The xylan decoration pattern in all lineages of gymnosperms permits hydrogen bonding of xylan to cellulose.
Abstract
The interaction between cellulose and xylan is important for the load-bearing secondary cell wall of flowering plants. Based on the precise, evenly spaced pattern of acetyl and glucuronosyl (MeGlcA) xylan substitutions in eudicots, we recently proposed that an unsubstituted face of xylan in a 2-fold helical screw can hydrogen bond to the hydrophilic surfaces of cellulose microfibrils. In gymnosperm cell walls, any role for xylan is unclear, and glucomannan is thought to be the important cellulose-binding polysaccharide. Here, we analyzed xylan from the secondary cell walls of the four gymnosperm lineages (Conifer, Gingko, Cycad, and Gnetophyta). Conifer, Gingko, and Cycad xylan lacks acetylation but is modified by arabinose and MeGlcA. Interestingly, the arabinosyl substitutions are located two xylosyl residues from MeGlcA, which is itself placed precisely on every sixth xylosyl residue. Notably, the Gnetophyta xylan is more akin to early-branching angiosperms and eudicot xylan, lacking arabinose but possessing acetylation on alternate xylosyl residues. All these precise substitution patterns are compatible with gymnosperm xylan binding to hydrophilic surfaces of cellulose. Molecular dynamics simulations support the stable binding of 2-fold screw conifer xylan to the hydrophilic face of cellulose microfibrils. Moreover, the binding of multiple xylan chains to adjacent planes of the cellulose fibril stabilizes the interaction further. Our results show that the type of xylan substitution varies, but an even pattern of xylan substitution is maintained among vascular plants. This suggests that 2-fold screw xylan binds hydrophilic faces of cellulose in eudicots, early-branching angiosperm, and gymnosperm cell walls.
The plant secondary cell wall is a complex network of various polysaccharides and phenolic compounds that act in concert to provide strength to the cell wall (Kumar et al., 2016). Cellulose, formed of strong crystalline fibrils of linear β1,4 glucan, comprises about 40% of dry plant biomass. The other secondary cell wall polysaccharides, largely xylan and glucomannan, comprise about 30% of dry plant biomass. The abundance and structure of these hemicelluloses vary with plant species and tissues, but they have in common that they are tightly associated with cellulose. It is believed that the most important biological role of hemicelluloses is their contribution to strengthening the cell wall by interaction with cellulose and, in some walls, with lignin (Scheller and Ulvskov, 2010). However, it is unclear how hemicelluloses interact with cellulose in the cell wall (Cosgrove and Jarvis, 2012). In this work, we were interested in the interaction of cellulose with xylan, one of the most abundant polysaccharides in nature.
To understand polysaccharide interactions in the cell wall, we need to know not only the hemicellulose primary structure, but also the conformation of the polysaccharide chains. The glucan chains in cellulose are similar to a flat ribbon known as a 2-fold helical screw and associate through lateral hydrogen bonding into sheets. The sheets of glucan chains stack on top of each other, resulting in highly ordered crystalline cellulose. It is still unclear how many glucan chains form a microfibril and whether the microfibril has a hexagonal or rectangular cross section. However, recent studies shed some light onto these questions (Fernandes et al., 2011; Newman et al., 2013; Thomas et al., 2013; Cosgrove, 2014; Oehme et al., 2015; Thomas et al., 2015; Wang and Hong, 2016; Vandavasi et al., 2016). Whichever model is favored, hydrophobic (e.g. 100 or 200) and hydrophilic (e.g. 110 or 010) crystal faces are exposed for interaction with other molecules such as hemicelluloses (Zhao et al., 2014; Cosgrove, 2014; Li et al., 2015).
The β1,4 xylan backbone is always further modified, often by acetyl (Ac), arabinosyl (Ara), and glucuronosyl (MeGlcA) side-chain substitutions. These substitutions are supposed to be necessary to maintain xylan solubility (Mikkelsen et al., 2015). Unsubstituted xylan forms crystalline fibers of chains adopting a 3-fold screw helix (Nieduszynski and Marchessault, 1971). Consequently, xylan substitutions are essential for xylan function and vascular plant viability (Mortimer et al., 2010; Xiong et al., 2013, 2015). In vitro experiments and in silico modeling suggest xylan interacts with cellulose, and it is widely accepted that this is partly through interactions on the hydrophobic faces of the cellulose fibrils (Bosmans et al., 2014; Köhnke et al., 2011; Kabel et al., 2007; Busse-Wicher et al., 2014). In contrast to the binding to the hydrophobic faces, the backbones of highly substituted hemicelluloses are thought to be unable to hydrogen bond effectively with the hydrophilic surfaces of cellulose fibrils because of steric hindrance. For example, hydrogen bonding of the xyloglucan backbone to cellulose would be blocked by steric restrictions of the side chains (Finkenstadt et al., 1995; Zhang et al., 2011). How then does the naturally occurring, highly substituted, xylan interact with cellulose? Our recent findings in the eudicot Arabidopsis (Arabidopsis thaliana) revealed that the majority of xylan bears substitutions solely on alternate xylosyl residues. Every second Xyl is acetylated (Busse-Wicher et al., 2014; Chong et al., 2014), and MeGlcA side chains reside on evenly spaced xylosyl residues, largely at 6-, 8-, 10-, or 12-residue intervals (Bromley et al., 2013). In this scenario, on a xylan backbone in the ribbon-like 2-fold helical screw conformation, all the decorations will face one side, creating an unsubstituted xylan surface. Therefore, in addition to forming stacking interactions on the hydrophobic surface, this xylan structure is compatible with hydrogen bonding to the hydrophilic surface of cellulose (Busse-Wicher et al., 2014; Busse-Wicher et al., 2016).
Both xylan and glucomannan are substantial components of vascular plant secondary cell walls (Timell, 1967; Willfor et al., 2005; McKee et al., 2016). In conifers (gymnosperms, Pinales), the main hemicellulose is glucomannan, but eudicots possess relatively little glucomannan (Scheller and Ulvskov, 2010; Huang et al., 2015), and the secondary cell walls are dominated by xylan, suggesting xylan might have adopted additional functions in these flowering plants that produce hardwoods (Dammström et al., 2009). Conifers, providing softwood for the paper, pulp, and construction industries, are of major ecological and economical value. Consequently, understanding the function and architecture of the cell wall components of softwoods and hardwoods is of great importance.
To investigate whether the precise arrangement of xylan decorations on evenly spaced xylosyl residues, as seen in eudicots, is a novel feature of hardwood xylan, we analyzed the pattern of xylan substitution in various gymnosperms and angiosperms. In addition to conifers, there are three further gymnosperm lineages: Cycad, Gingko and Gnetophyta (Fig. 1). There has been a debate whether Gnetophyta are the gymnosperm lineage most closely related to the angiosperms (Davis and Schaefer, 2011; Uddenberg et al., 2015). There are few studies across gymnosperm lineages to determine any divergence in the structure of xylans.
Figure 1.
Schematic representation of the phylogenetic relationship between gymnosperm and angiosperm species studied in this work. The distances do not correspond to phylogenetic distances.
Our work shows that some gymnosperm xylans have decorations and decoration patterns that are different to those of eudicot xylans. Nevertheless, these modifications largely reside on even xylosyl residues on the backbone. Molecular dynamics simulations support the hypothesis that this highly conserved organization of substitutions allows an unsubstituted surface of xylan to bind stably to hydrophilic faces of cellulose fibrils.
