Production of plant lysine-derived alkaloids originates with the convergent evolution of lysine decarboxylase.
Abstract
Lycopodium alkaloids (LAs) are derived from lysine (Lys) and are found mainly in Huperziaceae and Lycopodiaceae. LAs are potentially useful against Alzheimer’s disease, schizophrenia, and myasthenia gravis. Here, we cloned the bifunctional lysine/ornithine decarboxylase (L/ODC), the first gene involved in LA biosynthesis, from the LA-producing plants Lycopodium clavatum and Huperzia serrata. We describe the in vitro and in vivo functional characterization of the L. clavatum L/ODC (LcL/ODC). The recombinant LcL/ODC preferentially catalyzed the decarboxylation of l-Lys over l-ornithine (l-Orn) by about 5 times. Transient expression of LcL/ODC fused with the amino or carboxyl terminus of green fluorescent protein, in onion (Allium cepa) epidermal cells and Nicotiana benthamiana leaves, showed LcL/ODC localization in the cytosol. Transgenic tobacco (Nicotiana tabacum) hairy roots and Arabidopsis (Arabidopsis thaliana) plants expressing LcL/ODC enhanced the production of a Lys-derived alkaloid, anabasine, and cadaverine, respectively, thus, confirming the function of LcL/ODC in plants. In addition, we present an example of the convergent evolution of plant Lys decarboxylase that resulted in the production of Lys-derived alkaloids in Leguminosae (legumes) and Lycopodiaceae (clubmosses). This convergent evolution event probably occurred via the promiscuous functions of the ancestral Orn decarboxylase, which is an enzyme involved in the primary metabolism of polyamine. The positive selection sites were detected by statistical analyses using phylogenetic trees and were confirmed by site-directed mutagenesis, suggesting the importance of those sites in granting the promiscuous function to Lys decarboxylase while retaining the ancestral Orn decarboxylase function. This study contributes to a better understanding of LA biosynthesis and the molecular evolution of plant Lys decarboxylase.
Since plants are sessile organisms, they produce a diverse range of defense chemicals, known as specialized metabolites, that contribute to the adaptation to their ecological niches (Pichersky and Lewinsohn, 2011). Chemical compounds are important for plants, as they can serve as attractants for insect pollinators or as defense against pathogens and herbivores (Pichersky and Gang, 2000). Many plant species have been used in traditional medicines for the treatment of various human diseases (Tang and Eisenbrand, 1992). Almost one-fourth of modern medicines are derived from natural sources (De Luca et al., 2012). Alkaloids are one of the most important specialized metabolites and are mostly derived from amino acids. Alkaloids display a vast variety of biological activities, and many of them are currently used for clinical purposes; examples include morphine as an analgesic, artemisinin as an antimalarial, and camptothecin as an antineoplastic (De Luca et al., 2012).
Lycopodium alkaloids (LAs) are Lys-derived alkaloids that have quinolizine or pyridine and α-pyridine nuclei in their structures (Ma and Gang, 2004). LAs have been isolated primarily from the genera Lycopodium and Huperzia, which are clubmosses (Ma and Gang, 2004). Whole plants from the families Huperziaceae and Lycopodiaceae have been used in Chinese folk medicine for the treatment of various symptoms (Ma et al., 2007). Huperzia serrata produces huperzine A (HupA), a promising candidate drug for the treatment of Alzheimer’s disease, owing to its function as a potent acetylcholinesterase inhibitor (Wang et al., 2009; Qian and Ke, 2014). HupA and its derivative ZT-1 have been evaluated in clinical trials for the treatment of Alzheimer’s disease (Ma et al., 2007; Jia et al., 2013).
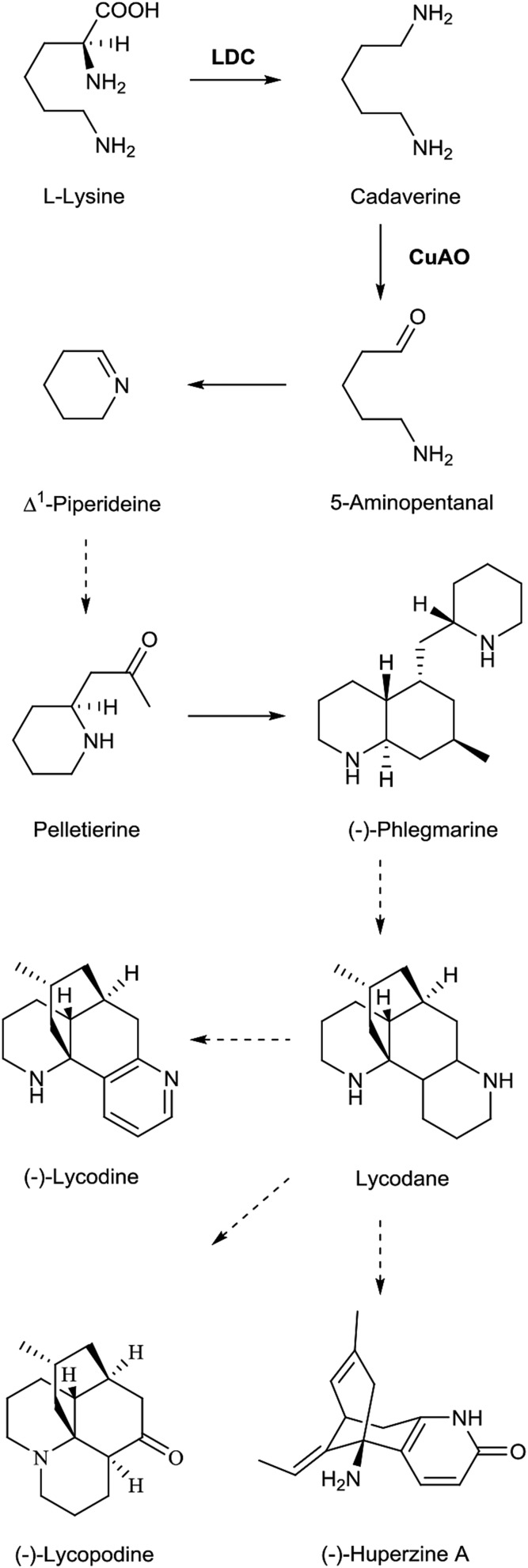
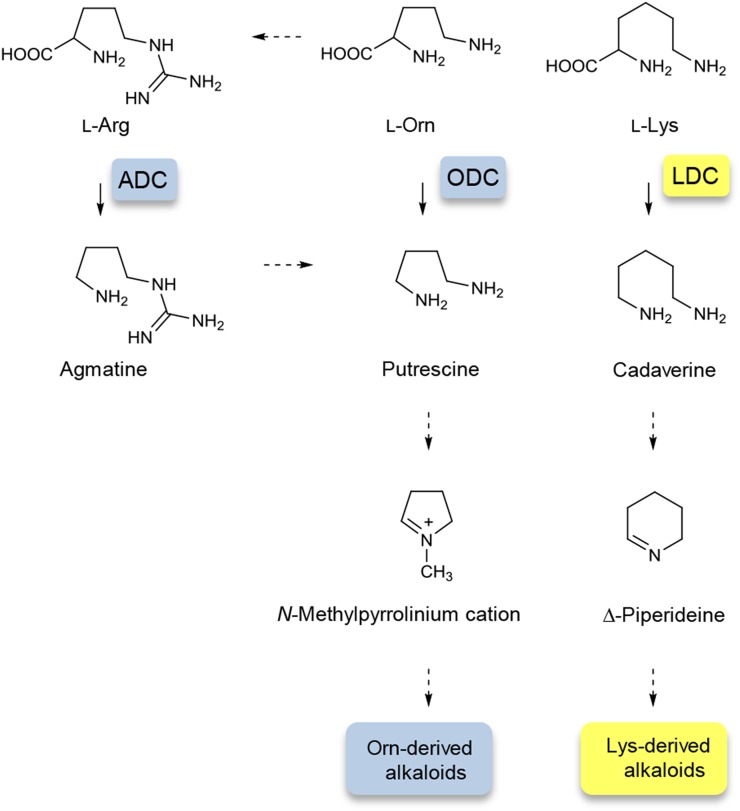
Owing to the difficulties in cultivation and in vitro propagation, the biosynthetic pathways for LAs are not well documented and have been proposed based on tracer experiments using labeled precursors and plants in their natural habitats (Ma and Gang, 2004, and refs. therein). Lysine decarboxylase (LDC) has been proposed as the entry-point enzyme in the LA biosynthetic pathway, which catalyzes the decarboxylation of Lys to yield cadaverine (Fig. 1). Cadaverine is then catalyzed by CuAO to produce 5-aminopentanal, which is spontaneously cyclized to the first intermediate for LA production, Δ1-piperideine (Ma and Gang, 2004). Based on analyses of the EST data from LA-producing plants, several candidate genes for LA biosynthesis have been proposed; however, no further investigation has been performed (Luo et al., 2010a, 2010b). Recently, the CuAO gene from H. serrata was cloned and characterized, using degenerate primers based on the conserved sequences of the known plant CuAO enzymes; however, the cloned CuAO showed a broad substrate specificity (Sun et al., 2012).
