Stilbene prenyltransferase that utilizes DMAPP from the plastidic terpenoid pathway shows a high substrate specificity.
Abstract
Prenylated stilbenoids synthesized in some legumes exhibit plant pathogen defense properties and pharmacological activities with potential benefits to human health. Despite their importance, the biosynthetic pathways of these compounds remain to be elucidated. Peanut (Arachis hypogaea) hairy root cultures produce a diverse array of prenylated stilbenoids upon treatment with elicitors. Using metabolic inhibitors of the plastidic and cytosolic isoprenoid biosynthetic pathways, we demonstrated that the prenyl moiety on the prenylated stilbenoids derives from a plastidic pathway. We further characterized, to our knowledge for the first time, a membrane-bound stilbenoid-specific prenyltransferase activity from the microsomal fraction of peanut hairy roots. This microsomal fraction-derived resveratrol 4-dimethylallyl transferase utilizes 3,3-dimethylallyl pyrophosphate as a prenyl donor and prenylates resveratrol to form arachidin-2. It also prenylates pinosylvin to chiricanine A and piceatannol to arachidin-5, a prenylated stilbenoid identified, to our knowledge, for the first time in this study. This prenyltransferase exhibits strict substrate specificity for stilbenoids and does not prenylate flavanone, flavone, or isoflavone backbones, even though it shares several common features with flavonoid-specific prenyltransferases.
A substantial part of nonhost defense responses in many plants is the pathogen-induced production of secondary metabolites, generally termed phytoalexins, that locally restrict disease progression due to bioactivities toxic to the pathogen (for review, see Ahuja et al., 2012). Peanut or groundnut (Arachis hypogaea) tissues mount a defense against infection by the soil fungus Aspergillus flavus and other pathogens by overproducing stilbene derivatives around sites of wounding and elicitor perception (Sobolev, 2013). Resveratrol, one of the most studied phytoalexin stilbenoids, has attracted great attention because of its bioactive properties shown through in vitro and in vivo assays to benefit human health. These include antiinflammatory (Das and Das, 2007) and antioxidant properties as well as antitumor and favorable cardiovascular effects (Gambini et al., 2015). However, the limited oral bioavailability of resveratrol due to its rapid absorption and metabolism restricts the future of this potentially valuable drug in clinical trials (Tomé-Carneiro et al., 2013; Gambini et al., 2015).
Prenylated stilbenoids naturally produced as phytoalexins in the peanut plant possess, for the most part, one isoprenyl moiety bound to the aromatic ring of the stilbene molecule (Fig. 1). When compared with resveratrol, these compounds exhibit similar or enhanced bioactivity in in vitro experiments. For instance, arachidin-1 and resveratrol showed similar antiinflammatory activity in lipid polysaccharide-treated RAW 264.7 macrophages, and this correlated with the inhibition of PG E2 production (Chang et al., 2006; Djoko et al., 2007). Arachidin-1, arachidin-2, and arachidin-3, also applied to macrophages, were more effective than resveratrol in inhibiting inducible nitric oxide production (Sobolev et al., 2011). In other antioxidant activity assays, arachidin-1 inhibited lipid oxidation more effectively than resveratrol (Abbott et al., 2010), and arachidin-2 and arachidin-3 showed greater potency over resveratrol in inhibiting the production of intracellular reactive oxygen species (Sobolev et al., 2011). Arachidin-1 further showed higher cytotoxicity than resveratrol to leukemia HL-60 cells (Huang et al., 2010) and other cancer cells (SK-MEL, KB, BT-549, and SK-OV-3; Sobolev et al., 2011). Interestingly, arachidin-1 and arachidin-3 were shown to bind to human cannabinoid receptors 2 (hCBR2s), while the affinity of their nonprenylated analogs, piceatannol and resveratrol, for hCBR2s was 5- to 10-fold lower. Molecular modeling studies with hCBR2s indicated that the prenyl moiety of the arachidins improved the binding affinity to the receptors (Brents et al., 2012).
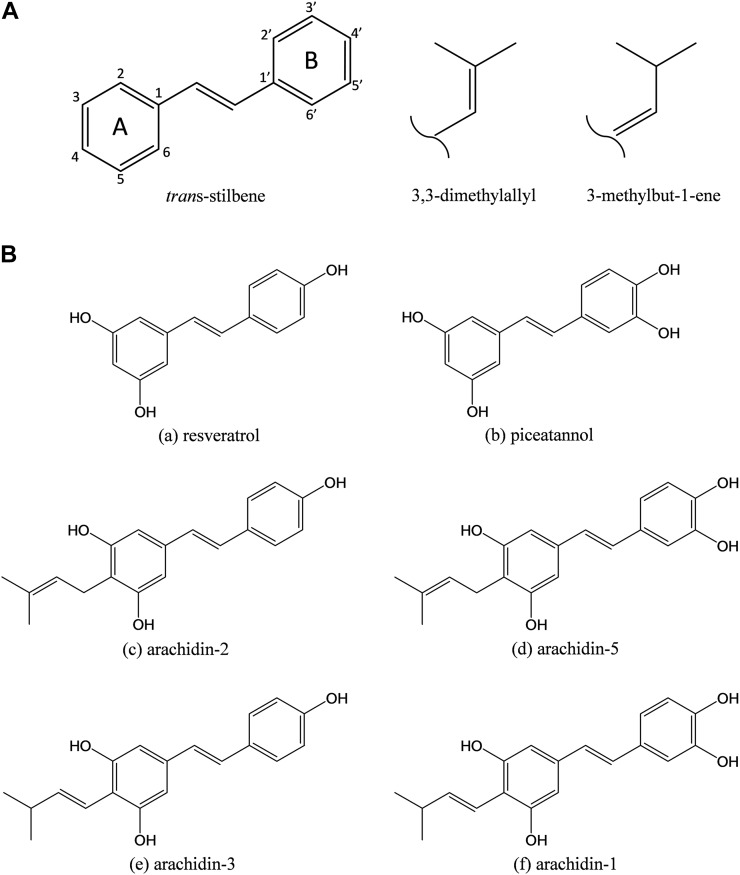
Figure 1.
A, Structure of the stilbene backbone and main prenylation patterns found on peanut prenylated stilbenoids. B, Chemical structures of stilbenoids identified in elicited peanut hairy roots: resveratrol (a), piceatannol (b), arachidin-2 (c), arachidin-5 (d), arachidin-3 (e), and arachidin-1 (f). All compounds are shown in their trans-isomers.
In addition to the above-mentioned stilbenoids (arachidin-1, arachidin-2, and arachidin-3), more than 20 other prenylated stilbenoids have been described in peanut tissues (Sobolev et al., 2006, 2016; Wu et al., 2011; Sobolev, 2013). The biosynthesis of stilbenoids derives from both the phenylpropanoid and acetate pathways. These merge to produce resveratrol by the action of resveratrol synthase, which catalyzes the cyclization of 4-coumaroyl-CoA and malonyl-CoA (Schöppner and Kindl, 1984). The prenylation step, in which one of two prenyl patterns (3,3-dimethylallyl and 3-methyl-but-1-enyl) is introduced to various positions of the stilbene backbone (Fig. 1), along with the oxidation, methylation, and cyclization steps plays a major role in the diversification of peanut prenylated stilbenoids. Although the enzymes involved in resveratrol biosynthesis have been elucidated (Chong et al., 2009), the enzymes involved in the prenylation steps of resveratrol or any other stilbenoid have not been described.
For other prenylated aromatic compounds, a prenyltransferase was found to be the critical activity for coupling the aromatic compound biosynthesis and terpenoid biosynthesis, the latter leading to the formation of the prenyl unit (Yazaki et al., 2009). Two pathways are known for the biosynthesis of prenylated compounds in plants, the mevalonic acid (MVA) pathway in the cytosol and the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway in the plastid (Lohr et al., 2012). Many studies have shown that dimethylallyl pyrophosphate (DMAPP) derived from the MEP pathway is used as a prenyl donor to form prenylated flavonoids or prenylated isoflavonoids in the plastid (Yamamoto et al., 2000; Yazaki et al., 2009). To determine the biosynthetic origin of these terpenoids, distinct metabolic inhibitors were applied to inhibit the key rate-limiting enzymes involved in either the MVA or MEP pathway. For instance, mevastatin, an inhibitor of 3-hydroxy-3-methylglutaryl-CoA reductase involved in the MVA pathway, was used in hairy root cultures of ginseng (Panax quinquefolius) to study the biosynthesis of ginsenosides (Zhao et al., 2014), while clomazone, an herbicide that inhibits 1-deoxy-d-xylulose-5-phosphate synthase during early steps of DMAPP biosynthesis in the plastid, was used to investigate the synthesis of monoterpenes in Catharanthus roseus (Han et al., 2013).
In order to elucidate the biosynthesis of peanut prenylated stilbenoids, we established hairy root cultures of peanut (Condori et al., 2010) and recently demonstrated that a sustainable production of the prenylated stilbenoids arachidin-1 and arachidin-3 can be achieved upon cotreatment of these cultures with methyl jasmonate (MeJA) and cyclodextrin (CD; Yang et al., 2015a). In this study, we took advantage of this bioproduction system and identified arachidin-2 and a new prenylated stilbenoid, named arachidin-5, as the prenylated products of the hairy root microsomal fraction using resveratrol and piceatannol as substrates, respectively. To determine the biosynthetic origin of the prenyl moiety of these prenylated stilbenoids, two metabolic inhibitors, mevastatin and clomazone were selected and applied to peanut hairy root cultures cotreated with MeJA and CD as elicitors. In the process, we identified and characterized a resveratrol 4-dimethylallyl transferase from the microsomal fraction of elicited peanut hairy roots. To our knowledge, this enzyme is the first stilbenoid-specific prenyltransferase that prenylates resveratrol and other specific stilbenoids at the 4-C position of the aromatic ring (Fig. 1).