RESULTS
Conifer Xylan Is Decorated with MeGlcA on Every Sixth Xylosyl Residue
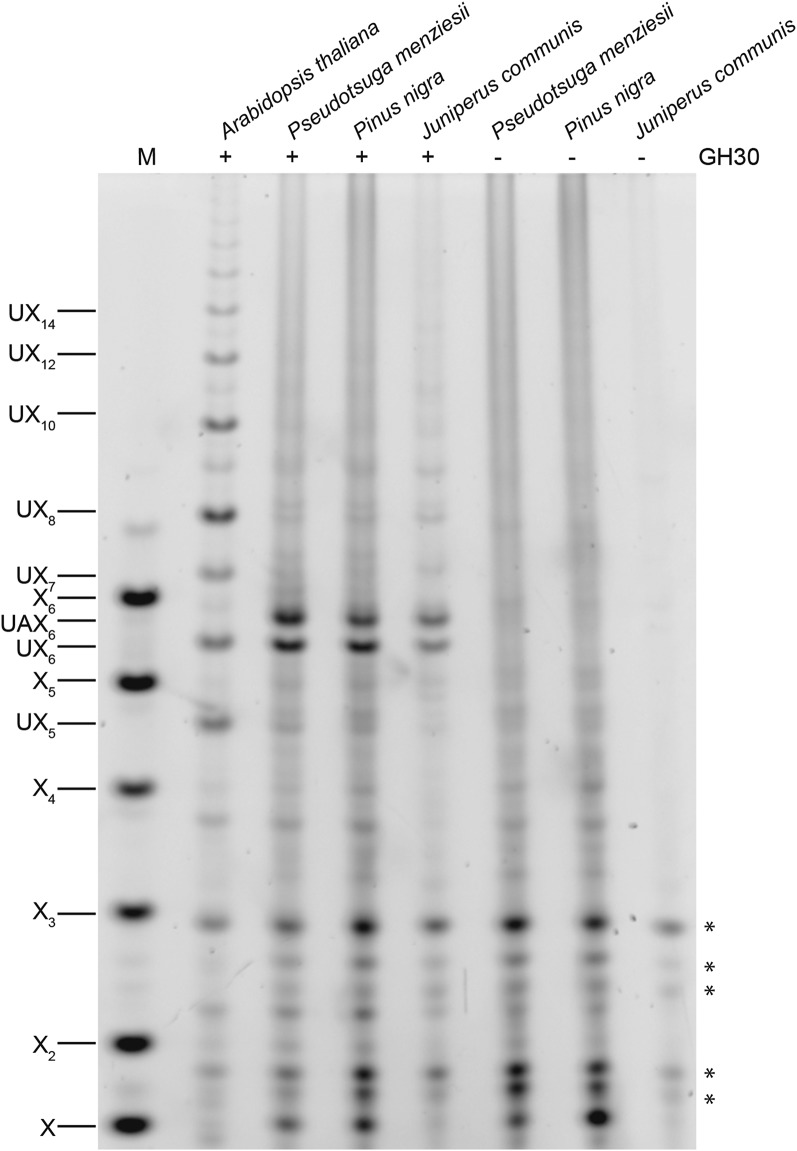
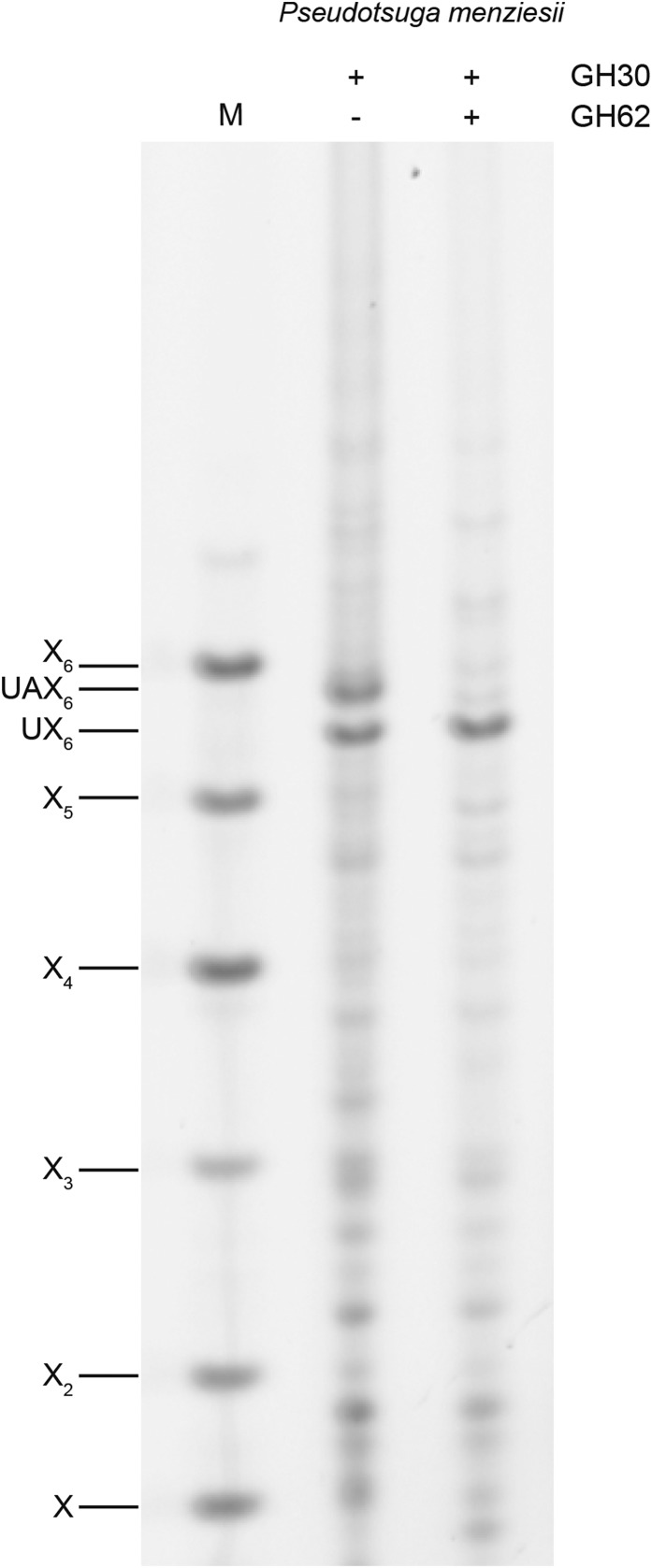
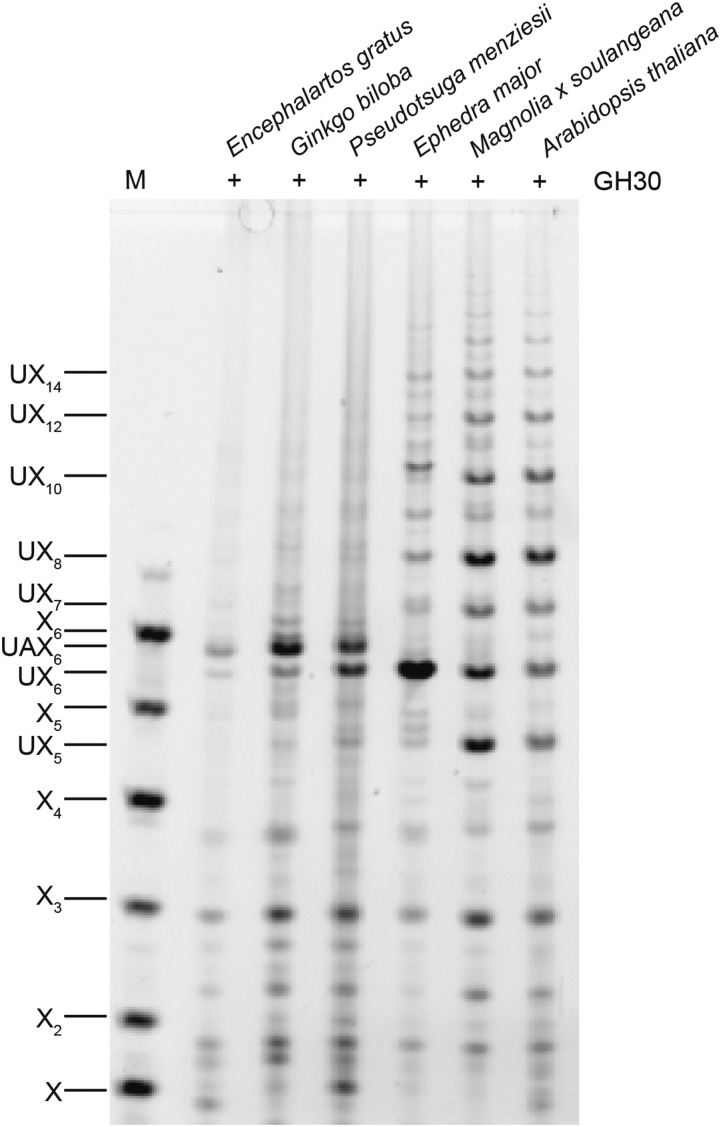
Conifer xylan is decorated by α1,3-arabinofuranose and α1,2-MeGlcA (Timell, 1967; Jacobs et al., 2001; Willfor et al., 2005; McKee, et al., 2016). Using partial acid hydrolysis and MALDI-ToF mass spectrometry (MS), Jacobs et al. (2001) analyzed the distribution of MeGlcA side chains on xylan from several conifers. They suggested that the majority of substitutions are regularly spaced, on every seventh or eighth xylosyl residue along the backbone. A spacing of seven residues would be incompatible with binding to the hydrophilic surface of cellulose fibrils (Busse-Wicher et al., 2016). To investigate the precise pattern of xylan MeGlcA substitution across plant lineages, we used a glucuronoxylanase in CAZy family GH30. This xylanase hydrolyses the xylan backbone adjacent to each MeGlcA, leaving MeGlcA at the O-2 linked position from the cleavage site (St John et al., 2011; Bromley et al., 2013). Alcohol insoluble residue (AIR) from debarked stems of three representative conifers (Pinales: Douglas fir [Pseudotsuga menziesii], black pine [Pinus nigra], and juniper [Juniperus communis]) were incubated with alkali to solubilize the hemicelluloses, and then digested with the GH30 xylanase. The resulting oligosaccharides were analyzed by PACE (polysaccharide analysis by carbohydrate gel electrophoresis). In contrast to the range of oligosaccharides released from Arabidopsis xylan, just two main oligosaccharides running between the standards Xyl5 and Xyl6 were produced from all three conifer xylans (Fig. 2). One of these oligosaccharides comigrated with MeGlcAXyl6 (UX6), a product from digestion of Arabidopsis xylan. The other oligosaccharide migrated slightly more slowly, suggesting an additional substitution. To investigate the nature of the additional substitution, we digested AIR from Douglas fir with GH30 followed by GH62 α-arabinofuranosidase. The oligosaccharide was sensitive to the arabinosidase, indicating arabinose decoration, and migrated with UX6 after digestion, so we named it UAX6 (Fig. 3). Taken together, the data show that the majority of MeGlcA substitutions are found on every sixth xylosyl residue of conifer xylan and the xylan is additionally modified by α-arabinosyl side chains.
Figure 2.
