Photoprotective proteins PsbS and LhcSR3 accumulate rapidly in the photosynthetic membrane of Chlamydomonas during highlight stress with PsbS activating nonphotochemical quenching.
Abstract
Photosynthetic organisms must respond to excess light in order to avoid photo-oxidative stress. In plants and green algae the fastest response to high light is non-photochemical quenching (NPQ), a process that allows the safe dissipation of the excess energy as heat. This phenomenon is triggered by the low luminal pH generated by photosynthetic electron transport. In vascular plants the main sensor of the low pH is the PsbS protein, while in the green alga Chlamydomonas reinhardtii LhcSR proteins appear to be exclusively responsible for this role. Interestingly, Chlamydomonas also possesses two PsbS genes, but so far the PsbS protein has not been detected and its biological function is unknown. Here, we reinvestigated the kinetics of gene expression and PsbS and LhcSR3 accumulation in Chlamydomonas during high light stress. We found that, unlike LhcSR3, PsbS accumulates very rapidly but only transiently. In order to determine the role of PsbS in NPQ and photoprotection in Chlamydomonas, we generated transplastomic strains expressing the algal or the Arabidopsis psbS gene optimized for plastid expression. Both PsbS proteins showed the ability to increase NPQ in Chlamydomonas wild-type and npq4 (lacking LhcSR3) backgrounds, but no clear photoprotection activity was observed. Quantification of PsbS and LhcSR3 in vivo indicates that PsbS is much less abundant than LhcSR3 during high light stress. Moreover, LhcSR3, unlike PsbS, also accumulates during other stress conditions. The possible role of PsbS in photoprotection is discussed.
Photosynthetic organisms can be frequently exposed to light intensities that surpass the photosynthetic electron transport capacity. In these conditions, the excess absorbed energy can be transferred from excited chlorophylls in the triplet state (3Chls*) to molecular O2 and cause the production of harmful reactive oxygen species (ROS). In plants and green algae most of the light is absorbed by membrane proteins of the Light harvesting complex (Lhc) family (Jansson, 1999; Ballottari et al., 2012; Buchel, 2015), which are associated with photosystem I and II (Dekker and Boekema, 2005; Caffarri et al., 2014) and participate in photoprotection (Duffy and Ruban, 2015).
The fastest photoprotective mechanisms are activated to harmlessly dissipate excess absorbed energy as heat in a process called non-photochemical quenching of the energy (NPQ). NPQ is a complex and multifactorial process that has been deeply investigated in recent years. Although the molecular mechanisms are only partially understood and still debated (Holt et al., 2005; Ruban et al., 2007; Bode et al., 2009; Holzwarth et al., 2009; Kruger et al., 2014), several key actors involved in NPQ have been discovered in plants and green algae (see Duffy and Ruban [2015] and Goss and Lepetit [2015] for recent reviews).
The fastest component of NPQ, called qE, is triggered by a low luminal pH that is detected by low pH-responsive proteins. So far, three proteins have been shown to have a major role in detecting and transducing the low pH signal: the low pH activated violaxanthin de-epoxidase enzyme (VDE; Arnoux et al., 2009) that catalyses the conversion of the xanthophyll violaxanthin into zeaxanthin, a pigment involved in NPQ and other photoprotection mechanisms (Demmig-Adams, 1990; Havaux and Niyogi, 1999); the pigment-binding LhcSR proteins that belong to the Lhc family and are very likely the active site of quenching after protonation of C-terminal residues (Peers et al., 2009; Bonente et al., 2011; Liguori et al., 2013); and the PsbS protein that is protonated on two luminal Glu residues (Li et al., 2000; Li et al., 2004) and is proposed to subsequently promote the reorganization of the photosynthetic apparatus in order to create quenching sites elsewhere (Bassi and Caffarri, 2000; Bonente et al., 2008a; Johnson et al., 2011). It should be noted that efficient energy quenching also requires the presence of Lhc proteins, but this does not require a specific Lhc isoform (Andersson et al., 2001, 2003; de Bianchi et al., 2008, 2011). Our present investigation is focused on PsbS and LhcSR3 expression in Chlamydomonas reinhardtii and the functionality of PsbS in the green alga.
PsbS belongs to the Lhc supergene family, but it has the unique property of being the only member with four transmembrane helices instead of the more common three-helix organization (Kim et al., 1992). Because no pigment binding has been detected either by biochemical analysis (Bonente et al., 2008a) or in in the recent crystal structure (Fan et al., 2015), it is very likely that PsbS is not the quenching site during NPQ. Therefore, it has been suggested that PsbS promotes a reorganization of the photosynthetic membranes in order to induce the formation of a quenching site in the Lhc antenna system (Bassi and Caffarri, 2000; Betterle et al., 2009; Johnson et al., 2011).
PsbS genes are present in organisms of the green lineage (plants and green algae) but are absent in other oxygenic photosynthetic organisms not belonging to this lineage (Buchel, 2015). The protein has been detected both in vascular (Tracheophytes) and nonvascular (Bryophytes) plants, as well in the closest algal relatives to plants (Charophytes) and in multicellular green algae (Chlorophytes; Bonente et al., 2008b). However, previous immunoblot analyses aimed at investigating the accumulation of PsbS in unicellular green algae such as Chlamydomonas reinhardtii showed that the protein did not appear to accumulate under several different growth conditions (Bonente et al., 2008b). Nevertheless, the presence of two highly homologous psbS genes in Chlamydomonas that code for proteins that are well conserved with respect to plant PsbS proteins (Bonente et al., 2008b) would suggest that it is unlikely that these genes are pseudogenes. However, it has been proposed that the Chlamydomonas PsbS does not play or plays only a minor role in NPQ (Bonente et al., 2008b). Consistent with this idea, a genetic screen based on NPQ induction capacity resulted in the isolation of a mutant (called npq4) with a very low level of NPQ (Peers et al., 2009). The mutation knocked out the LhcSR3 protein, which since the discovery of its role in NPQ, has been intensively studied. In Chlamydomonas, LhcSR proteins are encoded by three genes: lhcSR1, lhcSR3.1 and 3.2, the last two coding for identical proteins that are both knocked out in the npq4 mutant. Interestingly, LhcSR3 plays a predominant role in NPQ induction (Peers et al., 2009). LhcSR3 is homologous to the Lhc proteins: it has a similar structure with three transmembrane helices and it is also able to bind Chls and Cars (Bonente et al., 2011). It has been suggested that LhcSR3 itself is the energy quencher due to its pigment binding ability, the unusual fluorescence properties that show very short lifetime components, and its pH sensitivity (Bonente et al., 2011; Liguori et al., 2013; Tokutsu and Minagawa, 2013). A recent investigation suggests the LhcSR is localized both in grana membranes and stroma lamellae in the moss Physcomitrella patens (Pinnola et al., 2015), and investigations on Chlamydomonas suggest that LhcSR3 can bind both to PSI or PSII depending on illumination conditions (Allorent et al., 2013; Bergner et al., 2015). In contrast to psbS, lhcSR genes are widespread in a many photosynthetic organisms, including green algae, brown algae, diatoms, nonvascular plants, and even some vascular plants (Buchel, 2015). In the moss Physcomitrella patens, LhcSR-dependent and the PsbS-dependent NPQ operate independently and additively (Alboresi et al., 2008).
Even if PsbS proteins in Chlamydomonas have not been detected in previous analyses, the presence of conserved psbS genes, their expression under nitrogen deprivation conditions (Miller et al., 2010), and the expression of the genes during a dark to light shift as found in a very recent report (Zones et al., 2015) suggest a role for PsbS in the alga. Thus, in this work, we have reinvestigated the expression and accumulation of PsbS in Chlamydomonas during light stress and other stress conditions in order to determine whether this protein could play a role in NPQ under particular conditions and compared our results with those obtained on LhcSR3.
We found that PsbS rapidly and transiently accumulates during light stress, and that contrasts with LhcSR3 that accumulates over a much longer period. In order to demonstrate a role of the algal PsbS protein in NPQ, we created Chlamydomonas strains constitutively expressing the algal or the Arabidopsis psbS genes. Both the overexpressed PsbS proteins showed the ability to increase NPQ in Chlamydomonas.
A comparison with expression/accumulation of LhcSR suggests that LhcSR is more abundant and is involved in photoprotection under several stress conditions, while PsbS seems to respond very rapidly and transiently only to high light stress and has a minor role in photoprotection in Chlamydomonas.
RESULTS
Analysis of PsbS and LhcSR3 Accumulation in Chlamydomonas reinhardtii in High Light
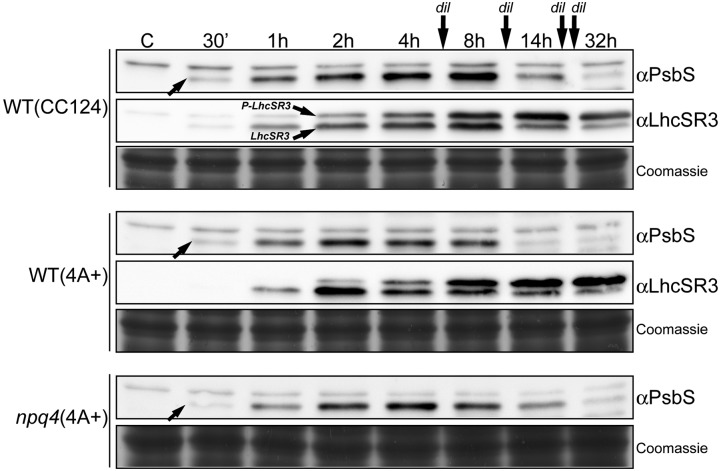
In previous investigations (Bonente et al., 2008b), PsbS was not detected upon high light acclimation. Therefore, we performed a new analysis of the kinetics PsbS accumulation during high light treatment by sampling cells at several time points starting from 30 min until 32 h after the switch from normal light (45 µmol photons m−2 s−1) to high light (Fig. 1; Supplemental Fig. S1). Experiments were performed in liquid minimal medium at 1200 µmol photons m−2 s−1 with four dilutions of the medium in order to avoid autoshading by the growing cells (Fig. 1) and were also repeated without dilutions at three different light intensities (400, 800, and 1200 µmol photons m−2 s−1; Supplemental Fig. S1). In all cases, the kinetics of PsbS induction were similar (Fig. 1; Supplemental Fig. S1): PsbS, undetectable in low light acclimated cells, showed a rapid increase and was already detectable after 30 min under high light reaching a maximum level after around 4 to 8 h. PsbS then decreased and was almost undetectable at 32 h. The decline in PsbS was independent of the increase in cell density. The maximal PsbS quantity appeared dependent on the light intensity (Supplemental Fig. S1). Interestingly, kinetics in wild-type (4A+) and npq4 cells were practically identical despite the absence of LhcSR3 in the npq4 mutant.