RESULTS
Purification and Structural Identification of a New Prenylated Stilbenoid from Peanut Hairy Root Culture
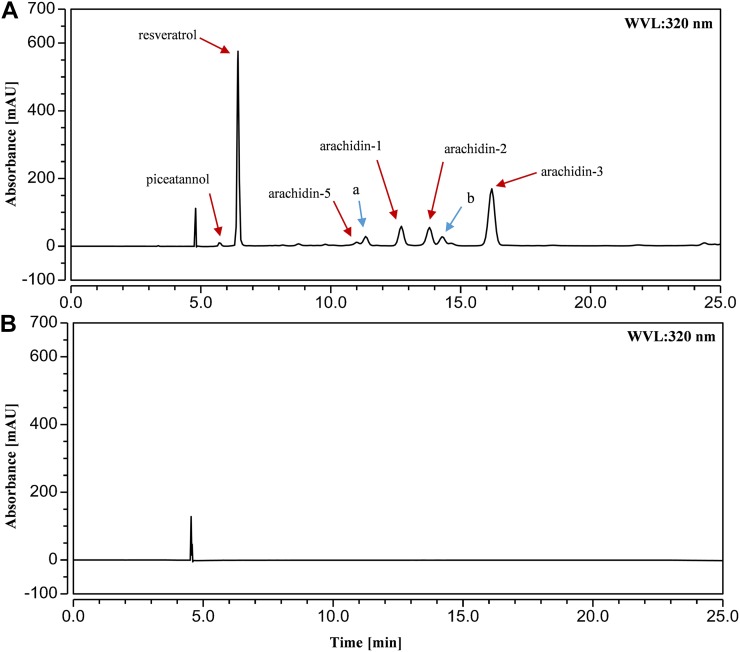
Our previous work had indicated that hairy roots of peanut have the capability to produce and secrete resveratrol (3,5,4′-trihydroxy-stilbene), piceatannol (3,5,3′,4′-tetrahydroxy-stilbene), and their prenylated analogs, arachidin-3 and arachidin-1, in the medium upon cotreatment with MeJA and CD as elicitors (Yang et al., 2015a). In this study, we have identified arachidin-2 in the ethyl acetate extract of the culture medium by comparing with the HPLC retention time, characteristic UV spectrum (λmax = 323 nm; Fig. 2), and mass spectrometric analysis of the purified arachidin-2 isolated from fungus-infected peanut seeds. The structure of arachidin-2 purified from peanut hairy root culture was confirmed subsequently by 1H- and 13C-NMR spectra (data not shown).
Figure 2.
HPLC results (UV light at 320 nm) of ethyl acetate extract from the medium of peanut hairy root culture elicited with 100 μm MeJA and 9 g L−1 CD for 72 h (A) and without elicitors for 72 h (B). a, Arachidin-5 derivative; b, arachidin-2 derivative, mAU, milliabsorbance units; WVL, wavelength.
A new prenylated stilbenoid with λmax = 327 nm in the HPLC mobile phase also was isolated from the peanut hairy root culture medium (Fig. 2). We named it arachidin-5. Mass spectrometry analysis (Table I) of arachidin-5 (m/z 313 [M+H]+) gave a main fragment with an m/z 257 [M+H-56]+ in MS2, which suggested the presence of a prenyl moiety. Arachidin-5 and arachidin-1 share very similar mass spectrometry, MS2, and MS3 spectra (Table I), indicating that the structural difference between these two compounds might be only in the position of olefinic bond on their prenylated moieties.
Table I. Stilbenoids detected in ethyl acetate extract from the medium of elicited peanut hairy root culture.
Analysis was done by HPLC-PDA-electrospray ionization-MS3.
| No. | Analyte | HPLC Retention Time | UV Light | [M+H]+ | MS2 Ionsa | MS3 Ions |
|---|---|---|---|---|---|---|
| min | nm | m/z | ||||
| 1 | Piceatannol | 5.41 | 240, 324 | 245 | 227, 199, 135 | 107 |
| 2 | Resveratrol | 6.08 | 237, 306, 317 | 229 | 211, 183 | 107 |
| 3 | Arachidin-5 | 10.29 | 240, 327 | 313 | 257 | 239, 229, 211 |
| 4 | Arachidin-5 derivative | 10.62 | 240, 355 | 311 | 283, 201, 135 | 241, 173, 135 |
| 5 | Arachidin-1 | 11.87 | 243, 341 | 313 | 257 | 239, 229, 211 |
| 6 | Arachidin-2 | 12.29 | 239, 311, 323 | 297 | 241 | 223, 213, 195 |
| 7 | Arachidin-2 derivative | 13.38 | 238, 350 | 295 | 267, 253, 201 | 239, 225, 172 |
| 8 | Arachidin-3 | 15.11 | 243, 335 | 297 | 241 | 223, 213, 195 |
MS2 ions in boldface were the most abundant ions and were subjected to MS3 fragmentation.
The 1H-NMR of arachidin-5 displayed two independent aromatic systems linked via a vinyl bridge. One of the aromatic systems had a characteristic ABX substitution pattern showing the H resonances at δ7.05 (d, 2.1 Hz; H-3′), δ6.87 (dd, 2.1 and 8.3 Hz; H-5′), and δ6.83 (d, 8.3 Hz; H-6′) ppm. The other aromatic system showed only one signal but integrating for two Hs (δ6.58 ppm, H-2 and H-6), indicating that four carbons of the benzene ring are substituted and that the two Hs are in an identical environment. The vinyl bridge H signals appeared at δ6.77 (d, 16.5 Hz; H-α) and δ6.86 (d, 16.5 Hz; H-β) ppm (Supplemental Fig. S1). These signals indicated that the stilbene core of arachidin-5 has the same substitution pattern as that of arachidin-1. The difference between these two compounds is in the signals of the prenyl group. The structure of the prenylated moiety on arachidin-5 was determined by 1H- and 13C-NMR. The presence of a five-carbon unit of the 3,3-dimethylallyl structure was evident from the peak at 5.3 ppm (t, 7.1 Hz) that is coupled to the peak at 3.36 ppm (d, 7.1) as well as the peaks at 1.77 and 1.65 ppm for the two methyl groups (H-5′′ and H-4′′, respectively; Supplemental Fig. S1). These proton resonances are the same as those published for arachidin-2 (Park et al., 2011). The 3,3-dimethylallyl structure also is supported by the presence of a quaternary carbon peak at 130.9 (C-3′′) in the 13C-NMR spectrum as well as peaks at 124.4 (C-2′′), 26 (C-4′′), 23.1 (C-1′′), and 17.9 (C-5′′; Supplemental Fig. S2). These resonances are in agreement with published resonances for the isoprene tail of arachidin-2 (Park et al., 2011). The NMR results showed that arachidin-5 has the same 3,3-dimethylallyl moiety as arachidin-2 but with an additional hydroxy group at the 3′ position, while arachidin-1 and arachidin-3 have the 3-methyl-but-1-enyl moieties instead.
In addition to arachidin-5, arachidin-1, arachidin-2, and arachidin-3, HPLC analysis of the culture medium showed the presence of two other compounds with similar characteristics of arachidin-5 and arachidin-2 based on UV light and mass spectrometry, MS2, and MS3 spectra (Fig. 2; Table I). These compounds were later designated as arachidin-5 derivative and arachidin-2 derivative, respectively.
Effects of Metabolic Inhibitors on the Yield of Prenylated Stilbenoids in Peanut Hairy Root Culture
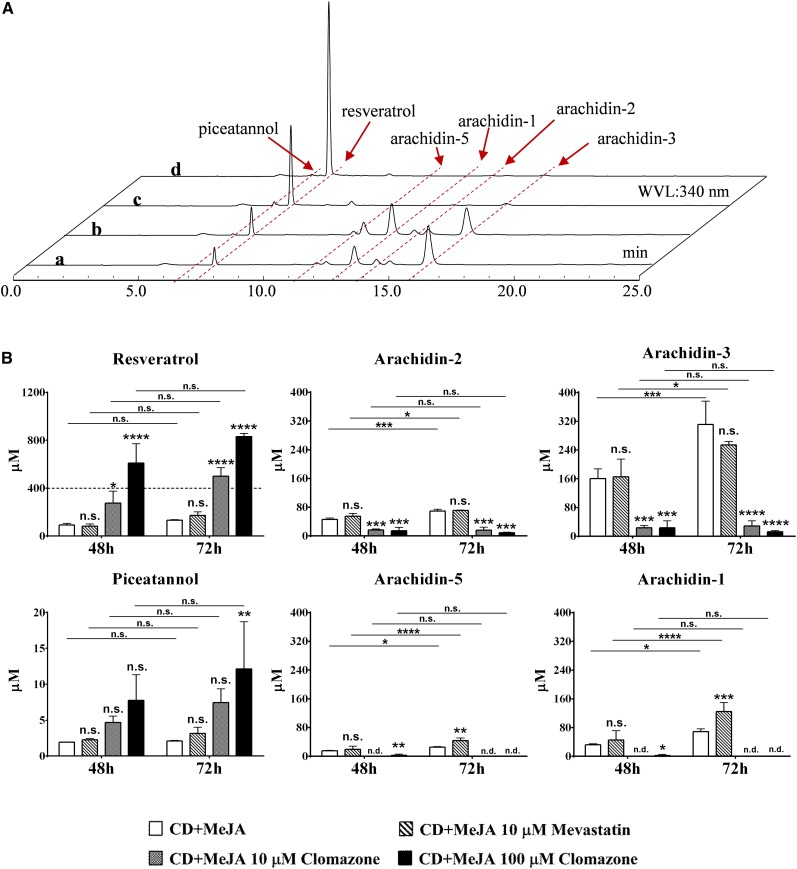
As expected for peanut phytoalexins, nonprenylated and prenylated stilbenoids were only present in the peanut hairy root cultures after elicitor treatment (Medina-Bolivar et al., 2007; Yang et al., 2015a). To study the metabolic origin of the prenyl moiety of the prenylated stilbenoids, mevastatin or clomazone was added to 9-d peanut hairy root cultures cotreated with 100 μm MeJA and 9 g L−1 CD. In preliminary experiments, we found that mevastatin at 1, 10, or 100 μm did not affect the levels of prenylated stilbenoids (Supplemental Fig. S3). On the other hand, the effects on the accumulation of resveratrol and the inhibition of prenylated stilbenoid accumulation were increased with the concentration of clomazone (1–100 μm; Supplemental Fig. S4). Hence, to quantify these effects, we determined the yields of resveratrol and prenylated stilbenoids after 48 and 72 h of elicitor treatment together with mevastatin (10 μm) or clomazone (10 and 100 μm). The yields of stilbenoids were expressed in micromoles to assess the molar contribution of resveratrol as a precursor of the prenylated stilbenoids.
Mevastatin had no significant effect on the yields of resveratrol, piceatannol, and prenylated stilbenoids with the exception of arachidin-5 and arachidin-1, which showed 27% and 41% increases in yield, respectively, after a 72-h treatment (Fig. 3). During the 24-h interval between the 48- and 72-h treatments, the noninhibitor and mevastatin-treated groups had significant increases in the yields of arachidin-5, arachidin-1, arachidin-2, and arachidin-3, indicating that mevastatin did not inhibit the accumulation of these prenylated stilbenoids (Fig. 3).
Figure 3.