MeGlcA is regularly distributed on xylan in conifers. AIR of Douglas fir, black pine, common juniper, and the eudicot Arabidopsis for comparison were hydrolyzed with glucuronoxylanase GH30 and analyzed by PACE. The major conifer products correspond to substituted xylohexoses UX6 and UAX6. Undigested material was used as a control (−). Marker (M), xylosyl oligosaccharides X1-X6; UXn, Xn substituted by MeGlcA. Bands marked with asterisks are nonspecific labeling products.
Figure 3.
α-Arabinofuranose decorations on conifer xylan. Glucuronoxylanase GH30 digestion products of Douglas fir were digested with α-arabinofuranosidase GH62 and analyzed by PACE. UAX6 was sensitive to the arabinosidase, yielding UX6. Marker (M), xylosyl oligosaccharides X1-X6.
Arabinose and MeGlcA Are Located Two Xylosyl Residues Apart on the Conifer Xylan Backbone
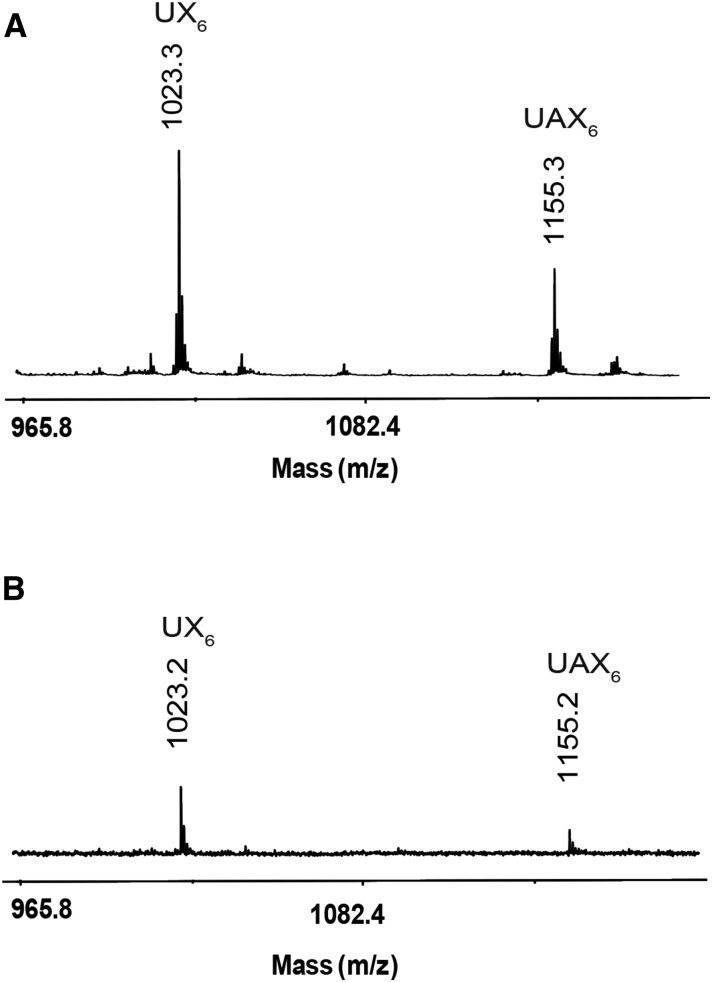
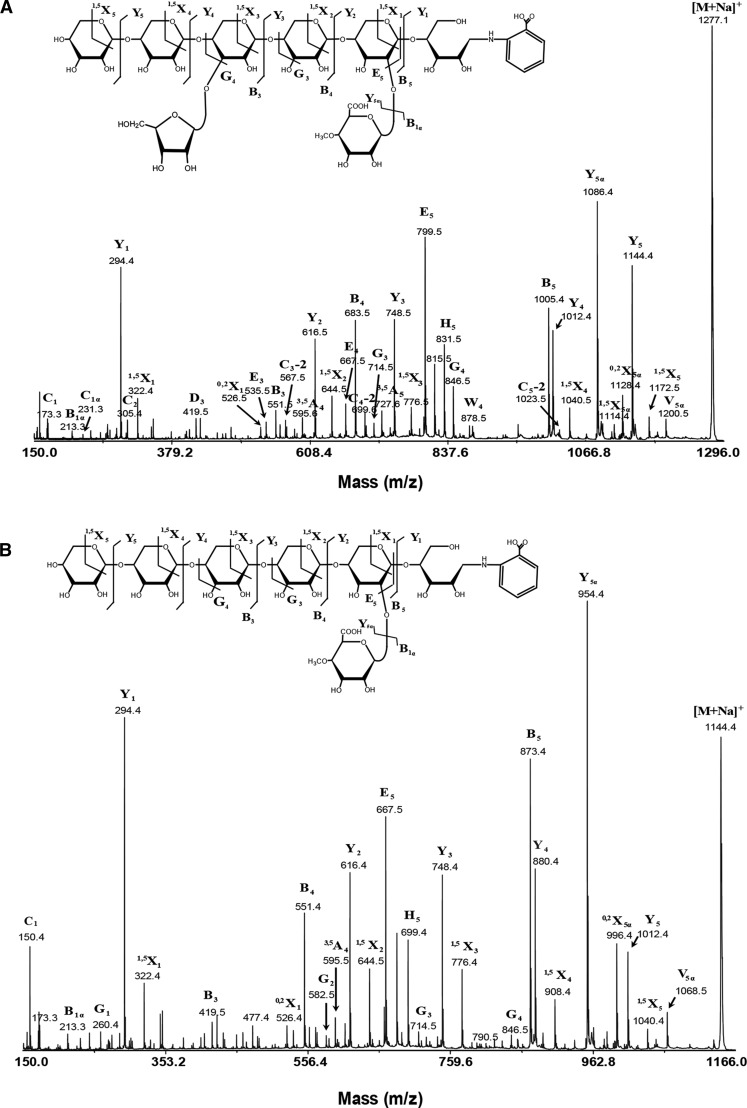
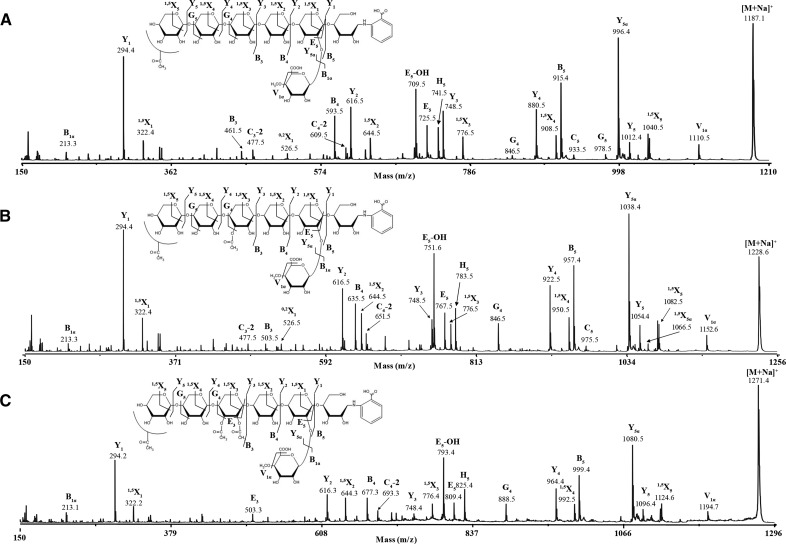
To determine any regularity in the position of the arabinosyl side chains on the backbone, we analyzed GH30 xylanase digestion products of alkali extracted (thus, all possible acetyl groups would have been removed) xylan from Douglas fir (conifer) by MS. MALDI-ToF MS revealed two main ions of mass corresponding to MeGlcAPent6 (m/z 1023.3) and MeGlcAPent7 (m/z 1155.3; Fig. 4), which is consistent with the PACE assignments of UX6 and UAX6. No substitutions with unmethylated GlcA were detected. MALDI-TOF CID MS/MS of 2-AB-labeled MeGlcAPent6 confirmed the oligosaccharide was UX6, a linear pentose backbone with 2-linked MeGlcA on the second xylosyl residue from the reducing end (Fig. 5). MS/MS of the MeGlcAPent7 peak showed similar fragmentation to UX6 but indicated that an additional pentosyl side chain substitution was located on the O-3 of the fourth Xyl from the reducing end (Fig. 5). Taken together with the α-arabinofuranosidase sensitivity (Fig. 3), we conclude that α-l-arabinofuranose is located at −2 position from MeGlcA on the conifer xylan backbone (UAX6), maintaining an evenly spaced pattern. PACE and MS analyses were repeated with xylan from black pine (conifer) and gave similar results (Supplemental Figs. S1 and S2).
Figure 4.
Mass spectrometry of glucuronoxylanase GH30 digestion products of conifer xylan. A, MALDI-ToF-MS of glucuronoxylanase GH30 digestion of NaOH-extracted, deacetylated Douglas fir xylan shows two species UX6 and UAX6. B, MALDI-ToF-MS of glucuronoxylanase GH30 digestion of DMSO-extracted conifer xylan shows no acetylation of UX6 or UAX6.
Figure 5.
MALDI-CID of glucuronoxylanase GH30 digestion products of conifer Douglas fir xylan shows arabinose and MeGlcA substitutions are spaced two residues apart. A, High-energy MALDI-CID of the conifer UAX6 oligosaccharide labeled with 2-AA and separated by HILIC. 1,3-Linked arabinose is present two xylosyl residues from the MeGlcA. B, High-energy MALDI-CID of the conifer UX6 oligosaccharide labeled with 2-AA and separated by HILIC.