Figure 1.
Kinetics of PsbS accumulation in Chlamydomonas wild-type (CC124 and 4A+) and npq4 cells during high light treatment. Cells grown in minimal medium were exposed to 1200 µmol photons m−2 s−1 for 32 h, and aliquots were harvested at different times as indicated. Two micrograms (in Chls) of total membranes were analyzed by SDS-PAGE and by immunoblot assays using antiPsbS and antiLhcSR3 antibodies. PsbS (arrow) showed a transient accumulation in all three genotypes with a peak around 4–8 h. LhcSR3 showed a slightly delayed induction and a strong accumulation until the end of the treatment. The position of phosphorylated LhcSR3 (P-LhcSR3) and non-phosphorylated LhcSR3 (LhcSR3) are indicated in the wild-type (CC124) immunoblot as example. Dilutions (dil) of the medium were done in order to avoid cell autoshading. Similar kinetics experiments at different light intensities and without dilutions are presented in Supplemental Figure S1. The Coomassie stained gels are shown as protein loading control.
In parallel, we also investigated the accumulation of the LhcSR3 protein, which has a key role in NPQ induction in Chlamydomonas (Peers et al., 2009). LhcSR3 is also induced by high light (Fig. 1; Supplemental Fig. S1), as previously shown (Peers et al., 2009; Bonente et al., 2011; Allorent et al., 2013). However, in contrast to PsbS, the maximum intensity was reached later (after 8 h) and accumulation remained rather stable and high until the end of our experiments (32 h). Moreover, LhcSR3 was detected at two positions in immunoblot assays, corresponding to the phosphorylated and unphosporylated proteins (Bonente et al., 2011), and the ratio of P-LhcSR3 over non-P-LhcSR3 increased during the stress period (Fig. 1).
It should be noted that in a previous work (Bonente et al., 2008b), the antibody used to check PsbS accumulation was raised against a recombinant barley PsbS protein. For this work, we used a new antibody raised against a synthetic peptide from the Chlamydomonas PsbS sequence that is also well conserved in the Arabidopsis PsbS (see “Materials and Methods”). The new antibody, despite some unspecific cross-reactivity, is able to detect the Chlamydomonas PsbS protein (Figs. 1 and 5) as well as the Arabidopsis one (Supplemental Fig. S2). Several immunoblot assays confirmed the ability of this antibody to correctly recognize the PsbS band: (1) the recombinant Chlamydomonas PsbS protein is detected (Supplemental Fig. S2A); (2) comparison of grana membranes from Arabidopsis wild-type and npq4 (koPsbS) plants showed a specific band at the MW of PsbS in the wild-type sample absent in the mutant (Supplemental Fig. S2A); (3) the putative PsbS bands disappear in samples from high light treated Chlamydomonas cells and Arabidopsis membranes in a competition assay where the antibody solution was preincubated with an excess of recombinant Chlamydomonas protein before the immunoblot analysis (Supplemental Fig. S2B); (4) PsbS was detected by mass spectrometry only in high light stressed thylakoids of Chlamydomonas (Supplemental Fig. S2C; Supplemental Table S2); (5) comparison between PsbS-overexpressor strains (see below) and high light treated cells showed that the transiently appearing band detected on stressed cells has exactly the same position as overexpressed PsbS (Fig. 5B).
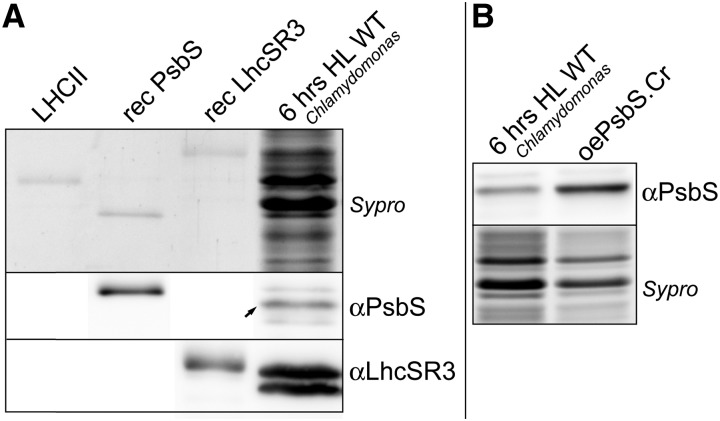
Figure 5.
Quantification of PsbS and LhcSR3 in vivo. A, Similar amounts (∼ 0.03 µg, see “Materials and Methods” for details) of recombinant Chlamydomonas PsbS, recombinant LhcSR3, native Arabidopsis LHCII, and a total extract (2 µg in Chls) of Chlamydomonas wild-type cells grown on minimal medium and treated for 6 h under high light were separated by SDS-PAGE and stained with Sypro or transferred onto nitrocellulose membranes for immunoblot analysis. The intensities of the chemiluminescent band of PsbS and LhcSR3 in the total extract were compared with the signals of the respective recombinant proteins in order to estimate the absolute and relative content in vivo. Note that the PsbS signal is much lower in the high light sample as compared with the recombinant protein, while for LhcSR3 it is the contrary. We found an LhcSR3/PsbS ratio in vivo of ∼ 8 ± 2 and a PsbS amount of 0.1 ± 0.05 protein per monomeric PSII. The values are the average and sd of four experiments using two biological replicates. Note that differences in mobility between recombinant and native proteins are due to the presence of more amino acids in the recombinant PsbS and LhcSR3. B, PsbS content in high light treated cells and oePsbS.Cr(wt) cells was compared by loading total cell extracts containing similar amounts of photosynthetic proteins (2 µg in Chls). We found a PsbS content in oePsbS strains of 0.5 ± 0.08 PsbS per monomeric PSII, about 5 times higher than in high light treated cells (average and sd of four experiments using two biological replicates).
Analysis of the Expression of psbS and lhcSR Genes
Quantitative real-time PCR experiments were performed on the same samples used for kinetic analysis of PsbS and LhcSR3 protein accumulation. For this analysis, we designed primers able to distinguish between the two psbS genes (JGI v. 5.5 locus Cre01.g016600 is psbS1 and locus Cre01.g016750 is psbS2; Zones et al., 2015), as well the two lhcSR3 genes (lhcSR3.1 and lhcSR3.2) and the lhcSR1 gene (Maruyama et al., 2014).
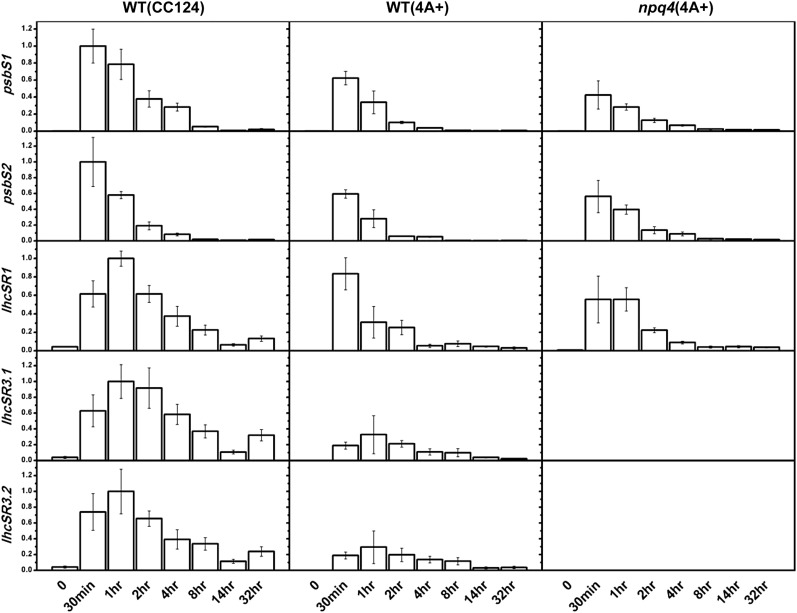
As shown in Figure 2, the results are in good agreement with the those obtained by immunoblot analysis on proteins (Fig. 1; Supplemental Fig. S1); the induction of psbS gene expression is extremely fast and transient, with a maximum in the very first part of the low light to high light transition. A similar behavior was found for lhcSR genes, but delayed compared to psbS and in agreement with the later appearance of the LhcSR3 protein.
Figure 2.
Kinetics of psbS and lhcSR expression during high light stress. Expression of the two psbS genes and the three lhcSR genes (lhcSR1, lhcSR3.2, and lhcSR3.2) was analyzed by quantitative real-time PCR on cells grown on minimal medium. In a similar manner to the immunoblot experiments, psbS expression was induced rapidly and transiently after the switch to high light in all three genotypes. LhcSR genes were also induced by high light, but the maximum expression was slightly delayed as compared to psbS, similar to the immunoblot results. Averages ± se are shown (three measurements from two biological replicates for wild-type [CC124] and npq4 cells; two replicates from one biological sample for wild-type [4A+] cells). Data are normalized to one for the maximum value obtained for a given transcript. A gene encoding the cytosolic 40S small ribosomal subunit RACK1, also known as Chlamydomonas G protein β-subunit-like polypeptide (CBLP), and the gene IDA5 were used as a control.