Effects of mevastatin and clomazone on the production of stilbenoids in elicited peanut hairy root culture. A, HPLC results (UV light at 340 nm) of ethyl acetate extracts from peanut hairy root cultures after 48 h of treatment with 100 μm MeJA and 9 g L−1 CD (a), 100 μm MeJA, 9 g L−1 CD, and 10 μm mevastatin (b), 100 μm MeJA, 9 g L−1 CD, and 10 μm clomazone (c), and 100 μm MeJA, 9 g L−1 CD, and 100 μm clomazone (d). WVL, Wavelength. B, Yields of resveratrol, arachidin-2, arachidin-3, piceatannol, arachidin-5, and arachidin-1 after 48 and 72 h of cotreatment of 10 μm mevastatin or 10 or 100 μm clomazone with elicitors (CD + MeJA). Values shown are average micromolar yields of three replicates, and error bars represent sd. Asterisks above the bars represent significant differences compared with the group treated with MeJA and CD alone (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; and ∗∗∗∗, P < 0.0001), and asterisks above the connecting lines represent significant differences between 48- and 72-h treatments (∗, P < 0.05; ∗∗∗, P < 0.001; and ∗∗∗∗, P < 0.0001). n.d., Not detected; n.s., not significant. Statistical analyses were performed by two-way ANOVA with Dunnett’s and Sidak’s multiple comparison test, respectively.
With an inherent limited production (about 100-fold lower than that of resveratrol) upon elicitor treatment, the piceatannol concentration increased only slightly in the 100 μm clomazone group after 72 h of treatment (Fig. 3). However, in both 10 and 100 μm clomazone-treated groups, there were significantly higher yields of resveratrol and significantly lower yields of prenylated stilbenoids when compared with the noninhibitor group. In particular, the accumulation of arachidin-5 and arachidin-1 was almost completely inhibited, and only trace amounts were observed in the 48-h treated groups containing 100 μm clomazone (Fig. 3). There were no significant increases in the levels of all prenylated stilbenoids during the 24-h interval between the 48- and 72-h time points, indicating that clomazone has an inhibitory effect on the accumulation of prenylated stilbenoids in peanut hairy root cultures. Since each of the inhibitors was applied to 9-d-old hairy root cultures together with elicitors, DMAPP or isopentenyl diphosphate (IPP) might already have been synthesized and stored in the tissue prior to the inhibition. Thus, even if the biosynthesis of DMAPP and IPP was blocked when clomazone was added into the medium, small amounts of arachidin-2 and arachidin-3 could be detected in the 48- and 72-h clomazone-treated samples (Fig. 3). Moreover, when different concentrations of clomazone were applied to the peanut hairy root culture without elicitors, none of these stilbenoids was detected in the ethyl acetate extracts of the culture medium, suggesting that clomazone was not able to induce the production of stilbenoids by itself (Supplemental Fig. S5). However, in the cotreated group of 100 μm clomazone with elicitors, the yield of resveratrol reached up to 830.73 ± 25.83 μm after 72 h, which was 6.3-fold higher than that in the noninhibitor group. The micromolar increase of resveratrol was about 1.54-fold greater than the overall decrease in the accumulation of arachidin-5, arachidin-1, arachidin-2, and arachidin-3, suggesting that these prenylated stilbenoids may have been derived from resveratrol.
Degradation of Exogenous Resveratrol in Peanut Hairy Roots
Upon cotreatment of peanut hairy root cultures with MeJA and CD as elicitors, most of the total resveratrol and prenylated stilbenoids produced were secreted into the culture medium and only trace amounts were found in the ethyl acetate extracts of the root tissue (Supplemental Fig. S6). Based on previous observations regarding the loss of resveratrol accumulation in medium of nonelicited peanut hairy root cultures (Yang et al., 2015a), we speculated that resveratrol could be metabolized by one or multiple enzymatic mechanisms in peanut hairy roots.
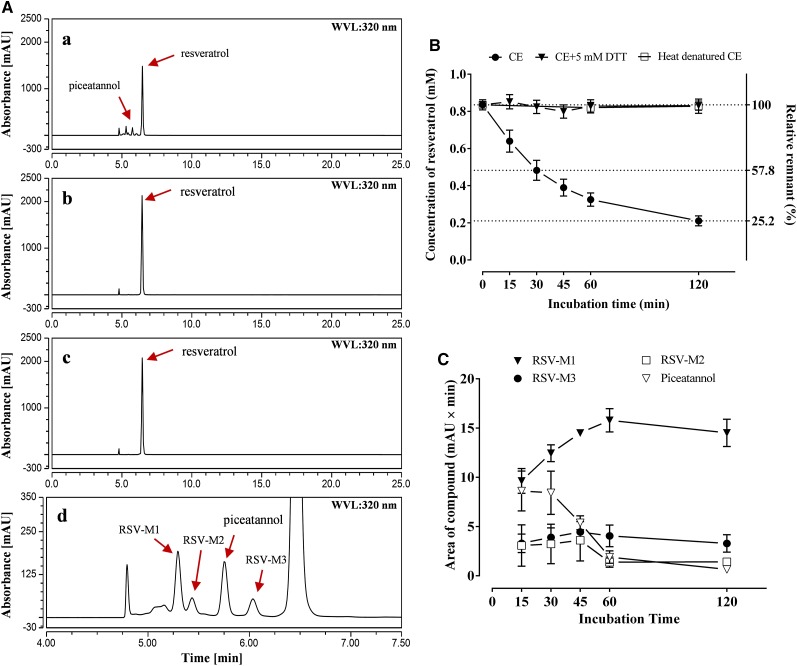
To confirm this hypothesis, 1 mm resveratrol was coincubated with a crude cell-free extract from nonelicited peanut hairy roots. Within a 2-h incubation, the color of the reaction mixtures was changed from colorless or pale yellow to dark yellow (Supplemental Fig. S7), and the concentration of resveratrol declined to 57.8% at 30 min and 25.2% at 120 min of initial levels. In the control group coincubated with heat-denatured crude cell-free extract, in contrast, resveratrol concentrations did not decrease significantly after 60- or 120-min incubations (Fig. 4B). Meanwhile, piceatannol, along with three other unidentified compounds detected as reaction products (Fig. 4A, a and d), were not found in the control group. One of the three reaction products, resveratrol metabolite-1, was found to increase within the first 60 min during the degradation of resveratrol, while the other two products, resveratrol metabolite-2 and metabolite-3, remained at similar levels during the incubation (Fig. 4C). Piceatannol concentrations started to decrease after 30 min and were undetectable after 120 min of incubation (Fig. 4C), suggesting that piceatannol along with resveratrol metabolite-2 and metabolite-3 might be intermediate products that are ultimately converted into resveratrol metabolite-1 during the degradation of exogenous resveratrol in peanut hairy roots.
Figure 4.
Enzymatic degradation of resveratrol by crude cell-free protein extract from nonelicited peanut hairy root. A, HPLC results (UV light at 320 nm) of ethyl acetate extracts from the incubation mixtures contained 1 mm resveratrol with 50 µg of crude cell-free extract for 30 min (a), 50 µg of heat-denatured crude cell-free protein extract for 30 min (b), or 50 µg of crude cell-free protein extract and an additional 5 mm DTT for 120 min (c). All reactions were done in a pH 7.6 Tris-HCl buffer. d shows an enlargement of the 4- to 7.5-min section of the chromatogram in a. B, Time course of resveratrol concentration remaining in reaction mixtures after coincubation with crude cell-free protein extract (CE), crude cell-free protein extract with an additional 5 mm DTT (CE+5 mm DTT), and heat-denatured protein (heat denatured CE) as a control. C, Time course of piceatannol and three other unidentified compounds, resveratrol metabolite-1 (RSV-M1), resveratrol metabolite-2 (RSV-M2), and resveratrol metabolite-3 (RSV-M3), produced during the coincubation of resveratrol with crude cell-free extract. mAU, Milliabsorbance units; WVL, wavelength.
In fact, according to our preliminary experiments, not only was resveratrol metabolized, but the accumulation of prenylated stilbenoids like arachidin-2 also was affected. Consequently, after 120 min of incubation, neither resveratrol nor its prenylation product, arachidin-2, was detected in the ethyl acetate extracts of the reaction mixtures (Supplemental Fig. S8). It is necessary, therefore, to address this degradation of stilbenoids before further investigation of prenyltransferase activity in peanut hairy roots. To avoid the oxidation of resveratrol into piceatannol, the reducing agent DTT was added to the reaction mixture of the crude cell-free extract from nonelicited cultures using 1 mm resveratrol as substrate. Upon 120 min of coincubation with 5 mm DTT, the original amount of resveratrol was retained and no piceatannol or other reaction products were detected in the mixture (Fig. 4Ac), suggesting that DTT prevented the initial oxidation and subsequent degradation of resveratrol.
Detection of Resveratrol Prenyltransferase Activity in Peanut Hairy Roots
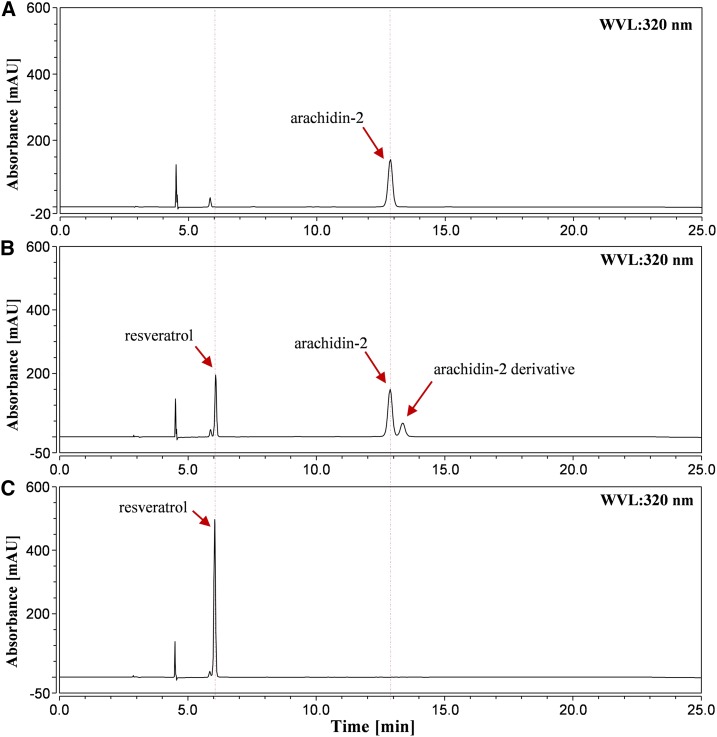
All flavonoid-specific prenyltransferases reported to date from other legume plants have been localized to plastid membranes, and the results described above indicate that the accumulation of prenylated stilbenoids in our hairy root culture system can be blocked in the presence of a plastid MEP pathway inhibitor. We hypothesized that the stilbenoid-specific prenyltransferase(s) in peanut also might be membrane localized and utilize DMAPP or IPP originating from the MEP pathway as the prenyl donor. Our clomazone inhibitor experiments also suggested resveratrol as a prenyl acceptor and a precursor of prenylated stilbenoids. Furthermore, according to our preliminary results, the prenyltransferase activities from the crude cell-free extracts of peanut hairy roots were inducible by elicitors and increased with incubation time (Supplemental Fig. S9). Considering that the yield of prenylated stilbenoids, including arachidin-1, arachidin-2, arachidin-3, and arachidin-5, increases from 9 to 72 h of elicitation (Supplemental Fig. S10), we selected a time point during this period for further studies. To this end, we prepared a microsomal fraction using ultracentrifugation from 48-h elicited cultures treated with resveratrol and DMAPP, along with DTT to reduce the degradation of resveratrol and its prenylated product. Two enzymatic products were synthesized in the reaction mixture. The predominant one was identified as arachidin-2 by comparison of its retention time, UV light spectrum, mass spectra, and fragmentation patterns obtained by tandem mass spectrometry (MS2 and MS3), each of which was identical to that of arachidin-2 purified from peanut hairy root culture (Fig. 5B). Another product (Fig. 5B) shared identical retention time, UV light spectrum, mass spectra, MS2, and MS3 with that of an unidentified compound found in peanut hairy root culture (Table I). Due to the presence in the enzymatic reaction with 100 μm arachidin-2 and crude cell-free extract (Supplemental Fig. S11), this product is considered as an enzymatic derivative of arachidin-2. None of the prenylated products were detected in the sample incubated with the microsomal fraction from nonelicited hairy root tissue (Supplemental Fig. S15), indicating the inducibility of this resveratrol prenyltransferase in peanut hairy roots. Moreover, the specific activity of resveratrol prenyltransferase in the microsomal fraction (421.16 ± 16.25 pkat mg−1 protein, based on the production of arachidin-2) was 9.3-fold higher than that in the crude cell-free extracts, suggesting that resveratrol prenyltransferase in peanut was bound to the membrane fraction of the root cells (Table II). Additional experiments using peanut membrane-bound protein as markers could verify these observations.