Xylan Decoration Is Not Identical in Different Lineages of Gymnosperms
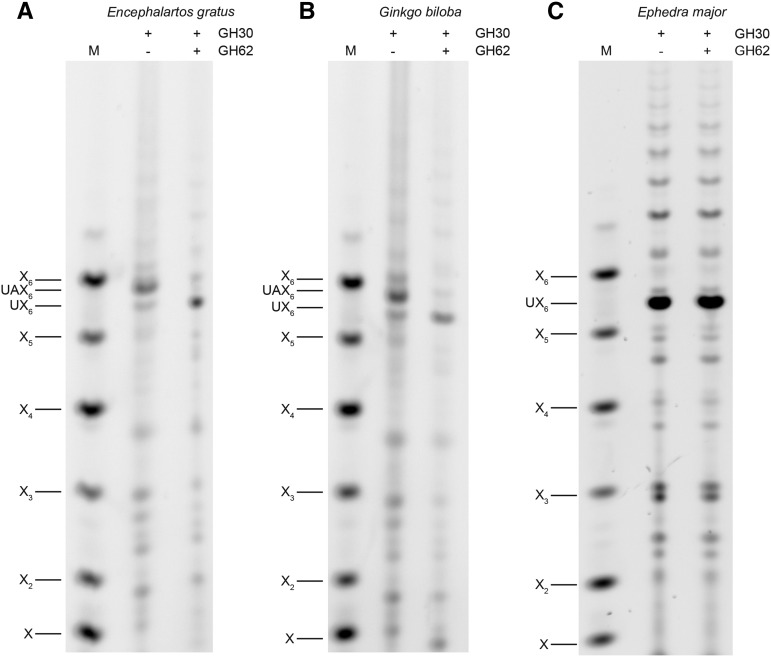
Xylan substitutions and substitution patterns are conserved in different conifer species. However, the xylan structure of the other gymnosperm lineages, Cycads, Gingko, and Gnetophyta, is unknown. To investigate their xylan structures, we produced AIR from mature woody petioles of a Cycad (Encephalartos gratus) and debarked stems from Ginkgo (Ginkgo biloba) and two representatives of the Gnetophyta (Ephedra major and Gnetum edule). Digestion of AIR with GH30 xylanase and analysis by PACE showed that two oligosaccharides were released from Cycad and Gingko xylan, which comigrate with the two oligosaccharides, UX6 and UAX6, released from the conifer Douglas fir (Fig. 6). Further digestion with GH62 arabinofuranosidase confirmed these products are identical to those from conifer xylan (Fig. 7). Interestingly, UAX6 appears to be relatively more abundant than the UX6, suggesting greater arabinosylation in Gingko and Cycads than in the conifer xylan. In contrast to this, UAX6 was completely absent in E. major (Gnetophyta) and UX6 was the predominant product (Fig. 6). The absence of arabinosyl decorations was confirmed by insensitivity of the products to digestion with GH62 arabinofuranosidase (Fig. 7). In addition to UX6, we also detected some degree of polymerization (DP) 8 and longer xylo-oligosaccharides in E. major (Gnetophyta) (Figs. 6 and 7), indicating that there is a low proportion of MeGlcA substitutions more widely spaced than every sixth xylosyl residue. Nevertheless, six xylosyl residues was the most frequent spacing of MeGlcA in Gnetophyta.
Figure 6.
Analysis of the MeGlcA distribution on xylan in the four gymnosperm lineages and Magnoliales. AIR prepared from representatives of each gymnosperm lineage: Cycad (E. gratus), Ginkgo (G. biloba), conifer (P. menziesii), and Gnetophyta (E. major) and a representative of Magnoliales (Magnolia x soulangeana), plus the eudicot Arabidopsis for comparison, were hydrolyzed by glucuronoxylanase GH30 and analyzed by PACE. The cycad, gingko, and conifer show UX6 and UAX6, but the Gnetophyta products are largely UX6. Marker (M), xylosyl oligosaccharides X1-X6.
Figure 7.
Xylan is decorated with α-arabinofuranose in cycad and Ginkgo but not Gnetophyta. Glucuronoxylanase GH30 digestion products of cycad (E. gratus; A), Ginkgo (G. biloba; B), and Gnetophyta (E. major; C) xylan were digested with α-arabinofuranosidase GH62 and analyzed by PACE. UAX6 present in cycad and Ginkgo GH30 digestion products is sensitive to the arabinosidase. Marker (M), xylosyl oligosaccharides X1-X6.
In Contrast to Other Gymnosperms, the Xylan of Gnetophyta Is Acetylated
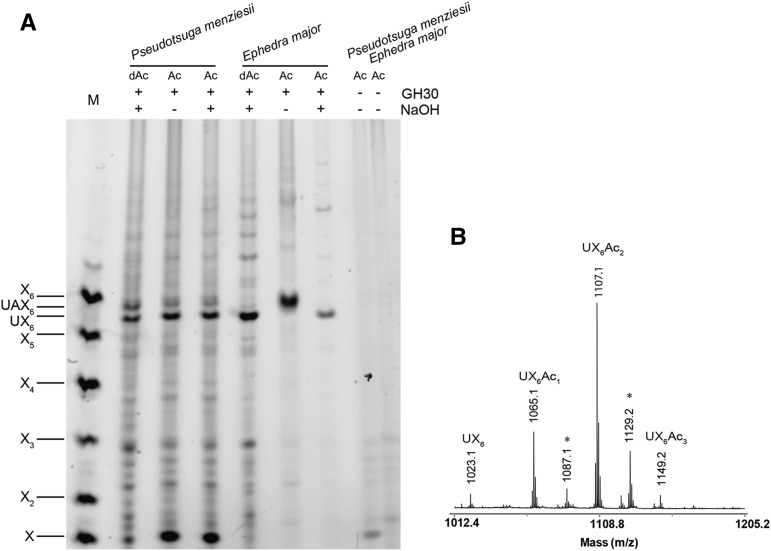
Acetylation of conifer xylan has not been reported. However, the Gnetophyta xylan is similar to secondary cell wall xylan in eudicots in that it lacks arabinose substitution. This raised the question whether acetylation might be present, perhaps in order to maintain solubility of the xylan during biosynthesis. To analyze the acetylation status of xylan in all four gymnosperm lineages, we extracted xylan using DMSO to preserve acetylation and digested it with GH30 xylanase. Douglas fir digestion products were not acetylated (Figs. 4 and 8) and neither were the products of ginkgo or cycads (Supplemental Fig. S3). In contrast to this, the GH30 xylanase digestion products of E. major (Gnetophyta) were sensitive to alkali treatment, suggesting possible acetylation (Fig. 8). MS of these xylo-oligosaccharides confirmed the presence of acetylation (Fig. 8). To investigate whether the acetylation is patterned on alternate xylosyl residues, MS/MS of the acetylated UX6 oligosaccharides was carried out. Three acetylated UX6 products were detectable, carrying the acetyl decorations precisely on alternate residues (Fig. 9). Interestingly, acetylation was not found on the xylosyl residues carrying MeGlcA, unlike eudicot xylan where almost all xylosyl residues bearing MeGlcA are acetylated (Koutaniemi et al., 2012). Moreover, analysis of xylan of G. edule showed that this second Gnetophyta species has similar MeGlcA and acetylation patterns (Supplemental Fig. S4). Therefore, xylan in Gnetophyta is acetylated and acetylation follows an evenly spaced pattern.
Figure 8.
Analysis of xylan acetylation in conifers and Gnetophyta. A, DMSO extracted acetylxylans denoted as “Ac,” from Douglas fir and E. major (Gnetophyta) were hydrolyzed with glucuronoxylanase GH30. After enzyme digestion and labeling with ANTS, some samples were subjected to alkali treatment (NaOH) to remove any ester-linked acetyl groups. A shift of the band mobility after NaOH treatment indicates the presence of acetylation. Digestion products of AIR deacetylated by alkali treatment, denoted as “dAc,” are shown for comparison. Undigested material was used as a control. Marker (M), xylosyl oligosaccharides X1-X6. B, MALDI-ToF-MS of glucuronoxylanase GH30 digested acetylated xylan from E. major reveals acetylation. Asterisk indicates doubly sodiated ions [M-H+2Na]+.
Figure 9.
Gnetophyta xylan oligosaccharides are acetylated on alternate xylosyl residues. Glucuronoxylanase GH30 products from E. major xylan were labeled with 2-AA and separated by HILIC. High-energy MALDI-CID of UX6Ac (A), UX6Ac2 (B), and UX6Ac3 (C). The position of the acetate on the nonreducing Xyl is unknown.
Xylan Substitution Patterning in the Magnoliales Is Similar to That of Eudicot Glucuronoxylan
The fact that in Gnetophyta we found eudicot-like acetylation substitution patterns, but MeGlcA patterns with characteristics of both conifers and eudicots, prompted us to analyze xylan substitution in Magnoliales, an early diverging angiosperm lineage. The oligosaccharides produced by GH30 xylanase digestion of magnolia (Magnolia x soulangeana; Fig. 6) and bay laurel (Laurus nobilis; Supplemental Fig. S5) xylan were highly similar to those from eudicots, with a spacing of 8 most frequent, but 6, 10, 12, and 14 spacing also substantial. The xylan was also acetylated and has no arabinose decorations (Supplemental Fig. S6). Therefore, the Magnoliales have a xylan decoration pattern very similar to eudicots (Bromley et al., 2013; Derba-Maceluch et al., 2015).