Figure 1.
Putative biosynthetic pathway for LAs. Dotted arrows indicate more than one catalytic conversion. CuAO, Copper amine oxidase.
Recently, we showed that bifunctional lysine/ornithine decarboxylases (L/ODCs) in the Lys-derived quinolizidine alkaloid (QA)-producing legumes were recruited by the ubiquitous enzyme ornithine decarboxylase (ODC; Bunsupa et al., 2012a). ODC catalyzes the decarboxylation of l-Orn to yield putrescine, which is the main precursor for the production of Orn-derived alkaloids. In plant cells, putrescine and its derivative polyamines, spermidine and spermine, are essential for a wide range of biological processes during plant growth and development (Fuell et al., 2010). In addition to its role in alkaloid biosynthesis, cadaverine has been implicated as a growth regulator and stress-response compound in several plant species (Tomar et al., 2013).
In this study, in order to elucidate the biosynthetic pathway of LAs and the evolution of plant LDC, we cloned L/ODC from Lycopodium clavatum and H. serrata. We provide results from both in vitro and in vivo experiments to confirm the functions of L/ODC in L. clavatum. Using the tests for positive selection and assays of enzyme function, we then show the convergent evolution of plant LDC in the Lys-derived alkaloid-producing plants. Furthermore, we were able to detect the substitution site that is under positive selection and is important for improving the LDC function.
RESULTS
Cloning of LDC from LA-Producing Plants
To identify the LDC-encoding cDNAs in L. clavatum and H. serrata, we used degenerate primers based on the sequence homology between the L/ODCs and other plant ODCs (Supplemental Fig. S1). The full-length cDNA clones of L. clavatum and H. serrata L/ODCs (hereafter referred to as LcL/ODC and HsL/ODC, respectively) were obtained using 5′- and 3′-RACE. The LcL/ODC contained a 1,500-bp open reading frame (ORF), encoding 500 amino acids. Two homologs of L/ODC from H. serrata, namely HsL/ODC1 and HsL/ODC2, were obtained. HsL/ODC1 and HsL/ODC2 contained 1,521- and 1,527-bp ORFs, encoding 507 and 509 amino acids, respectively. The deduced amino acid sequences of LcL/ODC, HsL/ODC1, and HsL/ODC2 were highly similar to one another (82% identity between LcL/ODC and HsL/ODCs, and 97% identity between HsL/ODC1 and HsL/ODC2). Lower sequence identities of about 55% with other plant L/ODCs and ODCs were observed. Sequence alignment of LcL/ODC with other eukaryotic ODCs and L/ODCs revealed that all amino acid residues responsible for substrate binding were completely conserved (Supplemental Fig. S1). The amino acid residue at position 344 of the narrow-leafed lupin (Lupinus angustifolius) L/ODC (LaL/ODC) was Phe. This Phe-344 residue is critical for enzymatic activities of both LDC and ODC in LaL/ODC (Bunsupa et al., 2012a). Interestingly, this position in LcL/ODC (position 374), HsL/ODC1 (position 379), and HsL/ODC2 (position 377) is Tyr (Supplemental Fig. S1). For comparison, we also cloned the partial sequence of L/ODC from Thermopsis lupinoides (TlL/ODC), which produces QAs. As expected, TlL/ODC had Phe at this position.
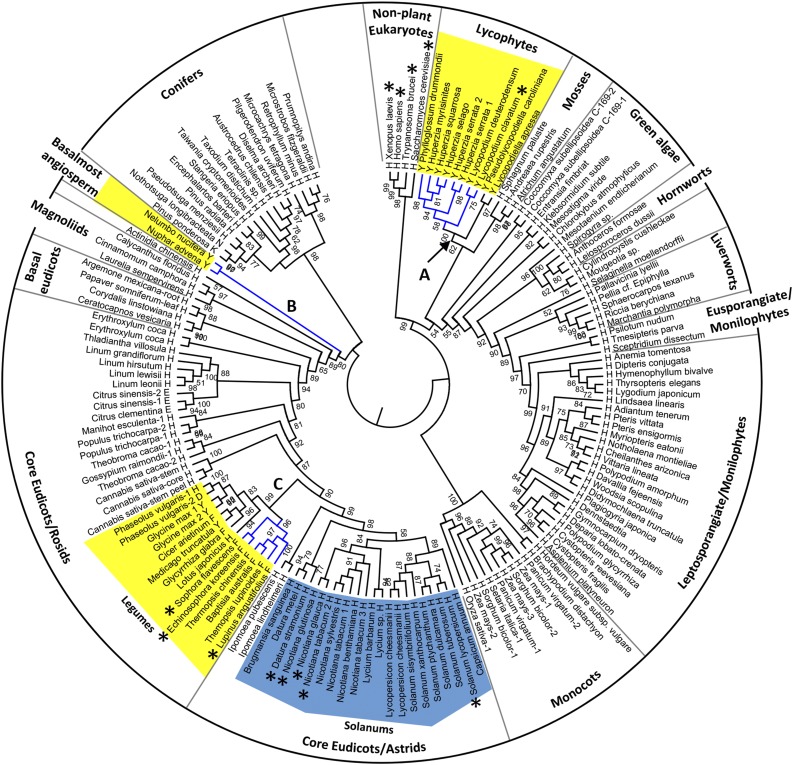
Phylogenetic analysis of the eukaryotic ODCs and LDCs provided good support for a monophyletic origin of the sequences belonging to their families (Fig. 2). LcL/ODC, HsL/ODC1, and HsL/ODC2 formed a clade that was distant from the Leguminosae L/ODCs, indicating a convergent evolution of the Lys-derived alkaloid production in distinct plant lineages.
Figure 2.
Rooted phylogenetic tree of ODC and LDC amino acid sequences from eukaryotes. From an alignment of highly conserved amino acids without gaps built using MEGA version 6 (Supplemental Data S1), the phylogenetic tree was constructed by PhyML3.0 using the best-fit mode. The divergence node derived from nonplant eukaryote genes is defined to be the root of the phylogenetic tree. Asterisks represent enzymes whose biochemical properties have been investigated. The blue branch lines indicate the Lys-derived alkaloid-producing plants. Uppercase letters next to the taxa represent the amino acid at position 344 (LaL/ODC numbering). The bootstrap values (1,000 replicates) are shown. Letters A, B, and C indicate the branches that are likely to be under positive selection for the production of Lys-derived alkaloids in plants: LAs (branch A), nuphar alkaloids (branch B), and QAs (branch C). Bootstrap values greater than 50% are shown. The accession numbers of the enzymes are listed in Supplemental Table S3.
In Vitro Activity Assays of Recombinant LcL/ODC Protein
To determine the biochemical functions of the identified sequences, the ORFs of LcL/ODC and HsL/ODC1 were heterologously expressed in Escherichia coli, which were then affinity purified and assayed for LDC and ODC activities. However, we were unable to purify the recombinant HsL/ODC1 because of its insoluble nature. A molecular mass of 54 kD, in good agreement with the predicted 54.21 kD, was observed upon SDS-PAGE of the tag-purified/cleaved LcL/ODC protein (Supplemental Fig. S2). This purified recombinant protein was used to test both LDC and ODC activities, at optimal pH values of 8 and 7, respectively. LcL/ODC exhibited both LDC and ODC activities to similar extents and at the same order of magnitude as the L/ODCs characterized previously from QA-producing plants (Table I). The kcat values were calculated as 3.17 and 2.13 s−1 for l-Lys and l-Orn, respectively, while the Km values were 1.69 and 5.48 mm for l-Lys and l-Orn, respectively. LcL/ODC preferentially catalyzed the decarboxylation of l-Lys over l-Orn by about 5 times the catalytic efficiency (kcat/Km).
Table I. Kinetic parameters of L/ODC and its mutant proteins.