Figure 5.
Resveratrol prenyltransferase activity in a microsomal fraction of elicited peanut hairy root. HPLC results (UV light at 320 nm) are shown for purified arachidin-2 (A), ethyl acetate extract of a 500-µL reaction mixture containing 30 µg of microsomal fraction, 100 μm resveratrol, 300 μm DMAPP, 10 mm MgCl2, and 5 mm DTT (B), and ethyl acetate extract of a 500-µL reaction mixture containing heat-denatured 30 µg of microsomal fraction, 100 μm resveratrol, 300 μm DMAPP, 10 mm MgCl2, and 5 mm DTT (C). All reactions were done for 60 min in a pH 9.2 Tris-HCl buffer. mAU, Milliabsorbance units; WVL, wavelength.
Table II. Resveratrol prenyltransferase activity from peanut hairy root protein fractions.
The preparation of fractions and the resveratrol prenyltransferase assay are described in “Materials and Methods.” Values are means ± sd for two replicates.
| Enzyme Solution | Total Activity | Total Protein | Specific Activity |
|---|---|---|---|
| pkat g−1 root tissue | mg g−1 root tissue | pkat mg−1 protein | |
| Crude cell-free extracts | 147.95 ± 16.71 | 3.27 ± 0.06 | 45.27 ± 4.35 |
| 156,000g supernatant | 23.48 ± 4.65 | 3.00 ± 0.22 | 7.80 ± 0.97 |
| Microsomal fraction | 50.38 ± 6.66 | 0.12 ± 0.01 | 421.16 ± 16.25 |
Biochemical Characterization of Resveratrol Prenyltransferase
In the resveratrol prenyltransferase reaction, the accumulation of the prenylated product arachidin-2 followed a linear relationship with input between 30 and 120 µg of microsomal protein. The reaction also was linear over the 120-min incubation time (Supplemental Fig. S12). The optimum pH for this prenyltransferase was 9.2 in a Tris-HCl buffer (Supplemental Fig. S13), which was close to the optimum pH values of 9 to 10 for flavonoid-specific prenyltransferases (Yamamoto et al., 2000). Divalent cations were absolutely required for resveratrol prenyltransferase activity, and Mg2+ was the most effective of these (100%), followed by Mn2+ (71.8%), Fe2+ (14.6%), and Ca2+ (0.9%; Supplemental Fig. S14). No prenyltransferase activity was detected when a divalent cation was absent from the reaction (Supplemental Fig. S15). The isoprenoid precursor IPP was tested as a prenyl donor for the prenyltransferase with resveratrol as the prenyl acceptor, and no activity was detected in the assay (Supplemental Fig. S15). The apparent Km values for resveratrol and DMAPP were calculated as 111.1 ± 40.44 and 91.89 ± 7.032 μm, respectively (Supplemental Fig. S16).
Substrate Specificity of Resveratrol Prenyltransferase
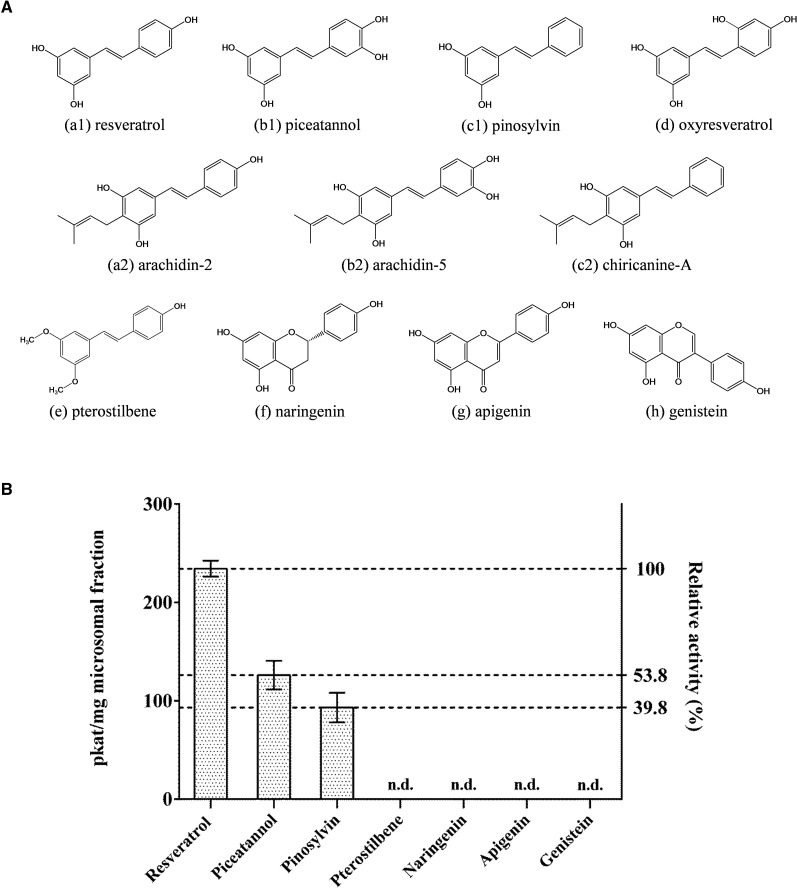
To analyze the substrate specificity of the resveratrol prenyltransferase from peanut hairy root, stilbenoids (piceatannol, pinosylvin, pterostilbene, and oxyresveratrol), flavanone (naringenin), flavone (apigenin), and isoflavone (genistein) were incubated with a microsomal fraction using DMAPP as a prenyl donor. When piceatannol was used as a prenyl acceptor, the microsomal fraction catalyzed the synthesis of arachidin-5 as the predominant prenylated product (Supplemental Fig. S17). Similar to the arachidin-2 derivative, another product considered as an enzymatic derivative of arachidin-5 also was detected in the reaction (Supplemental Fig. S18). In addition, the microsomal fraction catalyzed pinosylvin into chiricanine A, confirmed by comparison of its retention time, UV light spectrum, mass spectra, and fragmentation patterns with those of chiricanine A standard purified from fungus-challenged peanut seeds (Fig. 6; Supplemental Fig. S17).
Figure 6.
Substrate specificity of resveratrol prenyltransferase in a microsomal fraction of elicited peanut hairy root. A, Chemical structures of prenyl acceptors used for substrate specificity analysis and their prenylated products: stilbenoids (a1, resveratrol; a2 arachidin-2; b1, piceatannol; b2, arachidin-5; c1, pinosylvin; c2, chiricanine A; d, oxyresveratrol; and e, pterostilbene), flavanone (f, naringenin), flavone (g, apigenin), and isoflavone (h, genistein). B, Prenylation activity of a microsomal fraction with various prenyl acceptors. Values are averages of three replicates, and error bars represent sd. n.d., Not detected.
Using this microsomal fraction, an isoprene unit also was transferred to oxyresveratrol. Mass spectroscopic analysis of its reaction product (m/z 313 [M+H]+) showed a main fragment with m/z 257 [M+H-56]+ in MS2, which suggested the presence of a prenyl moiety (Supplemental Fig. S19). However, the position of the prenyl moiety on this prenylated oxyresveratrol remains undetermined due to the insufficient amount of compound for further structural elucidation.
Interestingly, pterostilbene, which has methoxy groups at the C-3 and C-5 positions, did not produce any product within the microsomal fraction (Fig. 6; Supplemental Fig. S17), suggesting that the additional methyl group on its structure might block prenylation and that either or both of the hydroxyl groups at C-3 and C-5 might be required for the prenylation reaction. Furthermore, neither prenylated flavanone, prenylated flavone, nor prenylated isoflavone was detected in the reaction mixtures (Fig. 6; Supplemental Fig. S17), indicating that this peanut prenyltransferase may be a stilbenoid-specific prenyltransferase.
DISCUSSION
A Membrane-Bound Prenyltransferase Specific for Stilbenoids from Peanut Hairy Root
As the limiting enzyme involved in the biosynthesis of prenylated flavonoids, flavonoid prenyltansferases have been a focus of previous research, particularly in plants of the Leguminosae family known to accumulate these type of specialized metabolites. The first flavonoid prenyltansferase, SfN8DT-1, was identified in Sophora flavescens cell cultures by Sasaki et al. (2008). Subsequently, SfG6DT and SfiLDT, which prenylate genistein to produce wighteone (6-dimethylallylgenistein) and isoliquiritigenin to produce dimethyllylisoliquiritigenin, respectively, were identified in the same species (Sasaki et al., 2011). While SfG6DT prenylates genistein on the A-ring of the isoflavone, LaPT1 isolated from another legume, white lupin (Lupinus albus), catalyzes the prenylation of the B-ring of genistein and 2′-hydroxygenistein to form isowighteone (3′-dimethylallylgenistein) and luteone (2′-hydroxy-3′-dimethylallyl-genistein), respectively (Shen et al., 2012). As a crucial prenyltransferase involved in glyceollin biosynthesis, GmG4DT was identified and characterized in soybean (Glycine max). This enzyme catalyzes the dimethylallylation of glycinol at position 4 to produce the precursor of the phytoalexin, glyceollin I (Akashi et al., 2009). More recently, GuA6DT, a flavone prenyltransferase, was identified in another legume species, liquorice (Glycyrrhiza uralensis; Li et al., 2014), and SfFPT characterized from S. flavescens displays high catalytic efficiency of C-8 prenylation on different types of flavonoids, including flavanone, flavone, and flavanonol (Chen et al., 2013).