Molecular Dynamics Simulations of Xylan Adsorption on Hexagonal Cross-Section Cellulose Microfibrils
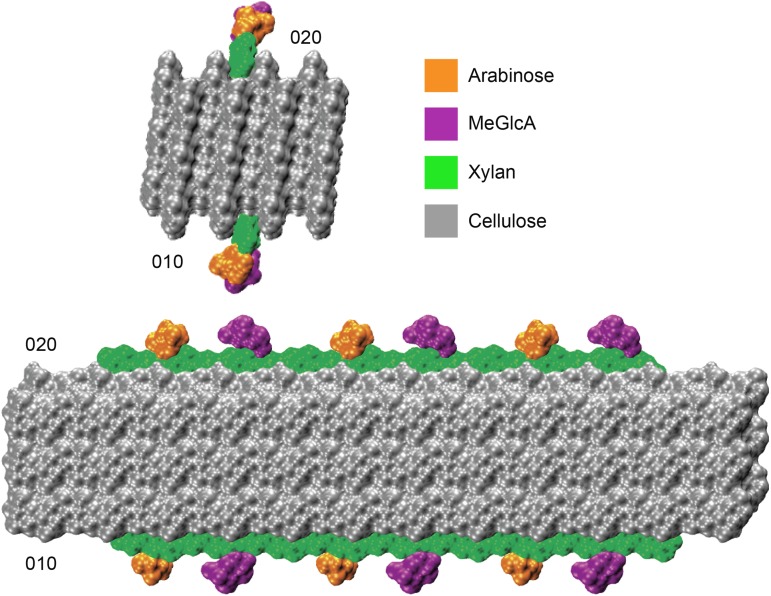
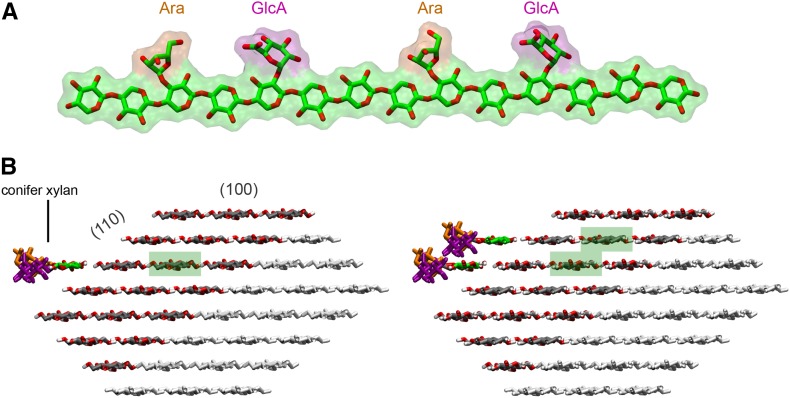
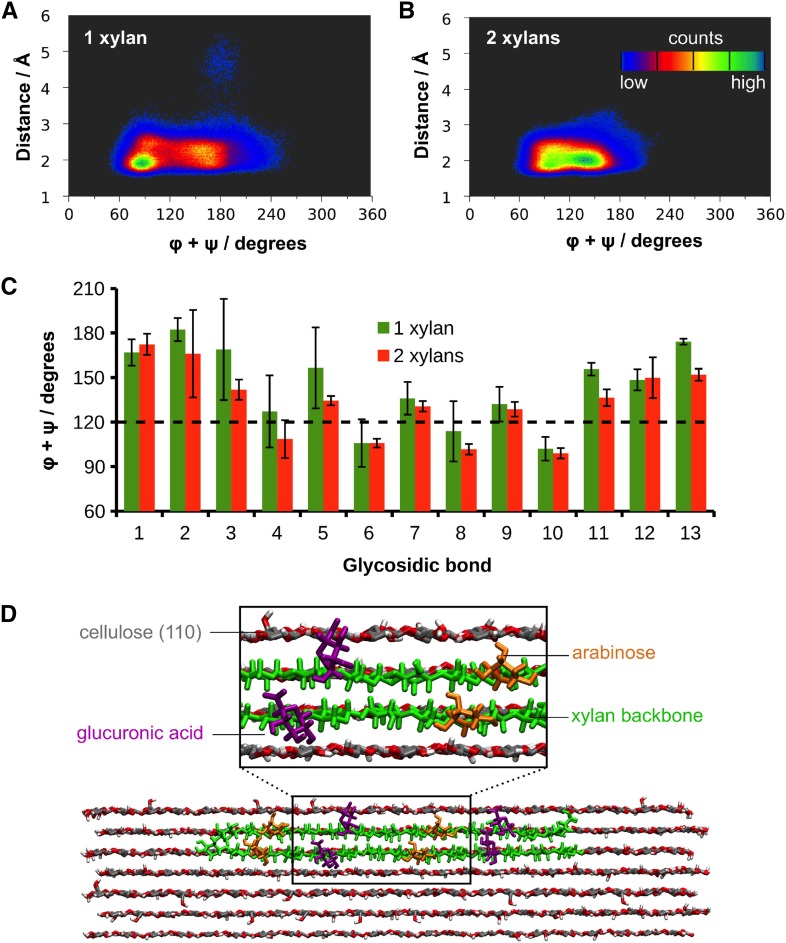
In silico docking of a model of gymnosperm xylan onto a rectangular 24-chain cellulose fibril showed that the even distribution of MeGlcA and arabinose allows xylan adsorption on the hydrophilic (010) and (020) surfaces without steric hindrance (Fig. 10). This indicates gymnosperm xylan could bind to this form of cellulose similarly to the glucuronoxylan from eudicots (Busse-Wicher et al., 2014). However, the structure of the cellulose fibrils in eudicots and gymnosperms is not resolved, and an alternative hexagonal structure cellulose fibril is also considered possible (Fernandes et al., 2011). The hydrophilic faces of such cellulose fibrils (e.g. 110) present glucan chains available for hydrogen bonding to xylan, but the absence of surface grooves could lead to fewer interplane glucan:xylan interactions and consequent instability in xylan chain binding. To investigate whether xylan could bind stably to cellulose fibrils with a hexagonal cross section, we performed molecular dynamics (MD) simulations of decorated gymnosperm xylan chains adsorbed on the 110 hydrophilic surface of an Iβ cellulose microfibril. We built a model of conifer xylan with backbone DP 14, decorated with two α-1,2-linked glucuronic acids spaced by 6 xylosyl residues, and two α-1,3-linked arabinoses located two xylosyl apart from each GlcA (Fig. 11). The cellulose microfibril was modeled as half of a hexagonal 36-chain elementary fibril of DP = 22 so that the (110) hydrophilic face was exposed to xylan adsorption. We considered the cases of one and two xylan chains adsorbed onto the cellulose surface (Fig. 11). The simulations showed that the conifer xylan stably binds to the hydrophilic cellulose surface (Supplemental Movies 1 and 2). Figure 12 show the two-dimensional distribution of xylan configurations defined by the distance of xylosyl residues from the cellulose surface and by the sum of the glycosidic torsion angles φ+ψ, which characterizes the xylan helical conformation. The first and last two residues were not considered in this analysis because they are considerably more flexible than the inner residues. Two-fold (21), right-handed 3-fold (31), and left-handed 3-fold screw conformations are characterized, respectively, by φ+ψ=120°, φ+ψ=50°, and φ+ψ=190°. Figure 12 show that the distance between the xylosyl residues and the cellulose surface remains mostly within 3 Å, indicating stable binding of the xylan chains. Xylan-xylan interactions stabilize the binding further, as seen by the distribution of φ+ψ, which are narrower for the two xylan chain case, indicating greater flexibility of the single chain. For the single conifer xylan, when the xylosyl residues are close to the (110) cellulose surface, the distribution of φ+ψ is spread around 120° with local maxima at 90° and 180° (Fig. 12). During the few times when some xylosyl residues move away from the cellulose surface, the helical conformation fluctuates strictly around the left-handed 31 screw (φ+ψ=190°), which is the conformation observed for xylans in solution (Nieduszynski and Marchessault, 1971). When two xylan chains are adsorbed in parallel with respect to one another and to the microfibril, the helical conformation also fluctuates around φ+ψ=120°, but with a more uniform distribution in the range φ+ψ=90° to 150° (Fig. 12). In Figure 12C, we show the average values of φ+ψ separately for each glycosidic bond of the xylan backbone. Except for the residues belonging to the extremities of the xylan chain, the sum φ+ψ alternates between values somewhat above and below 120° for neighboring residues, with the mean repeat distance of 10.4 Å (distance between bound xylobiose units), indicating that the xylan chains do not exhibit the idealized helical structure characterized by the same values of φ+ψ (120°) for all the residues of a segment. Instead, the values of φ+ψ from adjacent units average out to 120°, so that the adsorbed portions of the xylan chain adopts a 21 screw-like conformation. Thus, the alternating torsion angles provides a stretched conformation with all the substitutions pointing away from the cellulose surface, allowing the xylan backbone to establish hydrogen bonds with the cellulose fibril, as illustrated in Figure 12.
Figure 10.
Model of conifer hemicellulose interactions with hydrophilic surfaces (010 and 020) of a 24-chain cellulose microfibril. The reducing end view and side view are shown. DP 18 xylan chains, with evenly spaced alternate (α-1-3)-arabinofuranosyl and (α-1-2)-MeGlcA decorations at xylosyl residues, were modeled as 2-fold helical screws.