Both psbS genes are expressed, as previously found (Bonente et al., 2008b; Zones et al., 2015), as well all lhcSR genes. However, there are two major differences between the analysis on mRNA and on protein: (1) the maximum accumulation of mRNA occurs earlier than the maximum protein accumulation; and (2) while both psbS mRNA and PsbS protein decrease during the stress, this is not the case for lhcSR3 RNAs and LhcSR3 proteins, since lhcSR3 gene expression decreases significantly before the end of the 32 h kinetic experiment, while the protein remains abundant.
Overexpression of PsbS in Chlamydomonas
In order to test the functionality of the PsbS protein, we created overexpressor strains that we compared with wild-type cells that do not contain PsbS when grown under moderate light (Fig. 1).
Because nuclear transformation in Chlamydomonas is not straightforward and a previous assay to overexpress PsbS by nuclear transformation was unsuccessful (Bonente et al., 2008b), we generated PsbS-overexpressing strains by means of chloroplast transformation. We used two different constructs: (1) oePsbs.Cr, to create strains overexpressing the full length PsbS protein of Chlamydomonas (since the mature sequence is not known) and (2) oePsbS.At, to create strains overexpressing the mature PsbS protein of Arabidopsis. We transformed both wild-type (CC124) and npq4 (koLhcSR3, 4A+ background) cells, and the strains obtained are here after indicated as oePsbS.Cr(wt)/oePsbS.At(wt) and oePsbS.Cr(npq4)/oePsbS.At(npq4), where “Cr” and “At” indicated the presence of the Chlamydomonas or the Arabidopsis PsbS and the strain transformed is indicated in brackets. We used the npq4 (4A+) strain since it is the original koLhcSR3 mutant (Peers et al., 2009), and the wild type (CC124) as it is a very common strain used for genetic screening and physiological studies (Proschold et al., 2005). It should be noted that, even if we cannot directly compare oePsbS(npq4) to oePsbS(wt) cells, each oePsbS strain is compared with its own non-transformed control (WT-CC124 or npq4). In the case of the oePsbS(npq4) strains, for NPQ measurements (see next section), they are also compared with the wild type with the same genetic background (4A+).
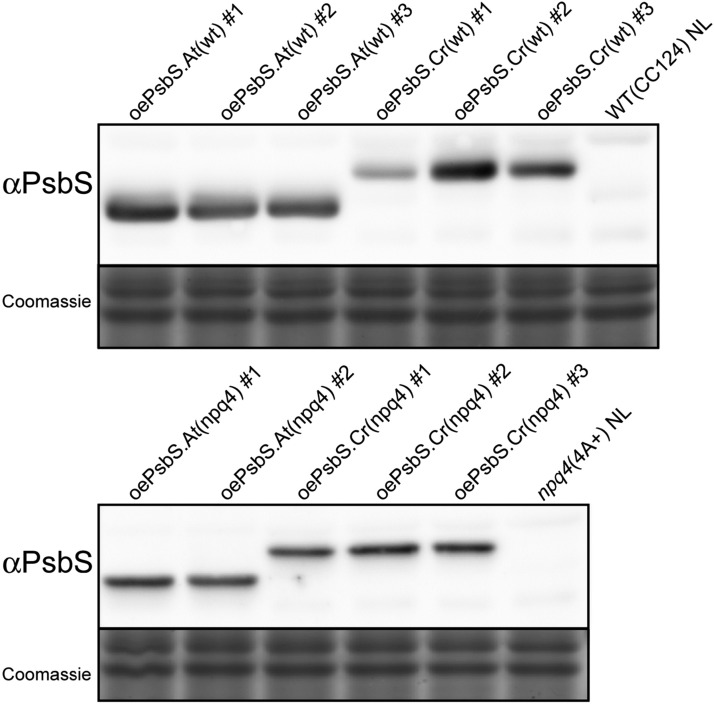
Immunoblot analysis performed on thylakoid membranes showed that both algal and plant PsbS accumulated in transplastomic Chlamydomonas strains (Fig. 3). Note also (Fig. 5B) that the full length Chlamydomonas PsbS protein overexpressed in the chloroplast is correctly processed, because the overexpressed protein is detected at a Mr identical to that of the nuclear-expressed PsbS in high light.
Figure 3.
PsbS overexpression in Chlamydomonas. PsbS overexpressing strains were created by chloroplast transformation using the Chlamydomonas psbS sequence (oePsbS.Cr) or the Arabidopsis psbS (oePsbS.At). Overexpression strains were made both in the wild-type (CC124) background (top) and in the npq4 (4A+) background (bottom). Immunoblot analysis on total cell extracts (2 µg in Chls) from independent transformants confirmed the presence of the recombinant PsbS proteins, absent in wild-type and npq4 cells grown under normal light (NL) on minimal medium. The strains oePsbS.Cr(wt)#1 and oePsbS.Cr(wt)#2 showed the lowest and highest PsbS content, respectively (results obtained on three biological replicates). Coomassie stained gels are shown as protein loading controls.
Similar PsbS accumulation levels were obtained for each construction/genotype, except for two oePsbS.Cr(wt) strains, where one line showed low expression and another high expression (oePsbS.Cr9Wt no. 1 and no. 2, respectively; Fig. 3). This was useful for correlating PsbS dose with the NPQ phenotype as described below.
Chlorophyll Fluorescence Properties of oePsbS Chlamydomonas Strains
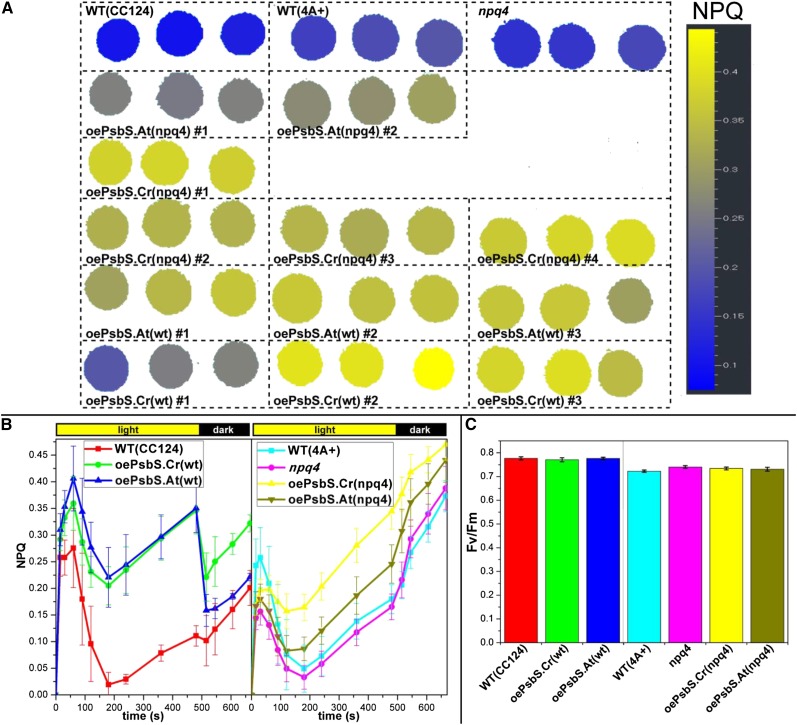
Twelve independent PsbS overexpressing lines [3 oePsbS.Cr(wt); 3 oePsbS.At(wt); 4 oePsbS.Cr(npq4); 2 oePsbS.At(npq4)] were tested for NPQ. Cells grown on a solid minimal medium at moderate light (45 µmol photons m−2 s−1) were exposed to 800 µmol photons m−2 s−1 of actinic light. NPQ induction was higher in all the oePsbS lines as compared with the respective control lines (WT-CC124 or npq4; Fig. 4 for an average of the lines with the same construction/genotype and Supplemental Fig. S3A for each independent line tested).
Figure 4.
Fluorescence phenotypes of PsbS overexpressing strains. A, Wild type (CC124), wild type (4A+), npq4, overexpressing strains of the Arabidopsis, and the Chlamydomonas PsbS in the npq4 (4A+) background [oePsbS.At(npq4) and oePsbS.Cr(npq4) respectively], and overexpressing strains of the Arabidopsis and the Chlamydomonas PsbS in the wild-type (CC124) background [oePsbS.At(wt) and oePsbS.Cr(wt), respectively] were plated on minimal medium, grown at ∼ 45 µmol photons m−2 s−1 and imaged with a fluorescence camera. The NPQ levels on a representative plate after 6 min under actinic light (800 µmol photons m−2 s−1) are indicated in false colors. All oePsbS lines showed a higher NPQ capacity as compared to their control. B, The full NPQ kinetics of the same strains as in A are shown. The values are the average ± sd for oePsbS lines with the same backgrounds/construction calculated from five replicate (n = 5) plates arranged as in A (three spots of each independent oePsbS strain per plate; six spots for wild-type and npq4 strains per plate). The kinetics of the single independent oePsbS strains are shown in Supplemental Figure S3. C, Maximum quantum yields (Fv/Fm) calculated over four replicate plates as in B.
Both the Chlamydomonas and the Arabidopsis PsbS were able to increase NPQ in the overexpressing strains, indicating that both algal and plant PsbS proteins are functional in Chlamydomonas.
The NPQ kinetics (Fig. 4B) showed a rapid induction in the first minute (likely due to a fast and elevated ΔpH formed at the beginning of the actinic light phase), followed by a decrease and then a second increase in NPQ levels after a few minutes.