All experiments were performed in 50 mm potassium phosphate buffer (at optimal pH for each enzyme). Kinetic parameters were calculated from mean values (n = 3–4). ND, Not detected.
| Protein |
Km |
Vmax |
kcat |
kcat/Km |
LDC/ODC Ratio of kcat/Km | ||||
|---|---|---|---|---|---|---|---|---|---|
| LDC | ODC | LDC | ODC | LDC | ODC | LDC | ODC | ||
| mm | nmol min−1μg−1 | s−1 | m−1 s−1 | ||||||
| LcL/ODC-wild type | 1.69 | 5.48 | 3.65 | 2.46 | 3.17 | 2.13 | 1,878 | 388 | 4.84 |
| LcL/ODC-Y344H | 22.39 | 8.21 | 0.51 | 2.47 | 0.44 | 2.14 | 20 | 261 | 0.08 |
| NtODC3-wild type | ND | 1.44 | ND | 30.30 | ND | 23.54 | ND | 16,351 | ND |
| NtODC3-H344Y | ND | 0.75 | ND | 14.33 | ND | 11.33 | ND | 14,794 | ND |
| NtODC3-H344F | ND | 0.61 | ND | 3.42 | ND | 2.66 | ND | 4,368 | ND |
A competition assay, performed by varying the concentration of l-Lys in the presence and absence of l-Orn and vice versa, showed a competitive reaction pattern (Supplemental Fig. S3, A and B). The inhibitor assay, using α-difluoromethyl-Orn, an ODC suicide inhibitor, showed a dose-dependent inhibition of both LDC and ODC activities (Supplemental Fig. S3, C and D). These results suggest that the catalytic sites of LcL/ODC were identical in l-Orn and l-Lys and similar to that of previously studied L/ODCs (Bunsupa et al., 2012a).
Overexpression of LcL/ODC in Tobacco Hairy Roots Significantly Increases Anabasine Biosynthesis
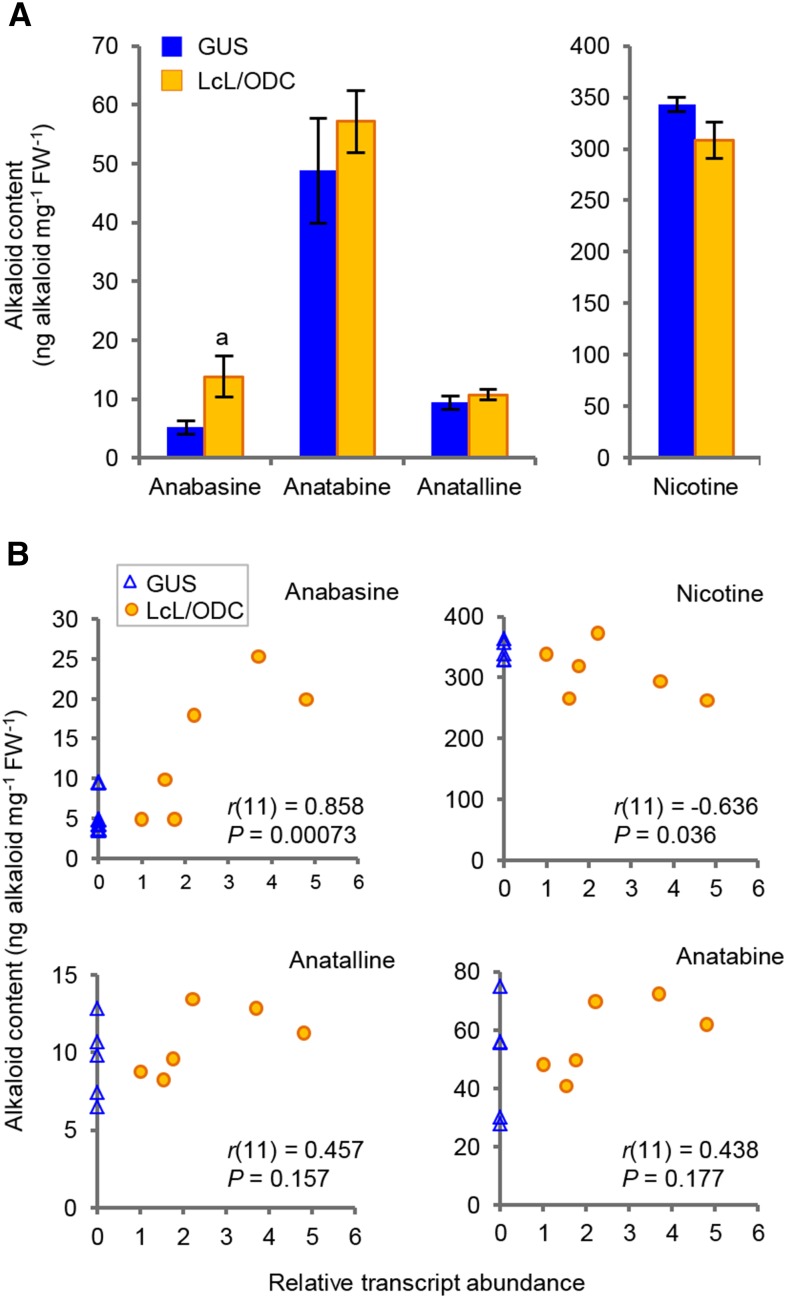
To show that LcL/ODC functions as an LDC for alkaloid biosynthesis, LcL/ODC was expressed under the control of the constitutive cauliflower mosaic virus 35S promoter in tobacco (Nicotiana tabacum) hairy roots as well as a control GUS. The expression of LcL/ODC transcript was confirmed using quantitative PCR. The alkaloid levels in the transgenic tobacco lines were analyzed using HPLC-photodiode array detection and liquid chromatography-mass spectrometry. The levels of anabasine, a Lys-derived alkaloid, in the LcL/ODC-transformed tobacco hairy roots increased significantly, showing an average 2.7-fold increase (P < 0.05). In contrast, the levels of other tobacco alkaloids did not change significantly compared with the control lines (P > 0.05; Fig. 3A).
Figure 3.
Major alkaloid levels and correlations between the relative abundance of LcL/ODC transcript and the alkaloid levels in tobacco hairy roots overexpressing LcL/ODC. A, Abundance of tobacco alkaloids in six and five independent hairy roots for LcL/ODC- and GUS-overexpressing lines, respectively (biological replicates, n = 4 for each line). Values are means ± se. Lowercase a indicates that the mean value is statistically different from the corresponding GUS control, based on a one-tailed Student’s t test: P < 0.05. B, Correlations between each of the tobacco alkaloids and the relative LcL/ODC transcripts in tobacco hairy roots overexpressing LcL/ODC (orange circles) and GUS (blue triangles) lines. Pearson correlation coefficients (r) with the number of tested samples in parentheses and the corresponding P values are shown. FW, Fresh weight.
Comparison of the LcL/ODC gene transcript levels and the tobacco alkaloid contents revealed a significant positive correlation between the LcL/ODC transcript levels and anabasine accumulation (Pearson’s correlation coefficient [r] = 0.858, P < 0.001; Fig. 3B). A significant negative correlation between the LcL/ODC transcript levels and the levels of nicotine, an Orn-derived alkaloid, was found (r = −0.636, P < 0.05; Fig. 3B). There was no significant correlation between the LcL/ODC transcript levels and the levels of other alkaloids (Fig. 3B).