Figure 11.
Cellulose-xylan complexes used for molecular dynamics simulations. A, Conifer xylan model. B, Cellulose microfibril with one and two conifer xylans docked onto the (110) face. Only the dark-gray part of the cellulose was considered in the simulations to save computational cost. Highlighted in green are the glucan chains that were replicated and translated to the cellulose surface to build the xylan chains.
Figure 12.
Molecular dynamics simulations of cellulose-xylan complexes. Two-dimensional distribution of the sum of dihedral angles φ+ψ of xylosyl residues against their distances from the cellulose surface for one (A) and two (B) conifer xylans. The two xylosyl residues at both ends of the xylan chain were not considered in the analysis. C, Average values of φ+ψ for each glycosidic bond of the xylan backbone. The error bars correspond to the standard deviations of the averages taken from the three independent simulations. D, Snapshot taken from the simulation of two xylans adsorbed on the (110) face of the cellulose fibril, showing the extended configuration of the chain.
DISCUSSION
Previously, little attention has been paid to the arrangement of substitutions along the xylan backbone and the resulting implications for the interaction of the polysaccharide with cellulose. Here, we show that precise patterning of xylan substitutions is widespread in vascular plants. This xylan patterning is not restricted to acetylation and MeGlcA substitutions, but extends to arabinosyl residues too. We propose that the patterning of xylan substitutions allows a compatible interaction between xylan and the cellulose microfibril hydrophilic surfaces in vascular plants (Busse-Wicher et al., 2014; Busse-Wicher et al., 2016).
The pattern of MeGlcA substitution of xylan is similar in the four different gymnosperm lineages (Conifers, Cycads, Ginkgo, and Gnetophyta), with a predominant MeGlcA periodicity of six. In contrast to the conserved MeGlcA pattern, acetylation and arabinosylation are different across the gymnosperm lineages. Conifer, Cycad, and Ginkgo xylans are arabinosylated, but acetylation was not detected. In contrast, Gnetophyta xylan is not arabinosylated but is acetylated, like the xylans of eudicots and the early diverging angiosperms such as magnolia. The relationship of the four gymnosperm and angiosperm lineages is to date still unresolved (Davis and Schaefer, 2011). The xylan structural similarities place the Gnetophyta cell walls closer to that of angiosperms than to the other gymnosperm lineages. Consistent with this, other cell wall components in Gnetophyta are also more similar to angiosperm than gymnosperm cell walls. Conifers possess largely G units in lignin, and the wall is dominated by glucomannan. In contrast, Gnetophyta walls have both S and G units in lignin (Grabber et al., 2004), and xylan has higher abundance than glucomannan (Melvin and Stewart, 1969). These results would tend to suggest that the cell walls of ancestors of angiosperms and gymnosperms had these characteristics. Xylan acetylation would be lost and arabinosylation gained in the conifer, ginkgo, and cycad gymnosperm lineages.
Patterning of arabinosyl substitution of cell wall xylan has not been previously reported. We showed that the conifer xylan arabinosyl substitution is precisely two xylosyl backbone residues from the MeGlcA. Furthermore, acetylation resides only on every second xylosyl residue in Gnetophyta. Taken together, we now know that the majority of substitutions are evenly spaced in the xylan of gymnosperms as well as in Magnoliales and eudicots even though the modifications vary between lineages. Therefore, the even spacing appears to be a highly conserved feature, strongly implying a biological role. It is unknown if a compatible pattern is present on the xylan of lower plant lineages such as mosses, Selaginella, and ferns. Moreover, the decoration pattern in monocot angiosperms has not yet been fully assessed. In the commelinoid monocots, including grasses, xylans have acquired additional features such as feruloylation (Carpita and Gibeaut, 1993; Scheller and Ulvskov, 2010). In order to understand xylan interaction with other cell wall components, it will be important to determine if cellulose-compatible xylan structures exist in these plant lineages too.
There are still not enough data to support any secondary plant cell wall model, and there are multiple possibilities how and to what extent the cellulose fibers may be covered and how cell wall components may interact (Harris and Stone, 2008). Having a compatible decoration pattern of MeGlcA and Ara, can the conifer xylan adsorb onto cellulose? In silico experiments (Fig. 10) showed that xylan fits to the (020)/(010) hydrophilic faces of the proposed rectangular 24-chain fibril structure, similarly to the undecorated and acetylated xylan tested previously (Busse-Wicher et al., 2014). This cellulose model, proposed by Fernandes et al. (2011) and Thomas et al. (2013), is supported by experimental data. There are also alternative models proposed, including microfibrils with irregular cross sections, as discussed by Cosgrove (2014) and Newman et al. (2013). Here, we performed MD simulations of the adsorption of xylans on the (110) hydrophilic face of a 36-chain hexagonal cellulose fibril. The main difference between the (110) and the (020)/(010) hydrophilic faces is that the former lacks the spatial grooves where the xylan chain may fit in well-defined binding sites and establish stacking interactions with neighboring glucan chains. In spite of the lack of such grooves in the hexagonal fibril, we showed that conifer xylan stably binds to the (110) cellulose surface. The backbone binds with a somewhat distorted 2-fold screw conformation through hydrogen bonds with the exposed cellulose hydroxyl groups. The 2-fold screw conformation allows all substitutions to point away from the cellulose surface. Also, we found that the presence of two xylan chains in adjacent planes of the cellulose stabilizes binding as a 2-fold helix. This suggests that xylans from conifers and others have the potential to bind multiple layers of glucan chains in the cellulose fibrils and therefore may coat the hydrophilic faces of cellulose. The MD simulations show that if the xylan diffuses from the cellulose surface, it relaxes into a 3-fold screw conformation. Therefore, adoption of the 2-fold screw conformation is dependent on interaction with the cellulose. Solid-state NMR lends support to the view that xylan adopts multiple conformations in the cell wall (Dupree et al., 2015) and will be important to test the models presented in this work.
Binding of an unsubstituted surface of the decorated xylan backbone to cellulose microfibrils in a 2-fold-screw conformation appears to have at least two advantages. First, the xylan chains can bind to the hydrophilic in addition to the more accessible hydrophobic faces of cellulose fibrils. In addition, when bound to the hydrophilic face, the xylan substitutions are exposed for interaction with other cell wall components. Each of the different xylan decorations likely adds different properties to the surface of the xylan-coated cellulose microfibril: Acetylated xylan would reduce water hydrogen bonding to cellulose or xylan hydroxyl groups and promote microfibril interactions with hydrophobic lignin and perhaps with other fibrils. The GlcA side chains confer a charge to the surface and might link to lignin (Reis and Vian, 2004; Oinonen et al., 2015) or bind metal cations. Arabinosyl side chains are hydrophilic. The surface of microfibrils coated with glucuronoarabinoxylan in conifers will therefore have different properties to fibrils coated with acetylated xylan in Gnetophyta, Magnoliales, or eudicots. Xylan modifications might have evolved to adapt to the composition of the specific cell walls, tuning the microfibril interaction between different lignin and hemicellulose cell wall components. In conifers, glucomannan is also in close proximity to cellulose (Fernandes et al., 2011; Salmen, 2015). We do not know if xylan and glucomannan on the cellulose microfibril surfaces provide a different function.
The position of Gnetophyta in gymnosperm evolution is not resolved. The walls of Gnetophyta show higher similarities to angiosperms than to the other gymnosperms. This could suggest that the Gnetophyta walls show ancestral features lost in the other gymnosperms. Alternatively, perhaps both the angiosperms and Gnetophyta convergently evolved similar types of walls to perform specific water conducting function. Most gymnosperm xylem is built of tracheids, which both conduct water and provide mechanical support. Interestingly, Gnetophyta possess vessel elements in the xylem similarly to eudicots. The more hydrophobic acetylated xylan occurs with the appearance of vessel cells specialized in water conduction.
CONCLUSION
Here, we focused on the spatial arrangement of the decorations on xylan and conclude that even spacing of substitutions is highly conserved in plants, suggesting an important role for xylan to fold as a 2-fold helical screw. We showed by means of MD simulations that the even spacing would allow xylan to bind to hydrophilic faces of cellulose microfibrils by hydrogen bonds. The binding of xylan as a 2-fold screw to cellulose is likely to be stable for hydrophilic faces of rectangular and hexagonal cellulose fibrils. These data underline the importance of patterning of xylan substitutions in plant cell wall architecture.
MATERIALS AND METHODS
Plant Material
Plant material was collected at the University of Cambridge Botanic Garden, Cambridge, as follows: The base of mature woody petioles of a Encephalartos gratus and Dioon edule (Cycads, grown at greenhouse), ∼1-cm-diameter branch of Ginkgo biloba (Ginkgo), ∼1-cm-diameter branch of Pinus nigra (black pine), ∼0.5-cm-diameter branch of Juniperus communis (juniper), ∼0.5-cm branch of Ephedra major (Gnetophyta), ∼0.5-cm branch of Gnetum edule (Gnetophyta, grown at greenhouse), ∼1-cm twig of Magnolia x soulangeana (magnolia), and ∼0.5-cm branch of Laurus nobilis (bay laurel). Sawdust of Pseudotsuga menziesii (Douglas fir) was obtained from Willemsen Naaldhout.
After collection, stems were boiled in 96% ethanol for 30 min to inactivate endogenous enzymes. Stems of all plants except E. major were debarked and all plant material was milled. AIR was prepared as described previously (Bromley et al., 2013).