Unexpectedly, we found differences in NPQ kinetics between strains in the CC124 and in the 4A+ backgrounds. In CC124 cells, both the plant and the algal PsbS overexpressors showed similar kinetics, and the maximum NPQ was much higher than the NPQ in the wild-type (CC124) control, and the NPQ was largely reversible during the first 30 s of the dark phase. In the 4A+ background, Chlamydomonas PsbS was more efficient than the Arabidopsis PsbS, the NPQ at the end of the actinic light phase was significantly higher in oePsbS (npq4) strains than in wild-type (4A+) and npq4 control cells, but the difference was smaller than that between oePsbS(wt) lines and its wild-type (CC124) control and the NPQ did not show reversibility. The NPQ measurements on liquid cultures with a classical PAM instrument (Supplemental Fig. S3B) are in accordance with measurements performed by video-imaging on cultures grown on solid medium. Moreover, in this case, we found reversibility of NPQ at the end of the actinic light for the oePsbS(npq4) strains, but not for npq4 and wild-type (4A+) cells despite some accumulation of LhcSR1 (Supplemental Fig. S4B). However, LhcSR1 accumulation was similar in all strains.
The level of NPQ showed a correlation with PsbS abundance, at least in the case of the Chlamydomonas PsbS where we obtained strains with different expression levels. oePsbS.Cr(wt) no. 1, which accumulates low amount of PsbS, (Fig. 3), showed a lower NPQ capability than oePsbS.Cr(wt) no. 2 that accumulates high amounts of PsbS (Fig. 4A; Supplemental Fig. S3A). This is similar to the situation in Arabidopsis, where PsbS abundance is also correlated to NPQ levels (Li et al., 2002a, 2002b).
It should be noted the increase of NPQ capability in oePsbS strains is very unlikely due to some major change in the organization of the photosynthetic complexes. Biochemical characterization of the strains showed that Chl a/b ratios of oePsbS and control strains are very similar, as well as the content of proteins representative of major photosynthetic complexes and protein involved in cyclic electron transfer, except for PGRL1 that was less abundant in oePsbS.At(wt) cells (Supplemental Fig. S4).
In conclusion, results from oePsbS lines strongly support that both the plant and the algal PsbS are functional and able to increase NPQ capability in Chlamydomonas cells.
In Vivo Quantification of PsbS and LhcSR
Knowledge about the absolute and relative amounts of PsbS and LhcSR3 in vivo might be important to provide further information about their role in photoprotection.
Because PsbS and LhcSR3 in vivo stoichiometry is not directly measurable by immunoblot analyses, we used purified recombinant proteins as standards for parallel SDS-PAGE and immunoblot analyses. Similar amounts of recombinant PsbS and LhcSR (based on Sypro staining in a SDS-PAGE) were loaded on a gel together with a known amount of photosynthetic proteins from high light treated Chlamydomonas cells (Fig. 5A). We also compared PsbS content between high light treated cells with oePsbS.Cr strains (Fig. 5B). In this manner, we were able to compare the chemiluminescent immunoblot signals between the lanes with the recombinant proteins (for which the protein amount is quantifiable, see “Materials and Methods” for details) and the lanes of the cellular extracts, where PsbS and LhcSR3 are mixed with several other proteins and thus undetectable in a Sypro stained gel. This allowed the approximate estimation of the relative and absolute amounts of the two proteins in vivo (Fig. 5A), as well the relative amount between high light treated cells and oePsbs.Cr strains (Fig. 5B).
Our result showed that the amount of PsbS in vivo around the time of maximum accumulation (6 h of high light) is about 8 ± 2 (sd) times less than LhcSR3, for which we estimated a LhcSR3/monomeric-PSII ratio of 0.9 ± 0.4 (sd). Our results therefore indicate that PsbS in vivo is present in much lower amounts than LhcSR3 and in substoichiometric amounts compared to PSII (∼ 0.1 PsbS per monomeric PSII).
Accumulation of native PsbS during a light stress was also compared with the accumulation of PsbS in oePsbS strains, and we found that PsbS in oePsbS lines is about five times more than in high light stressed cells (0.50 ± 0.08 sd protein per monomeric-PSII, Fig. 5B).
PsbS and LhcSR3 Accumulation in Nitrogen-Depleted Medium and Other Growth Conditions
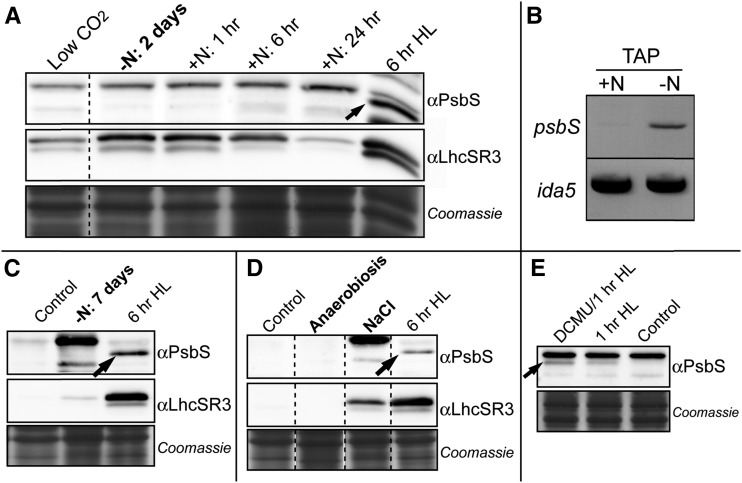
It has been reported that under nitrogen deprivation, psbs mRNA accumulates to high levels (Miller et al., 2010). Thus, we performed immunoblot experiments to check the accumulation of the PsbS protein in cells grown in these conditions. As shown in Figure 6A, PsbS protein was practically undetectable after 2 d under nitrogen deprivation. On the contrary, the psbs mRNA accumulated to a much larger extent under nitrogen starvation as compared to normal conditions (Fig. 6B), in agreement with previous findings (Miller et al., 2010). Addition of nitrogen into the medium did not promote accumulation of PsbS protein during a time course experiment of 24 h (Fig. 6A).
Figure 6.
PsbS and LhcSR3 expression under different stress conditions. A, SDS-PAGE on total extracts of Chlamydomonas cells incubated in TAP medium as in Miller et al. (2010) in nitrogen starvation conditions for 2 d, and after replenishment with nitrogen. PsbS protein was not detected at any time. On the contrary, LhcSR3 protein accumulated after 2 d of starvation and decreased after nitrogen addition. LhcSR3, but not PsbS, is also present in low CO2 conditions (cells grown without air bubbling, left lane). Cells at 6 h of a high light stress are the positive control. B, An increase in psbS mRNA abundance after 2 d of nitrogen starvation in TAP medium was detected by semiquantitative PCR, in accordance with Miller et al. (2010). C, A faint band of LhcSR3 was still visible after 7 d of nitrogen starvation in TAP medium. D, Anaerobiosis and 25 mm NaCl stress were also tested in TAP medium: PsbS was never detected, while LhcSR3 was clearly induced under a salt stress. Coomassie gels are shown as loading control for all immunoblot experiments. E, Cells grown on minimal medium at 45 µmol photons m−2 s−1 have been treated for 1 h at 800 µmol photons m−2 s−1 in the presence or not of DCMU (10 µm). Cells kept at low light and without DCMU are also shown (control). PsbS accumulation was higher in presence of DCMU.
Contrarily to PsbS, LhcSR3 accumulated during a nitrogen starvation period, and its content diminished when the nitrogen was added to the medium (Fig. 6A). After 7 d of nitrogen starvation, PsbS remained undetectable, while LhcSR3 was still detectable (Fig. 6C).
We also tested other growth conditions, some of which were previously tested with a different antibody (Bonente et al., 2008b). In particular, we tested the accumulation of PsbS during dark anaerobic growth and under salt stress: in each case, we did not detect the PsbS protein (Fig. 6D). On the contrary, LhcSR3 significantly accumulated during salt stress although, as for PsbS, not during dark anaerobic growth.
It is worth noting that lhcSR3 genes are expressed in low CO2 conditions (Maruyama et al., 2014), and we detected LhcSR3 protein when cultures were not bubbled with air (Fig. 6A), in agreement with previous results showing LhcSR3 accumulation during a shift form high CO2 to air (Dang et al., 2014). On the contrary, PsbS was not detected in these conditions (Fig. 6A).
Finally, when we treated low light acclimated cells to a high light stress (800 µmol photons m−2 s−1) in the presence of DCMU, an inhibitor of electron transport at PSII impairing plastoquinone reduction, we found that the abundance of PsbS after 1 h of high light was higher in cells incubated with DCMU than in cells treated in the same light condition but without DCMU (Fig. 6E).
PsbS and Photoprotection
In order to investigate the ability of PsbS in increasing photoprotection capability, we tested the responses of the oePsbS strains and their controls (wild type and npq4) to high light. In particular, we compared the decrease in the maximum quantum yield of PSII (Fv/Fm) after high light stress under constant or fluctuating illumination. Under these conditions, the values of Fv/Fm were similar or even lower in the majority of the oePsbS strains as compared with control strains (Table I). Only the Arabidopsis oePsbS strains in the wild-type background showed a slightly higher Fv/Fm after the different light stresses. We therefore concluded that overexpression of the Chlamydomonas PsbS protein had no positive effect on photoprotection under these high light growth conditions and using Fv/Fm as an output.
Table I. Maximum PSII quantum yield after high light treatment.