In this study, we have described, to our knowledge, the first plant stilbenoid prenyltransferase activity from the microsomal fraction of elicited peanut hairy roots. This prenyltransferase catalyzes the dimethylallylation of resveratrol at C-4 and shows several common features with other prenyltransferases, such as those described above. For instance, all prenylation activities mentioned above and demonstrated here require a divalent cation as cofactor and a basic buffer for optimal reaction rate (except for GmG4DT, whose reaction rate optimum is pH 7.5 buffer for the reaction; Akashi et al., 2009). Because the activity of this enzyme was concentrated in the microsomal fraction and the accumulation of prenylated stilbenoid in peanut hairy roots was inhibited by clomazone, we suggest that this peanut resveratrol prenyltransferase is a membrane-bound protein located in the plastid. This further suggests that the DMAPP used in the prenylation reaction was derived from the MEP plastidic pathway. Our hypothesis is in agreement with the plastidic subcellular location of flavonoid prenyltransferases identified in other legume species.
During the biosynthesis of stilbenoids and flavonoids, both resveratrol synthase and chalcone synthase use 4-coumaroyl-CoA as a substrate and perform three condensation reactions with malonyl-CoA to form a linear tetraketide, which is later folded into new ring structures. These two enzymes are distinguished by a special property of stilbene synthases, which loses the terminal carboxyl group as CO2, resulting in the release of four CO2 molecules. In contrast, each reaction catalyzed by chalcone synthase releases three CO2 molecules. It has been suggested that stilbene synthase may have developed from chalcone synthase via gene duplication and mutation, rendering new and improved functions (Tropf et al., 1994).
Among the prenylation products of resveratrol, piceatannol, pinosylvin, and oxyresveratrol catalyzed by the microsomal fraction containing resveratrol prenyltransferase, only arachidin-2 and arachidin-5 were detected in the spent medium of peanut hairy root cultures cotreated with MeJA and CD. Interestingly, oxyresveratrol and prenylated oxyresveratrol reported in the bark of Artocarpus dadah (Su et al., 2002) along with pinosylvin isolated from pine (Pinus spp.) hardwood have not been identified in any peanut tissue, while the prenylated product of pinosylvin, chiricanine A, that was first described in the roots of Lonchocarpus chiricanus (Ioset et al., 2001), has been described in peanut seeds after infection with the fungus A. flavus (Sobolev et al., 2009).
Due to the variety of enzymes that could be present in the microsomal fraction, our studies described here address neither whether any stilbenoid prenyltransferases act with strict substrate specificities nor whether multiple stilbenoid substrates are recognized by a single prenyltransferase in peanut. Characterization of the genes encoding these prenyltransferases and purification of the specific prenyltransferases will be necessary to address these activities.
Recently, the genome sequences and gene annotation of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut, were described (Bertioli et al., 2016). We used the amino acid sequences of six well-characterized flavonoid prenyltransferases (Supplemental Table S1) to perform BLASTP against the translated coding sequence (CDS) databases of both A. duranensis and A. ipaensis genomic annotations. In brief, 22 and 37 prenyltransferase candidates were selected from the A. duranensis and A. ipaensis genomes, respectively. Among these 59 sequences, 58 of them were found to possess at least one transmembrane domain, according to predictions of the TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/). Plastidial targeting signals were predicted by at least one of three subcellular localization prediction programs, ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/), TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/), and iPSORT (http://ipsort.hgc.jp/; Table III), in 42 of the candidate sequences. The prenyltransferase candidates from A. duranensis and A. ipaensis with the most significant alignments share from 50% to 63% sequence identity with different flavonoid prenyltransferases, and both types of enzymes are predicted to contain nine transmembrane α-helices (Supplemental Table S2). In addition to newly available genomic information, numerous transcript references from three different species of Arachis, A. duranensis, A. ipaensis, and peanut (Supplemental Table S3), now provide us with resources to investigate resveratrol prenyltransferases in peanut. As with current genomic candidates, prenyltransferase candidates from Arachis spp. transcriptome BLASTN searches also show sequence identities of 50% to 63% to various flavonoid prenyltransferase sequences (Supplemental Table S4). All of them are predicted to possess nine transmembrane α-helices and plastidial targeting signals (Supplemental Table S5). We believe these abundant genomic and transcriptomic resources will help us identify the stilbenoid prenyltransferase genes from peanut in the near future.
Table III. Screening for stilbenoid prenyltransferase candidate sequences from A. duranensis and A. ipaensis genomes.
BLASTP was performed using flavonoid prenyltransferase protein sequences against translated CDS databases of A. duranensis and A. ipaensis genomes. Nonredundant candidates were further analyzed by the ChloroP 1.1, TargetP 1.1, and iPSORT programs for the prediction of chloroplast transit peptides. The transmembrane α-helices in the candidate sequences were predicted by the TMHMM 2.0 program.
| Prediction with Various Programs | A. duranensis | A. ipaensis | Total |
|---|---|---|---|
| Hits of nonredundant candidate sequences from BLASTP search | 22 | 37 | 59 |
| ChloroP | 12 | 17 | 29 |
| TargetP | 11 | 14 | 25 |
| iPSORT | 11 | 14 | 25 |
| Number of candidates predicted by at least one of the above programs | 17 | 25 | 42 |
| TMHMM | 22 | 36 | 58 |
Biosynthesis of Prenylated Stilbenoids in Peanut Hairy Root Cultures
The use of abiotic elicitors to induce the biosynthesis of stilbenoid in hairy root cultures provides for an axenic sustainable and controlled production platform for these specialized metabolites that can be leveraged to study their biosynthetic pathway. The metabolic steps to produce resveratrol from Phe, including Phe ammonia lyase, cinnamate-4-hydroxylase, 4-coumarate:CoA ligase, and resveratrol synthase, already have been elucidated (Watts et al., 2006). In this study, a stilbenoid prenyltransferase that prenylates resveratrol to form arachidin-2 was characterized from the microsomal fraction of elicited peanut hairy roots. Since resveratrol accumulated in the medium while clomazone blocked the plastid MEP pathway in our inhibitor feeding experiments, it may be that prenylated stilbenoids in addition to arachidin-2 also derive from resveratrol. A study of phytoalexins induced in peanut kernels by soil fungal exposure also suggests that resveratrol might be the precursor for other prenylated stilbenoids in peanut (Sobolev, 2008). Interestingly, after inhibition of the accumulation of prenylated stilbenoids by clomazone, the increase in the levels of resveratrol was much higher than the decrease in the concentration of the prenylated stilbenoids (arachidin-1, arachidin-2, arachidin-3, and arachidin-5), suggesting that the prenylated stilbenoids themselves may feedback inhibit the production of resveratrol. In addition, other stilbenoids that derive from resveratrol also may be produced. Further experiments will be needed to address these issues.
The main prenylated stilbenoids produced by peanut hairy root can be categorized into two groups according to the structure of their prenyl side chains. One group includes arachidin-2 and arachidin-5 (identified in this study), having a 3,3-dimethylallyl moiety. This is the most common type of prenylation found in prenylated stilbenoids from various plant families. For instance, longistylines C, longistylines D, and chiricanine A found in L. chiricanus (Leguminosae; Ioset et al., 2001) also have the same dimethylallyl moiety. Artoindonesianin N with one dimethylallyl moiety and 4-dimethylallyl-oxystilbene were reported in Artocarpus integer (Moraceae; Boonlaksiri et al., 2000) and Artocarpus gomezianus (Moraceae; Hakim et al., 2002), respectively. Mappain found in Macaranga mappa (Euphorbiaceae; van der Kaaden et al., 2001) has one dimethylallyl moiety and one geranyl moiety, while schweinfurthin C, with two geranyl moieties, was isolated from Macaranga alnifolia (Euphorbiaceae; Yoder et al., 2007). Moreover, the dimethylallyl moiety also is the most common prenylation pattern present in prenylated flavonoids, which are mostly found in the following families: Cannabaceae, Guttiferae, Leguminosae, Moraceae, Rutaceae, and Umbelliferae. The longer form of the dimethylallyl moiety, geranyl and lavandulyl moiety, also has been reported in prenylated flavonoids (Botta et al., 2005; Yang et al., 2015b). During the biosynthesis of these prenylated stilbenoids and prenylated flavonoids, prenyltransferases are responsible for dimethylallylation and geranylation, attaching DMAPP and GPP, respectively, to different positions of the stilbenoid and flavonoid skeletons.
One major group of prenylated stilbenoids in peanut hairy roots is represented by arachidin-1 and arachidin-3, which harbor a 3-methyl-but-1-enyl moiety. When compared with the dimethylallyl moieties commonly present in other prenylated stilbenoids and flavonoids, this unique prenylated form has been reported in peanut and very few other species. To our knowledge, the 3-methyl-but-1-enyl-oxystilbene isolated from A. integer (Moraceae; Boonlaksiri et al., 2000) is the only prenylated stilbenoid with this moiety reported in a species other than peanut. Due to the difference in the position of the olefinic bond on the prenylated moieties, 3-methyl-but-1-enyl stilbenoids such arachidin-3 and arachidin-1 have higher lipophilicity, as evident from a later retention time in reverse-phase HPLC when compared with their 3,3-dimethylallyl analogs, arachidin-2 and arachidin-5, respectively. The λmax of UV light absorbance of 3-methyl-but-1-enyl stilbenoids has an apparent shift to 335 to 340 nm when compared with that shown at 323 to 327 nm for 3,3-dimethylallyl stilbenoids (Table I). However, the effects of these two moieties on the bioactivity of peanut stilbenoids is still unclear, because arachidin-1 and arachidin-3 are not commercially available, limiting their studies, and because of the relatively low yields of arachidin-2 and arachidin-5 produced in peanut hairy roots.
The yields of arachidin-1 and arachidin-3 in our peanut hairy root culture system were much higher than those of their dimethylallyl analogs, arachidin-2 and arachidin-5. As described in the activity assays above, both arachidin-2 and arachidin-5 were further modified to other derivatives, suggesting that dimethylallyl stilbenoids are important intermediates for the biosynthesis of other peanut prenylated stilbenoids. As a substrate recognized by stilbenoid prenyltransferase, piceatannol was considered to be the putative precursor of arachidin-5 in peanut. However, the yield of piceatannol in our peanut hairy root culture is very limited and far less than that of resveratrol. Even after blocking the biosynthesis of the prenylated moiety by 100 μm clomazone, the cultures only produced 9.02 μm piceatannol compared with 845.7 μm resveratrol (Fig. 3). These observations suggests that arachidin-5 and arachidin-1 might not be synthesized from piceatannol due to its limited amounts; instead, arachidin-1 may derive from arachidin-3, which is relatively abundant in the medium, by hydroxylation at the C-3′ position.