Replication of Experiments
Five species of Pinales obtained from the Botanic Garden were used in this study, two species of Gnetophyta, two Cycadales, and one Ginkgoales. Technically, each experiment was repeated at least three times.
PACE
To release xylan for digestion, 400 μg to 1 mg of AIR was treated with 20 to 50 μL of 4 m NaOH for 1 h before neutralizing with HCl. Alternatively, DMSO extracted xylan was used (to retain potential acetylation, procedure as described in (Busse-Wicher et al., 2014). Xylan was hydrolyzed with Erwinia chrysanthemi GH30 (at concentration 0.1 –0.5 μm) in 0.1 m ammonium acetate buffer, pH 6.0, for 60 min. EcGH30 (Urbániková et al., 2011) was obtained from Novozymes.
For sequential digestion, samples were boiled to heat inactivate the enzyme and redigested with Penicillium aurantiogriseum GH62, a gift from Novozymes. The digestion conditions were 0.1 m ammonium acetate buffer, pH 6.0, for 12 h. After digestion, samples were taken to dryness in vacuo. For the visualization of oligosaccharides, the labeling of digestion products and standards (Xyl to xylohexaose, obtained from Megazyme) was performed with 8-aminonaphthalene-1,3,6- trisulfonic acid (ANTS; Invitrogen; Goubet et al., 2002), and gel running and visualization were carried using a G-box (Syngene), as described by Bromley et al. (2013).
Preparation of Oligosaccharide Samples for Mass Spectrometry
For the preparation of oligosaccharides, 100 μg of AIR was treated as described above. Enzymes were then deactivated by boiling for 30 min, and the hydrolysates were dried in a centrifugal evaporator. The samples were filtered using Nanosep system (Mr cutoff 10 kD; Pall) and dried in vacuo.
For the reductive amination of xylooligosaccharides, HyperSep Hypercarb cartridges (Thermo Scientific) were used for desalting as previously described (Maslen et al., 2007). Purified oligosaccharides were reductively aminated with 2-aminobenzoic acid (2-AA; Sigma-Aldrich) using optimized labeling conditions and further purified using GlykoClean S cartridges (Prozyme) as described previously (Tryfona and Stephens, 2010).
HILIC-MALDI-ToF/ToF-MS/MS
Capillary HILIC was carried out using an LC-Packings Ultimate system (Dionex), which was used to generate the gradient that flowed at 3 μL min−1 as previously described (Tryfona et al., 2012). Solvent A was 50 mm ammonium formate adjusted to pH 4.4 with formic acid. Solvent B was 20% solvent A in acetonitrile. The labeled oligosaccharides, dissolved in 80% acetonitrile, were loaded onto an amide-80 column (300 μm × 25 cm; 3-μm particle size; Dionex) and eluted with increasing aqueous concentration: column was equilibrated with 5% A. The gradient was initiated 5 min after injection and increased linearly to 52% A over 96 min. The column was then equilibrated for 10 min with 5% solvent A prior to the next injection (Tryfona and Stephens, 2010). The column eluent passed through a capillary UV detector (254 nm) to the MALDI sample spotter. For HILIC-MALDI-ToF/ToF-MS/MS, a Probot sample fraction system (Dionex) was employed for automated spotting of the HPLC eluent onto the MALDI target at 20-s intervals. Following air drying, the samples were overlayed with DHB matrix and analyzed by MALDI-ToF-MS (4700 Proteomics Analyzer; Applied Biosystems). The MS spectra were obtained in automatic mode with an average of 1500 laser shots/spectrum (mass range: 350–2,000 m/z). The oligosaccharide molecular ions [M+Na]+ were identified in the MALDI data and their HILIC elution positions were determined by carrying out an extracted ion chromatogram. High-energy (1 kV) MALDI-CID spectra were acquired with an average 10,000 shots/spectrum. The oligosaccharides were allowed to collide in the CID cell with argon at a pressure of 2 × 10−6 Torr.
Molecular Dynamics Simulations
To model conifer hemicellulose (arabino-glucurono-xylan) chains onto cellulose Iβ hydrophilic surfaces 010 and 020, a set of Cartesian coordinates (in PDB format) for atoms in a large cellulose crystal were obtained using cellulose-builder (Gomes and Skaf, 2012). Some cellulose chains were then removed by editing the PDB file describing this larger crystal, in order to obtain a 24-chain square cellulose microfibril and two additional cellulose chains: one on its 010 face, the other on its 020 face. These two additional cellulose chains were then transformed into arabino-glucurono-xylan chains by removing the exocyclic (C6) moieties and by adding (α-1-3)-arabinofuranosyl and (α-1-2)-4-O-methyl-glucuronosyl decorations. To add such decorations, psfgen (Phillips et al., 2005) and CHARMM topology files (Guvench et al., 2008) were used. The resulting PDB file was then displayed using VMD (Humphrey et al., 1996) as a Van der Waals solid surface model using a 1 Å probe radius.
To build complexes of cellulose with one and two conifer xylan chains, we first used cellulose-builder (Gomes and Skaf, 2012) to obtain the structure of a hexagonal cellulose elementary fibril containing 36 glucan chains of DP = 22 in the Iβ polymorph (Ding and Himmel, 2006; Nishiyama et al., 2002). We then replicated glucan chains (see the glucan chains highlighted in green in Fig. 11) and transposed them to the (110) hydrophilic cellulose surface. From such glucan chains, the hydroxymethyl groups were removed to create xylan chains docked onto the cellulose surface. Only the 14 central residues of the xylan chains were considered to reduce edge effects coming from the cellulose fibril. GlcA (in the nonprotonated state) and arabinose (Ara) substitutions were introduced according to the following pattern: Ara(3), GlcA(5), Ara(9), and GlcA(11) (the numbers in parentheses represent the position along the xylan chain; Fig. 11). This pattern is representative of a conifer xylan, and the substitutions were added according to the CHARMM36 force field internal coordinates (Guvench et al., 2009; Raman et al., 2010). Subsequently, the cellulose fibril was cut in the middle, parallel to the plane (110), so as to consider only the chains close to the face where the xylan is adsorbed (Fig. 11). The system was then immersed in a box of water such that a solvent layer at least 16 Å thick from the cellulose-xylan complex was formed. One Ca2+ ion was added to the simulation box for simulations with one xylan to keep the system electrically neutral. Two Ca2+ ions were added for the case of two xylans.
The simulations were performed under periodic boundary conditions. Electrostatic interactions were treated with the particle mesh Ewald method (Darden et al., 1993) and short-range interactions were truncated at a cutoff radius of 12 Å. A time step of 2 fs was used to numerically solve the equations of motion and all the chemical bonds involving hydrogen atoms were fixed at their equilibrium bond lengths. The temperature was kept at 300K and the pressure at 1 atm by Langevin thermostat and piston, respectively (Phillips et al., 2005). To prevent distortions from the Iβ cellulose crystal structure, the C1, C2, C3, C4, C5, and O5 atoms of the cellulose Glc rings were kept harmonically restrained with a force constant of 50 kcal/mol⋅Å2. The hydroxymethyl groups (C6) were allowed to move freely.
Before the production runs, we submitted the system to the following steps: (1) with only the substitution and solvent atoms free, 100 steps of energy minimization using conjugate gradients followed by 500 ps of MD; (2) the same as (1) but with only the cellulose atoms restrained (as described above). After these steps, we carried out 200 ns of MD simulation, over which the analyses were performed. Three independent 200-ns simulations were performed starting from step (1). The simulations were executed using NAMD (Phillips et al., 2005) with the CHARMM36 force field (Guvench et al., 2009; Raman et al., 2010) and the TIP3P water model (Jorgensen et al., 1983).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PACE analysis of xylan of black pine (Pinus nigra).
Supplemental Figure S2. MALDI-CID analysis of xylan of Pinus nigra.
Supplemental Figure S3. Analysis of xylan acetylation in Gingko biloba and Dioon edule.
Supplemental Figure S4. MALDI-CID analysis of xylan of Gnetum edule.
Supplemental Figure S5. Analysis of the MeGlcA distribution on xylan in Laurus nobilis (Magnoliales).
Supplemental Figure S6. Xylan acetylation in Magnolia x soulangeana and Arabidopsis thaliana.
Supplemental Movie 1. Molecular dynamics simulations of cellulose-xylan complexes.
Supplemental Movie 2. Molecular dynamics simulations of cellulose-xylan complexes.
Acknowledgments
The enzymes used in this work were a kind gift from Novozymes, Dr. Dave Bolam, and Prof. Harry Gilbert (University of Newcastle, UK). We thank Dr. Peter Michna for help collecting the samples from the Cambridge University Botanic Garden. We also thank Dr. Nadine Anders and Dr. Samuel Brockington for helpful discussions and critical reading of the manuscript.
Glossary
- AIR
alcohol insoluble residue
- PACE
polysaccharide analysis by carbohydrate gel electrophoresis
- MD
molecular dynamics
- 2-AA
2-aminobenzoic acid
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
This work was supported by the Leverhulme Trust Centre for Natural Material Innovation (M.B.-W. and P.D.), by The Low Carbon Energy University Alliance (A.L.), by Grant BB/G016240/1 from the BBSRC Sustainable Bioenergy Centre Cell Wall Sugars Programme (T.T. and P.D.), and the Sao Paulo Research Foundation (R.L.S., C.S.P., M.S.S., and T.C.F.G.; Grants 2013/08293-7, 2014/10448-1, and 2015/25031-1).