Cells were plated on minimal medium similarly as in Figure 4 and treated for 5 h under continuous high light (800 µmol photons m−2 s−1) or 2 h of fluctuating high light (3 min at 1200 µmol photons m−2 s−1, 3 min at 20 µmol m−2 s−1). Values are averages ± sd of all oePsbS strains with the same backgrounds/construction (three spots per strain per plate; two replicates) and of wild-type (WT) and npq4 cells (six spots per box, two replicates). Only oePsbS.At(wt) strains showed a small but significantly higher Fv/Fm (*P value < 0.05, Student's t test).
| 5 h Continuous HL Fv/Fm | 2 h Fluctuating HL Fv/Fm | |
|---|---|---|
| WT(CC124) | 0.571 ± 0.010 | 0.487 ± 0.006 |
| oePsbS.Cr(wt) | 0.581 ± 0.011 | 0.475 ± 0.018 |
| oePsbS.At(wt) |
0.593 ± 0.008* |
0.507 ± 0.011* |
| WT(4A+) | 0.451 ± 0.010 | 0.436 ± 0.005 |
| npq4 | 0.433 ± 0.008 | 0.486 ± 0.009 |
| oePsbS.Cr(npq4) | 0.419 ± 0.013 | 0.440 ± 0.026 |
| oePsbS.At(npq4) | 0.420 ± 0.006 | 0.442 ± 0.025 |
We also compared the growth rates of strains overexpressing the Chlamydomonas PsbS protein with their control strains under fluctuating high light conditions. In these experiments, growth rates were similar between all strains (Table II). This suggests, in agreement with the Fv/Fm experiments, that overexpression of the endogenous PsbS does not provide a significant advantage under these growth conditions as compared to control strains, which, however, accumulate some endogenous PsbS under high light, even if in a lower amount (Fig. 5B).
Table II. Growth rates of oePsbS.Cr strains versus their control strains in minimal medium.
Growth rates at low light (45 µmol photons m−2 s−1) and fluctuating high light (3 min at 1200 µmol photons m−2 s−1, 3 min at 45 µmol photons m−2 s−1) were not significantly different between wild-type (WT) and oePsbS strains. Values are averages ± sd of three biological replicates (wild-type background) or two biological replicates (npq4 background) using two different oePsbS.Cr strains and three flasks per strain per replicate.
| Low Light Doubling Time (Days) | Fluct. High Light Doubling Time (Days) | |
|---|---|---|
| WT(CC124) | 0.450 ± 0.018 | 1.369 ± 0.053 |
| oePsbS.Cr(wt) |
0.436 ± 0.026 |
1.403 ± 0.054 |
| npq4 | 0.444 ± 0.000 | 1.382 ± 0.029 |
| oePsbS.Cr(npq4) | 0.420 ± 0.046 | 1.447 ± 0.206 |
DISCUSSION
PsbS Accumulates Transiently and Is Functional in Chlamydomonas
So far, the role of PsbS in Chlamydomonas has been elusive. On one hand, PsbS was not detected under several growth conditions (Bonente et al., 2008b), and the npq4 mutant (koLhcSR3) suggests that almost all algal NPQ capability depends on LhcSR3 (Peers et al., 2009). However, on the other hand, the Chlamydomonas genome contains two psbS genes that are conserved with one another and with the plant psbS (Merchant et al., 2007), and some investigations have shown that psbS RNA accumulates in particular conditions (Miller et al., 2010; Zones et al., 2015).
Thus, we (re)investigated PsbS gene expression and protein accumulation in Chlamydomonas. Here, we show that the PsbS protein accumulates rapidly and transiently during high light stress, peaking around 4/8 h of the treatment (Fig. 1; Supplemental Fig. S1); PsbS accumulation and the expression of both psbS genes follow a similar but slightly delayed trend during high light stress; psbS gene expression kinetics are independent of the presence of LhcSR3 and the high light intensity (at least in the conditions we tested, which all saturate photosynthesis), but PsbS protein amounts seem dependent on the high light intensity (Supplemental Fig. S1).
LhcSR3 accumulation was delayed as compared to PsbS and the amount remained elevated during the entire 32 h light stress. Moreover, the ratio of the phosphorylated LhcSR3 over the nonphosphorylated LhcSR3 (upper and lower band, respectively, in Fig. 1 and Supplemental Fig. S1; Bonente et al., 2011) increased during the light stress. Phosphorylation of LhcSR3 could be involved in the localization of the protein (Allorent et al., 2013; Bergner et al., 2015). Minor and major LHCII antenna can be phosphorylated in Chlamydomonas to move from PSII to PSI during state transition (Iwai et al., 2008; Rochaix, 2014), and this could also be the case for LhcSR3. Although clear evidence showing interactions between LhcSR3 and PSI are still lacking in Chlamydomonas, in the moss Physcomitrella patens, LhcSR proteins have been shown to interact and protect both photosystems (Pinnola et al., 2015).
The results showing a delay between maximum gene expression and maximum protein accumulation can be explained by a relatively slow turnover of the proteins, which thus accumulate during the first hours when the RNA is still abundant. However, in the case of LhcSR3, the turnover appears to be particularly slow, since the accumulation of the protein (Fig. 1) continues for several hours after the maximum gene expression (Fig. 2). Alternatively, the relatively low abundant lhcSR3 transcripts in the late stage of the high light period could be very efficiently transcribed.
PsbS in Photoprotection
Our results suggest that PsbS expression is very rapidly activated in response to light stress (30 min are enough to detect PsbS accumulation), but that its role seems to be only transient, in contrast to the situation in plants and mosses. Instead, LhcSR3 might have a much more important role in acclimation to high light, and possibly some change at the level of photosystems would require phosphorylation of LhcSR3 in a late stage of the light stress. Therefore, in addition to the observed temporal separation between PsbS and LhcSR3 expression, it is reasonable to think that the two proteins are differentially localized within the thylakoid membranes as recently observed in Physcomitrella patens (Pinnola et al., 2015). Physcomitrella is the first organism where both PsbS and LhcSR have been found and for which KO mutants have been created (Alboresi et al., 2010). Interestingly, while in Chlamydomonas, PsbS and LhcSR are stress-inducible proteins that accumulate transiently during stress conditions; in plants, PsbS is usually detected under non-stress conditions; and in Physcomitrella, both PsbS and LhcSR proteins can be detected in low light grown plants (Gerotto et al., 2011). This phenomenon occurs as well in the multicellular green alga Ulva linza (Zhang et al., 2013). This might support the idea of Allorent et al. (2013) proposing that an inducible and less efficient NPQ system in Chlamydomonas is compensated by a large state transition capacity.
In order to investigate the ability of the Chlamydomonas PsbS to induce NPQ and increase photoprotection, we created PsbS overexpressor strains by chloroplast transformation. We also generated strains accumulating the Arabidopsis PsbS protein to determine whether the plant protein is functional in Chlamydomonas cells. OePsbS lines, which constitutively accumulate PsbS, were compared with wild-type and npq4 cells grown in conditions where PsbS is undetectable (low light) and LhcSR3 has no or negligible accumulation (low light and high CO2/air bubbled cultures) (Figs. 1 and 6; Supplemental Fig. S4B). As visible in Figure 4 and Supplemental Figure S3, all oePsbS lines showed a significantly higher NPQ capacity as compared with their respective controls. In the case of oePsbS lines in the wild-type (CC124) background, the NPQ was rapidly reversible in the dark, as it is the case in plants, due to the rapid neutralization of the low luminal pH that is necessary for activating PsbS. NPQ measurements on liquid cultures (Supplemental Fig. S3B) showed reversibility also for the oePsbS strains in the npq4 (4A+) background, but not for wild-type (4A+) and npq4 controls. The fact that wild-type strains and npq4 cells did not show a significant NPQ reversibility at the end of the actinic light is in agreement with fact that LhcSR3 has no or negligible accumulation in the growth conditions used (Figs. 1 and 6; Supplemental Fig. S4B). However, we noticed that LhcSR1 was accumulated in our growth conditions (Supplemental Fig. S4B), but since the amount is similar in oePsbS and control strains, and considering the less important role of LhcSR1 in NPQ (Peers et al., 2009), NPQ differences between oePsbS(wt) strains and their controls almost certainly have to be ascribed to the activity of PsbS.
In both genotypes (CC124 and 4A+), PsbS-dependent NPQ seems superimposed on another phenomenon that is detected as NPQ and seems stronger in the 4A+ genotype. This could be state transitions, as investigated in high light treated cells of Chlamydomonas (Allorent et al., 2013). Considering the different NPQ responses of oePsbS strains in the two genetic backgrounds, it is likely that significant photosynthetic differences exist between the two genotypes, but investigation of this is beyond the scope of our present research.
The fact that the heterologous expression of the Arabidopsis PsbS had a similar effect as the Chlamydomonas PsbS (Fig. 3) suggests that the mechanism of PsbS, which is still debated, is at least partially conserved between plants and the green algae despite some differences in photosystem II organization between the organisms. In particular, LHCII proteins in Chlamydomonas have biochemical/spectroscopic properties similar to those of plants (Natali and Croce, 2015), but they cannot be classified in the Lhcb1-3 isoforms as in plants (Jansson, 1999; Elrad and Grossman, 2004). In Chlamydomonas, the CP24 subunit, which is next to the LHCII-M trimer (Dekker and Boekema, 2005) and that has been suggested to detach from PSII during NPQ induction in a PsbS-dependent phenomenon (Betterle et al., 2009), is lacking and at its place a third LHCII trimer is bound to PSII (Tokutsu et al., 2012; Drop et al., 2014). Even if it is possible that PsbS in Chlamydomonas interacts somehow differently with PSII than in plants, we found that PsbS accumulation has a positive correlation with NPQ capacity as in plants (Li et al., 2002a, 2002b), further supporting a conserved PsbS-dependent NPQ mechanism.
