ishizaki Y, Hagiwara K, Komatsu K, Shiwa Y, Izumikawa K, Yoshikawa H, Umezawa T, Sakata Y, Takezawa D (2015) Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc Natl Acad Sci USA 112: E6388–E6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK (2010) Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Jha SK, Bagri J, Pandey GK (2015) ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One 10: e0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ram H, Abbas N, Chattopadhyay S (2012) Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J Biol Chem 287: 25995–26009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Kurata N, Ohyanagi H, Hake S (2014) Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26: 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. (2011) LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6: 1975–1983 [DOI] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang YN, Xue LJ, Zou MJ, Liu JY, Chen F, Xue HW (2011) Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol 156: 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin CC, Ma B, Collinge DP, Pogson BJ, He SJ, Xiong Q, Duan KX, Chen H, Yang C, Lu X, et al. (2015) Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell 27: 1061–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Du H, Hu H, Xiong L (2014) A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol Biol 84: 693–705 [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Li X, Ning J, Hu H, Li X, Xiao J, Xiong L (2013) The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J Exp Bot 64: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Xing L, Liu X, Hou YJ, Chinnusamy V, Wang P, Duan C, Zhu JK (2013) The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res 23: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Zhong X, You J, Xiong L (2013) Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol 81: 175–188 [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683 [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360: 307–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.