References
- Bosmans TJ, Stépán AM, Toriz G, Renneckar S, Karabulut E, Wågberg L, Gatenholm P (2014) Assembly of debranched xylan from solution and on nanocellulosic surfaces. Biomacromolecules 15: 924–930 [DOI] [PubMed] [Google Scholar]
- Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P (2013) GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J 74: 423–434 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M, Gomes TCF, Tryfona T, Nikolovski N, Stott K, Grantham NJ, Bolam DN, Skaf MS, Dupree P (2014) The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J 79: 492–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse-Wicher M, Grantham NJ, Lyczakowski JJ, Nikolovski N, Dupree P (2016) Xylan decoration patterns and the plant secondary cell wall molecular architecture. Biochem Soc Trans 44: 74–78 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Chong SL, Virkki L, Maaheimo H, Juvonen M, Derba-Maceluch M, Koutaniemi S, Roach M, Sundberg B, Tuomainen P, Mellerowicz EJ, Tenkanen M (2014) O-acetylation of glucuronoxylan in Arabidopsis thaliana wild type and its change in xylan biosynthesis mutants. Glycobiology 24: 494–506 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2014) Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol 22: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Jarvis MC (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammström S, Salmén L, Gatenholm P (2009) On the interactions between cellulose and xylan, a biomimetic simulation of the hardwood cell wall. Bioresources 4: 3–14 [Google Scholar]
- Darden T, York D, Pedersen L (1993) Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J Chem Phys 98: 10089–10092 [Google Scholar]
- Davis CC, Schaefer H (2011) Plant evolution: pulses of extinction and speciation in gymnosperm diversity. Curr Biol 21: R995–R998 [DOI] [PubMed] [Google Scholar]
- Derba-Maceluch M, Awano T, Takahashi J, Lucenius J, Ratke C, Kontro I, Busse-Wicher M, Kosik O, Tanaka R, Winzéll A, et al. (2015) Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol 205: 666–681 [DOI] [PubMed] [Google Scholar]
- Ding SY, Himmel ME (2006) The maize primary cell wall microfibril: a new model derived from direct visualization. J Agric Food Chem 54: 597–606 [DOI] [PubMed] [Google Scholar]
- Dupree R, Simmons TJ, Mortimer JC, Patel D, Iuga D, Brown SP, Dupree P (2015) Probing the molecular architecture of Arabidopsis thaliana secondary cell walls using two- and three-dimensional (13)C solid state nuclear magnetic resonance spectroscopy. Biochemistry 54: 2335–2345 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkenstadt VL, Hendrixson TL, Millane RP (1995) Models of xyloglucan binding to cellulose microfibrils. J Carbohydr Chem 14: 601–611 [Google Scholar]
- Gomes TCF, Skaf MS (2012) Cellulose-builder: a toolkit for building crystalline structures of cellulose. J Comput Chem 33: 1338–1346 [DOI] [PubMed] [Google Scholar]
- Goubet F, Jackson P, Deery MJ, Dupree P (2002) Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem 300: 53–68 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. C R Biol 327: 455–465 [DOI] [PubMed] [Google Scholar]
- Guvench O, Hatcher ER, Venable RM, Pastor RW, Mackerell AD (2009) CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J Chem Theory Comput 5: 2353–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD Jr (2008) Additive empirical force field for hexopyranose monosaccharides. J Comput Chem 29: 2543–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PJ, Stone BA (2008) Chemistry and molecular organization of plant cell walls. In Himmel ME, ed, Biomass Recalcitrance: Deconstructing the Plant Cell Wall for Bioenergy. Blackwell Publishing, Oxford, UK, pp 61–93 [Google Scholar]
- Huang Y, Wang LCY, Nawawi DS, Akiyama T, Yokoyama T, Matsumoto Y (2015) Relationships between hemicellulose composition and lignin structure in woods. J Wood Chem Technol 36: 9–15 [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- Jacobs A, Larsson PT, Dahlman O (2001) Distribution of uronic acids in xylans from various species of soft- and hardwood as determined by MALDI mass spectrometry. Biomacromolecules 2: 979–990 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935 [Google Scholar]
- Kabel MA, van den Borne H, Vincken JP, Voragen AGJ, Schols HA (2007) Structural differences of xylans affect their interaction with cellulose. Carbohydr Polym 69: 94–105 [Google Scholar]
- Köhnke T, Ostlund A, Brelid H (2011) Adsorption of arabinoxylan on cellulosic surfaces: influence of degree of substitution and substitution pattern on adsorption characteristics. Biomacromolecules 12: 2633–2641 [DOI] [PubMed] [Google Scholar]
- Koutaniemi S, Guillon F, Tranquet O, Bouchet B, Tuomainen P, Virkki L, Petersen HL, Willats WG, Saulnier L, Tenkanen M (2012) Substituent-specific antibody against glucuronoxylan reveals close association of glucuronic acid and acetyl substituents and distinct labeling patterns in tree species. Planta 236: 739–751 [DOI] [PubMed] [Google Scholar]
- Kumar M, Campbell L, Turner S (2016) Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67: 515–531 [DOI] [PubMed] [Google Scholar]
- Li L, Pérré P, Frank X, Mazeau K (2015) A coarse-grain force-field for xylan and its interaction with cellulose. Carbohydr Polym 127: 438–450 [DOI] [PubMed] [Google Scholar]
- Maslen SL, Goubet F, Adam A, Dupree P, Stephens E (2007) Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydr Res 342: 724–735 [DOI] [PubMed] [Google Scholar]
- McKee LS, Sunner H, Anasontzis GE, Toriz G, Gatenholm P, Bulone V, Vilaplana F, Olsson L (2016) A GH115 α-glucuronidase from Schizophyllum commune contributes to the synergistic enzymatic deconstruction of softwood glucuronoarabinoxylan. Biotechnol Biofuels 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin JF, Stewart CM (1969) Chemical composition of wood of gnetum gnemon. Holzforschung 23: 51–56 [Google Scholar]
- Mikkelsen D, Flanagan BM, Wilson SM, Bacic A, Gidley MJ (2015) Interactions of arabinoxylan and (1,3)(1,4)-β-glucan with cellulose networks. Biomacromolecules 16: 1232–1239 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, Dupree P (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski I, Marchessault RH (1971) Structure of beta-D-(1-->4′)xylan hydrate. Nature 232: 46–47 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124: 9074–9082 [DOI] [PubMed] [Google Scholar]
- Oehme DP, Downton MT, Doblin MS, Wagner J, Gidley MJ, Bacic A (2015) Unique aspects of the structure and dynamics of elementary Iβ cellulose microfibrils revealed by computational simulations. Plant Physiol 168: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinonen P, Zhang L, Lawoko M, Henriksson G (2015) On the formation of lignin polysaccharide networks in Norway spruce. Phytochemistry 111: 177–184 [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman EP, Guvench O, MacKerell AD Jr (2010) CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J Phys Chem B 114: 12981–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis D, Vian B (2004) Helicoidal pattern in secondary cell walls and possible role of xylans in their construction. C R Biol 327: 785–790 [DOI] [PubMed] [Google Scholar]
- Salmen L. (2015) Wood morphology and properties from molecular perspectives. Ann For Sci 72: 679–684 [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- St John FJ, Hurlbert JC, Rice JD, Preston JF, Pozharski E (2011) Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase. J Mol Biol 407: 92–109 [DOI] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Martel A, Grillo I, Altaner CM, Jarvis MC (2015) Diffraction evidence for the structure of cellulose microfibrils in bamboo, a model for grass and cereal celluloses. BMC Plant Biol 15: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timell TE. (1967) Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol 1: 45–70 [Google Scholar]
- Tryfona T, Stephens E (2010) Analysis of carbohydrates on proteins by offline normal-phase liquid chromatography MALDI-TOF/TOF-MS/MS. Methods Mol Biol 658: 137–151 [DOI] [PubMed] [Google Scholar]
- Uddenberg D, Akhter S, Ramachandran P, Sundström JF, Carlsbecker A (2015) Sequenced genomes and rapidly emerging technologies pave the way for conifer evolutionary developmental biology. Front Plant Sci 6: 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T, Liang HC, Kotake T, Tsumuraya Y, Stephens E, Dupree P (2012) Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol 160: 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbániková L, Vršanská M, Mørkeberg Krogh KB, Hoff T, Biely P (2011) Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J 278: 2105–211621501386 [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, et al. (2016) A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers. Plant Physiol 170: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Hong M (2016) Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J Exp Bot 67: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willfor S, Sundberg A, Pranovich A, Holmbom B (2005) Polysaccharides in some industrially important hardwood species. Wood Sci Technol 39: 601–617 [Google Scholar]
- Xiong G, Cheng K, Pauly M (2013) Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6: 1373–1375 [DOI] [PubMed] [Google Scholar]
- Xiong G, Dama M, Pauly M (2015) Glucuronic acid moieties on xylan are functionally equivalent to O-acetyl-substituents. Mol Plant 8: 1119–1121 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brumer H, Ågren H, Tu Y (2011) The adsorption of xyloglucan on cellulose: effects of explicit water and side chain variation. Carbohydr Res 346: 2595–2602 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Crespi VH, Kubicki JD, Cosgrove DJ, Zhong LH (2014) Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: effects of surface hydrophobicity and side-chain variation. Cellulose 21: 1025–1039 [Google Scholar]