Metabolism of Prenylated Stilbenoids in Peanut Hairy Root Cultures
Under treatment with MeJA and CD, arachidin-1, arachidin-2, arachidin-3, and arachidin-5 are the major prenylated stilbenoids secreted and accumulated in the spent culture together with resveratrol. When CD is not added in the hairy root cultures, only limited amounts of resveratrol and stilbenoids are detected in the medium. These observations were reported previously when peanut hairy roots were treated with NaOAc, H2O2, or MeJA alone as elicitors (Yang et al., 2015a). Importantly, when resveratrol was added in nonelicited peanut hairy root cultures, 47% and 97% reductions of the initial resveratrol concentration in the culture medium were observed after 0.5 and 1 h of coincubation, respectively (Yang et al., 2015a). In this study, we confirmed that the degradation of extrinsic resveratrol in peanut hairy root culture occurs due to activities of the root tissue. Unlike resveratrol synthase, which is induced and involved in stilbenoid biosynthesis, the enzymes involved in the degradation of resveratrol appear to be constitutively expressed in nonelicited root tissue, leading to the dramatic decline of extrinsic resveratrol. Results from the experiments described here suggest that the peanut enzymes present initiate this process by oxidizing resveratrol into piceatannol and then by converting this into other derivatives. This enzymatic degradation process also may apply to other prenylated stilbenoids, such as arachidin-2 (Supplemental Fig. S8), and may explain the observation that only trace amounts of stilbenoids are detected in ethyl acetate extracts of root tissue (Supplemental Fig. S6). The constitutive degradation of stilbenoids may provide the plant with the ability to manage potential toxic effect of stilbenoids when they accumulate at high levels within the cell. Interestingly, other species such as grape (Vitis vinifera) accumulate resveratrol as glucosides (i.e. piceid). However, this conjugate was not identified in the peanut hairy roots and has not been described in fungus-elicited peanut kernels. Altogether, these findings suggest that peanut may have evolved a distinct mechanism to metabolize resveratrol-type phytoalexins after they are produced.
CONCLUSION
Harnessing the inducible bioproduction capabilities of the peanut hairy root culture system, we have newly identified a prenylated stilbenoid, arachidin-5, and have demonstrated that the prenyl moiety on peanut prenylated stilbenoids is derived from a plastidic biosynthesis pathway. We have characterized, to our knowledge for the first time, a plant membrane-bound stilbenoid-specific prenyltransferase activity from the microsomal fraction of peanut hairy roots. The characteristics of this enzyme provide important information for subsequent cloning and comprehensive definition of the prenyltransferase genes of peanut. Moreover, we have observed the enzymatic degradation of exogenous resveratrol by peanut hairy root tissue, an observation that will lead to the elucidation of further mechanisms governing phytoalexin accumulation in plants.
MATERIALS AND METHODS
Chemical Reagents
Authentic standards of resveratrol and piceatannol were obtained from Biophysica and Axxora, respectively; arachidin-1, arachidin-2, arachidin-3, and arachidin-5 standards were purified from elicited peanut (Arachis hypogaea) hairy root as described below. Pinosylvin, oxyresveratrol, pterostilbene, naringenin, apigenin, and genistein used in this study were purchased from Sigma-Aldrich. Chiricanine A was purified from fungus-challenged peanut seeds as described below. DMAPP was obtained from Isoprenoids. Stock solutions of the inhibitors mevastatin (100 mm; Sigma-Aldrich) and clomazone (100 mm; Sigma-Aldrich) were prepared in ethanol and stored at 4°C.
Isolation and Purification of Chiricanine A from Fungus-Challenged Peanut Seeds
One kilogram of seeds of the 31-1314 peanut runner breeding line from the National Peanut Research Laboratory, stored at 4°C for 2 years after harvest, were allowed to imbibe distilled water for 18 h at 4°C. They were then chopped with a custom-made hand cutter into 3- to 6-mm pieces, thoroughly washed with distilled water, blotted with a paper towel, air dried to the condition where sliced peanuts did not leave water spots on filter paper, and placed on aluminum trays so that the thickness of the layer did not exceed 1 cm. The trays were evenly sprayed with the fungal spores of Aspergillus caelatus NRRL 25528 (105 mL−1), placed into autoclave bags, and incubated at 30°C for 96 h. The bags were opened every 24 h to allow fresh air to the peanuts and growing fungus.
The incubated peanut seeds were extracted with 3 L of methanol overnight at room temperature without agitation. This procedure was repeated two more times. The combined mixture was filtered through a paper filter in a Büchner-type funnel under reduced pressure. The combined filtrates were defatted three times with 0.5 L of n-hexane. The methanol layer was evaporated to dryness. The residue was redissolved in CHCl3 and applied to a chromatographic column (34 mm i.d.) packed with silica gel (Silica Gel 60, 0.063−0.200 mm; EM Science) to the height of 400 mm. The column was eluted subsequently with 0.5 L of CHCl3, 1 L of EtOAc, 1 L of acetone, and 1 L of methanol (all solvents were purchased from Fisher). Eight fractions were collected from the column and analyzed by HPLC. Fractions containing chiricanine A were combined, evaporated to dryness with a rotary evaporator, and subjected to further purification on a similar silica gel column. The column was subsequently eluted with 0.4 L of CHCl3, 1.5 L of CHCl3/EtOAc (1:1), 1.3 L of EtOAc, and 1 L of acetone. Twenty-four fractions were collected. Combined fractions containing chiricanine A were evaporated to dryness on a rotary evaporator, redissolved in methanol, filtered, and subjected to final purification with a preparative 100-mm × 19-mm i.d., 5-μm XTerra Prep RP18 OBD HPLC column (Waters). The flow rate was 9.5 mL min−1, and column temperature was maintained at 40°C. The following mobile phase was used: 73% CH3CN, 3% of 1% HCOOH in water, and 24% of water. Pure fractions of chiricanine A obtained from HPLC were evaporated with a rotary evaporator to a point where almost all of the organic solvent was removed. Then, the target compound was extracted four times with EtOAc (water:EtOAc ratio 1:1, v/v). The combined EtOAc layers were evaporated nearly to dryness with a rotary evaporator. The residue was transferred into a 15-mL vial with EtOAc and evaporated nearly to dryness with a stream of N2. The residue was redissolved in EtOAc, filtered, transferred into 4-mL vials, and evaporated to dryness with a stream of N2. Then, the vial was placed into a lyophilizer for 2 h at room temperature to remove traces of the solvents. Chiricanine A was obtained as a slightly yellowish oil (6.5 mg).
Analyses of Stilbenoids from Peanut Hairy Root Cultures
Hairy root cultures of peanut ‘Hull’ line 3 established by the Medina-Bolivar laboratory (Condori et al., 2010) were maintained in 50 mL of modified Murashige and Skoog medium (Condori et al., 2010) with 3% Suc in 250-mL flasks. In order to induce the synthesis and secretion of stilbenoids, the spent medium of 9-d-old hairy root cultures was discarded and replaced with 50 mL of fresh Murashige and Skoog medium containing 3% Suc with 100 μm MeJA and 9 g L−1 CD (Cavasol) as elicitors and incubated in the dark at 28°C for an additional 72 h as described previously (Yang et al., 2015a). After the elicitation period, 1 mL of spent medium was partitioned with ethyl acetate twice. The combined organic phase was evaporated under nitrogen gas and dissolved in methanol for subsequent HPLC and mass spectrometry analyses.
Quantitative analysis of stilbenoids in peanut hairy root culture extracts was performed in an UltiMate 3000 LC system (Dionex; Thermo Scientific) equipped with a photodiode array detector. The separation was performed on a SunFire C18 5-µm, 4.6- × 250-mm column (Waters) at 40°C with a flow rate at 1 mL min−1. The mobile phase consisted of 2% formic acid in water (A) and methanol (B). The column was initially equilibrated with 100% A for 1 min. Then, a linear gradient was performed from 40% A and 60% B to 35% A and 65% B (1–20 min), followed by a linear gradient from 35% A and 65% B to 100% B (20–25 min). Then, the column was washed with 100% A for 5 min (25–30 min). Calibration curves of various stilbenoids were established using A320 for resveratrol, piceatannol, arachidin-2, and arachidin-5 and at A340 for arachidin-1 and arachidin-3.
For liquid chromatography-mass spectrometry qualitative analysis of stilbenoids, an UltiMate 3000 rapid separation LC system (Dionex; Thermo Scientific) was used for the chromatographic separation. The separation method was similar to the HPLC conditions described above with the following modification: 0.02% formic acid in water was used instead 2% in mobile phase A, and the LTQ XL linear ion trap mass spectrometer (Thermo Scientific) with an electrospray ionization source was used to obtain structural information of stilbenoids following the method described previously (Marsh et al., 2014). Briefly, all mass spectra were performed in the positive ion mode with ion spray voltage at 4 kV, sheath gas (high-purity nitrogen) at 45 arbitrary units, auxiliary gas (high-purity nitrogen) at 15 arbitrary units, capillary voltage at 9 V, capillary temperature at 300°C, and tube lens offset at 45 V. Full mass scans were recorded in the range m/z 100 to 2,000. Ultra-high-purity helium was used as the collision gas, and 35% collision energy was applied in collision-induced dissociation. The data were recorded and analyzed by Xcalibur software (Thermo Scientific).