Investigation of the absolute and relative PsbS and LhcSR3 stoichiometry in vivo indicated that the level of PsbS is sub-stoichiometric as compared to PSII (we found about 0.1 PsbS per monomeric PSII). Even at its maximum expression (∼6 h of high light, Fig. 1) PsbS accumulates to much lower levels than LhcSR3, being about eight times less (it should be noted that our estimation of LhcSR3 amount is higher than the value of ∼0.2 previously proposed (Bonente et al., 2011, possibly due to the different approach or growth conditions). Since the mode of action of PsbS is still not well understood, and since PSII particles are densely packed within the grana, it might be possible that one PsbS per ten PSII is enough to efficiently protect the photosynthetic apparatus. However, PsbS in Arabidopsis accumulates at higher and likely stoichiometric amounts with PSII (see, for instance, 2D gels in Li et al. [2004] and Caffarri et al. [2009]) where PsbS is present in amounts comparable to those of monomeric Lhc proteins). Due to the fact that PsbS in algae and plants likely works in a similar manner and that there is a correlation between the amount of PsbS and NPQ level, we suggest that the low amount of PsbS in vivo might contribute to only a small part of the NPQ capacity of Chlamydomonas.
Our tests to determine whether PsbS overexpression affects photoprotection did not reveal a significant difference in oePsbS strains versus control strains. It should be noted, however, that the control strains are also able to accumulate the endogenous PsbS (Fig. 1) and LhcSR1 (and LhcSR3 in the wild-type background), which could attenuate the differences between control and oePsbS strains. It should be also noted that the Chlamydomonas npq4 mutant lacking LhcSR3 and unable to significantly increase NPQ capability when grown at high light (Peers et al., 2009) also did not show significant differences for either Fv/Fm or growth in high light as compared with wild-type cells (Peers et al., 2009). Similarly, Arabidopsis mutants lacking PsbS also show relatively small differences in comparison with wild-type plants under high light conditions, due to the enhancement of other photoprotective mechanisms (Golan et al., 2006). It is likely that photoprotection strategies are partially redundant and, thus, that other photosynthetic controls such as state transitions (Allorent et al., 2013) and cyclic electron transport (Chaux et al., 2015; Saroussi et al., 2016), which seem particularly important in Chlamydomonas photoprotection, could mask the photoprotection ability of PsbS in laboratory conditions. Thus, PsbS may have roles in increasing fitness in the simultaneous presence of high light and other natural stress conditions.
PsbS Accumulates in Response to Light; LhcSR3 Accumulates under Several Stress Conditions
We determined PsbS accumulation under different stresses to obtain more information about the possible role of PsbS under unfavorable growth conditions. As previously found (Miller et al., 2010), we detected psbS gene transcription under nitrogen starvation (Fig. 6). However, we did not detect protein accumulation, a result that suggests the existence of some control on mRNA translation. Besides the absence of the PsbS protein under nitrogen deprivation conditions, we did not detect PsbS after acclimation to any of the other stresses tested, including low CO2, high salt conditions, and anaerobiosis. On the contrary, LhcSR3 was detected after acclimation to low CO2, in agreement with previous investigations (Dang et al., 2014; Maruyama et al., 2014) and also in response to high salt and nitrogen starvation (and its quantity decreased after the addition of nitrogen in the medium; Fig. 6). In contrast to anaerobiosis, these three stresses lead to production of reactive oxygen species that could be involved in the activation of the lhcSR genes. In summary, after acclimation to various stresses, only LhcSR was found in cells, but not PsbS, indicating that psbS gene expression is under the control of a signal appearing very rapidly in the cells after a transition to high light. Notably, high light specifically provokes a rapid and significant production of singlet oxygen (1O2*; Flors et al., 2006) due to energy transfer from triplet excited chlorophylls to oxygen, especially at the level of PSII. We cannot exclude a role of the redox state of the plastoquinone (PQ) on PsbS accumulation. However, this possibility is less likely since the PQ redox state is mainly involved in optimizing photosynthesis under not saturating light conditions, such as during state transitions, or during long-term acclimation (Fey et al., 2005; Adamiec et al., 2008). Moreover, our result (Fig. 6E) showing an increase of PsbS accumulation during high light treatment in presence of DCMU, which blocks plastoquinone reduction, further supports a minor role of the PQ redox state in psbS gene expression in the first phase of a light stress. In later stages of stress or in the deficiency of the final electron acceptor CO2, other reactive oxygen species, such as superoxide (O2−) and hydrogen peroxide (H2O2) formed at PSI by the Mehler reaction, or the redox state of the chloroplast, may be more important in triggering a change in the genetic expression necessary for acclimation to the stress. We therefore argue that the rapid accumulation of PsbS is likely triggered by a 1O2* derived signal and is important for a fast and transient need of energy dissipation at the level of PSII. This action would be performed in parallel with LhcSR or even earlier, due to the more rapid appearance of PsbS. In later stages of the stress, LhcSR, but not PsbS, would have a major role in photoprotection of both photosystems.
CONCLUSION
We investigated the expression of two key proteins in NPQ induction, PsbS and LhcSR. We showed that Chlamydomonas psbS gene transcripts and the PsbS protein accumulate very rapidly and transiently during high light stress. We demonstrate that the endogenous PsbS protein is able to activate NPQ in Chlamydomonas, as well as the Arabidopsis PsbS, suggesting a conserved mechanism of action. PsbS accumulation in Chlamydomonas seems strongly dependent on high light stress, while LhcSR3 accumulation is detected under several different stress conditions that can lead to general oxidative stress even under moderate light. We also showed that PsbS in vivo stoichiometry is significantly lower than that of LhcSR3. We cannot exclude that other stress conditions (not tested) are able to induce PsbS, but our results, together with those indicating that LhcSR3 accumulation can explain almost all fast induced NPQ in high light acclimated Chlamydomonas cells (Peers et al., 2009), suggest that in Chlamydomonas PsbS plays a less important role in the protection of the photosynthetic apparatus than in plants. However, the conservation of functional PsbS proteins in the green alga and their very rapid accumulation (faster than LhcSR3) indicate that PsbS must be important for the fitness of Chlamydomonas in natural conditions during a transient response to high light.
MATERIALS AND METHODS
Strains and Culture Conditions
The Chlamydomonas reinhardtii wild-type strain CC124 (mt+ nit1 nit2), the 4A+ wild type and the mutant npq4 (Peers et al., 2009) were grown at 22°C under continuous illumination in a Percival AR-41L3 incubator (CLF PlantClimatics, Germany). Batch cultures were grown on a rotary shaker in Erlenmeyer flasks (75 mL) under 45 µmol photons m−2 s−1. TAP or minimal medium (E. Harris, The Chlamydomonas Sourcebook) were used. For high light treatments, cells were cultured in 1 L flasks on a rotary shaker and supplied with continuous filtered air bubbling generated by an aquarium pump. For light stress, illumination was supplied by two white LED panels (Photon System Instruments, Czech Republic) placed radially along the flasks to reach an intensity up to 1,200 µmol photons m−2 s−1. Sampling for immunoblot and qRT-PCR were performed after 30 min, 1 h, 2 h, 4 h, 8 h, 14 h, and 32 h. A volume of 12 mL of cells was harvested by centrifugation (3,500 g, 10 min, 4°C), supernatant was removed, and the pellet was immediately frozen in liquid nitrogen. Cell pellet was stored at −80°C until used.
Nitrogen depletion experiments were carried out as described by Miller et al. (2010) at 100 µmol m−2 s−1. Anaerobic conditions were created in the dark as described by Bonente et al. (2008b). Sodium chloride stress was created growing cells in TAP with the addition of 25 mm NaCl for 3 d under 45 µmol photons m−2s−1. Prolonged nitrogen stress was created maintaining cells without a nitrogen source for 7 d under 45 µmol photons m−2 s−1. A nitrogen replenishing experiment was carried out as described by Miller et al. (2010). After 2 d of nitrogen depletion, NH4Cl was added to the medium and sampling was performed after 1 h, 6 h, and 24 h.
DCMU experiment was performed on cells grown on minimal medium at 45 µmol photons m−2 s−1. The cell culture was split in two flasks before high light treatment and DCMU (10 µm) was added to one flask, and then cells were exposed to 800 µmol photons m−2 s−1 for 1 h
Fluctuating light conditions for Fv/Fm analysis (3 min at 1,200 µmol m−2 s−1 + 3 min at 20 µmol m−2 s−1) were created in the Fluorcam FC 800-O using the blue led actinic panel (Photon System Instruments). Fluctuating light conditions for growth rate experiments (3 min at 1,200 µmol m−2 s−1 + 3 min at 45 µmol m−2 s−1) were created with a white Fytoled system equipped with a LC100 controller (Photon System Instrument, Czech Republic). Growth rates were calculated on the chlorophyll absorption in the red and corrected for the light scattering (two measurements per day, at least 2 d for each light conditions).
Overexpression of PsbS in C. reinhardtii
Synthetic psbS genes from C. reinhardtii and Arabidopsis were designed to optimize the chloroplast codon usage of C. reinhardtii. We used the mature sequence for the Arabidopsis PsbS and the full sequence in the case of Chlamydomonas PsbS (the mature Chlamydomonas PsbS is not known; however, the PsbS precursor is correctly processed inside the chloroplast, Fig. 5B). Note that the two psbS genes from C. reinhardtii encode for identical proteins (Bonente et al., 2008b), except for one conservative Ser ⁄Thr substitution at position 28, which is very likely in the signal peptide sequence. The synthetic genes were integrated in the pLM21 vector using the BglII restriction site using the In-Fusion kit (Clontech). The pLM21 vector was originated from the IR-int vector from Michelet et al. (2011), designed for gene insertion into the chloroplast genome by homologous recombination. The IR-int vector was digested by ClaI and PmlI to excise VapA and its promoter. A PCR was performed on atpB-int vector to amplify the psaA promoter and 5′UTR using Advantage HD (Clontech) using the following primers: 5′-ATGGAGCTGTACCACGTTTAAACAGATCTCCATGGATTTCTCCTTATAA-3′ and 5′-CGGATCCGATATCGATAAGCTTTCTTAATTCAAC-3′. These primers added BglII and PmeI as multicloning sites. Then, the PCR product was cloned by In-fusion kit (Clontech) between ClaI and PmlI within the IR-int vector.