Transgenic Arabidopsis Plants Expressing LcL/ODC Showed a Significant Increase in Cadaverine Production
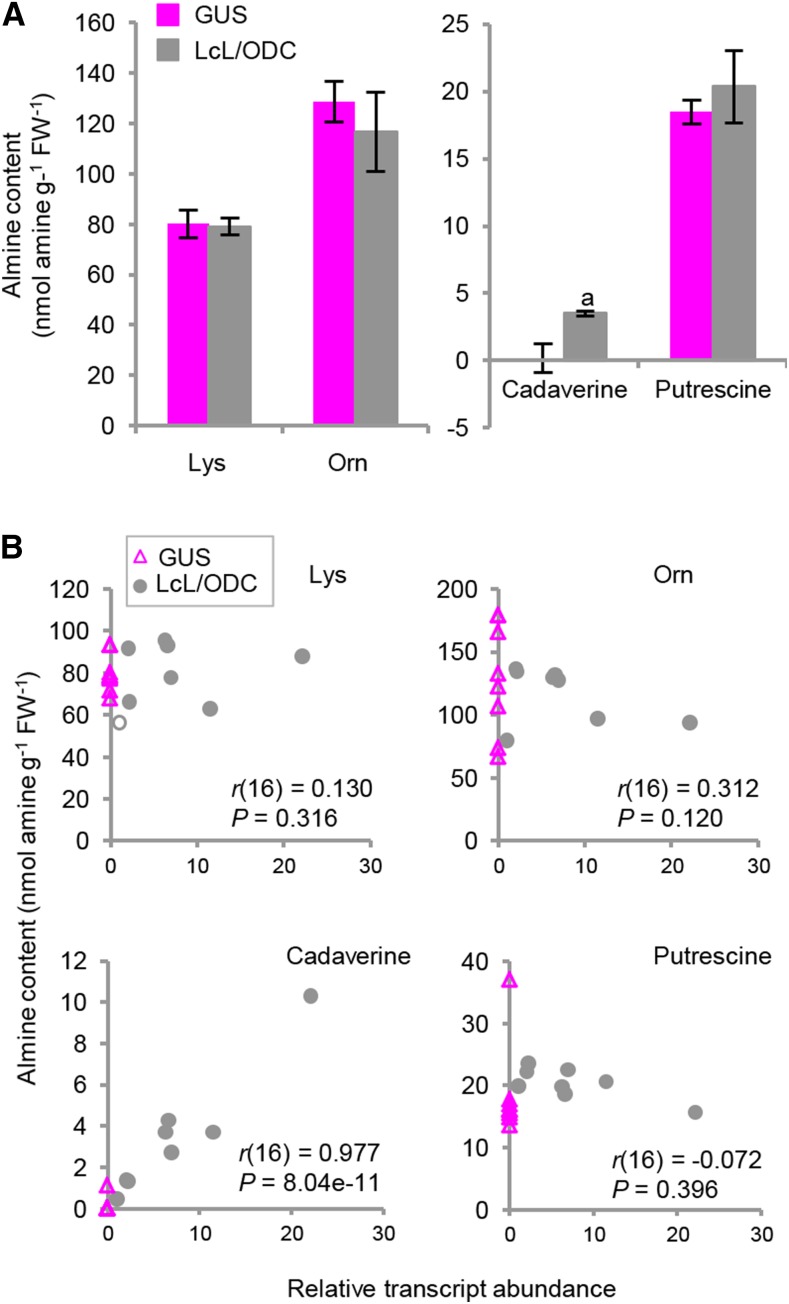
The levels of amines, including l-Lys, l-Orn, cadaverine, and putrescine, in the LcL/ODC- and control (GUS)-transformed Arabidopsis (Arabidopsis thaliana) plants were analyzed by capillary electrophoresis-mass spectrometry. The LcL/ODC-expressing Arabidopsis plants displayed significantly increased levels of cadaverine, which were, on average, 22-fold higher (P < 0.01) compared with the control plants. In contrast, l-Lys, l-Orn, and putrescine levels did not change significantly (P > 0.05; Fig. 4A). Only the cadaverine levels showed a significant positive correlation with the LcL/ODC transcript levels (r = 0.977, P < 0.001; Fig. 4B).
Figure 4.
Amine levels and correlations between the relative abundance of LcL/ODC transcript and the amine levels in Arabidopsis plants expressing LcL/ODC. A, Abundance of amines in pooled samples (six to eight plants) of Arabidopsis plants expressing LcL/ODC or GUS, with eight independent lines for each. Values are means ± se. Lowercase a indicates that the mean value is statistically different from the corresponding GUS control, based on a one-tailed Student’s t test: P < 0.01. B, Correlations between each of the amines and the relative LcL/ODC transcripts in tobacco hairy roots overexpressing LcL/ODC (gray circles) and GUS (magenta triangles) lines. Pearson correlation coefficients (r) with the number of tested samples in parentheses and P values are shown. FW, Fresh weight.
Localization of LcL/ODC Protein
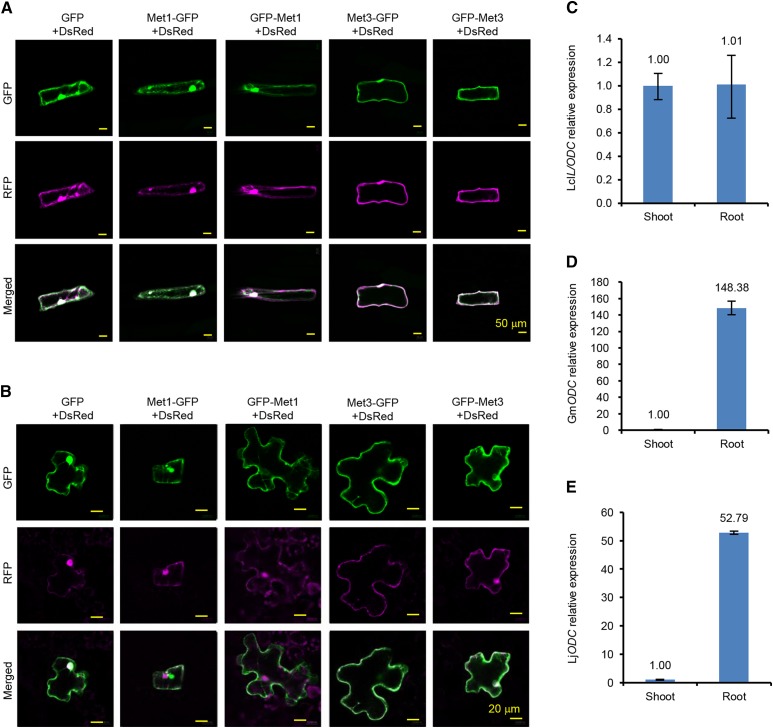
The analysis of LcL/ODC nucleotide sequence indicated alternative translational initiation sites, 1AUG (LcL/ODC-Met-1) and 47AUG (LcL/ODC-Met-3; http://www.cbs.dtu.dk/services/NetStart/). The iPSORT program predicted that LcL/ODC-Met-3 has a chloroplast transit peptide (http://ipsort.hgc.jp/).
In order to determine the subcellular localization sites of the alternatively translated products of LcL/ODC, the full-length (LcL/ODC-Met-1) and truncated (LcL/ODC-Met-3) sequences of LcL/ODC were fused to GFP at either the N or the C terminus under the control of the 35S cauliflower mosaic virus promoter. As a control, a vector for the expression of only GFP and red fluorescent protein (RFP) from Discosoma sp. (DsRed) was used for cytosolic localization. Each resulting construct was expressed simultaneously with DsRed in onion (Allium cepa) epidermal cells and Nicotiana benthamiana leaves using particle gun bombardment. The overlay of the green and red fluorescent images for all constructs localized the detected signal to the cytosol in both onion epidermal cells and N. benthamiana leaves (Fig. 5, A and B). These localization patterns were identical to the cytosol localization references.
Figure 5.
Subcellular localization of LcL/ODC fused with GFP in onion epidermal cells and N. benthamiana leaves, and the relative abundance of LcL/ODC, GmODC, and LjODC transcript levels. A and B, LcL/ODC from the first (Met-1) and the third (Met-3) start codons were fused with GFP at the N-terminal (GFP-Met-1 and GFP-Met-3) or C-terminal (Met-1-GFP and Met-3-GFP) end. The resultant constructs were simultaneously and transiently expressed with Discosoma sp. (DsRed). RFP from DsRed and GFP were used as references for cytosolic localization. GFP (green; top row), RFP (red; middle row), and merged (green and red; bottom row) fluorescence observed in onion epidermal cells (A) and N. benthamiana leaves (B) expressing the indicated target constructs are shown. Bars = 50 μm and 20 μm for the onion epidermal cells (A) and the N. benthamiana leaves (B), respectively. C to E, Quantitative real-time PCR analysis of ODC or L/ODC transcript levels in the shoots and roots of L. clavatum (C), soybean (D), and L. japonicus (E). Values shown are means ± sd of analytical replicates; n = 3 to 4.
L/ODC and ODC Transcript Levels and Metabolite Profiles of Alkaloid-Producing and Nonproducing Plants
To determine the tissues where LcL/ODC is expressed, quantitative real-time PCR was performed with the shoots and roots of L. clavatum, and the transcript levels in the roots were normalized to that of the shoots. LcL/ODC expression levels were similar for both the tested organs (Fig. 5C).