Purification and Identification of Arachidin-5 from Peanut Hairy Root Cultures
For the purification of arachidin-5 and other prenylated stilbenoids, 1 L of spent medium obtained from a pool of about 20 flasks of 72-h elicited peanut hairy root cultures was collected and extracted with an equal volume of ethyl acetate twice in a 2-L separatory funnel. The organic phase was recovered and dried in a rotavapor (Buchi), and the crude extract (approximately 800 mg) was stored at 4°C for subsequent high-performance counter current chromatography (HPCCC) fractionation. A two-phase HPCCC solvent system (hexane:ethyl acetate:methanol:water, 4:5:3:3, v/v/v/v) was equilibrated at room temperature in a 2-L separatory funnel. The upper phase of this solvent mixture was used as a stationary phase and the lower one was used as a mobile phase for the preparative HPCCC system (Dynamic Extractions). The multilayer coil was filled with the stationary phase at a flow rate of 8 mL min−1 and then started spinning at 1,600 rpm. Then, the hydrodynamic equilibrium was established by pumping 6 mL min−1 mobile phase into the column until a clear mobile phase was eluted at the outlet. Crude extract (300 mg) was dissolved in 5 mL of the two-phase solvent and injected manually. The effluent was monitored at UV light of 340 nm. The fractions were collected every 30 s, dried in a SpeedVac, and analyzed by HPLC as described above. According to the HPLC profiles, the fractions collected between 27 and 32 min contained arachidin-1 and arachidin-5, whereas those collected between 53 and 62 min contained arachidin-2 and arachidin-3. Fractions containing arachidin-1 and arachidin-5 were combined as one sample, whereas those containing arachidin-2 and arachidin-3 were combined as a separate sample. Then, the organic solvent in the samples was removed using a rotavapor. The remaining aqueous mixtures containing the target compounds were extracted with ethyl acetate (1:1, v/v), evaporated nearly to dryness with a rotavapor, and redissolved in methanol. Then, these combined fractions were applied to thin-layer chromatography plates (TLC Silica Gel 60 RP-18F254s; Millipore) and separated using as a developing solvent a system composed of methanol:water:acetic acid (15:45:40, v/v/v). After separation, the purified prenylated stilbenoids on the adsorbent were scraped off, redissolved in methanol, and dried under nitrogen gas for subsequent 1H- and 13C-NMR spectra analysis. In summary, 4.6 mg of arachidin-5, 20.3 mg of arachidn-1, 5.2 mg of arachidin-2, and 17.8 mg of arachidin-3 were purified from the peanut hairy root culture medium using the HPCCC and preparative thin-layer chromatography method.
For NMR analysis of purified arachidin-5 and arachidin-2, 1H-NMR was recorded at 400 MHz and 13C-NMR was recorded at 100 MHz in acetone-d6 on a Bruker AV-400 NMR spectrometer.
Inhibitor Feeding Experiment
Nine-day-old peanut hairy root cultures were used in the inhibitor feeding experiment. Prior to treatment, the spent medium from each culture was removed and replaced with 50 mL of fresh Murashige and Skoog medium containing 3% Suc and 100 μm MeJA and 9 g L−1 CD as elicitors. Then, mevastatin (10 μm) or clomazone (10 and 100 μm) was applied to the culture medium. For the control group, 50 µL of absolute ethanol (solvent of mevastatin and clomazone) was added to the medium. All treatments were performed at 28°C and continuous darkness with three biological replicates per treatment. One milliliter of spent medium was collected from each treatment at 48 and 72 h after treatment. The 1-mL medium aliquots were extracted with 1 mL of ethyl acetate, then the organic phase was collected, dried under nitrogen gas, and redissolved in methanol for HPLC analysis as described above.
Enzyme Preparation
Enzyme solutions from peanut hairy roots for the prenyltransferase assay were prepared using 9-d-old roots elicited with 100 μm MeJA and 9 g L−1 CD for 48 h. Ten grams (fresh weight) of elicited hairy root tissues was ground and homogenized in 20 mL of extraction buffer composed of 100 mm Tris-HCl buffer (pH 7.6), 10 mm DTT, and 2.5% (w/v) polyvinylpyrrolidone (average Mr, 40,000; Sigma-Aldrich) using a mortar and pestle. The homogenate was centrifuged at 12,000g for 15 min at 4°C to remove the cell debris. Crude cell-free extracts were obtained by passing 2.5 mL of the 12,000g supernatant through a PD-10 desalting column (GE Healthcare) using 100 mm Tris-HCl (pH 9.2) containing 10 mm DTT as equilibration buffer. About 13.5 mL of the remaining 12,000g supernatant was centrifuged subsequently at 156,000g for 45 min at 4°C. The 156,000g supernatant was collected and cleaned up through a PD-10 desalting column (GE Healthcare) equilibrated with 100 mm Tris-HCl buffer (pH 9.2) containing 10 mm DTT. The microsomal pellet was resuspended in 100 mm Tris-HCl buffer (pH 9.2) containing 10 mm DTT, recentrifuged (156,000g for 45 min), and finally resuspended in 1 mL of the same buffer for subsequent enzyme reaction.
For the degradation of resveratrol assay, crude cell-free extract was prepared from 12-d-old nonelicited peanut hairy roots following a similar procedure to that described for elicited roots above except that no DTT was included in the extraction and equilibration buffers.
Protein Quantification
Protein contents in the various enzyme solutions were determined using the Coomassie protein assay (Thermo Scientific) using bovine serum albumin as standard.
Degradation of Resveratrol Assay
The standard assay condition contained 1 mm resveratrol with 50 µg of crude cell-free extract from 12-d-old nonelicited peanut hairy root in 500 µL of 100 mm Tris-HCl buffer (pH 7.6). To study the effect of the reducing agent on the degradation of resveratrol, an additional 5 mm DTT was added to the standard assay mixture. The reactions were performed at 28°C under varying incubation times (15, 30, 45, 60, and 120 min) and stopped by heating at 99°C for 20 min. After 12,000g centrifugation and filtration with a 0.2-μm nylon filter, the amount of resveratrol remaining in each reaction mixture was analyzed by HPLC. In the control group, crude cell-free extract was heated at 99°C for 20 min.
Prenyltransferase Assay
The standard assay was performed in a total volume of 500 µL containing 100 μm resveratrol, 300 μm DMAPP as a prenyl donor, 10 mm MgCl2, 5 mm DTT, and 30-µg microsomal fractions in 100 mm Tris-HCl buffer (pH 9.2). After 60 min of incubation at 28°C, the reaction mixture was terminated by adding 20 µL of 6 m HCl and extracted with 500 µL of ethyl acetate. The extracts were dried under nitrogen gas and dissolved in methanol, and the reaction product was quantified by HPLC analysis. The prenylation activity for each reaction was quantified by the molar concentration of the generated prenylated product per second with 1-mg microsomal fractions (kat mg−1).
In the linearity study, the prenyltransferase activities under varying incubation times (30, 60, 90, and 120 min) with a 30-µg microsomal fraction and varying amounts of microsomal faction (30, 60, 90, and 120 µg) incubated for 60 min were measured. For the pH dependency study, the activities of prenyltransferase were measured using 100 mm Tris-HCl buffer at pH 7, 8, 8.4, 8.6, 8.8, 9, 9.2, 9.4, 9.6, and 10. For the divalent cation dependency study, 10 mm MnCl2, FeCl2, CaCl2, CoCl2, ZnCl2, NiCl2, or CuCl2 was added to the reaction mixture instead of MgCl2 as described above, and the enzyme activity was compared with the reaction containing MgCl2. For kinetic studies, varying concentrations (10, 20, 40, 80, 160, 320, and 640 μm) of resveratrol with a fixed concentration of DMAPP (640 μm) and varying concentrations (10, 20, 40, 80, 160, 320, and 640 μm) of DMAPP with a fixed concentration of resveratrol (640 μm) were incubated with microsomal fractions of peanut hairy root in a total volume of 250 µL at 28°C for 60 min. These reactions were used to calculate Vmax and Km values using nonlinear regression analysis of the Michaelis-Menten equation using GraphPad Prism 6 software. For the substrate specificity assay, 100 μm stilbenoids (resveratrol, piceatannol, pinosylvin, pterostilbene, and oxyresveratrol), flavanone (naringenin), flavone (apigenin), and isoflavone (genistein) with 300 μm DMAPP as a prenyl donor were incubated with microsomal fractions of peanut hairy root in a total volume of 250 µL at 28°C for 60 min. Arachidin-2 and arachidin-5 purified from peanut hairy root culture and chiricanine A purified from fungus-challenged peanut seed were diluted to various concentrations, and calibration curves of their A320 were used for quantitative analysis.
Statistical Analysis
Two-way ANOVA with multiple comparison tests was conducted for the data in Figure 3. Analyses were done with GraphPad Prism 6 software, version 6.02.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. 1H-NMR analysis of arachidin-5.
Supplemental Figure S2. 13C-NMR analysis of arachidin-5.
Supplemental Figure S3. Effect of mevastatin on the production of stilbenoids in elicited peanut hairy root culture.
Supplemental Figure S4. Effect of clomazone on the production of stilbenoids in elicited peanut hairy root culture.
Supplemental Figure S5. Effect of clomazone on the production of stilbenoids in nonelicited peanut hairy root culture.
Supplemental Figure S6. Comparison of stilbenoid yields from the culture medium and root tissue.
Supplemental Figure S7. Colors of resveratrol degradation reaction mixtures at various incubation time points.
Supplemental Figure S8. Time course of resveratrol prenyltansferase assay without DTT and subsequent degradation of resveratrol and arachidin-2 in the reaction.
Supplemental Figure S9. Time course of prenyltransferase activity in crude cell-free extract of peanut hairy roots upon treatment with 100 μm MeJA and 9 g L−1 CD.
Supplemental Figure S10. Accumulation of prenylated stilbenoids, arachidin-1, arachidin-3, arachidin-5, and arachidin-2, in the medium of peanut hairy root culture upon treatment with 100 μm MeJA and 9 g L−1 CD.
Supplemental Figure S11. Biotransformation of arachidin-2 by protein extracts from elicited peanut hairy root culture.
Supplemental Figure S12. Resveratrol prenyltransferase activity.
Supplemental Figure S13. pH dependency of resveratrol prenyltransferase activity.
Supplemental Figure S14. Divalent cation requirement for resveratrol prenyltransferase activity.
Supplemental Figure S15. Biochemical characterization of resveratrol prenyltransferase in microsomal fraction of elicited peanut hairy root.
Supplemental Figure S16. Effects of resveratrol and DMAPP concentrations on resveratrol prenyltransferase activity.
Supplemental Figure S17. Substrate specificity of resveratrol prenyltransferase in microsomal fraction of elicited peanut hairy root.
Supplemental Figure S18. Biotransformation of arachidin-5 by protein fractions from elicited peanut hairy root culture.
Supplemental Figure S19. Substrate specificity of resveratrol prenyltransferase in microsomal fraction of elicited peanut hairy root.
Supplemental Table S1. Protein sequences used to perform BLAST analysis against Arachis spp. genomic and transcriptomic databases and their accession numbers.
Supplemental Table S2. Alignment results of various flavonoid prenyltransferases with translated CDS of A. duranensis and A. ipaensis gene models.
Supplemental Table S3. Available transcriptome databases of Arachis spp.
Supplemental Table S4. Alignment results of various flavonoid prenyltransferases with translated transcripts of A. duranensis, A. ipaensis, and peanut.
Supplemental Table S5. Chloroplast transit peptide predictions of sequences identified from Arachis spp. transcripts.
Supplementary Material
Glossary
- MVA
mevalonic acid
- MEP
2-C-methyl-d-erythritol-4-phosphate
- DMAPP
dimethylallyl pyrophosphate
- MeJA
methyl jasmonate
- CD
cyclodextrin
- IPP
isopentenyl diphosphate
- CDS
coding sequence
- HPCCC
high-performance counter current chromatography
Footnotes
This work was supported by the U.S. Department of Agriculture-NIFA (grant no. 2014–67014–21701), the National Science Foundation-EPSCoR (grant no. EPS 0701890, Center for Plant-Powered Production), the Arkansas ASSET Initiative, the Arkansas Science and Technology Authority, and the Arkansas Biosciences Institute.