In the pLM21-CpPSBSCr and in the pLM21-CpPSBSAt, both psbs genes are under the control of the psaA 5′untranslated region (UTR) promoter region and the rbcL 3′UTR region, while the aadA gene (spectinomycin resistance) is under control of atpA 5′UTR promoter region and the rbcL 3′UTR region. For plastid transformation experiments, cells were grown in TAP medium at 45 µmol photons m−2 s−1 until reaching 2.5 × 106 cells mL−1 and then plated onto minimal medium agar plate containing 50 µg mL−1 spectinomycin. Biolistic transformation and DNA extraction for genotyping were performed as described in Baltz et al. (2014). The chloroplast DNA loading control has been done on 16S using fw primer 5′-TCCATGGAGAGTTTGATCCTG-3′ and rev primer 5′-TCCTCTCAGACCAGCTACTGC-3′, Tm: 50°C. Homoplasmy was verified by PCR (forward primer 5′-CGTTATTAGCCTTTCGTCGCT-3′ and reverse primer 5′-CCGAAACGGTGGTTATTCCAGGCC-3′) by the lack of amplification of the insertion region in the chloroplast genome, since it is broken. The presence of PSBSCr or PSBSAt was verified using specific primers (PSBSAt fw primer 5′-GGTACATCAGGTGGTATTGG-3′, PSBSAt rev primer 5′-CCTAATTGAGCTAAACGACC-3′, PSBSCr fw primer 5′-TCAAAAACAGAAGTTGAACG-3′, PSBSCr rev primer 5′-CGAATTGTGCTAATGCACCT-3′). PCR was performed using the Taq‘Ozime Purple Mix.
Thylakoid Membranes, Total Membranes, and Total Protein Purification
Thylakoid membranes were isolated as described by Bonente et al. (2008b). Total membranes were prepared breaking 10–20 mL of harvested cells in the buffer containing 25 mm HEPES (pH 7.5), 5 mm MgCl2, 0.3 m Suc, and a protease inhibitor cocktail (EasyPack EDTA free, Roche) using glass beads. Glass beads and unbroken cells were removed by centrifugation (600 g, 2 min), and from the supernatant fraction, membranes were pelleted by centrifugation at 12,000 g for 5 min. Membranes were suspended and stored in 5 mm HEPES-KOH (pH 7.5), 10 mm EDTA and 50% glycerol and stored at −80°C. For total protein extraction, a volume of 10–20 mL of exponential grown culture was harvested by centrifugation (3,500 g, 10 min) and suspended in SDS-PAGE sample buffer (62.5 mm Tris [pH 6.8], 10% glycerol, 5% SDS, 5% β-mercaptoethanol) containing a protease inhibitor cocktail (EasyPack EDTA free, Roche). An amount of 200 mg of glass beads (425–600 µm) were then added, and cells were broken using vigorous vortexing. Glass beads and cell debris were removed by centrifugation (21,000 g, 10 min), and the chlorophyll amount was quantified using a fitting procedure of the absorption spectrum after acetone extraction (Croce et al., 2002).
Immunoblot
About 1.5–2 µg of chlorophyll were loaded on 12% (w/v) SDS-PAGE with 6 m urea and transferred onto a nitrocellulose membrane. Immublotting analyses were performed using an antibody raised in rabbit against the synthetic peptide (C)-TGKGALAQFDIETG, corresponding to the first part of the second luminal loop of the Chlamydomonas PsbS. This sequence has been chosen for the good predicted antigenicity and for the strong conservation with the first homolog loop of Chlamydomonas PsbS and with the corresponding loops of Arabidopsis PsbS, in order to have a wide cross-reactivity (see also Supplemental Fig. S2). αLhcSR3, αLhcSR1, αPsbO, αPsaC, and αPetA antibodies were purchased from Agrisera. αPGRL1 and αNDA2 are described in Tolleter et al. (2011) and Desplats et al. (2009), respectively. αPsbC was raised in rabbit against a recombinant peptide corresponding to the loop E. An antirabbit IgG peroxidase-conjugated antibody (CellSignaling) and the Fusion Fx7 (Vilber Lourmat) was used for detection.
Peptide Competition Assay
For the competition assay, 7.5 µg of PsbS antibody was incubated with the recombinant His-tagged PsbS of C. reinhardtii (25 µg of protein) (Bonente et al., 2008b) in 15 mL of PBS buffer containing 2% (w/v) milk powder and 0.25% (v/v) TritonX-100 for 1 h at room temperature on a rotary shaker. This solution was used to hybridize a membrane as described in the Immunoblot section.
Quantification of PsbS and LhcSR3
Similar visual amount of recombinant PsbS (Bonente et al., 2008b), recombinant LhcSR3 (Bonente et al., 2011) and native LHCII were loaded on a SDS-PAGE together with 2 µg (in Chls) of a total extract of high light treated Chlamydomonas wild-type cells and oePsbS.Cr cells. SDS-PAGE were either stained with Sypro or transferred to nitrocellulose membranes for immunoblot analysis with the antiPsbS and antiLhcSR3 antibodies. Amount of recombinant PsbS and LhcSR3 were calculated by densitometry of the Sypro stained SDS-PAGE and comparison with the signal of LHCII, for which 0.03 µg of apoprotein were loaded (calculated on the Chls absorption, where 1 ODQy/cm is equal to 40 µg/ml of apoprotein). The values obtained by densitometry were also corrected for the number of basic residues in each protein since Sypro is mainly bound by His, Arg and Lys residues. Apoprotein amounts in vivo were obtained by densitometry and comparison of the ECL signals between lanes containing the recombinant proteins and lanes containing total cell extracts. For absolute quantification, the same assumption as in (Bonente et al., 2011) about PSII/PSI ratio (1.18) and photosystem antenna size (PSI = 240 Chls, PSII = 222 Chls) were considered.
RNA Extraction, Synthesis of cDNA, and Semi-quantitative and Quantitative RT-PCR Analysis
A volume of 12 mL of pelleted cells was used for RNA extraction using the TRIreagent and subjected to DNase treatment with the RNAase-free DNase I (Thermo scientific). A total of 250 ng of DNA-free RNA was used for the cDNA synthesis using PrimeScript RT reagent kit (Perfect Real Time) from Takara in accordance to the manufacturer’s instructions. Primers for quantitative real-time (qRT)-PCR are listed in Supplemental Table S1. Each reaction contained the master mix, 200 ng of each primer, and cDNA corresponding to 12.5 ng input RNA in the reverse transcriptase reaction. Real-time (RT)-PCR was performed on the light Cycler Instrument (CFX96 Real-Time PCR Detection System, Bio Rad) using SYBR Green I as a fluorescent dye. The reaction conditions were as follow: 95°C for 3 min, followed by cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 30 s up to a total of 44 cycles. Controls without template or reverse transcriptase were included. A gene encoding the cytosolic 40S small ribosomal subunit RACK1 also known as Chlamydomonas G protein β-subunit-like polypeptide (CBLP) and the gene IDA5 were used as a control. Data analysis was performed using Bio-Rad CFX Manager software. Gene expression ratios were normalized to controls using the 2-ΔCt. For semiquantitative RT-PCR, cDNA was synthetize using the Superscript III Reverse transcriptase (Invitrogen) following manufacture’s instruction. PCR cycle conditions were 3 min of initial denaturation at 94°C, following 34 cycles of 30 s of denaturation, 30 s of annealing at 60°C and 3 min of elongation at 72°C. Final elongation was performed for 10 min at 72°C. Primers were the same used in (Miller et al., 2010).
Fluorescence Measurements
Fluorescence measurements on cells grown on solid minimal medium have been performed using an imaging fluorometer (Fluorcam FC 800-O; Photon System Instruments). An amount of cells corresponding to about 1.5 µg of chlorophyll were spotted onto minimal medium agar plate and let grow for 5-7 d at 45 µmol photons m−2 s−1 before measurements. Cells were dark-acclimated for at least 30 min and then subjected to NPQ measurement with 800 µmol photons m−2 s−1 of blue light. Fluorescence measurements on cells grown on liquid minimal medium have been performed with a DUAL-PAM-100 (DUAL-PAM/F, Walz) on dark adapted cultures concentrated ∼ 0.5 µg Chls/ml. Actinic light was provided by a red LED set at 1200 µmol photons m−2 s−1. Sedimentation was avoided by using a miniature magnetic stirrer. NPQ was calculated as (Fm - Fm’)/Fm’ where Fm is the maximum fluorescence of dark-adapted tissues and Fm’ is the maximum fluorescence of light adapted tissues. Maximum quantum yield of PSII (Fv/Fm) was calculated as (Fm-Fo)/Fm.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Kinetics of PsbS accumulation in Chlamydomonas WT(4A+) and npq4 cells during a high light treatment at three different light intensities.
Supplemental Figure S2. Test of the antibody against PsbS.
Supplemental Figure S3. NPQ of the individual oePsbS strains and on liquid cultures.
Supplemental Figure S4. Biochemical characterization of the oePsbS and control strains.
Supplemental Materials and Methods S1. Mass spectrometry analysis.
Supplemental Table S1. Primers used for the quantitative real-time PCR experiments.
Supplemental Table S2. Mass spectrometry results on Chlamydomonas thylakoids from low light and high light treated cells.
Supplementary Material
Acknowledgments
Christophe Laloi, Ben Field, and Patrice Crete at the LGBP are thanked for technical help and fruitful discussion on qRT-PCR. Ben Field is further thanked for manuscript revision. Sabrina Lignon, Rémy Puppo, Pascal Mansuelle, and Régine Lebrun, from the Proteomic Platform of the Mediterranean Institute of Microbiology, Marseille Proteomique IBiSA labeled, are thanked for mass spectrometry analyses. Roberto Bassi is thanked for the LhcSR3 recombinant protein. Riot Claire at the LB3M is thanked for technical assistance in chloroplast transformation.