In order to investigate the metabolite profiles and the gene expression patterns of plant L/ODCs and ODCs, we determined the metabolite profiles of L. clavatum and H. serrata. In addition, we assessed the transcript levels and metabolite profiles of two alkaloid-free legumes: soybean (Glycine max) and Lotus japonicus. In contrast with the transcript levels of LcL/ODC, which were expressed equally in the shoots and the roots, soybean ODC2 (GmODC2) and L. japonicus ODC (LjODC) transcripts were expressed at higher levels in the roots (Fig. 5, D and E). l-Lys and l-Orn were found mainly in the shoots of the tested plants. On the other hand, cadaverine was detected only in soybean, mainly in the roots (Supplemental Tables S1 and S2). LAs, such as lycodine and HupA, were higher in the shoots than in the roots (Supplemental Table S1).
Evolutionary Analyses of Plant ODCs and L/ODCs Detect a Positive Selection Site at Amino Acid Position 344
There were three evolutionary events that led to the production of Lys-derived alkaloids in plants: LAs (branch A), nuphar alkaloids (branch B), and QAs (branch C; Fig. 2). If these events were advantageous, the branches representing them (branches A, B, and C) would likely be under positive selection (Fig. 2). To examine whether these branches were positively selected, we first performed a codon site test. However, there were no positive selection sites found (Table II). Since the positively selected site(s) might be found in only the three evolutionary events that led to LAs in plants (Fig. 2), we simultaneously performed the branch-site test by selecting the branches A, B, and C as the foreground and the other branches as the background (Bielawski and Yang, 2005; Zhang et al., 2005). The ratio of nonsynonymous (amino acid replacing) substitution rate (Ka) over the synonymous (silent) substitution rate (Ks; ω = Ka/Ks) in the foreground (branches A, B, and C) was 1.4 (Table II). We used the likelihood ratio test (LRT) to test the statistical significance of the detection of positive selection (Zhang et al., 2005). The LRTs for positive selection in the selected foreground branches yielded statistically significant results (P < 0.05, χ2 test, degrees of freedom = 1; Table II). In the three branches, two amino acid residues, 112 and 344, were positively selected, as shown using the Bayes empirical Bayes method (posterior probability > 0.95; Table II; Bielawski and Yang, 2005).
Table II. Molecular evolutionary analysis of eukaryotic ODCs and LDCs.
| Model | ts/tva | No. of Genes | No. of Codon Sites | ωb in the Background Branches | ωb in the Foreground Branches | Log Likelihood | P | Positively Selected Sitesc |
|---|---|---|---|---|---|---|---|---|
| Branch-site model | 1.764 | 156 | 675 | 0.094 | 1 | −48,716.1203 | Not applicable | |
| 1.763 | 156 | 675 | 0.094 | 1.43 | −48,714.1402 | 0.028 | 112 (0.980) | |
| 344 (0.951)d |
Transversion:transition ratio.
ω value is the ratio of nonsynonymous (amino acid replacing) substitution rate (Ka) over the synonymous (silent) substitution rate (Ks), Ka/Ks.
The amino acid position is based on LaL/ODC amino acid numbering. The sites that have posterior probabilities > 0.9, by Bayes empirical Bayes analysis, are shown with the posterior probabilities in parentheses.
Boldface numbers indicate positively selected sites (posterior probabilities > 0.95).
The homology modeling-based methods revealed that only the amino acid 344 was located near the enzyme active site (Supplemental Fig. S4). Therefore, the amino acid substitutions at site 344 were predicted to enlarge the active site cavity of LDC in QA-producing plants to allow access to l-Lys, which has one more carbon than l-Orn (Bunsupa et al., 2012a).
Substitutions at Amino Acid 344 Are Important for a Shift of ODC to LDC Activity
Since the amino acid at position 112 was not located near the active site and was not conserved across LA-producing plants, we focused on the substitutions at amino acid 344 (Supplemental Table S4). To investigate the catalytic importance of the substitutions at amino acid 344, an LcL/ODC-Y344H mutant was constructed. In addition, tobacco ODC-3 (NtODC3) and its mutants, NtODC3-H344F and NtODC3-H344Y, were cloned and prepared (Supplemental Fig. S2). The LcL/ODC-Y344H mutant exhibited a reduced catalytic efficiency (Kcat/Km) of LDC over ODC activities, ranging from 4.84- to 0.08-fold, compared with the LcL/ODC-wild type (Table I). The NtODC3-H344F and NtODC3-H344Y mutants exhibited a reduced Kcat/Km toward ODC activity by 1.1- and 3.7-fold, respectively, compared with the NtODC3-wild type (Table I). However, LDC activity was not detected in either the wild type or the NtODC3 mutants.
These results strongly suggest that the amino acid substitution of His to Tyr in clubmosses, or from His to Phe/Tyr in legumes, is an important event that allows LDC activity, although further substitutions are required to optimize the LDC activity. In addition, putative ODCs from Nuphar avena and Nelumbo nucifera, which produce LAs, have Tyr at position 344 (Forrest and Ray, 1971). In chickpea (Cicer arietinum), putrescine and cadaverine are accumulated and degraded in a similar manner during seed germination and seedling development, and the presence of LDC in this plant was confirmed by feeding experiments using labeled [14C]Lys (Torrigiani and Scoccianti, 1995). The chickpea L/ODC has Phe at position 344. Taken together, these results support the importance of amino acid substitutions from His to Tyr or Phe at position 344.
DISCUSSION
Lys-derived alkaloids are widely distributed throughout the plant kingdom, from clubmosses to flowering plants (Bunsupa et al., 2012b). Based on the skeleton structure, the Lys-derived alkaloids can be subdivided into four main groups: quinolizidine, lycopodium, piperidine, and indolizidine alkaloids. With the exception of indolizidine alkaloids, LDC is the enzyme involved in the first step of Lys-derived alkaloid biosynthesis (Bunsupa et al., 2012b). In previous studies, we reported the cloning and characterization of LDC, which is responsible for the production of QAs (Bunsupa et al., 2012a). In this study, we isolated the LcL/ODC gene from LA-producing plants, thus supporting the important role of LDC in the production of alkaloids. Our results also provide a better understanding of the evolution of plant LDC.
Physiological Importance of LDC for Alkaloid Production
The recombinant LcL/ODC preferentially catalyzed the decarboxylation of l-Lys over l-Orn, with a 5-fold increase in efficiency in vitro, unlike LaL/ODC, which catalyzes both substrates nearly equally (Bunsupa et al., 2012a). The cellular abundance of Lys is expected to play an important role in the production Lys-derived alkaloids. The l-Lys level was about 15 times higher than that of l-Orn in L. clavatum and 45 times higher in the narrow-leafed lupin (Supplemental Table S1; Bunsupa et al., 2012a). LaL/ODC is localized in the chloroplast, where the last step of Lys biosynthesis is thought to take place, whereas LcL/ODC is localized in the cytosol (Mazelis et al., 1976; Bunsupa et al., 2012a). These results suggest that the subcellular trafficking of Lys to the cytosol may play a role in the efficient production of LAs. However, it is difficult to differentiate between a cytosolic localization and localization in the plasma membrane or endoplasmic reticulum. Further experiments, such as those employing fluorescence recovery after photobleaching and colocalization studies with membrane markers, are needed to provide additional evidence of cytosolic localization, to further address this issue of compartmentation of biosynthesis. The similar transcript levels of L/ODC observed in the shoots and roots of L. clavatum were inconsistent with the fact that major accumulation of the LAs happens in the stems and leaves. Thus, the downstream enzymes in LA biosynthesis might be localized in the shoots, or the transportation of the alkaloids produced might play a role in the differential accumulation of LAs.
The functions of LcL/ODC in vivo were characterized using a stable transformation in tobacco hairy roots and Arabidopsis plants, because of the difficulty in the transformation of L. clavatum. Analysis of the transgenic Arabidopsis plants and tobacco hairy roots expressing LcL/ODC showed a significant increase in cadaverine and the Lys-derived alkaloid, anabasine, respectively (Figs. 3 and 4). Furthermore, the correlation analysis showed a significant correlation between the expression of LcL/ODC and the cadaverine levels in transgenic Arabidopsis as well as between LcL/ODC and anabasine in the transgenic tobacco hairy roots. Anabasine is composed of two rings, a piperidine ring derived from Lys and a pyridine ring derived from nicotinic acid. The two rings in nicotine are a pyrrolidine ring derived from Orn and a pyridine ring (Bunsupa et al., 2014). A negative correlation between LcL/ODC transcript and nicotine was found, but the nicotine levels did not decrease significantly. This result suggests a tight regulation of nicotine biosynthesis in tobacco. Taken together, these results clearly support the function of LcL/ODC in plants.