Articles can be viewed without a subscription.
References
- Abbott JA, Medina-Bolivar F, Martin EM, Engelberth AS, Villagarcia H, Clausen EC, Carrier DJ (2010) Purification of resveratrol, arachidin-1, and arachidin-3 from hairy root cultures of peanut (Arachis hypogaea) and determination of their antioxidant activity and cytotoxicity. Biotechnol Prog 26: 1344–1351 [DOI] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Akashi T, Sasaki K, Aoki T, Ayabe S, Yazaki K (2009) Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol 149: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EKS, Liu X, Gao D, Clevenger J, Dash S, et al. (2016) The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet 48: 438–446 [DOI] [PubMed] [Google Scholar]
- Boonlaksiri C, Oonanant W, Kongsaeree P, Kittakoop P, Tanticharoen M, Thebtaranonth Y (2000) An antimalarial stilbene from Artocarpus integer. Phytochemistry 54: 415–417 [DOI] [PubMed] [Google Scholar]
- Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem 12: 717–739 [DOI] [PubMed] [Google Scholar]
- Brents LK, Medina-Bolivar F, Seely KA, Nair V, Bratton SM, Ñopo-Olazabal L, Patel RY, Liu H, Doerksen RJ, Prather PL, et al. (2012) Natural prenylated resveratrol analogs arachidin-1 and -3 demonstrate improved glucuronidation profiles and have affinity for cannabinoid receptors. Xenobiotica 42: 139–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Lai YH, Djoko B, Wu PL, Liu CD, Liu YW, Chiou RYY (2006) Biosynthesis enhancement and antioxidant and anti-inflammatory activities of peanut (Arachis hypogaea L.) arachidin-1, arachidin-3, and isopentadienylresveratrol. J Agric Food Chem 54: 10281–10287 [DOI] [PubMed] [Google Scholar]
- Chen R, Liu X, Zou J, Yin Y, Ou B, Li J, Wang R, Xie D, Zhang P, Dai J (2013) Regio- and stereospecific prenylation of flavonoids by Sophora flavescens prenyltransferase. Adv Synth Catal 355: 1817–1828 [Google Scholar]
- Chong J, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177: 143–155 [Google Scholar]
- Condori J, Sivakumar G, Hubstenberger J, Dolan MC, Sobolev VS, Medina-Bolivar F (2010) Induced biosynthesis of resveratrol and the prenylated stilbenoids arachidin-1 and arachidin-3 in hairy root cultures of peanut: effects of culture medium and growth stage. Plant Physiol Biochem 48: 310–318 [DOI] [PubMed] [Google Scholar]
- Das S, Das DK (2007) Anti-inflammatory responses of resveratrol. Inflamm Allergy Drug Targets 6: 168–173 [DOI] [PubMed] [Google Scholar]
- Djoko B, Chiou RYY, Shee JJ, Liu YW (2007) Characterization of immunological activities of peanut stilbenoids, arachidin-1, piceatannol, and resveratrol on lipopolysaccharide-induced inflammation of RAW 264.7 macrophages. J Agric Food Chem 55: 2376–2383 [DOI] [PubMed] [Google Scholar]
- Gambini J, Inglés M, Olaso G, Lopez-Grueso R, Bonet-Costa V, Gimeno-Mallench L, Mas-Bargues C, Abdelaziz KM, Gomez-Cabrera MC, Vina J, et al. (2015) Properties of resveratrol: in vitro and in vivo studies about metabolism, bioavailability, and biological effects in animal models and humans. Oxid Med Cell Longev 2015: 837042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim EH, Ulinnuha UZ, Syah YM, Ghisalberti EL (2002) Artoindonesianins N and O, new prenylated stilbene and prenylated arylbenzofuran derivatives from Artocarpus gomezianus. Fitoterapia 73: 597–603 [DOI] [PubMed] [Google Scholar]
- Han M, Heppel SC, Su T, Bogs J, Zu Y, An Z, Rausch T (2013) Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS ONE 8: e62467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CP, Au LC, Chiou RYY, Chung PC, Chen SY, Tang WC, Chang CL, Fang WH, Lin SB (2010) Arachidin-1, a peanut stilbenoid, induces programmed cell death in human leukemia HL-60 cells. J Agric Food Chem 58: 12123–12129 [DOI] [PubMed] [Google Scholar]
- Ioset JR, Marston A, Gupta MP, Hostettmann K (2001) Five new prenylated stilbenes from the root bark of Lonchocarpus chiricanus. J Nat Prod 64: 710–715 [DOI] [PubMed] [Google Scholar]
- Li J, Chen R, Wang R, Liu X, Xie D, Zou J, Dai J (2014) GuA6DT, a regiospecific prenyltransferase from Glycyrrhiza uralensis, catalyzes the 6-prenylation of flavones. ChemBioChem 15: 1673–1681 [DOI] [PubMed] [Google Scholar]
- Lohr M, Schwender J, Polle JE (2012) Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae. Plant Sci 185-186: 9–22 [DOI] [PubMed] [Google Scholar]
- Marsh Z, Yang T, Nopo-Olazabal L, Wu S, Ingle T, Joshee N, Medina-Bolivar F (2014) Effect of light, methyl jasmonate and cyclodextrin on production of phenolic compounds in hairy root cultures of Scutellaria lateriflora. Phytochemistry 107: 50–60 [DOI] [PubMed] [Google Scholar]
- Medina-Bolivar F, Condori J, Rimando AM, Hubstenberger J, Shelton K, O’Keefe SF, Bennett S, Dolan MC (2007) Production and secretion of resveratrol in hairy root cultures of peanut. Phytochemistry 68: 1992–2003 [DOI] [PubMed] [Google Scholar]
- Park BH, Lee HJ, Lee YR (2011) Total synthesis of chiricanine A, arahypin-1, trans-arachidin-2, trans-arachidin-3, and arahypin-5 from peanut seeds. J Nat Prod 74: 644–649 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Mito K, Ohara K, Yamamoto H, Yazaki K (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol 146: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Tsurumaru Y, Yamamoto H, Yazaki K (2011) Molecular characterization of a membrane-bound prenyltransferase specific for isoflavone from Sophora flavescens. J Biol Chem 286: 24125–24134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöppner A, Kindl H (1984) Purification and properties of a stilbene synthase from induced cell suspension cultures of peanut. J Biol Chem 259: 6806–6811 [PubMed] [Google Scholar]
- Shen G, Huhman D, Lei Z, Snyder J, Sumner LW, Dixon RA (2012) Characterization of an isoflavonoid-specific prenyltransferase from Lupinus albus. Plant Physiol 159: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev VS. (2008) Localized production of phytoalexins by peanut (Arachis hypogaea) kernels in response to invasion by Aspergillus species. J Agric Food Chem 56: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Sobolev VS. (2013) Production of phytoalexins in peanut (Arachis hypogaea) seed elicited by selected microorganisms. J Agric Food Chem 61: 1850–1858 [DOI] [PubMed] [Google Scholar]
- Sobolev VS, Khan SI, Tabanca N, Wedge DE, Manly SP, Cutler SJ, Coy MR, Becnel JJ, Neff SA, Gloer JB (2011) Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J Agric Food Chem 59: 1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev VS, Krausert NM, Gloer JB (2016) New monomeric stilbenoids from peanut (Arachis hypogaea) seeds challenged by an Aspergillus flavus strain. J Agric Food Chem 64: 579–584 [DOI] [PubMed] [Google Scholar]
- Sobolev VS, Neff SA, Gloer JB (2009) New stilbenoids from peanut (Arachis hypogaea) seeds challenged by an Aspergillus caelatus strain. J Agric Food Chem 57: 62–68 [DOI] [PubMed] [Google Scholar]
- Sobolev VS, Potter TL, Horn BW (2006) Prenylated stilbenes from peanut root mucilage. Phytochem Anal 17: 312–322 [DOI] [PubMed] [Google Scholar]
- Su BN, Cuendet M, Hawthorne ME, Kardono LBS, Riswan S, Fong HHS, Mehta RG, Pezzuto JM, Kinghorn AD (2002) Constituents of the bark and twigs of Artocarpus dadah with cyclooxygenase inhibitory activity. J Nat Prod 65: 163–169 [DOI] [PubMed] [Google Scholar]
- Tomé-Carneiro J, Larrosa M, González-Sarrías A, Tomás-Barberán FA, García-Conesa MT, Espín JC (2013) Resveratrol and clinical trials: the crossroad from in vitro studies to human evidence. Curr Pharm Des 19: 6064–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G (1994) Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol 38: 610–618 [DOI] [PubMed] [Google Scholar]
- van der Kaaden JE, Hemscheidt TK, Mooberry SL (2001) Mappain, a new cytotoxic prenylated stilbene from Macaranga mappa. J Nat Prod 64: 103–105 [DOI] [PubMed] [Google Scholar]
- Watts KT, Lee PC, Schmidt-Dannert C (2006) Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Song L, Huang D (2011) Food grade fungal stress on germinating peanut seeds induced phytoalexins and enhanced polyphenolic antioxidants. J Agric Food Chem 59: 5993–6003 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Senda M, Inoue K (2000) Flavanone 8-dimethylallyltransferase in Sophora flavescens cell suspension cultures. Phytochemistry 54: 649–655 [DOI] [PubMed] [Google Scholar]
- Yang T, Fang L, Nopo-Olazabal C, Condori J, Nopo-Olazabal L, Balmaceda C, Medina-Bolivar F (2015a) Enhanced production of resveratrol, piceatannol, arachidin-1, and arachidin-3 in hairy root cultures of peanut co-treated with methyl jasmonate and cyclodextrin. J Agric Food Chem 63: 3942–3950 [DOI] [PubMed] [Google Scholar]
- Yang X, Jiang Y, Yang J, He J, Sun J, Chen F, Zhang M, Yang B (2015b) Prenylated flavonoids, promising nutraceuticals with impressive biological activities. Trends Food Sci Technol 44: 93–104 [Google Scholar]
- Yazaki K, Sasaki K, Tsurumaru Y (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70: 1739–1745 [DOI] [PubMed] [Google Scholar]
- Yoder BJ, Cao S, Norris A, Miller JS, Ratovoson F, Razafitsalama J, Andriantsiferana R, Rasamison VE, Kingston DGI (2007) Antiproliferative prenylated stilbenes and flavonoids from Macaranga alnifolia from the Madagascar rainforest. J Nat Prod 70: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Wang L, Liu L, Liang Y, Sun Y, Wu J (2014) Both the mevalonate and the non-mevalonate pathways are involved in ginsenoside biosynthesis. Plant Cell Rep 33: 393–400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.