References
- Adamiec M, Drath M, Jackowski G (2008) Redox state of plastoquinone pool regulates expression of Arabidopsis thaliana genes in response to elevated irradiance. Acta Biochim Pol 55: 161–173 [PubMed] [Google Scholar]
- Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T (2008) In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS One 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G, Tokutsu R, Roach T, Peers G, Cardol P, Girard-Bascou J, Seigneurin-Berny D, Petroutsos D, Kuntz M, Breyton C, Franck F, Wollman FA, et al. (2013) A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Walters RG, Horton P, Jansson S (2001) Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: implications for the mechanism of protective energy dissipation. Plant Cell 13: 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S (2003) Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Arnoux P, Morosinotto T, Saga G, Bassi R, Pignol D (2009) A structural basis for the pH-dependent xanthophyll cycle in Arabidopsis thaliana. Plant Cell 21: 2036–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M, Girardon J, Dall’osto L, Bassi R (2012) Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim Biophys Acta 1817: 143–157 [DOI] [PubMed] [Google Scholar]
- Baltz A, Dang KV, Beyly A, Auroy P, Richaud P, Cournac L, Peltier G (2014) Plastidial expression of type II NAD(P)H dehydrogenase increases the reducing state of plastoquinones and hydrogen photoproduction rate by the indirect pathway in Chlamydomonas reinhardtii. Plant Physiol 165: 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R, Caffarri S (2000) Lhc proteins and the regulation of photosynthetic light harvesting function by xanthophylls. Photosynth Res 64: 243–256 [DOI] [PubMed] [Google Scholar]
- Bergner SV, Scholz M, Trompelt K, Barth J, Gäbelein P, Steinbeck J, Xue H, Clowez S, Fucile G, Goldschmidt-Clermont M, Fufezan C, Hippler M (2015) STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol 168: 615–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betterle N, Ballottari M, Zorzan S, de Bianchi S, Cazzaniga S, Dall’osto L, Morosinotto T, Bassi R (2009) Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J Biol Chem 284: 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode S, Quentmeier CC, Liao PN, Hafi N, Barros T, Wilk L, Bittner F, Walla PJ (2009) On the regulation of photosynthesis by excitonic interactions between carotenoids and chlorophylls. Proc Natl Acad Sci USA 106: 12311–12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Formighieri C, Mantelli M, Catalanotti C, Giuliano G, Morosinotto T, Bassi R (2011) Mutagenesis and phenotypic selection as a strategy toward domestication of Chlamydomonas reinhardtii strains for improved performance in photobioreactors. Photosynth Res 108: 107–120 [DOI] [PubMed] [Google Scholar]
- Bonente G, Howes BD, Caffarri S, Smulevich G, Bassi R (2008a) Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J Biol Chem 283: 8434–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Passarini F, Cazzaniga S, Mancone C, Buia MC, Tripodi M, Bassi R, Caffarri S (2008b) The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem Photobiol 84: 1359–1370 [DOI] [PubMed] [Google Scholar]
- Büchel C. (2015) Evolution and function of light harvesting proteins. J Plant Physiol 172: 62–75 [DOI] [PubMed] [Google Scholar]
- Caffarri S, Kouril R, Kereïche S, Boekema EJ, Croce R (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S, Tibiletti T, Jennings RC, Santabarbara S (2014) A comparison between plant photosystem I and photosystem II architecture and functioning. Curr Protein Pept Sci 15: 296–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux F, Peltier G, Johnson X (2015) A security network in PSI photoprotection: regulation of photosynthetic control, NPQ and O2 photoreduction by cyclic electron flow. Front Plant Sci 6: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce R, Canino G, Ros F, Bassi R (2002) Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41: 7334–7343 [DOI] [PubMed] [Google Scholar]
- Dang KV, Plet J, Tolleter D, Jokel M, Cuiné S, Carrier P, Auroy P, Richaud P, Johnson X, Alric J, Allahverdiyeva Y, Peltier G (2014) Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26: 3036–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bianchi S, Betterle N, Kouril R, Cazzaniga S, Boekema E, Bassi R, Dall’Osto L (2011) Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bianchi S, Dall’Osto L, Tognon G, Morosinotto T, Bassi R (2008) Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20: 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B. (1990) Carotenoids and photoprotection in plants: A role for the xanthophyll zeaxanthin. Biochim Biophys Acta 1020: 1–24 [Google Scholar]
- Desplats C, Mus F, Cuiné S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in chlamydomonas chloroplasts. J Biol Chem 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Drop B, Webber-Birungi M, Yadav SK, Filipowicz-Szymanska A, Fusetti F, Boekema EJ, Croce R (2014) Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim Biophys Acta 1837: 63–72 [DOI] [PubMed] [Google Scholar]
- Duffy CD, Ruban AV (2015) Dissipative pathways in the photosystem-II antenna in plants. J Photochem Photobiol B 152(Pt B): 215–226 [DOI] [PubMed] [Google Scholar]
- Elrad D, Grossman AR (2004) A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr Genet 45: 61–75 [DOI] [PubMed] [Google Scholar]
- Fan M, Li M, Liu Z, Cao P, Pan X, Zhang H, Zhao X, Zhang J, Chang W (2015) Crystal structures of the PsbS protein essential for photoprotection in plants. Nat Struct Mol Biol 22: 729–735 [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Bräutigam K, Pfannschmidt T (2005) Photosynthetic redox control of nuclear gene expression. J Exp Bot 56: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Flors C, Fryer MJ, Waring J, Reeder B, Bechtold U, Mullineaux PM, Nonell S, Wilson MT, Baker NR (2006) Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J Exp Bot 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Giacometti GM, Bassi R, Morosinotto T (2011) Role of PSBS and LHCSR in Physcomitrella patens acclimation to high light and low temperature. Plant Cell Environ 34: 922–932 [DOI] [PubMed] [Google Scholar]
- Golan T, Müller-Moulé P, Niyogi KK (2006) Photoprotection mutants of Arabidopsis thaliana acclimate to high light by increasing photosynthesis and specific antioxidants. Plant Cell Environ 29: 879–887 [DOI] [PubMed] [Google Scholar]
- Goss R, Lepetit B (2015) Biodiversity of NPQ. J Plant Physiol 172: 13–32 [DOI] [PubMed] [Google Scholar]
- Havaux M, Niyogi KK (1999) The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci USA 96: 8762–8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt NE, Zigmantas D, Valkunas L, Li XP, Niyogi KK, Fleming GR (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Holzwarth AR, Miloslavina Y, Nilkens M, Jahns P (2009) Identification of two quenching sites active in the regulation of photosynthetic light-harvesting studied by time-resolved fluorescence. Chem Phys Lett 483: 262–267 [Google Scholar]
- Iwai M, Takahashi Y, Minagawa J (2008) Molecular remodeling of photosystem II during state transitions in Chlamydomonas reinhardtii. Plant Cell 20: 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Johnson MP, Goral TK, Duffy CD, Brain AP, Mullineaux CW, Ruban AV (2011) Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sandusky P, Bowlby NR, Aebersold R, Green BR, Vlahakis S, Yocum CF, Pichersky E (1992) Characterization of a spinach psbS cDNA encoding the 22 kDa protein of photosystem II. FEBS Lett 314: 67–71 [DOI] [PubMed] [Google Scholar]
- Krüger TP, Ilioaia C, Johnson MP, Ruban AV, van Grondelle R (2014) Disentangling the low-energy states of the major light-harvesting complex of plants and their role in photoprotection. Biochim Biophys Acta 1837: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem 279: 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Niyogi KK (2002a) Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J Biol Chem 277: 33590–33597 [DOI] [PubMed] [Google Scholar]
- Li XP, Muller-Moule P, Gilmore AM, Niyogi KK (2002b) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA 99: 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori N, Roy LM, Opacic M, Durand G, Croce R (2013) Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: the C-terminus of LHCSR is the knob of a dimmer switch. J Am Chem Soc 135: 18339–18342 [DOI] [PubMed] [Google Scholar]
- Maruyama S, Tokutsu R, Minagawa J (2014) Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol 55: 1304–1310 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Lefebvre-Legendre L, Burr SE, Rochaix JD, Goldschmidt-Clermont M (2011) Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol J 9: 565–574 [DOI] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, Bullard B, Sears BB, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natali A, Croce R (2015) Characterization of the major light-harvesting complexes (LHCBM) of the green alga Chlamydomonas reinhardtii. PLoS One 10: e0119211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Pinnola A, Cazzaniga S, Alboresi A, Nevo R, Levin-Zaidman S, Reich Z, Bassi R (2015) Light-harvesting complex stress-related proteins catalyze excess energy dissipation in both photosystems of Physcomitrella patens. Plant Cell 27: 3213–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Harris EH, Coleman AW (2005) Portrait of a species: Chlamydomonas reinhardtii. Genetics 170: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65: 287–309 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Berera R, Ilioaia C, van Stokkum IHM, Kennis JTM, Pascal AA, van Amerongen H, Robert B, Horton P, van Grondelle R (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Saroussi SI, Wittkopp TM, Grossman AR (2016) The type II NADPH dehydrogenase facilitates cyclic electron flow, energy-dependent quenching, and chlororespiration metabolism during acclimatiation of Chlamydomonas reinhardtii to nitrogen deprivation. Plant Physiol 170: 1975–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, Cuiné S, Plet J, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ye N, Mou S, Xu D, Fan X (2013) Occurrence of the PsbS and LhcSR products in the green alga Ulva linza and their correlation with excitation pressure. Plant Physiol Biochem 70: 336–341 [DOI] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.