Evolution of Plant LDC for the Production of Alkaloids
ODC, L/ODC, arginine decarboxylase (ADC), and diaminopimelate decarboxylase are pyridoxal-5′-phosphate (PLP)-dependent enzymes that belong to the Ala racemase family (Christen and Mehta, 2001). The functional specialization of most PLP-dependent enzymes occurred more than 1,500 million years ago, before the divergence of eukaryotes, archaebacteria, and eubacteria; their substrate specificities were altered by the substitution of specific amino acids in the enzyme active site (Christen and Mehta, 2001). Plants are the only eukaryotes that possess the Arg pathway that are not dependent on ODC (Fig. 6; Fuell et al., 2010). Interestingly, the protozoa Trypanosoma cruzi lacks ODC activity and cannot grow in a medium without putrescine (Algranati, 2010).
Figure 6.
Biosynthetic pathways for Lys- and Orn-derived alkaloids in plants.
In this study, we addressed the evolution of promiscuous functions, focusing on the activities of two enzymes: ODC and LDC. Our data suggest that promiscuous activities existed in an ancestral gene, because most of the functionally characterized ODC genes exhibited both ODC and LDC activities, although a majority of them had a minor (promiscuous) LDC activity and a major ODC activity. In the two distant lineages, legumes and clubmosses, the LDC activity was reinforced independently via at least one event of positive selection at amino acid position 344. The independent occurrence of the same event is likely to be a consequence of natural selection rather than genetic drift.
Bifunctional L/ODC could be advantageous for both lineages, because both primary (putrescine for polyamine production) and specialized (cadaverine for alkaloid production) metabolisms are important for cell growth and differentiation and for protection against pathogens and herbivores (Pichersky and Gang, 2000), respectively. In contrast, the orthologous ODC gene disappeared in other plant lineages, such as Arabidopsis and the moss Physcomitrella patens (Fuell et al., 2010). Plants possess an Arg pathway consisting of enzymes derived from a cyanobacterial ancestor (Illingworth et al., 2003) for the complementation of putrescine production (Fig. 6). Therefore, it is likely that ODC is not truly required in plants.
The eukaryotic ODC forms a homodimer, the subunits of which interact in a head-to-tail manner, producing two active sites at their interphase (Lee et al., 2007). The fact that only ODC or LDC is found in plants could be explained by dominant-negative mutations, which lead to mutant enzymes that disrupt the original activity (Veitia, 2007). Thus, the spatial expression of duplicated copies, ancestral and novel/improved LDC functions of ODC, might release these two copies from molecular constraints, which was reported during the evolution of homospermidine synthase for the production of pyrrolizidine alkaloids (Kaltenegger et al., 2013). The proteins encoded by ODC and LDC might form heterodimers that are less efficient or even inactive. Therefore, either the native or the evolved enzymes could become fixed in the population via natural selection.
We used the amino acid sequences of LaL/ODC and LcL/ODC as the query sequences to perform BLAST searches against the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi), Phytozome (http://www.phytozome.net/), and OneKP (Johnson et al., 2012; Matasci et al., 2014; Wickett et al., 2014; Xie et al., 2014; https://www.bioinfodata.org/Blast4OneKP/) databases. We identified two copies of ODC from foxtail millet (Setaria italica) on the same scaffold with a distance of approximately 24 kb (hereafter referred to as SiODC1 and SiODC2). SiODC1 and SiODC2 have His and Tyr at position 344, respectively. Although SiODC1 had no intron in its genomic sequence, SiODC2 contained one intron and lacked 64 nucleotides in the coding region, which resulted in the loss of 22 amino acids (Supplemental Fig. S5). This was probably accomplished via a pseudoexonization mechanism, during which an exon sequence becomes intronic (Xu et al., 2012; Supplemental Figs. S1 and S5). These specific amino acids are very important for the ODC and LDC enzymes to bind to their cofactor, PLP (Lee et al., 2007). Therefore, it is likely that SiODC2 is a pseudogene. However, functional analysis of SiODC1 and SiODC2 is needed to support this hypothesis.
In addition, recent draft genome sequence studies on narrow-leafed lupin revealed the presence of a single LDC gene (Conant and Wolfe, 2008; Yang et al., 2013). As active copies of both ODC and LDC were not found in the same plant, divergence in the regulatory regions due to changes in the expression patterns of the LDC and ODC copies to reduce the dominant-negative effect might not have occurred during plant LDC evolution. Therefore, either ODC or LDC was selected during evolution and were maintained in the population.
The results presented here indicate that an adaptive change from ODC to LDC occurred in plants that produce Lys-derived alkaloids and cadaverine, via their promiscuous functions. The LDC activity could have been gained independently within the Leguminosae and clubmoss lineages. There are several models that could explain the possible routes by which the plant ODC diverged to obtain an LDC function. First, the promiscuous LDC activity from the ancestral ODC, which was involved mainly in the primary metabolism, could have evolved gradually via several mutations and selections because of its physiological advantage for the production of Lys-derived alkaloids. This would have increased the LDC function without drastically altering the original ODC function (i.e. a bifunctional enzyme). Additionally, the alternative metabolic ADC pathway also could produce putrescine; thus, the ancestral ODC was likely not constrained to maintain its original function. Therefore, the process of LDC evolution could have started prior to the gene duplication of ODC. This kind of evolutionary process has been termed a weak negative tradeoff, where the divergence of a novel enzyme function occurs via a generalist intermediate (Khersonsky and Tawfik, 2010). Second, when environmental changes made the promiscuous LDC function beneficial for plants, gene duplication would have been advantageous to increase the dose of the ancestral ODC gene, thus resulting in increased protein levels. This process would have allowed a wider variety of function-altering mutations to accumulate, including potentially beneficial mutations that increase the LDC function, and get fixed in the population. In contrast, the less functional copies and those containing deleterious mutations, including the parental gene, could have been lost. This evolutionary process has been proposed as the innovation, amplification, and divergence model (Bergthorsson et al., 2007). This model is supported by the identification of ODC-like and LDC-like sequences from the Lys-derived alkaloid-free foxtail millet; however, the LDC-like sequences show signatures of pseudogenization. In both models, subsequent gene duplication could have helped resolve the adaptive conflict between the ODC and LDC activities by allowing the optimization of each activity in two separate copies. However, our data suggest that the divergence path toward a newly specialized LDC enzyme has not been completed; therefore, present-day enzymes exhibit only ODC or L/ODC (bifunctional) activity.
CONCLUSION
Overall, our results describe a clear case of the evolutionary innovation that uses promiscuous activities as the starting point for the divergence of novel enzymes. In addition, the occurrence of an alternative metabolic pathway might increase the evolutionary adaptability of the related enzymes. These findings contribute to a better understanding of how the enzymes in the primary metabolism, which are under a strong purifying selection, could evolve to have a novel function for the specialized metabolism. The molecular cloning and characterization of LcL/ODC shed light on LA biosynthesis and can serve as a basis for further biotechnological production of LAs for human benefit.
MATERIALS AND METHODS
Plant Materials
Soybean (Glycine max; B01151) and Lotus japonicus (Gifu) seeds were obtained from the National BioResource Project and Dr. Hiroshi Sudo (Hoshi University), respectively. Thermopsis lupinoides (syn. Thermopsis fabacea), a QA-producing plant, was obtained from the Medicinal Plant Gardens of the Graduate School of Pharmaceutical Sciences at Chiba University. Lycopodium clavatum and Huperzia serrata were purchased from plant markets in Japan. Tobacco (Nicotiana tabacum ‘Petit Havana’ line SR1) was obtained from Ghent University.
Metabolite Profiling
Soybean and L. japonicus were cultured on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc with 0.8% agar in a growth chamber at 25°C under 16-h/8-h light (approximately 3,000 lx)/dark cycles for 30 d before metabolite analysis. L. clavatum and H. serrata were maintained in a growth chamber in the same conditions as soybean and L. japonicus. Alkaloids, amines, and amino acids were extracted from the different organs of L. clavatum, H. serrata, soybean, and L. japonicus and analyzed using capillary electrophoresis-mass spectrometry as described previously (Oikawa et al., 2011). The (±)-HupA standard was purchased from Sigma-Aldrich.
Measurement of RNA Levels
The total RNA was extracted (RNeasy kit; Qiagen) and reverse transcribed into cDNA as described elsewhere (Bunsupa et al., 2012a). Real-time PCR was performed using the SYBR Green master mix (Applied Biosystems) at a final volume of 25 μL, including the appropriate primer pair for each target (Supplemental Table S4). Assays were run in quadruplicate in a StepOnePlus real-time PCR system (Applied Biosciences). The amplification program consisted of 40 cycles of 95°C for 15 s followed by 60°C for 1 min. The relative quantification of gene expression was performed using the comparative threshold cycle method. β-TUBULIN was used as an endogenous reference (Supplemental Table S4).
Cloning ODC and L/ODCs from Plants
The cDNAs encoding LcL/ODC, HsL/ODC1, HsL/ODC2, and TlL/ODC were isolated using degenerate primers as described elsewhere (Bunsupa et al., 2012a). The full-length cDNAs were obtained using 5′- and 3′-RACE (TaKaRa Bio). However, only a partial sequence for TlL/ODC was obtained. The full-length sequence for NtODC3 was isolated from tobacco using specific primers designed from NtODC (GenBank accession no. AB031066).
Heterologous Expression of Recombinant Proteins
The LcL/ODC and NtODC3 ORFs were amplified using gene-specific primers with overhangs containing restriction sites (Supplemental Table S4). The mutants were then prepared by PCR-based mutagenesis (Higuchi et al., 1988) using the primers listed in Supplemental Table S4. The amplified fragments were inserted in frame into the same restriction sites within the pGEX-6P-2 expression vector (GE Healthcare), which yielded recombinant gene products with N-terminal glutathione S-transferase protein tags. The complete constructs were sequenced to confirm the correct orientation, expressed in Escherichia coli, and purified as described elsewhere (Bunsupa et al., 2012a). We also cloned and expressed HseL/ODC1 in E. coli; however, we were unable to purify the recombinant HseL/ODC1 protein because of its insoluble nature. The ratio of the targeted recombinant protein to other coeluted proteins, quantified by densitometry using ImageJ software (http://imagej.nih.gov/ij/), was used to calculate the protein concentration.
LDC and ODC Activity Assays
LDC and ODC enzyme activities were determined by measuring the CO2 released from [14C]l-Lys and [14C]l-Orn, respectively (Gaines et al., 1988). The decarboxylase activities were assayed in 50 mm potassium phosphate, 5 mm EDTA, 4 mm dithiothreitol, 0.3 mm PLP, 0.5 to 3 mm l-[1-14C]Lys (40 μCi) or l-[1-14C]Orn (40 μCi), and 0.5 to 1 μg of purified enzyme, at pH 7.5 (except for the LDC activity assay for LcL/ODC, which was performed at pH 7), in a final volume of 500 μL. The released labeled CO2 was trapped on Whatman 3MM filter paper soaked in Soluene 350 (Perkin-Elmer), which was put to the top of a glass tube and closed with a rubber cap. Each reaction was performed at 37°C for 30 min. The ODC and LDC activities were then determined by measuring 14CO2 released from l-[1-14C]Orn and l-[1-14C]Lys, respectively, by liquid scintillation counting. The kinetics of decarboxylation of both l-Lys and l-Orn were analyzed by measuring the initial velocities over a range of substrate concentrations (0.5–2 mm). The competition assays were performed using 2 and 4 mm l-Orn or l-Lys, while 10 and 20 μm α-difluoromethyl-Orn were used for inhibitor assays.
Molecular Modeling
The three-dimensional model structures of LcL/ODC were predicted using SWISS-MODEL (Arnold et al., 2006) and the published human-ODC-putrescine complex (Protein Data Bank entry 2000) as the template (Dufe et al., 2007). The modeled protein was visualized using PyMOL (www.pymol.org).
Phylogenetic Analysis
LaL/ODC and LcL/ODC were used as queries and blasted with TBLASTN against the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi), Phytozome (http://www.phytozome.net/), and OneKP (Johnson et al., 2012; Matasci et al., 2014; Wickett et al., 2014; Xie et al., 2014; https://www.bioinfodata.org/Blast4OneKP/) databases. The ODCs and L/ODCs from plants that had E values < 10 e−06 and had important catalytic residues that are important for both ODC and LDC activities (Supplemental Fig. S1), especially Asp at position 343 (LaL/ODC numbering), were selected (Grishin et al., 1999; Kern et al., 1999). Other eukaryotic ODCs, such as those from yeast and human, also were included in the phylogenetic tree. The accession numbers of each ODC and L/ODC are shown in Supplemental Table S3. Amino acid alignments were performed using MEGA version 6 and manually adjusted to improve the reliability of the alignment (Supplemental Data S2; Tamura et al., 2013). If a codon site has at least a gap in the generated alignment, the codon site with a gap was not used to generate the phylogenetic tree. Only highly conserved amino acids without gaps were used for further analysis (Supplemental Data S1). To generate a phylogenetic tree, the best-fit model for the amino acid replacements was searched using ProtTest (Abascal et al., 2005); chosen for the best-fit model is LG (an improved general amino acid replacement matrix) + I (invariable sites) + G (γ shape). The γ shape is 1.116 in four rate categories. The proportion of invariable sites was 0.08. Using the best-fit mode, we generated the phylogenetic tree in PhyML3.0 (Guindon et al., 2010).
Test for Positive Selection
The ORFs corresponding to all available ODC and L/ODC amino acid sequences, as described above, were aligned. The resulting alignments were used for further analyses. We performed two analyses, the codon-site test and the branch-site test, in codeml of the PAML (version 4) package (Yang, 2007). In the codon site test, we performed two analyses, using models M7 (model = 0 and NSsites = 7) and M8 (model = 0 and NSsites = 8). The LRT was used to compare the two models, assuming that twice the log-likelihood difference between the two models (2∆L) follows a χ2 distribution with a number of degrees of freedom. In the branch-site model, we selected two branches that led to Lys-derived alkaloids as foreground branches and searched for the positively selected sites (model = 2, NSsites = 2, and fix_omega = 0 [Ka/Ks = free]). For the null hypothesis, we used the branch-site model with following parameters: model = 2, NSsites = 2, and fix_omega = 1 [Ka/Ks = 1]. The LRT was used to compare the two models, assuming that twice the log-likelihood difference between the two models (2∆L) follows a χ2 distribution with a number of degrees of freedom.
Protein Localization Analysis
The chimeric gene constructs of 35Spro:LcL/ODC-Met-1 and 35Spro:LcL/ODC-Met-3 fused with GFP at the N or C terminus were created using the primers presented in Supplemental Table S4 and subsequently cloned into the pTH2 vector (Chiu et al., 1996). An empty vector fused with RFP from Discosoma sp. (DsRed), 35Spro:DsRed, was used as a reference for the localization to cytosol (Kitajima et al., 2009). The resulting plasmids were expressed transiently in onion (Allium cepa) epidermal cells and Nicotiana benthamiana leaves (8-week-old plants) using a Helios gene gun (Bio-Rad) as described elsewhere (Bunsupa et al., 2012a). The GFP and RFP signals were observed using a confocal laser-scanning microscope (LSM700; Zeiss). For GFP, we used an argon laser with excitation at 488 nm with FSet38 wf filter. An argon laser with excitation at 555 nm with FSet43 wf filter was used for RFP. All images were acquired from single optical sections and were merged using the ZEN 2012 lite imaging software (Zeiss).
Plasmid Construction and Plant Transformation
To construct pGWB2-LcL/ODC (35Spro:LcL/ODC), the full-length sequence for LcL/ODC was cloned into the binary vector pGWB2 (Nakagawa et al., 2007; for primer sequences, see Supplemental Table S4) via Gateway technology (Invitrogen). Transgenic tobacco (‘Petit Havana’ line SR1) hairy roots and Arabidopsis (Arabidopsis thaliana) were generated as described elsewhere (Bunsupa et al., 2012a). Tobacco alkaloids and amines were measured as described elsewhere (Bunsupa et al., 2012a).
Statistical Analysis
Student’s one-tailed t test was used to identify statistically significant differences in the metabolite levels of transgenic Arabidopsis plants and tobacco hairy roots. Pearson correlation analysis was performed to calculate the correlation between the metabolite levels and the expression levels of LcL/ODC in transgenic Arabidopsis plants and tobacco hairy roots. For all statistical tests, significance was determined at P < 0.05.
Accession Numbers
The new DNA sequences reported here are deposited in the DNA Data Bank of Japan under accession numbers AB915695 (LcL/ODC), AB915696 (HsL/ODC1), AB915697 (HsL/ODC2), AB915698 (TlL/ODC), and LC030209 (NtODC3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of selected eukaryotic ODC and L/ODC amino acid sequences.
Supplemental Figure S2. SDS-PAGE of the recombinant LcL/ODC, NtODC3, and their mutant proteins purified from E. coli.
Supplemental Figure S3. Competition and inhibition studies of LcL/ODC.
Supplemental Figure S4. Overview of the predicted protein structure of the LcL/ODC complex homology model with the Schiff base intermediate of putrescine and PLP at the active site.
Supplemental Figure S5. Alignment of genomic sequences of putative ODCs from foxtail millet.
Supplemental Table S1. Levels of amines and LAs in each organ of L. clavatum and H. serrata.
Supplemental Table S2. Levels of amines in each organ of soybean and L. japonicus.
Supplemental Table S3. Accession numbers of the sequences used for phylogenetic analysis.
Supplemental Table S4. List of primers used in this study.
Supplemental Data S1. Highly conserved amino acid alignment without gaps for the construction of the phylogenetic tree in Figure 2.
Supplemental Data S2. Original amino acid alignment using ClustalW in the MEGA6 program.
Supplementary Material
Acknowledgments
We thank Dr. Ryo Nakabayashi (RIKEN Center for Sustainable Resource Science) for preliminary analyses of LAs by liquid chromatography-mass spectrometry; Satoko Sugawara (RIKEN Center for Sustainable Resource Science) for excellent technical support in the preparation of transgenic Arabidopsis plants; Tsuyoshi Nakagawa (Shimane University) for providing the destination vector pGWB2; Toshiaki Mitsui (Niigata University) for providing pWx-TP-DsRed vector; and all 1000 Plants Project contributors for gene sequencing data.
Glossary
- LA
lycopodium alkaloid
- QA
quinolizidine alkaloid
- ORF
open reading frame
- LRT
likelihood ratio test
- PLP
pyridoxal-5′-phosphate
Footnotes
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, JST, Strategic International Collaborative Research Program (SICORP), and by the Strategic Priority Research Promotion Program, Chiba University.
Articles can be viewed without a subscription.
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Algranati ID. (2010) Polyamine metabolism in Trypanosoma cruzi: studies on the expression and regulation of heterologous genes involved in polyamine biosynthesis. Amino Acids 38: 645–651 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR (2007) Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci USA 104: 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z (2005) Maximum likelihood methods for detecting adaptive protein evolution. In Nielsen R, eds, Statistical Methods in Molecular Evolution. Springer, New York, pp 103–124 [Google Scholar]
- Bunsupa S, Katayama K, Ikeura E, Oikawa A, Toyooka K, Saito K, Yamazaki M (2012a) Lysine decarboxylase catalyzes the first step of quinolizidine alkaloid biosynthesis and coevolved with alkaloid production in Leguminosae. Plant Cell 24: 1202–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunsupa S, Komastsu K, Nakabayashi R, Saito K, Yamazaki M (2014) Revisiting anabasine biosynthesis in tobacco hairy roots expressing plant lysine decarboxylase gene by using 15N-labeled lysine. Plant Biotechnol 31: 511–518 [Google Scholar]
- Bunsupa S, Yamazaki M, Saito K (2012b) Quinolizidine alkaloid biosynthesis: recent advances and future prospects. Front Plant Sci 3: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Christen P, Mehta PK (2001) From cofactor to enzymes: the molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Chem Rec 1: 436–447 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- De Luca V, Salim V, Atsumi SM, Yu F (2012) Mining the biodiversity of plants: a revolution in the making. Science 336: 1658–1661 [DOI] [PubMed] [Google Scholar]
- Dufe VT, Ingner D, Heby O, Khomutov AR, Persson L, Al-Karadaghi S (2007) A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane. Biochem J 405: 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest TP, Ray S (1971) Nuphar alkaloids: 3-epinupharamine. Can J Chem 49: 1774–1775 [Google Scholar]
- Fuell C, Elliott KA, Hanfrey CC, Franceschetti M, Michael AJ (2010) Polyamine biosynthetic diversity in plants and algae. Plant Physiol Biochem 48: 513–520 [DOI] [PubMed] [Google Scholar]
- Gaines DW, Friedman L, McCann PP (1988) Apparent ornithine decarboxylase activity, measured by 14CO2 trapping, after frozen storage of rat tissue and rat tissue supernatants. Anal Biochem 174: 88–96 [DOI] [PubMed] [Google Scholar]
- Grishin NV, Osterman AL, Brooks HB, Phillips MA, Goldsmith EJ (1999) X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with α-difluoromethylornithine. Biochemistry 38: 15174–15184 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16: 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C, Mayer MJ, Elliott K, Hanfrey C, Walton NJ, Michael AJ (2003) The diverse bacterial origins of the Arabidopsis polyamine biosynthetic pathway. FEBS Lett 549: 26–30 [DOI] [PubMed] [Google Scholar]
- Jia JY, Zhao QH, Liu Y, Gui YZ, Liu GY, Zhu DY, Yu C, Hong Z (2013) Phase I study on the pharmacokinetics and tolerance of ZT-1, a prodrug of huperzine A, for the treatment of Alzheimer’s disease. Acta Pharmacol Sin 34: 976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, Chase MW, Clarke ND, Covshoff S, Edger PP, et al. (2012) Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PloS ONE 7: e50226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenegger E, Eich E, Ober D (2013) Evolution of homospermidine synthase in the Convolvulaceae: a story of gene duplication, gene loss, and periods of various selection pressures. Plant Cell 25: 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Oliveira MA, Coffino P, Hackert ML (1999) Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure 7: 567–581 [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79: 471–505 [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Michael AJ, Martynowski D, Goldsmith EJ, Phillips MA (2007) Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases. J Biol Chem 282: 27115–27125 [DOI] [PubMed] [Google Scholar]
- Luo H, Li Y, Sun C, Wu Q, Song J, Sun Y, Steinmetz A, Chen S (2010a) Comparison of 454-ESTs from Huperzia serrata and Phlegmariurus carinatus reveals putative genes involved in lycopodium alkaloid biosynthesis and developmental regulation. BMC Plant Biol 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Sun C, Li Y, Wu Q, Song J, Wang D, Jia X, Li R, Chen S (2010b) Analysis of expressed sequence tags from the Huperzia serrata leaf for gene discovery in the areas of secondary metabolite biosynthesis and development regulation. Physiol Plant 139: 1–12 [DOI] [PubMed] [Google Scholar]
- Ma X, Gang DR (2004) The lycopodium alkaloids. Nat Prod Rep 21: 752–772 [DOI] [PubMed] [Google Scholar]
- Ma X, Tan C, Zhu D, Gang DR, Xiao P (2007) Huperzine A from Huperzia species: an ethnopharmacolgical review. J Ethnopharmacol 113: 15–34 [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. (2014) Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelis M, Miflin BJ, Pratt HM (1976) A chloroplast-localized diaminopimelate decarboxylase in higher plants. FEBS Lett 64: 197–200 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway Binary Vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Matsuda F, Kikuyama M, Mimura T, Saito K (2011) Metabolomics of a single vacuole reveals metabolic dynamism in an alga Chara australis. Plant Physiol 157: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Qian ZM, Ke Y (2014) Huperzine A: is it an effective disease-modifying drug for Alzheimer’s disease? Front Aging Neurosci 6: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Morita H, Chen G, Noguchi H, Abe I (2012) Molecular cloning and characterization of copper amine oxidase from Huperzia serrata. Bioorg Med Chem Lett 22: 5784–5790 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Eisenbrand G (1992) Chinese Drugs of Plant Origin: Chemistry, Pharmacology, and Use in Traditional and Modern Medicine. Springer-Verlag, Berlin [Google Scholar]
- Tomar PC, Lakra N, Mishra SN (2013) Cadaverine: a lysine catabolite involved in plant growth and development. Plant Signal Behav 8: doi/10.4161/psb.25850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrigiani P, Scoccianti V (1995) Regulation of cadaverine and putrescine levels in different organs of chick-pea seed and seedlings during germination. Physiol Plant 93: 512–518 [Google Scholar]
- Veitia RA. (2007) Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19: 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BS, Wang H, Wei ZH, Song YY, Zhang L, Chen HZ (2009) Efficacy and safety of natural acetylcholinesterase inhibitor huperzine A in the treatment of Alzheimer’s disease: an updated meta-analysis. J Neural Transm (Vienna) 116: 457–465 [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, et al. (2014) SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30: 1660–1666 [DOI] [PubMed] [Google Scholar]
- Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci USA 109: 1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Tao Y, Zheng Z, Zhang Q, Zhou G, Sweetingham MW, Howieson JG, Li C (2013) Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. PLoS ONE 8: e64799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z (2005) Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.